Abstract
The three-dimensional structure of adeno-associated virus (AAV) serotype 6 (AAV6) was determined using cryo-electron microscopy and image reconstruction and using X-ray crystallography to 9.7- and 3.0-Å resolution, respectively. The AAV6 capsid contains a highly conserved, eight-stranded (βB to βI) β-barrel core and large loop regions between the strands which form the capsid surface, as observed in other AAV structures. The loops show conformational variation compared to other AAVs, consistent with previous reports that amino acids in these loop regions are involved in differentiating AAV receptor binding, transduction efficiency, and antigenicity properties. Toward structure-function annotation of AAV6 with respect to its unique dual glycan receptor (heparan sulfate and sialic acid) utilization for cellular recognition, and its enhanced lung epithelial transduction compared to other AAVs, the capsid structure was compared to that of AAV1, which binds sialic acid and differs from AAV6 in only 6 out of 736 amino acids. Five of these residues are located at or close to the icosahedral 3-fold axis of the capsid, thereby identifying this region as imparting important functions, such as receptor attachment and transduction phenotype. Two of the five observed amino acids are located in the capsid interior, suggesting that differential AAV infection properties are also controlled by postentry intracellular events. Density ordered inside the capsid, under the 3-fold axis in a previously reported, conserved AAV DNA binding pocket, was modeled as a nucleotide and a base, further implicating this capsid region in AAV genome recognition and/or stabilization.
Adeno-associated viruses (AAVs) are nonpathogenic single-stranded DNA (ssDNA) parvoviruses that belong to the Dependovirus genus and require helper viruses, such as Adenovirus or Herpesvirus, for lytic infection (4, 8, 22, 67). These viruses package a genome of ∼4.7 kb inside an icosahedral capsid (∼260 Å in diameter) with a triangulation number equal to 1 assembled from a total of 60 copies of their overlapping capsid viral protein (VP) 1 (VP1), VP2, and VP3 in a predicted ratio of 1:1:8/10 (10). The VPs are encoded from a cap open reading frame (ORF). VP3 is 61 kDa and constitutes 90% of the capsid's protein composition. The less abundant VPs, VP1 (87 kDa) and VP2 (73 kDa), share the same C-terminal amino acid sequence with VP3 but have additional N-terminal sequences. A rep ORF codes for four overlapping proteins required for replication and DNA packaging.
To date, more than 100 AAV isolates have been identified (21). Among the human and nonhuman primate AAVs isolated, 12 serotypes (AAV serotype 1 [AAV1] to AAV12) have been described and are classified into six phylogenetic clades on the basis of their VP sequences and antigenic reactivities, with AAV4 and AAV5 considered to be clonal isolates (21). AAV1 and AAV6, which represent clade A, differ by only 6 out of 736 VP1 amino acids (5 amino acids within VP3) and are antigenically cross-reactive. Other clade representatives include AAV2 (clade B), AAV2-AAV3 hybrid (clade C), AAV7 (clade D), AAV8 (clade E), and AAV9 (clade F) (21).
The AAVs are under development as clinical gene delivery vectors (e.g., see references 5, 9, 12, 13, 24, 25, 53, and 61), with AAV2, the prototype member of the genus, being the most extensively studied serotype for this application. AAV2 has been successfully used to treat several disorders, but its broad tissue tropism makes it less effective for tissue-specific applications and the prevalence of preexisting neutralizing antibodies in the human population (11, 43) limits its utilization, especially when readministration is required to achieve a therapeutic outcome. Efforts have thus focused on characterizing the capsid-associated tissue tropism and transduction properties conferred by the capsid of representative serotypes of other clades (21). Outcomes of these studies include the observation that AAV1 and AAV6, for example, transduce liver, muscle, and airway epithelial cells more efficiently (e.g., up to 200-fold) than AAV2 (27, 28, 30). In addition, the six residues (Table 1) that differ between the VPs of AAV1 and AAV6 (a natural recombinant of AAV1 and AAV2 [56]) confer functional disparity between these two viruses. For example, AAV6 shows ∼3-fold higher lung cell epithelium transduction than AAV1 (27), and AAV1 and AAV6 bind terminally sialylated proteoglycans as their primary receptor, whereas AAV6 additionally binds to heparan sulfate (HS) proteoglycans with moderate affinity (70, 71). Therefore, a comparison of the AAV1 and AAV6 serotypes and, in particular, their capsid structures can help pinpoint the capsid regions that confer differences in cellular recognition and tissue transduction.
