Abstract
Positive transcription elongation factor b (P-TEFb) plays an important role in stimulating RNA polymerase II elongation for viral and cellular gene expression. P-TEFb is found in cells in either an active, low-molecular-weight (LMW) form or an inactive, high-molecular-weight (HMW) form. We report here that human T-lymphotropic virus type 1 (HTLV-1) Tax interacts with the cyclin T1 subunit of P-TEFb, forming a distinct Tax/P-TEFb LMW complex. We demonstrate that Tax can play a role in regulating the amount of HMW complex present in the cell by decreasing the binding of 7SK snRNP/HEXIM1 to P-TEFb. This is seen both in vitro using purified Tax protein and in vivo in cells transduced with Tax expression constructs. Further, we find that a peptide of cyclin T1 spanning the Tax binding domain inhibits the ability of Tax to disrupt HMW P-TEFb complexes. These results suggest that the direct interaction of Tax with cyclin T1 can dissociate P-TEFb from the P-TEFb/7SK snRNP/HEXIM1 complex for activation of the viral long terminal repeat (LTR). We also show that Tax competes with Brd4 for P-TEFb binding. Chromatin immunoprecipitation (ChIP) assays demonstrated that Brd4 and P-TEFb are associated with the basal HTLV-1 LTR, while Tax and P-TEFb are associated with the activated template. Furthermore, the knockdown of Brd4 by small interfering RNA (siRNA) activates the HTLV-1 LTR promoter, which results in an increase in viral expression and production. Our studies have identified Tax as a regulator of P-TEFb that is capable of affecting the balance between its association with the large inactive complex and the small active complex.
Human T-lymphotropic virus type 1 (HTLV-1) is the etiological agent of adult T-cell leukemia (ATL) (29, 58) and the chronic inflammatory disease HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) (18, 19, 24, 30, 35). HTLV-1 encodes a 40-kDa protein, Tax, which is critical for viral replication, transformation, and gene regulation (3, 4, 8, 32). Numerous experiments have shown that Tax associates with CREB (1, 16, 44, 57, 59) and recruits CBP (25, 45, 53), p300 (14, 17, 40), and PCAF (21) to the Tax-responsive element (TRE) within the HTLV-1 long terminal repeat (LTR) for transcriptional activation. Recently, we demonstrated that P-TEFb is an essential transcription factor for transactivation of the HTLV-1 LTR by Tax (10, 60).
Functionally, P-TEFb, which is composed of CDK9 and cyclin T1, T2, or K, is essential for transcription of most major histocompatibility complex (MHC) class II protein-coding genes and plays an important role in transcriptional elongation (13, 37). Productive transcription elongation requires hyperphosphorylation at the Ser 2 site of the RNA polymerase II (pol II) C-terminal domain (CTD) by the CDK9 subunit of P-TEFb (34, 39). This phosphorylation is important for coupling of pre-mRNA synthesis with splicing and polyadenylation (2, 7, 9, 11, 27). P-TEFb also phosphorylates negative elongation factors DSIF and NELF, which cooperatively block RNA pol II processivity, to stimulate the transcription elongation (38, 41, 46, 47, 50, 51, 61). Glycerol gradient sedimentation and size exclusion chromatography have demonstrated that P-TEFb is found primarily in two distinct complexes within the cell (20, 33). The kinase-active P-TEFb is associated with the low-molecular-weight (LMW) complex (20, 54). A larger high-molecular-weight (HMW) complex is composed of P-TEFb associated with 7SK snRNP and HEXIM1 (31, 33). In cells, the ratio between the active and inactive forms of P-TEFb is tightly regulated (20, 38, 54). Inhibition of transcription or stress-inducing conditions such as treatment with actinomycin D, 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB), or UV irradiation result in the disruption of 7SK snRNP/HEXIM1-bound HMW P-TEFb complex and increase the LMW Brd4/P-TEFb complex, resulting in an increase of mRNA and protein synthesis (33, 55, 56).
In a previous report, we have demonstrated that the HTLV-1 Tax protein binds to cyclin T1 and can compete with Brd4 for P-TEFb binding (10). Here, we present evidence that the Tax/P-TEFb complex sediments in the LMW fractions in a complex, which is distinct from the LMW Brd4/P-TEFb complex. Interestingly, when Tax was expressed in cells, we observed a decrease in HMW P-TEFb complexes and an increase in LMW P-TEFb. The increase of P-TEFb in the LMW complex did not coincide with an increase in Brd4 levels in the LMW complex. We also show that Tax can decrease the interaction of 7SK snRNP and HEXIM1 with P-TEFb in the HMW complex. Dissociation of 7SK snRNP/HEXIM1 involves the direct interaction of Tax with cyclin T1, since a cyclin T1 peptide (amino acids [aa] 383 to 426) containing the Tax binding site inhibits the dissociation. Chromatin immunoprecipitation (ChIP) analysis of the HTLV-1 LTR demonstrates that Brd4 is associated with the inactive basal promoter, but upon Tax expression, Brd4 is dissociated from the transcription complex. Dissociation of Brd4 corresponds with Tax-activated transcription. Furthermore, the knockdown of Brd4 results in HTLV-1 promoter activation and viral production. These studies are the first to demonstrate that P-TEFb exists in HTLV-1-transformed cells in distinct complexes that are regulated by Tax expression. Our studies not only provide information on HTLV-1 transcription regulation but provide insight into mechanisms of P-TEFb regulation in eukaryotic cells.
MATERIALS AND METHODS
Cell culture and adenovirus transduction.
C8166 cells (Tax-expressing HTLV-1 transformed cells) were cultured in RPMI medium with 10% fetal bovine serum, 2 mM glutamine, and penicillin-streptomycin (Quality Biological Inc.). HeLa and pA-18G-BHK-21 cells were maintained in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum, 2 mM glutamine, and penicillin-streptomycin (Quality Biological Inc.). Transduction of adenovirus expressing green fluorescent protein (AdGFP) or Tax (AdTax) into the HeLa or pA-18G-BHK-21 cells was performed as described previously (10). Briefly, cells at 80% confluence were incubated with virus at a multiplicity of infection (MOI) of 100 (HeLa cells) or 200 (BHK-21 cells) infectious units/cell for 2 h in the absence of serum. The cells were incubated for 12 or 24 h in DMEM supplemented with 10% serum.
Nuclear fraction purification and glycerol gradient sedimentation.