TABLE 1.
Amino acid differences between AAV1 and AAV6 and their reported mutants
| AAV | Amino acid at positiona: |
Glycan targetb | Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 129 | 418 | 531 | 532 | 584 | 598 | 642 | |||
| AAV1 | L | E | E | D | F | A | N | S | 70 |
| AAV1-E/K | L | E | K | D | F | A | N | HS+ (and S)c | 70 |
| AAV6 | F | D | K | D | L | V | H | HS and S | 70 |
| AAV6.1 | F | D | E | D | L | V | H | HS− (and S)c | 40, 70 |
| AAV6.2 | L | D | K | D | L | V | H | HS (and S)c | 40, 70 |
| AAV6R2 | L | D | E | D | L | V | H | HS− (and S)c | 40 |
| HAE1 | L | E | E | D | L | V | N | (HS− and S)d | 39 |
| HAE2 | L | D | K | D | L | V | N | (HS and S)d | 39 |
| shH10 | F | D | K | N | L | V | N | HS (and S-inde) | 33 |
Mutant residues in boldface have an AAV6 parental original; those underlined have an AAV1 parental origin.
S, sialic acid; HS, heparan sulfate; HS+, HS positive.
The sialic acid binding phenotypes of these mutants were not discussed in the respective publications but are assumed to be still present.
The glycan targets for these mutants were not discussed in this publication; thus, the phenotypes indicated are assumed.
This mutant is sialic acid independent (S-ind) for cellular transduction.
The structures of AAV1 to AAV5 and AAV8 have been determined by X-ray crystallography and/or cryo-electron microscopy and image reconstruction (cryo-EM) (23, 36, 47, 52, 66, 73; unpublished data), and preliminary characterization of crystals has also been reported for AAV1, AAV5, AAV7, and AAV9 (15, 45, 46, 55). The capsid VP structures contain a conserved eight-stranded (βB to βI) β-barrel core and large loop regions between the strands that form the capsid surface. The capsid surface is characterized by depressions at the icosahedral 2-fold axes of symmetry, finger-like projections surrounding the 3-fold axes, and canyon-like depressions surrounding the 5-fold axes. A total of nine variable regions (VRs; VRI to VRIX) were defined when the two most disparate structures, AAV2 and AAV4, were compared (23). The VRs contain amino acids that contribute to slight differences in surface topologies and distinct functional phenotypes, such as in receptor binding, transduction efficiency, and antigenic reactivity (10, 23, 37, 47).
The structure of virus-like particles (VLPs) of AAV6, produced in a baculovirus/Sf9 insect cell expression system, has been determined by two highly complementary approaches, cryo-EM and X-ray crystallography. The AAV6 VP structure contains the general features already described for the AAVs and has conformational differences in the VRs compared to the VRs of other AAVs. The 9.7-Å-resolution cryoreconstructed structure enabled the localization of the C-α positions of five of the six amino acids that differ between highly homologous AAV6 and AAV1 but did not provide information on the positions of the side chains or their orientations. The X-ray crystal structure determined to 3.0-Å resolution enabled us to precisely map the atomic positions of these five residues at or close to the icosahedral 3-fold axes of the capsid. Reported mutagenesis and biochemical studies had functionally annotated the six residues differing between AAV1 and AAV6 with respect to their roles in receptor attachment and differential cellular transduction. Their disposition identifies the 3-fold capsid region as playing essential roles in AAV infection.
MATERIALS AND METHODS
Production and purification of AAV6 VLPs.
A recombinant baculovirus encoding VP2 and VP3 of AAV6 was constructed using the Bac-to-Bac system (Gibco/Invitrogen Corporation), as described previously for AAV1 and AAV5 but with an inserted AAV6 cap ORF. DH10Bac competent cells containing the baculovirus genome were transformed with pFastBac transfer plasmids containing the AAV component insert. Bacmid DNA purified from recombination-positive white colonies was transfected into Sf9 cells using Cellfectin reagent (Gibco/Invitrogen Corporation). At 3 days posttransfection, media containing recombinant AAV6 baculovirus were harvested and plaque assays were conducted to prepare independent plaque isolates. Several individual plaques were propagated to passage one (P1) to evaluate the levels of VP2 and VP3 expression. The clone with the highest level of protein expression was propagated to P2 and then P3 for VLP production.