Cells (2 × 107) were harvested for lysis with 1 ml of a cytoplasmic fraction preparation buffer containing 10 mM HEPES (pH 7.4), 10 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), and a protease inhibitor mixture (Roche). Following centrifugation at 12,000 rpm, the pellet extracted with 500 μl of a nuclear extraction buffer containing 20 mM HEPES (pH 7.4), 420 mM NaCl, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 1 mM PMSF, and protease inhibitors. Following extraction, the buffer was adjusted to a final concentration of 150 mM NaCl. Four hundred or 500 μg of nuclear extracts was fractionated on a 10 to 40% glycerol gradient by ultracentrifugation in an SW41 Ti rotor (Beckman) at 35,000 rpm for 24 h. Twenty-four fractions (500 μl each) were collected and immunoblotted with specific antibodies against Tax, cyclin T1 (Santa Cruz), CDK9 (Santa Cruz), or Brd4 (kindly provided by K. Ozata, NCI).
Coimmunoprecipitation and reverse transcription-PCR (RT-PCR).
Nuclear extracts were fractionated by glycerol gradient sedimentation, followed by immunoprecipitation. Fractions containing the low-molecular-weight P-TEFb complex were pooled and immunoprecipitated with CDK9, cyclin T1, Tax, or Brd4 antibody for 4 h at 4°C. High-molecular-weight complex fractions were immunoprecipitated with CDK9 (Santa Cruz) antibody. Magnetic protein G (Invitrogen) was added to the mixture and incubated for an additional 2 h. The beads were washed five times using buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl, 10% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, and 1 mM PMSF, and subjected to Western blotting as described above. To detect 7SK snRNA bound to CDK9 in the HMW complex, the HMW complex fraction immunoprecipitated with CDK9 was used for isolation of total RNA, followed by cDNA synthesis. PCR was performed with 25 to 30 cycles using the Taq DNA polymerase (Applied Biosystems).
In vitro competition assay.
Glycerol fractions containing HMW P-TEFb complexes obtained from HeLa or BHK-21 nuclear extracts transduced with AdGFP were incubated with 0.5 μg of purified His-Tax, baculovirus-purified Brd4, or bovine serum albumin (BSA) in binding buffer (25 mM HEPES [pH 7.4], 1 mM DTT, 20 mM NaCl, 0.2 mM EDTA, 0.1% Triton X-100, 20% glycerol, protease inhibitor cocktail [Roche], and RNase inhibitor [Promega]). Six hours later, CDK9 antibody was added and incubated for 4 h. The CDK9 immunocomplex was precipitated with magnetic protein G, and 7SK snRNA was detected by RT-PCR as described above. For peptide competitive inhibition, a 5-fold molar excess of cyclin T1 peptide (aa 383 to 426, PSAKVSLKEYRAKHAEELAAQKRQLENMEANVKSQYAYAAQNLL) or control peptide (AYAAQNLLSHHDSHSSVILKM; the nuclear localization signal is in bold) was incubated with His-Tax protein in binding buffer for 30 min before being mixed with HMW complex fractions. Six hours later, the reaction mixtures were subjected to immunoprecipitation with CDK9 antibody followed by real-time RT-qPCR (Stratagene Mx3000P system).
ChIP assay.
The ChIP assay was carried out using 10 μg of antibody to CDK9, Brd4, or RNA pol II (8WG16) (Covance) or CREB, CBP, or p300 (Santa Cruz) as described previously (60). BHK-21-HTLV-1 LTR-lacZ cells (5 × 106) were transduced with AdGFP or AdTax for 2 h at an MOI of 200. After 12 h of incubation, cells were cross-linked and sheared by sonication to get 200- to 800-bp DNA fragments. The chromatin extracts were precleared with salmon sperm DNA and magnetic protein G beads. The supernatants were diluted 10-fold with ChIP dilution buffer, and the antibodies were added and incubated overnight at 4°C. The immune complexes were collected by the addition of magnetic protein G beads and washed stepwise. The DNA was purified by proteinase K (Sigma) treatment, phenol extraction, and ethanol precipitation. PCR was performed using primers specific for the HTLV-1 LTR (nucleotides −160 to −139 [5′-CCACAGGCGGGAGGCGGCAGAA-3′] and nucleotides −102 to −79 [5′-TCATAAGCTCAGACCTCCGGGAAG-3′]) or LacZ-coding region (nucleotides −1010 to 1026 [5′-AAAATGGTCTGCTGCTG-3′] and nucleotides −1243 to −1227 [5′-TGGCTTCATCCACCACA-3′]).
siRNA transfection, reporter assay, and HTLV p19 Gag quantification assay.
For the reporter assay, C8166 cells were seeded in 24-well plates at 2 × 105 cells/well, and small interfering RNAs (siRNAs) (150 nM) were transfected using HyperFect transfection reagent (Qiagen) as described by the manufacturer. Forty-eight hours later, the reporter plasmid pGL3.HTLV-1-LTR-Luc (0.5 μg) was transfected using TransFast (Promega). The plasmid pRSV.LacZ (0.5 μg) was used to monitor transfection efficiency. After 24 h of incubation, cells were harvested for luciferase and beta-galactosidase activities. Cell lysates were examined for protein expression levels using Western blotting with anti-Brd4, -RNA pol II, -CDK9, or -Tax antibody. 293T cells were transfected with siRNA (150 nM) and 0.4 μg of pACH-WT, an HTLV-1 molecular clone kindly provided by L. Ratner (Washington University), using 6 μl HiPerFect transfection reagent as described by the manufacturer (Qiagen, Valencia, CA). Seventy-two hours later, cell supernatant was collected and HTLV p19 Gag levels quantitated using an enzyme-linked immunosorbent assay (ELISA) (ZeptoMetrix Corporation, Buffalo, NY) according to the manufacturer's protocol.
RESULTS
Tax and Brd4 are present in active LMW P-TEFb complex.
While P-TEFb has been shown to exist in a HMW inactive complex containing 7SK snRNP and HEXIM1 and in an LMW active complex containing Brd4 (20, 54), the distribution of P-TEFb in HTLV-1-transformed cells is not known. To characterize the Tax/P-TEFb complex in transformed cells, we first performed glycerol gradient sedimentation analysis on nuclear extracts from the HTLV-1-transformed C8166 cell line. Following centrifugation, gradient fractions were analyzed by Western blot analysis with CDK9, cyclin T1, Brd4, and Tax antibodies. Similar to the case for uninfected cells, the results of Western blot analysis for CDK9 and cyclin T1 were consistent with the presence of both a HMW complex (fractions 13 to 17) and a LMW complex (fractions 7 to 11) in the HTLV-1-transformed cells (Fig. 1A). Western blot analysis also showed that both Tax and Brd4 sediment in overlapping fractions with CDK9 in LMW complex regions of the glycerol gradient. Similar sedimentation profiles were observed when extracts from HeLa cells infected with a Tax-expressing adenovirus were examined (Fig. 1B). Although the apparent level of Brd4 remains unchanged in AdTax- versus AdGFP-transduced cells, CDK9 levels shift from the HMW to the LMW complex when Tax is expressed. This is clearly seen when corresponding fractions from Tax-positive versus Tax-negative extracts are run side by side (Fig. 1C). Together these results suggest that Tax alters the distribution of P-TEFb in vivo and binds to P-TEFb, forming LMW complexes.
FIG. 1.
Tax and Brd4 are present in the LMW complex. (A) Nuclear extracts from C8166 cells were fractionated on a 10% to 40% glycerol gradient by ultracentrifugation. After 24 h, 24 fractions were collected and analyzed by Western blot analysis with CDK9, Brd4, or Tax antibody. I indicates 1/50 of the extract used on the gradient. (B) HeLa cells (80% confluent) were transduced with adenovirus expressing GFP (AdGFP) or Tax (AdTax) at an MOI of 100. Nuclear extracts were prepared at 24 h posttransduction and analyzed by glycerol gradient sedimentation and Western blot analysis as described above. (C) Glycerol gradient fractions for adenovirus-transduced HeLa cells AdGFP (−Tax) or AdTax (+Tax) were separated on 4 to 20% Tris-glycine gels and Western blot analysis performed for CDK9 (top panel). Corresponding gradient fractions, indicated above the panels, from LMW (middle) and HMW (bottom) fractions of this gradient were separated on 4 to 20% Tris-glycine gels and Western blot analysis performed for CDK9 or Tax.
Tax interacts with P-TEFb and forms complexes distinct from those for Brd4.
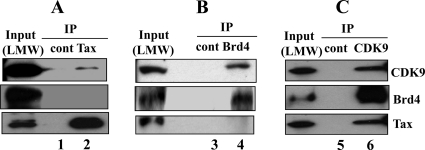
The results from glycerol gradient sedimentation demonstrate that Tax and Brd4 sediment in overlapping fractions with the LMW P-TEFb complex. Therefore, we determined whether Tax and Brd4 are in distinct LMW P-TEFb complexes. The LMW pool fractions from C8166 cells were immunoprecipitated with Tax, CDK9, or Brd4 antibody, followed by Western blot analysis with anti-Tax, -CDK9, and -Brd4 antibodies to determine the composition of the complexes (Fig. 2). As expected, when the LMW P-TEFb fraction was immunoprecipitated with CDK9 antibody, both Tax and Brd4 coimmunoprecipitated (Fig. 2C, lane 5 versus lane 6). In contrast, when the LMW fractions were immunoprecipitated with antibody to Tax (Fig. 2A, lane 2, top panel versus middle panel), only CDK9 and not Brd4 (bottom panel) was detected. Likewise, when antibody to Brd4 was used to isolate complexes from the LMW fractions, only CDK9 was detected (Fig. 2B, lane 4, top panel versus bottom panel). These results are consistent with the presence of two distinct LMW P-TEFb complexes in the HTLV-1-transformed cells, one composed of Tax/P-TEFb and the other composed of Brd4/P-TEFb.
FIG. 2.
Tax forms a distinct Tax/P-TEFb complex. Low-molecular-weight (LMW) P-TEFb complex fractions (fractions 7 to 11) of C8166 nuclear extracts were immunoprecipitated with control antibody (lanes 1, 3, and 5), Tax (A) (lane 2), Brd4 (B) (lane 4), or CDK9 (C) (lane 6) antibody and analyzed by Western blotting with CDK9 (upper panels), Brd4 (middle panels), or Tax (lower panels) antibody. Input lanes represent 1/20 of the extract used for immunoprecipitation.
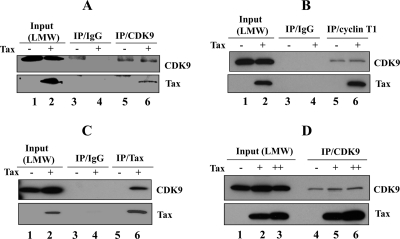
These results were further confirmed using the LMW fraction purified from HeLa nuclear extracts transduced with either AdGFP or AdTax. The results presented in Fig. 3A and B demonstrate that Tax is present in the anti-CDK9 or anti-cyclin T1 immunoprecipitates, consistent with the association of Tax with P-TEFb (Fig. 3A and B, lanes 6). In cells transduced with the control GFP virus, no Tax was detected in either CDK9 or cyclin T1 immunoprecipitates (Fig. 3A and B, lanes 5). In addition, the LMW P-TEFb pool was immunoprecipitated with anti-Tax antibody and then analyzed by Western blotting with anti-CDK9 antibody. The results presented in Fig. 3C demonstrate that CDK9 was detected in the Tax immunoprecipitates.
FIG. 3.
P-TEFb/Tax complex. (A to C) LMW P-TEFb complex fractions from nuclear extracts of HeLa cells transduced with AdGFP (−) or AdTax (+) were immunoprecipitated with control IgG antibody or antibody to CDK9 (A), cyclin T1 (B), or Tax (C). (D) LMW P-TEFb complex fractions of HeLa cells transduced with AdGFP at an MOI of 100 (−) or with AdTax at an MOI of 20 (+) or 100 (++) were immunoprecipitated with anti-CDK9 antibody and immunoblotted with CDK9 (upper panel) or Tax (lower panel) antibody.
To determine whether the level of Tax expression affects the formation of the Tax-P-TEFb complex, HeLa cells were transduced with AdTax virus at an MOI of 20 or 100 and incubated for 24 h. Nuclear extracts were fractionated on glycerol gradients, and the LMW P-TEFb complexes were immunoprecipitated with CDK9 antibody. The results shown in Fig. 3D demonstrate that as expression of Tax is increased, the interaction between Tax and P-TEFb quantitatively increases (Fig. 3D, lanes 4 to 6), suggesting that the level of Tax/P-TEFb in the cell is proportional to the amount of Tax expression. This is the first demonstration that Tax is associated with the LMW active P-TEFb fraction and may be immunoprecipitated with antibody to either the cyclin T1 or CDK9 subunit of P-TEFb. Moreover, it appears that the amount of Tax/P-TEFb complex in the cell is controlled, in part, by the overall level of Tax expression.
Tax decreases the HMW P-TEFb complex through inhibition of binding of 7SK snRNP/HEXIM1 to P-TEFb.