Sf9 insect cells cultured in Sf900 II serum-free medium (Gibco/Invitrogen Corporation) were infected at a multiplicity of infection of 5.0 PFU/cell with a recombinant AAV6 baculovirus expressing VP2 and VP3 whose titer had been determined (P3 stock). AAV6 VLPs were released from infected cells by three freeze-thaw cycles in lysis buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.2% Triton X-100), with benzonase (Merck, Germany) being added after the second cycle. The sample was clarified by centrifugation at 12,100 × g at 4°C for 15 min. Next, the cell lysate was pelleted through a 20% (wt/vol) sucrose cushion in TNET buffer (25 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.06% Triton X-100) by ultracentrifugation at 149,000 × g at 4°C for 3 h. The pellet from the sucrose cushion was resuspended in TNTM buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.06% Triton X-100, 30 mM MgCl2) overnight at 4°C. The sample was clarified by several rounds of centrifugation at 10,000 × g to remove insoluble material. The clarified sample was loaded onto a sucrose step gradient (5 to 40% [wt/vol]) and spun at 151,000 × g at 4°C for 3 h. A visible blue virus band was extracted from the 20/25% sucrose layer and dialyzed into 20 mM Tris-HCl (pH 7.5), 2 mM MgCl2, and 350 mM NaCl at 4°C. The approximate VLP concentration (in mg/ml) was calculated on the basis of optical density measurements at 280 nm, assuming an extinction coefficient of 1.7. The purity and integrity of the VLPs were monitored using SDS-PAGE and negative-stain electron microscopy (EM), respectively.
Structure determination of AAV6 VLPs by cryo-EM.
Small (3.5-μl) aliquots of purified VLPs (∼10 mg/ml) were vitrified via standard rapid freeze-plunging procedures [2, 17]). Samples were applied to glow-discharged (∼15 s in an Emitech K350 glow-discharge unit) Quantifoil holey grids, blotted for ∼5 s, plunged into liquid ethane, and transferred to liquid nitrogen and then into a precooled Gatan 626 cryospecimen holder. Images of vitrified specimens were recorded in an FEI Sphera microscope (200 keV, equipped with a LaB6 electron gun) at ×50,000 nominal magnification. Eighteen micrographs with minimal astigmatism and specimen drift recorded with the objective lens underfocused by 1.0 to 2.5 μm were digitized at 7-μm intervals (representing 1.4-Å pixels) on a Zeiss SCAI scanner. A total of 1,870 particles were extracted and preprocessed, and their defocus levels were estimated using the RobEM program (http://cryoEM.ucsd.edu/programs.shtm). The AUTO3DEM program (74) was used as previously described (26) to combine these particle images and reconstruct the structure of the AAV6 VLPs. To compensate for the effects of phase reversals in the contrast-transfer function of the images, phase corrections were performed but no amplitude corrections were applied. A conservative Fourier shell correlation of 0.5 was used as the threshold for estimating the resolution of the cryoreconstructed structure (65). The available coordinates for an AAV1 VP3 polyalanine capsid model (amino acids [aa] 218 to 736, VP1 numbering), extracted from a recently determined X-ray crystal structure (L. Govindasamy and M. Agbandje-McKenna, unpublished data), were docked into the AAV6 cryoreconstruction using the Situs package (68) for pseudoatomic model interpretation of the density map. The cryoreconstructed map and docked model were visualized in the Chimera program (54).
Structure determination of AAV6 VLPs by X-ray crystallography.
Crystals were grown from purified VLPs in 100 mM HEPES, pH 7.3, at a concentration of 10.0 mg/ml using the method of hanging-drop vapor diffusion with 60 mM MgCl2 and 100 mM NaCl as additives, 4% polyethylene glycol 6000 as a precipitant, and 25% glycerol as the cryoprotectant. X-ray diffraction data were collected from a single crystal at the Cornell High-Energy Synchrotron Source (CHESS) with a crystal-to-detector distance of 350 mm, an oscillation angle of 0.3° per image, and an exposure time of 50 s per image. The data were indexed, processed, scaled, and reduced using the HKL-2000 package (51) (Table 2). The crystal diffracted X-rays to a 3.0-Å resolution and was determined to be in the rhombohedral crystal system and space group R32 with unit-cell parameters of a equal to 262.6 Å and c equal to 609.9 Å (in the hexagonal setting). The data collection and processing statistics are given in Table 2.