We have previously shown that Tax expression decreases the amount of 7SK snRNA associated with P-TEFb in vivo, suggesting that Tax may function to regulate the HMW P-TEFb complex (10). We next investigated whether Tax could directly affect the stability of the HMW complex and dissociate the 7SK snRNP and HEXIM1 inhibitory subunits. The HMW P-TEFb fractions were isolated from HeLa (Fig. 4A) or BHK-21 (Fig. 4B) cells. The HMW fraction was then incubated with purified Tax, Brd4, or control BSA protein. Although equivalent amounts of CDK9 were immunoprecipitated from all reactions, Tax incubation with the HMW fraction resulted in a 50 to 60% decrease in the level of 7SK snRNA bound to P-TEFb (Fig. 4A [compare lane 1 and lane 2] and B [compare lane 2 and lane 3). A similar decrease was seen when Brd4 protein, which is known to decrease 7SK snRNP binding to P-TEFb (20), was used (Fig. 4A [lane 3] and B [lane 4]). Quantification of the reduction in 7SK snRNP binding to P-TEFb is shown graphically (Fig. 4A and B, right panels). This suggests that Tax, like Brd4, can dissociate P-TEFb from the 7SK snRNP and HEXIM1 complex.
FIG. 4.
Tax destabilizes the HMW P-TEFb complex through inhibition of the binding of 7SK snRNP/HEXIM1 with CDK9. (A and B) One microgram of purified His-Tax, Brd4, or BSA was incubated with the HMW P-TEFb complex fractions isolated from HeLa (A) or BHK-21 (B) cells. The reaction mixtures were then immunoprecipitated (IP) with CDK9 antibody and subjected to RT-PCR for 7SK snRNA detection (upper panel). The level of CDK9 immunoprecipitated was assessed by immunoblotting with anti-CDK9 antibody (lower panel). Quantitation of 7SK RNA and CDK9 levels was done using FluorChem (Alpha Innotech). The values for 7SK RNA were adjusted for CDK9 levels and the BSA control set to 1. The values were graphed as fold difference. Standard deviations from at least three independent experiments are shown. (C) Purified Tax protein was preincubated with a 5-fold molar excess of a cyclin T1 competing or control peptide. The reaction mixtures were then incubated with purified HMW P-TEFb complex from HeLa nuclear extracts. The incubation mixtures were immunoprecipitated with anti-CDK9 antibody. Half of the CDK9-immunoprecipitated complex was analyzed for CDK9 levels by immunoblotting. The other half was subjected to RNA purification and cDNA synthesis. The cDNA was used for real-time PCR (Stratagene Mx3000p) with 7SK snRNA-specific primers. the value for the HMW fraction alone was set to 1 (100% binding to CDK9). The relative percent binding for three independent experiments is shown.
We have previously demonstrated that Tax interacts with two independent sites of cyclin T1, one within amino acids 383 to 426 and the other between amino acids 432 and 533 (10). If the direct interaction of Tax with cyclin T1 is responsible for the dissociation of 7SK snRNP and HEXIM1 from the HMW complex, one would predict that a peptide covering the Tax binding domain would inhibit the dissociation. Since Brd4 binding to cyclin T was mapped to amino acids 432 to 533, we choose the peptide spanning amino acids 383 to 426, the second Tax binding region of cyclin T. Tax and the HMW P-TEFb fractions were incubated in the absence or presence of a 5-fold excess of control or competitor peptide spanning amino acids 383 to 426 of cyclin T1. Following incubation, the reaction mixtures were immunoprecipitated with anti-CDK9 antibody and analyzed for 7SK snRNA (Fig. 4C). Western blot analysis showed that similar levels of CDK9 were precipitated from each reaction mixture. Consistent with the results presented above, Tax decreased the amount of 7SK snRNA associated with P-TEFb by approximately 90% (Fig. 4C, compare lane 1 and lane 2). Control peptide had little effect on 7SK RNA levels in either the absence or presence of Tax (Fig. 4C, compare lanes 1 and 5 and lanes 2 and 3, respectively). In contrast, in the presence of the peptide spanning aa 383 to 426, dissociation of 7SK snRNA by Tax was decreased from 90% dissociation to only 40% dissociation (Fig. 4C). These results suggest that Tax interaction with cyclin T1 is important for the dissociation of P-TEFb from the 7SK snRNP and HEXIM1 complex.
Interaction of Brd4/P-TEFb and Tax/P-TEFb with the HTLV-1 LTR.
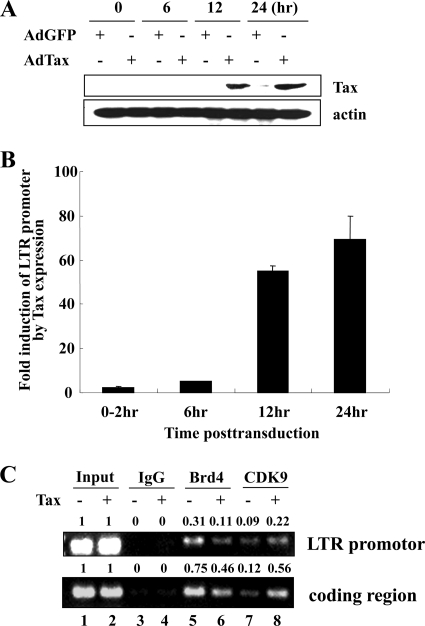
To investigate the role of Brd4 and Tax interaction with P-TEFb in HTLV-1 transcription, we performed ChIP assays using BHK-21-HTLV-1 LTR-lacZ cells, which have a single integrated copy of the HTLV-1 LTR promoter linked to the beta-galactosidase gene. The cells were transduced with either AdTax or control AdGFP virus for 0 to 24 h, harvested, and analyzed for Tax expression and reporter gene activity. As shown in Fig. 5A, Tax expression was detected as early as 12 h postinfection and increased slightly at 24 h. Concomitant with the expression of Tax, we observed a 50- to 60-fold increase in the level of HTLV-1 LTR activity (Fig. 5B). Based on the results of these studies, BHK-21-HTLV-1 LTR-lacZ cells were transduced with AdGFP or AdTax for 12 h and then fixed with 1% formaldehyde for ChIP analysis. Equivalent amounts of cross-linked chromatin were then immunoprecipitated with control IgG (Fig. 5C, lanes 3 and 4) or anti-Brd4 (Fig. 5C, lanes 5 and 6) and -CDK9 (Fig. 5C, lanes 7 and 8) antibody and subjected to semiquantitative RT-PCR amplification with specific primers for the LTR-promoter or the LacZ-coding region. In the absence of Tax expression, we found that Brd4 and CDK9 were bound to the basal promoter (Fig. 5C, top panel, lanes 5 and 7). Interestingly, when Tax was expressed in the cell, the level of Brd4 bound to the promoter region decreased significantly (Fig. 5C, top panel, compare lanes 5 and 6), suggesting that part of Tax transactivation involves dissociation of the Brd4/P-TEFb complex in favor of the Tax/P-TEFb complex. Consistent with the decrease in Brd4 at the promoter, Brd4 association in the coding region of the gene was also decreased when Tax was present (Fig. 5C, bottom panel, compare lanes 5 and 6). In parallel with the increase in HTLV-1 LTR transcription activity, we observed a significant increase in binding of CDK9 to the Tax-transactivated template (Fig. 5C, compare lanes 7 and 8). The levels of CREB, CBP, p300, and RNA pol II bound to the HTLV-1 LTR increased approximately 2- to 3-fold in the presence of Tax (data not shown). Together these results suggest that the basal HTLV-1 promoter is occupied by a transcription complex including Brd4/P-TEFb. In the presence of Tax, Brd4 is dissociated from the promoter and transcription factors CREB, CBP, Tax/P-TEFb, and pol II are recruited to the LTR to form the Tax-transactivated transcription complex.
FIG. 5.
ChIP analysis of the integrated HTLV-1 promoter in the absence and presence of Tax. (A and B) BHK-21-HTLV-1 LTR-lacZ cells, which contain a single integrated copy of the HTLV-1 LTR promoter linked with beta-galactosidase gene (lacZ), were transduced with AdGFP or AdTax. Cell lysates were prepared at 0, 6, 12, and 24 h posttransduction and analyzed by Western blot with anti-Tax antibody to check the level of expression in the cell. Beta-galactosidase activity was also measured to determine promoter activity. (C) BHK-21-HTLV-1 LTR-lacZ cells were transduced with AdGFP (−) or AdTax (+) at an MOI of 200 and subjected to ChIP assays at 12 h posttransduction. Antibodies specific for Brd4 and CDK9 were used for immunoprecipitation. PCR was used to analyze immunoprecipitated DNA using primers specific for the HTLV-1 LTR (upper panel) or LacZ-coding region (lower panel). Quantitation of PCR band intensity was done using FluorChem (Alpha Innotech). The values were determined from input set to 1.
Knockdown of Brd4 enhances Tax transactivation of the viral LTR and viral production.
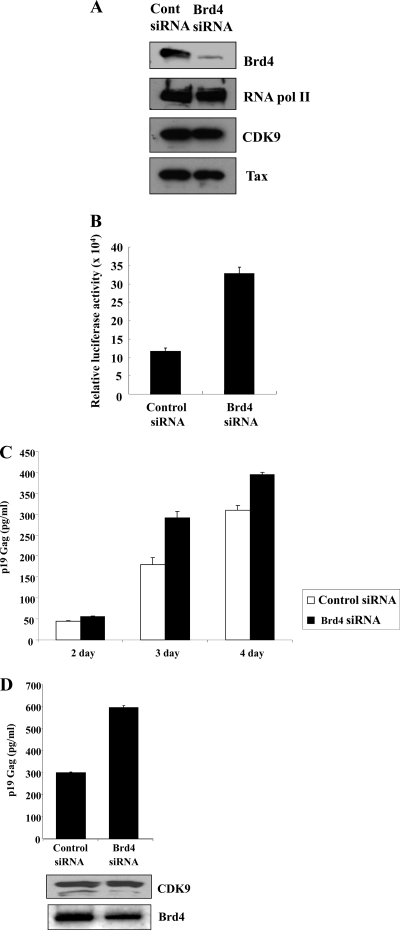
We reasoned that if Brd4/P-TEFb complexes have low LTR activity, then knockdown of Brd4 protein in HTLV-1 transformed cells should increase the viral LTR promoter activity. To test this, C8166 cells were transfected with control or Brd4 siRNA. Western blot analysis showed that when cells were transfected with siRNA to Brd4, the level of Brd4 protein decreased significantly (approximately 90%) compared to that in cells transfected with control siRNA (Fig. 6A). Brd4 siRNA was specific for Brd4 knockdown, since no significant decreases in expression of Tax, CDK9, or RNA pol II were found compared to that with the siRNA control (Fig. 6A). If Brd4 competes with Tax for P-TEFb, then reducing Brd4 protein should increase the Tax/P-TEFb complexes and thus increase promoter activity. Indeed, when viral promoter activity is measured using an HTLV-1 LTR reporter construct in transfection experiments, we observe a 3-fold increase in LTR activity in cells cotransfected with siRNA specific for Brd4 compared to control siRNA (Fig. 6B). Although we see a 90% reduction in Brd4 protein, we propose that only a modest increase in promoter activity is observed because active Tax/P-TEFb complexes already exist in these cells.
FIG. 6.
Brd4 knockdown increases viral promoter activity and virus production. C8166 cells were transfected with 150 nM control or Brd4 siRNA using HiPerFect transfection reagent. Forty-eight hours later, the cells were transfected with the reporters HTLV-1-LTR-Luc (0.5 μg) plasmid and RSV-LacZ (0.5 μg) plasmid. Cell lysates were prepared 24 h later. (A) Cell lysates were analyzed by immunoblotting using antibody to Brd4, RNA pol II, CDK9, or Tax. (B) Luciferase activity from the HTLV-1-LTR reporter plasmid was measured, and values were adjusted for transfection efficiency using beta-galactosidase activity. The values were graphed as relative luciferase activity and represent the mean activity from at least three independent experiments. (C) 293T cells were cotransfected with control or Brd4 siRNA and ACH-WT (HTLV-1 molecular clone) plasmid using HyperFect. Culture supernatants were harvested at 2, 3, and 4 days after transfection and p19 Gag antigen measured by ELISA. Values are given in pg/ml and are the average from three independent transfections. (D) 293T cells were cotransfected with control or Brd4 siRNA and ACH-WT (HTLV-1 molecular clone) plasmid using HyperFect. After 72 h, the cells were harvested for Western blot analysis of Brd4 or CDK9 and the supernatants used for quantitation of HTLV-1 p19 levels (pg/ml) using an ELISA.
Next, we investigated whether Brd4 levels affect virus production. To address this, we used a molecular clone of HTLV-1 (ACH-WT). When ACH-WT is transfected into cells, viral proteins are expressed and mature virus released into the supernatant. 293T cells were transfected with ACH-WT and either control siRNA or siRNA to Brd4. As shown in Fig. 6C, virus production increased with time. A clear difference in virus production between siRNA control and Brd4 siRNA cultures can be seen at day 3 and day 4 (Fig. 6C). Western blot analysis confirmed that cells cotransfected with Brd4 siRNA had a 35% reduction in Brd4 protein levels and no change in CDK9 protein levels (Fig. 6D, bottom panel). HTLV-1 p19 (Gag) ELISA results from culture supernatants show that reduced Brd4 expression enhances HTLV-1 virus production (1.6- to 2-fold) (Fig. 6C and D, top panel). Together with the ChIP data, these results suggest that Brd4 has low transactivation function on the LTR and acts as a negative regulator of HTLV-1 transcription, competing with Tax for P-TEFb. Although basal LTR activity is very low, siRNA to Brd4 does increase basal activity 4-fold (data not shown), suggesting that Brd4 may be a repressor. However, more studies are required to determine if Brd4 is indeed an active repressor or is insufficient to activate the LTR alone.