TABLE 2.
Data collection and processing statisticsa
| Data collection | CHESS F1 |
|---|---|
| Wavelength (λ, Å) | 0.917 |
| Space group | R32:H |
| Unit cell parameters (Å) | a = 262.6, c = 609.9 |
| Resolution (Å) | 40.0-3.0 (3.1-3.0) |
| No. of unique reflections | 119,617 (8,285) |
| Completeness (%) | 72.3 (50.5) |
| Average I/sigma | 6.5 (2.5) |
| Rmerge (%) | 15.4 (44.3) |
| Refinement | CNS, version 1.2 |
| No. of atoms (protein/solvent/DNA) | 4,117/12/25 |
| Average B factors (Å2) | 61.87 |
| Rcryst/Rfree (%) | 27.5/28.8 |
| RMSD bonds (Å) and angles (degrees) | 0.009 and 1.48 |
| Ramachandran plot | |
| Most favorably allowed (%) | 92.5 |
| Additionally allowed (%) | 7.5 |
Values in parentheses are for the highest resolution. CNS, crystallography and NMR system (6). Rmerge = Σ|Ihkl − <Ihkl>|/Σ|Ihkl| × 100, where Ihkl is the intensity of an individual hkl reflection and <Ihkl> is the mean intensity for all measured values of this reflection; the summation is over all equivalent intensities. Rcryst = Σ||Fobs| − |Fcalc||/Σ|Fobs| × 100, where Fobs and Fcalc are the amplitudes for the observed and calculated reflections, respectively. Rfree was calculated with the 5% of reflections excluded from the data set during refinement.
The orientation of the AAV6 VLPs in the crystal unit cell was determined using the self-rotation function in the General Lock Rotation Function (GLRF) program (63) with κ equal to 72°, 120°, and 180° in the search for icosahedral 2, 3-, and 5-fold-symmetry axes, respectively, with observed data being in the 10.0- to 5.0-Å resolution range. The crystallographic 2- and 3-fold-symmetry operators were shown to be coincident with icosahedral symmetry operators, resulting in 10 VPs (noncrystallographic symmetry [NCS] operators) per crystallographic asymmetric unit of the R32 space group.
The diffraction data were phased using the molecular replacement method in the AMoRe program (48). The orientation and position of the AAV6 10-mer in the crystal unit cell were determined by cross-rotation and translation searches, respectively, using atomic coordinates for 10 AAV1 VP3 monomers (a 10-mer of residues 218 to 736, VP1 numbering) from a recently determined crystal structure (Govindasamy and Agbandje-McKenna, unpublished), with the amino acids within VP3 that differ from those in AAV6, i.e., E418, E531, F584, A598, and N642, being computationally changed to alanine to eliminate model phase bias. This VP3 10-mer model was oriented and positioned into the AAV6 crystal unit cell, on the basis of the output rotation angles, and positioned at (0, 0, 0), on the basis of space group packing considerations, to calculate a set of initial phases. These phases were improved by refinement using the Crystallography and NMR (nuclear magnetic resonance) system (CNS) package (6, 7), using simulated annealing, energy minimization, atomic position, and temperature factor refinement and with the application of strict NCS operators. A single cycle of electron-density Fourier map (2Fo − Fc and Fo − Fc, in which Fo represents the observed structure factors and Fc represents those calculated from the model) averaging was carried out in CNS, while maintaining strict NCS operators (6, 7), using the experimentally measured amplitudes and the improved phases following each model refinement cycle. The refinement and averaging procedures were alternated with model building, using the Coot program (18), into averaged electron-density maps. To improve the quality of the maps between refinement steps, density map modification was carried out using the Density Modification subroutine in the CCP4 program (3), which performed histogram matching, solvent flattening, and NCS averaging.
Following the building of VP3 common amino acids 218 to 736 into the averaged density maps, two regions of unassigned densities were observed in the Fo − Fc density map (at a contour threshold of 3.0 standard deviations [σ]) in the interior of the capsid. A purine nucleotide (NT) and a pyrimidine base were modeled into these unassigned densities. The resolution of the icosahedrally averaged map (3.0 Å) was insufficient to determine unambiguously the identities of the purine and pyrimidine bases. However, there were no amino side group density observed at the C-2 position of the purine ring and no methyl group density observed at the C-5 position of the pyrimidine ring; thus, the base densities were interpreted as adenosine and cytosine, respectively. An occupancy of 0.5 was determined for each of the two bases by means of an empirical approach in which values (1.0 to 0.3) were applied to achieve temperature factors that were comparable with those of the average VP3 amino acid atoms (Table 2). Finally, 12 solvent molecules were built into remaining averaged positive Fo − Fc density at 3.0σ.
The refined structure had Rcryst (where Rcryst = Σ||Fobs| − |Fcalc||/Σ|Fobs| × 100, where Fobs and Fcalc are the amplitudes for the observed and calculated reflections, respectively) and Rfree values of 27.5% and 28.8%, respectively, and final root mean square deviations (RMSDs) of 0.009 Å for bond lengths and 1.48° for bond angles. These values are within the range of those for structures reported at a comparable resolution, as calculated by the Polygon subroutine (62, 64) in the program PHENIX (Python-based Hierarchical Environment for Integrated Xtallography) (1). The quality of the refined structure was analyzed using the Coot (18) and MOLPROBITY (42) programs. A Ramachandran plot showed values of 92.5% and 7.5% in the most favorably and additionally allowed regions, respectively. The refined VP3 coordinates were retrospectively fitted into the cryoreconstructed AAV6 density map without any main-chain adjustments, as was the case for the AAV1-based model.
Structural comparison of AAV6 VLP to other AAV structures.
The refined AAV6 VP3 structure was compared to the VP3 structures of AAV1 (Govindasamy and Agbandje-McKenna, unpublished), AAV2 (Protein Data Bank [PDB] accession no. 1LP3), AAV3b (PDB accession no. 3KIC), AAV4 (PDB accession no. 2G8G), and AAV8 (PDB accession no. 2QA0) by structural alignment with the secondary structure matching (SSM) subroutine in the Coot program (18). The program superimposes C-α positions and provides information on residues that are structurally equivalent, inserts gaps when the atoms are too far apart to superimpose, and provides the distances (in Å) between the C-α positions. VRs were identified, as previously defined (23), as containing stretches of two or more than two amino acids with C-α positions that are >1.0 Å apart between two serotypes.
Protein structure accession number.
The refined coordinates and structure factors for the AAV6 VP3 structure at 3.0-Å resolution have been deposited in the RCSB PDB with accession number 3OAH.
RESULTS AND DISCUSSION
Structure of AAV6 VLP.
The structure of the AAV6 VLP was determined to 9.7-Å resolution by cryo-EM. The capsid surface of the reconstructed AAV6 structure exhibits the previously defined characteristic features of AAV capsids, with a depression at each 2-fold axis, protrusions surrounding each 3-fold axis, and a canyon-like depression surrounding the channel at each 5-fold axis (Fig. 1A and B). Consistent with the high degree of structural similarity between AAV1 and AAV6, a correlation coefficient of 0.94 was calculated for the fit between the cryoreconstructed density map and a map generated from structure factors calculated from the docked AAV1 crystal structure-based polyalanine model using the Mapman program (31). The docked model provided information on the C-α positions of five of the six amino acids (418, 531, 584, 598, and 642) that differ between AAV1 and AAV6 in the C-terminal regions of VP3 (Fig. 1C and D). The side chain orientations and potential interactions of these residues could not be determined from the cryoreconstructed structure but were obtained from the X-ray crystal structure.
FIG. 1.