DISCUSSION
Studies have shown that P-TEFb is found in two major complexes in the cell, an active, low-molecular-weight (LMW) complex and an inactive, high-molecular-weight (HMW) complex. However, these studies were not performed with HTLV-1-transformed cells. Our studies are the first to demonstrate that P-TEFb is also found in two major forms, an HMW complex and an LMW complex, in Tax-expressing cells. This profile was found when nuclear extracts were made using detergent with either high or low salt concentrations. Further, our studies demonstrate that Tax deregulates the P-TEFb pathway in two ways. One mechanism involves competition with Brd4. Tax interacts with the cyclin T1 subunit of P-TEFb, competing or blocking Brd4 binding, and forms a Tax/P-TEFb complex. Because overexpression of Brd4 decreases HTLV-1 LTR transcription activity, the transition to the Tax/P-TEFb complex likely plays an important role in transcription activation. The results presented in this paper demonstrate that the Tax/P-TEFb cosediments with the LMW P-TEFb complex. Whether other proteins in addition to Tax and P-TEFb are present in the complex is under investigation.
We have utilized ChIP assays to analyze the composition of HTLV-1 LTR transcription complexes in the presence and absence of Tax. Our results demonstrate that Brd4 is bound to the inactive LTR basal promoter, presumably as a part of the Brd4/P-TEFb complex. In addition, lower levels of CREB, CBP, p300, and RNA pol II are found on the basal promoter. To date our report is the first demonstrating that Brd4 is present on a promoter in an inactive form and does not contribute positively to transcription. We interpret these findings to suggest that while a transcription complex is formed on the basal promoter, the composition and/or architecture of the complex is not conducive to transcription. Indeed, Easley and colleagues (12) have also recently reported the requirement for chromatin-remodeling factors in the activation of the latent HTLV-1 LTR. Using conventional chromatography, they found association of Tax with the chromatin remodeling factors BRG1 and SWI/SNF. They went on to show that in latent cells, this association occurs on the HTLV-1 LTR upon TNF-α stimulation and contributes to viral gene expression. Unfortunately, Easley et al. did not examine the level of Brd4 on the latent versus the activated LTR. Our results show that in the presence of Tax, binding of the Brd4 to the HTLV-1 promoter decreases. Concomitant with the decrease in Brd4 binding, we see an increase in the level of CDK9 binding, suggesting an influx of the Tax/P-TEFb complex. Consistent with numerous studies on Tax transactivation which have demonstrated that Tax increases the amount and stability of CREB, CBP, p300, and RNA pol II binding, we also observed an increase in the levels of these factors binding to the promoter (15, 26). Interestingly, the modest changes we find in transcription factor binding lead to a 60-fold activation of transcription. It appears, therefore, that the formation of the Tax/P-TEFb complex may play an important role in the ability of Tax to activate the viral promoter in a specific manner. It will be of interest to determine if the Tax/P-TEFb complex also plays a role in the activation of Tax-responsive cellular promoters.
In addition to recruiting P-TEFb to activate the viral LTR, a second way in which Tax regulates P-TEFb is to destabilize the HMW 7SK snRNP/HEXIM1/P-TEFb complex, leading to the dissociation of the inhibitory 7SK snRNP and HEXIM1 subunits. This effect of Tax has been observed in vivo and in vitro with purified Tax protein. The latter results suggest that Tax directly dissociates the HMW complex. The ability of Tax to dissociate the HMW complex does not appear to be through direct interaction with HEXIM1, as in vitro binding assays failed to detect an interaction between the proteins (data not shown). Work by several groups has recently demonstrated that LARP7 and MEPCE proteins are important components of the 7SK snRNP complex, regulating its stability and in turn P-TEFb function (23, 28, 49). Thus, it will be important to determine if Tax utilizes LARP7 and/or MEPCE to alter the regulation of P-TEFb in the cell.
It is interesting to compare the activities of the viral transcription activators HTLV-1 Tax and HIV-1 Tat in P-TEFb regulation. Sedore et al. (42) have reported that Tat expression decreases the amount of HMW P-TEFb complex in the cell. Tat competed with HEXIM1 for binding to 7SK RNA, blocked the formation of the P-TEFb/HEXIM1/7SK snRNP complex, and caused a release of P-TEFb from the P-TEFb/HEXIM1/7SK snRNP complex. In a separate study, Barboric et al. (6) reported that Tat could dissociate, and prevent the formation of, the P-TEFb/HEXIM1/7SK snRNP complex. This activity was dependent upon the interaction between Tat and cyclin T1. The dissociation of the HMW P-TEFb complex by Tax described in this paper is through binding of Tax to cyclin T1. Thus, the binding sites for Tat and Tax on cyclin T1 are distinct.
As noted above, in cells the ratio between the active and inactive forms of P-TEFb is tightly regulated (20, 38, 54). Inhibition of transcription or stress-inducing conditions such as treatment with actinomycin D, DRB, or UV irradiation result in the disruption of 7SK snRNP/HEXIM1-bound HMW P-TEFb complex and an increase in the LMW P-TEFb fraction, resulting in an increase of mRNA and protein synthesis (33, 55, 56). The exact mechanism by which the HMW complex is disrupted in the cell is unknown. We have shown that Tax interacts with the cyclin T1 subunit of P-TEFb. Our data suggest that dissociation of 7SK snRNP and HEXIM1 is the result of Tax interaction with cyclin T1. Indeed, a cyclin T1 peptide spanning the Tax binding site inhibits Tax dissociation of the HMW complex. It is possible that the interaction of Tax with cyclin T1 causes a conformational change in the complex, leading to disruption of the interaction of 7SK snRNP and HEXIM1. We have also observed that Brd4, which interacts with a domain of cyclin T1 that overlaps or is in close proximity to the Tax binding domain, is also able to dissociate binding of HEXIM1 and 7SK snRNP to P-TEFb. What regulates the redistribution of P-TEFb is unclear. Our results suggest that Tax competes with 7SK snRNP/HEXIM1 and Brd4 for P-TEFb binding. The Tax/P-TEFb complex is more efficient at transcriptional activation on the viral LTR than the Brd4/P-TEFb complex and thus converts basal to activated transcription. This model is supported by work showing that Myc, RelA, and AIRE depend on P-TEFb for activated transcription (5, 22, 36). Our studies with Tax and Brd4 provide key insight into the regulation of P-TEFb in the cell. It would be of interest to identify other cellular proteins which interact with this domain of cyclin T1 to determine if cellular proteins utilize a similar mechanism for regulating P-TEFb activity.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
We thank the members of the Brady lab for thoughtful discussions.