AAV6 structure. (A) Surface representation of the AAV6 cryoreconstructed image at 9.7-Å resolution. The capsid surface density is shown as a radially colored, depth-cued image (low to high radii, pink to gray). Selected icosahedral 2-fold (2f), 3-fold (3f), and 5-fold (5f) axes of the capsid are indicated by arrows. (B) Cross-sectioned slab from the cryo-EM density map (gray isosurface) with the docked C-α backbone of the polyalanine model (residues 218 to 736, pink) derived from the AAV1 crystal structure. Dashed arrows indicate the approximate locations of icosahedral axes of symmetry. (C) Coil representation of an AAV6 VP backbone trace (pink) showing the locations of the five amino acids (pink spheres) within VP3 that differ between AAV1 and AAV6. The first and last letters in each residue label refer to AAV6 and AAV1, respectively. (D) A trimer of AAV6 VPs (pink, green, and blue) showing the symmetry-related clustering of the differing residues (spheres) shown in panel C and colored according to the monomer in which they reside. These residues cluster near the icosahedral 3-fold axes in both the interior (residues 418 and 642) and exterior (residues 531, 584, and 598) surfaces of the capsid. The view is approximately down the icosahedral 3-fold axis. Approximate positions of icosahedral 2-, 3-, and 5-fold-symmetry axes of the capsid are depicted as filled ovals, triangles, and pentagons, respectively, in panels C and D. Panels A and B were generated using the Chimera program (53), and panels C and D were generated using the PyMol program (14).
The AAV6 crystal structure was determined to 3.0-Å resolution (Table 2). The refinement and molecular geometry statistics are consistent with those reported for other members of the Parvoviridae as well as structures determined for other virus families at comparable resolution, as reported on the VIPERdb website (http://viperdb.scripps.edu/). As has been previously reported for other AAV capsid structures, the N-terminal region of VP2 and the first 15 amino acids of VP3 were unresolved in both the cryo-EM and X-ray structures (23, 36, 37, 47, 52, 66, 73). In all of these AAV structures, only the overlapping C-terminal VP (∼520-aa) region common to the VP1, VP2, and VP3 sequence is unambiguously resolved. The lack of ordered density for the VP N termini in the AAV6 VLPs used in this study likely results from the low copy number of VP2 (∼5 copies) and the fact that the VP3 N terminus likely adopts different conformations, two factors which are incompatible with the icosahedral symmetry imposed during the structure determination procedures. The C-terminal 519 amino acids (amino acids 218 to 736, VP1 numbering) common to VP2 and VP3 (hereafter referred to as VP3) were unambiguously assigned in the averaged AAV6 electron-density map (Fig. 2A and B). This same stretch of amino acids was fitted into the cryoreconstructed density map (Fig. 1B to D).
FIG. 2.
Crystal structure of AAV6. (A and B) Sections of the 2Fo − Fc electron-density map (gray mesh) of AAV6, contoured at 1.0σ, for two of the residues (residue 584 in panel A and residue 642 in panel B) that differ between AAV6 and AAV1. The AAV1 (purple) and AAV6 (atom type) coordinates are shown in stick form. (C) Ribbon diagram representation of AAV6 VP3 monomer (ordered residues 218 to 736), with labels highlighting the conserved β-barrel core motif (βBIDG-βCHEF, pink) and the αA helix (blue). Loop regions (orange) between the core β strands also contain small stretches of β-strand structure (pink). The DE and HI loops are labeled. Approximate positions of icosahedral 2-, 3-, and 5-fold-symmetry axes in the capsid are depicted with filled symbols, as described in the legend to Fig. 1. Positions of the purine (Pu) and cytosine (Cyt) NT bases that are ordered in the averaged density map are shown and labeled. (D) The conserved nucleotide binding pocket, showing the ordered densities interpreted as a deoxyadenylate and cytosine (labeled), close to the 3-fold axis (see panel C). The deoxyadenylate and cytosine molecules are shown inside the Fo − Fc difference density map (dark gray mesh) and contoured at 3.0σ. AAV6 amino acids within 2.4 Å to 5.0 Å of the ordered density are shown and labeled; residue D609 has been omitted for clarity. (E) Comparison of nucleotide binding pocket in AAV3 (yellow), AAV4 (black), and AAV6 (pink) crystal structures. This region (structure and DNA) is also conserved in AAV8 (data not shown). AAV6 amino acid positions are labeled. This figure was generated using the PyMol program (14).