Footnotes
Published ahead of print on 6 October 2010.
REFERENCES
- 1.Adya, N., and C. Z. Giam. 1995. Distinct regions in human T-cell lymphotropic virus type I tax mediate interactions with activator protein CREB and basal transcription factors. J. Virol. 69:1834-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67-76. [DOI] [PubMed] [Google Scholar]
- 3.Alexandre, C., P. Charnay, and B. Verrier. 1991. Transactivation of Krox-20 and Krox-24 promoters by the HTLV-1 Tax protein through common regulatory elements. Oncogene 6:1851-1857. [PubMed] [Google Scholar]
- 4.Alexandre, C., and B. Verrier. 1991. Four regulatory elements in the human c-fos promoter mediate transactivation by HTLV-1 Tax protein. Oncogene 6:543-551. [PubMed] [Google Scholar]
- 5.Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. [DOI] [PubMed] [Google Scholar]
- 6.Barboric, M., J. H. Yik, N. Czudnochowski, Z. Yang, R. Chen, X. Contreras, M. Geyer, B. Matija Peterlin, and Q. Zhou. 2007. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 35:2003-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bird, G., D. A. Zorio, and D. L. Bentley. 2004. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol. Cell. Biol. 24:8963-8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brauweiler, A., J. E. Garrus, J. C. Reed, and J. K. Nyborg. 1997. Repression of bax gene expression by the HTLV-1 Tax protein: implications for suppression of apoptosis in virally infected cells. Virology 231:135-140. [DOI] [PubMed] [Google Scholar]
- 9.Cho, E. J., T. Takagi, C. R. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, W. K., M. Zhou, M. K. Jang, K. Huang, S. J. Jeong, K. Ozato, and J. N. Brady. 2007. Modulation of the Brd4/P-TEFb interaction by the human T-lymphotropic virus type 1 tax protein. J. Virol. 81:11179-11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cramer, P., A. Srebrow, S. Kadener, S. Werbajh, M. de la Mata, G. Melen, G. Nogues, and A. R. Kornblihtt. 2001. Coordination between transcription and pre-mRNA processing. FEBS Lett. 498:179-182. [DOI] [PubMed] [Google Scholar]
- 12.Easley, R., L. Carpio, I. Guendel, Z. Klase, S. Choi, K. Kehn-Hall, J. N. Brady, and F. Kashanchi. 2010. Human T-lymphotropic virus type 1 transcription and chromatin-remodeling complexes. J. Virol. 84:4755-4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu, T. J., J. Peng, G. Lee, D. H. Price, and O. Flores. 1999. Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription. J. Biol. Chem. 274:34527-34530. [DOI] [PubMed] [Google Scholar]
- 14.Geiger, T. R., N. Sharma, Y. M. Kim, and J. K. Nyborg. 2008. The human T-cell leukemia virus type 1 tax protein confers CBP/p300 recruitment and transcriptional activation properties to phosphorylated CREB. Mol. Cell. Biol. 28:1383-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giebler, H. A., J. E. Loring, K. van Orden, M. A. Colgin, J. E. Garrus, K. W. Escudero, A. Brauweiler, and J. K. Nyborg. 1997. Anchoring of CREB binding protein to the human T-cell leukemia virus type 1 promoter: a molecular mechanism of Tax transactivation. Mol. Cell. Biol. 17:5156-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goren, I., E. Tavor, and A. Honigman. 1999. Gene regulation mediated by interaction between HTLV-1 promoter elements and transcription factors Tax and CREB. Virology 256:303-312. [DOI] [PubMed] [Google Scholar]
- 17.Harrod, R., Y. Tang, C. Nicot, H. S. Lu, A. Vassilev, Y. Nakatani, and C. Z. Giam. 1998. An exposed KID-like domain in human T-cell lymphotropic virus type 1 Tax is responsible for the recruitment of coactivators CBP/p300. Mol. Cell. Biol. 18:5052-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ijichi, S., N. Eiraku, M. Osame, S. Izumo, R. Kubota, I. Maruyama, M. Matsumoto, T. Niimura, and S. Sonoda. 1989. Activated T lymphocytes in cerebrospinal fluid of patients with HTLV-I-associated myelopathy (HAM/TSP). J. Neuroimmunol. 25:251-254. [DOI] [PubMed] [Google Scholar]
- 19.Izumo, S., F. Umehara, N. Kashio, R. Kubota, E. Sato, and M. Osame. 1997. Neuropathology of HTLV-1-associated myelopathy (HAM/TSP). Leukemia 11(Suppl. 3):82-84. [PubMed] [Google Scholar]
- 20.Jang, M. K., K. Mochizuki, M. Zhou, H. S. Jeong, J. N. Brady, and K. Ozato. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19:523-534. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, H., H. Lu, R. L. Schiltz, C. A. Pise-Masison, V. V. Ogryzko, Y. Nakatani, and J. N. Brady. 1999. PCAF interacts with tax and stimulates tax transactivation in a histone acetyltransferase-independent manner. Mol. Cell. Biol. 19:8136-8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanazawa, S., L. Soucek, G. Evan, T. Okamoto, and B. M. Peterlin. 2003. c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene 22:5707-5711. [DOI] [PubMed] [Google Scholar]
- 23.Krueger, B. J., C. Jeronimo, B. B. Roy, A. Bouchard, C. Barrandon, S. A. Byers, C. E. Searcey, J. J. Cooper, O. Bensaude, E. A. Cohen, B. Coulombe, and D. H. Price. 2008. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 36:2219-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubota, R., Y. Furukawa, S. Izumo, K. Usuku, and M. Osame. 2003. Degenerate specificity of HTLV-1-specific CD8+ T cells during viral replication in patients with HTLV-1-associated myelopathy (HAM/TSP). Blood 101:3074-3081. [DOI] [PubMed] [Google Scholar]
- 25.Kwok, R. P., M. E. Laurance, J. R. Lundblad, P. S. Goldman, H. Shih, L. M. Connor, S. J. Marriott, and R. H. Goodman. 1996. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature 380:642-646. [DOI] [PubMed] [Google Scholar]