The structural topology of the AAV6 VP3 is similar to that previously reported for other parvoviruses, with a conserved eight-stranded β-barrel (βBIDG-βCHEF sheets) core that forms the contiguous capsid and an α helix (αA, residues 290 to 302, VP1 numbering) located on the wall of the depression surrounding the icosahedral 2-fold axes (23, 37, 47, 73) (Fig. 2C). A tubular density feature consistent with helix αA was also observed in the cryoreconstructed density map (data not shown). The capsid surface is formed by loop structures inserted between the β strands (Fig. 2C). These loops also contain small stretches of β structure (Fig. 2C). Comparisons of the previously determined AAV crystal structures identified regions of variable conformation (VRI to VRIX) in these loops (23). The VRs are spread throughout the VP3 structure but are clustered on the capsid surface (23). These regions also differ between AAV6 and the other AAVs, in particular, AAV4 (Fig. 3A; see Table S1 in the supplemental material). These VRs contribute to phenotypic differences, such as receptor attachment, transduction efficiency, and antigenic reactivity, between the AAVs (10, 23, 37, 47, 52, 66, 73). The conserved DE and HI loops (Fig. 2C and 3A), between the βDE and βHI strands, respectively, play essential structural and functional roles in the life cycle of the AAVs and other parvoviruses. The DE loops in five, symmetry-related monomers interact and form the channel at the 5-fold axis through which genomic ssDNA is postulated to be packaged (35). This is also where a phospholipase A2 (PLA2) domain, located within the VP1 unique N termini, is proposed to be externalized during cellular trafficking (reviewed in reference 10) (35). Structural variation is observed at the top of the DE loop (Fig. 3A, VRII), consistent with dynamics which might be required for genome packaging or the PLA2 externalization (34, 35). The HI loop lines the floor of the depression around the icosahedral 5-fold axes and is implicated in capsid assembly as well as capsid dynamics associated with receptor attachment (16, 38).
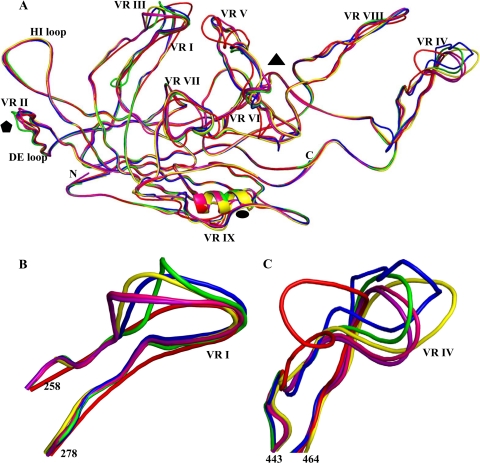
FIG. 3.
Comparison of available AAV crystal structures. (A) Superposition of the VP3 monomer structures of AAV1 (purple), AAV2 (blue), AAV3 (yellow), AAV4 (red), AAV6 (pink), and AAV8 (green). Common variable regions VRI to VRIX (as defined in reference 23) are labeled with Roman numerals. The DE and HI loops are labeled. Approximate positions of icosahedral 2-, 3-, and 5-fold axes are depicted as described in the legend to Fig. 1. (B and C) Close-up views of VRI and VRIV, respectively. This figure was generated using the PyMol program (14).
In addition to the VP3 structure and solvent molecules, densities consistent with a purine NT and a pyrimidine base were observed in the AAV6 VLP capsid structure, despite the fact that these particles should be empty (devoid of DNA). The purine base, assigned as an adenine due to the lack of a C-2 amino group density, is located in a conserved DNA binding region and occupies the same position as the bases previously reported in the crystal structures of AAV3, AAV4, and AAV8 (23, 37, 47) (Fig. 2D and E; the structure of AAV8 is not shown). The conserved binding region contains amino acids E417, V419, P420, D609, H630, P631, and S632, which are capable of forming polar and hydrophobic interactions with the NT and sandwich the bases between the two prolines (Fig. 2D and E; residue D609 is not shown). However, in AAV6, the orientation of the base as modeled into the Fo − Fc map is rotated 180° about the plane of the base relative to the bases built into the other AAV structures due to the position of the density interpreted as the deoxyribose sugar (see Fig. S1A in the supplemental material). In the 2Fo − Fc Fourier map (calculated before initial NT model building), a dual position of the density interpretable as the deoxyribose was observed, whereas the densities interpretable as the base and phosphate groups overlap with positions observed for the other AAV NTs (see Fig. S1B in the supplemental material). The dual orientation of the sugar portion of this ordered NT suggests that both orientations can occur with equal probability. The overlapping position of the base within the conserved binding pocket suggests that its interactions with the surrounding amino acids dictate the ordering of this nucleotide. The observation of a single sugar conformation in the Fo − Fc Fourier density map (see Fig. S1A in the supplemental material) suggests a higher propensity for the orientation modeled inside the AAV6 VLPs.