- 26.Lenzmeier, B. A., H. A. Giebler, and J. K. Nyborg. 1998. Human T-cell leukemia virus type 1 Tax requires direct access to DNA for recruitment of CREB binding protein to the viral promoter. Mol. Cell. Biol. 18:721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, S., G. Coutinho-Mansfield, D. Wang, S. Pandit, and X. D. Fu. 2008. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 15:819-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markert, A., M. Grimm, J. Martinez, J. Wiesner, A. Meyerhans, O. Meyuhas, A. Sickmann, and U. Fischer. 2008. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 9:569-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuoka, M. 2005. Human T-cell leukemia virus type I (HTLV-I) infection and the onset of adult T-cell leukemia (ATL). Retrovirology 2:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meekings, K. N., J. Leipzig, F. D. Bushman, G. P. Taylor, and C. R. Bangham. 2008. HTLV-1 integration into transcriptionally active genomic regions is associated with proviral expression and with HAM/TSP. PLoS Pathog. 4:e1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michels, A. A., V. T. Nguyen, A. Fraldi, V. Labas, M. Edwards, F. Bonnet, L. Lania, and O. Bensaude. 2003. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell. Biol. 23:4859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munoz, E., and A. Israel. 1995. Activation of NF-kappa B by the Tax protein of HTLV-1. Immunobiology 193:128-136. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 34.Ni, Z., A. Saunders, N. J. Fuda, J. Yao, J. R. Suarez, W. W. Webb, and J. T. Lis. 2008. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol. Cell. Biol. 28:1161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osame, M., M. Nakagawa, F. Umehara, S. Ijichi, T. Moritoyo, I. Higuchi, K. Usuku, K. Arimura, and S. Izumo. 1997. Recent studies on the epidemiology, clinical features and pathogenic mechanisms of HTLV-I associated myelopathy (HAM/TSP) and other diseases associated to HTLV. J. Neurovirol. 3(Suppl. 1):S50-S51. [PubMed] [Google Scholar]
- 36.Oven, I., N. Brdickova, J. Kohoutek, T. Vaupotic, M. Narat, and B. M. Peterlin. 2007. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol. Cell. Biol. 27:8815-8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng, J., Y. Zhu, J. T. Milton, and D. H. Price. 1998. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 12:755-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterlin, B. M., and D. H. Price. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23:297-305. [DOI] [PubMed] [Google Scholar]
- 39.Prelich, G. 2002. RNA polymerase II carboxy-terminal domain kinases: emerging clues to their function. Eukaryot. Cell 1:153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramirez, J. A., and J. K. Nyborg. 2007. Molecular characterization of HTLV-1 Tax interaction with the KIX domain of CBP/p300. J. Mol. Biol. 372:958-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renner, D. B., Y. Yamaguchi, T. Wada, H. Handa, and D. H. Price. 2001. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 276:42601-42609. [DOI] [PubMed] [Google Scholar]
- 42.Sedore, S. C., S. A. Byers, S. Biglione, J. P. Price, W. J. Maury, and D. H. Price. 2007. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 35:4347-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, J. A., E. A. White, M. E. Sowa, M. L. Powell, M. Ottinger, J. W. Harper, and P. M. Howley. 2010. Genome-wide siRNA screen identifies SMCX, EP400, and Brd4 as E2-dependent regulators of human papillomavirus oncogene expression. Proc. Natl. Acad. Sci. U. S. A. 107:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tie, F., N. Adya, W. C. Greene, and C. Z. Giam. 1996. Interaction of the human T-lymphotropic virus type 1 Tax dimer with CREB and the viral 21-base-pair repeat. J. Virol. 70:8368-8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vendel, A. C., S. J. McBryant, and K. J. Lumb. 2003. KIX-mediated assembly of the CBP-CREB-HTLV-1 tax coactivator-activator complex. Biochemistry 42:12481-12487. [DOI] [PubMed] [Google Scholar]
- 46.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, C. H., Y. Yamaguchi, L. R. Benjamin, M. Horvat-Gordon, J. Washinsky, E. Enerly, J. Larsson, A. Lambertsson, H. Handa, and D. Gilmour. 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 17:1402-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, S. Y., A. Y. Lee, S. Y. Hou, J. K. Kemper, H. Erdjument-Bromage, P. Tempst, and C. M. Chiang. 2006. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 20:2383-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xue, Y., Z. Yang, R. Chen, and Q. Zhou. 2010. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 38:360-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamaguchi, Y., N. Inukai, T. Narita, T. Wada, and H. Handa. 2002. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 22:2918-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 52.Yan, J., Q. Li, S. Lievens, J. Tavernier, and J. You. 2010. Abrogation of the Brd4-positive transcription elongation factor B complex by papillomavirus E2 protein contributes to viral oncogene repression. J. Virol. 84:76-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan, J. P., J. E. Garrus, H. A. Giebler, L. A. Stargell, and J. K. Nyborg. 1998. Molecular interactions between the coactivator CBP and the human T-cell leukemia virus Tax protein. J. Mol. Biol. 281:395-400. [DOI] [PubMed] [Google Scholar]
- 54.Yang, Z., J. H. Yik, R. Chen, N. He, M. K. Jang, K. Ozato, and Q. Zhou. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19:535-545. [DOI] [PubMed] [Google Scholar]
- 55.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317-322. [DOI] [PubMed] [Google Scholar]
- 56.Yik, J. H., R. Chen, R. Nishimura, J. L. Jennings, A. J. Link, and Q. Zhou. 2003. Inhibition of P-TEFb (CDK9/cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12:971-982. [DOI] [PubMed] [Google Scholar]
- 57.Yin, M. J., and R. B. Gaynor. 1996. HTLV-1 21 bp repeat sequences facilitate stable association between Tax and CREB to increase CREB binding affinity. J. Mol. Biol. 264:20-31. [DOI] [PubMed] [Google Scholar]
- 58.Yoshida, M., J. Fujisawa, J. Inoue, and M. Seiki. 1986. Mechanism of the gene expression of HTLV-I and its association with ATL. AIDS Res. 2(Suppl. 1):S71-S78. [PubMed] [Google Scholar]
- 59.Zhao, L. J., and C. Z. Giam. 1992. Human T-cell lymphotropic virus type I (HTLV-I) transcriptional activator, Tax, enhances CREB binding to HTLV-I 21-base-pair repeats by protein-protein interaction. Proc. Natl. Acad. Sci. U. S. A. 89:7070-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, M., H. Lu, H. Park, J. Wilson-Chiru, R. Linton, and J. N. Brady. 2006. Tax interacts with P-TEFb in a novel manner to stimulate human T-lymphotropic virus type 1 transcription. J. Virol. 80:4781-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu, W., T. Wada, S. Okabe, T. Taneda, Y. Yamaguchi, and H. Handa. 2007. DSIF contributes to transcriptional activation by DNA-binding activators by preventing pausing during transcription elongation. Nucleic Acids Res. 35:4064-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]