Five angstroms from the purine base, a second base, cytosine (assigned on the basis of the absence of methyl group density at the C-5 position of the pyrimidine ring), is ordered in a position proximal to the 3-fold axis (Fig. 2C). No sugar or phosphate groups were observed for this base, which interacts with the main chain of H630. This histidine is conserved in representative clade members of the AAVs (Fig. 2E), but despite this conservation, the density for this base was not reported in the AAV3 crystal structure (37), nor was it observed in our structures of AAV4 and AAV8 (23, 47). There is no indication that this cytosine base and the purine NT are components of a single DNA chain.
Significantly, like the AAV6 structure, that of AAV8 was also determined from baculovirus/Sf9-expressed VLPs produced in the absence of the rep ORF. These observations support a proposal that the AAVs are able to package fragments of host cellular DNA in the absence of Rep proteins (38). A similar packaging of cellular genomic material is commonly observed for RNA viruses expressed in a heterologous system, most likely due to a requirement for interaction with nucleic acid for capsid assembly (20). For bacteriophage φX174, a small ssDNA virus that packages a genome similar in size to those of parvoviruses, in virions, subgenomic pieces of DNA are also observed in empty capsids and are also likely required to facilitate capsid assembly (44). DNA packaging is not required for the assembly of autonomous parvovirus VLPs, as reported for minute virus of mice (29, 34), but the AAV observations suggest that it may play a role for the dependoviruses, though this remains to be verified.
The lower occupancy (0.5) of the AAV6 bases, relative to the surrounding protein (as was reported for AAV8 [47]), is consistent with the expected lack of icosahedral symmetry for NTs ordered within VLPs. Indeed, only a single copy of the ssDNA genome is packaged into wild-type virions, and thus, the same NT/base cannot be ordered in all 60 sites within the capsid, unless it is part of a conserved DNA sequence repeated 60 times and forming specific interactions with the capsid. Such a conserved DNA sequence has not been reported for the AAVs. Given occupancy of less than one, the strict NCS utilized for electron-density averaging during the structure determination would be expected to result in reduced sigma for NT/base density at each averaging cycle and eventual loss of signal. Thus, the unexpected observation of ordered DNA density inside AAV6 and other AAVs suggests the presence of a common DNA recognition motif inside the capsid directly under the 3-fold axes (Fig. 2C). The fact that the recognition site amino acids are conserved in most AAV sequences and all the structures determined to date suggests an important function for this DNA interaction in the life cycle of the viruses, such as a role in capsid stabilization or assembly.
Comparison of the AAV6 structure to structures of other AAVs pinpoints capsid regions that control receptor attachment and transduction efficiency.
Superposition of the AAV6 structure with those available for AAV1 to AAV4 and AAV8 using the SSM application in the Coot program (18) shows differences (stretches of two or more amino acids with C-α positions that are >1.0 Å apart between two serotypes [23]) with (i) AAV2 at VRI, VRII, VRIV, VRV, and VRVII; (ii) AAV3 at VRI, VRII, VRIV, VRVI, VRVII, and VRIX; (iii) AAV4 at VRI to VRIX; and (iv) AAV8 at VRI, VRII, VRIV, VRV, and VRVII (Fig. 3A; see Table S1 in the supplemental material). Variable regions I and IV were commonly divergent in conformation between AAV6 and these four AAVs (Table 3; Fig. 3B and C). The amino acids that form these two surface loops are also highly divergent between the representative members of the AAV clades (52). AAV1 and AAV6 are 99% identical and superimpose with an RMSD of 0.33 Å, and they exhibit the lowest difference between the C-α positions of residues in their VRIV regions (Table 3; Fig. 3A and C). AAV6 is the most structurally diverse from AAV4, with which it shares the lowest degree of sequence homology (59% compared to 80 to 99% with the other AAVs; Table 3). For example, the C-α positions of amino acids in VRIV differ from 1.0 to 14.6 Å, though the structures superimpose with an overall RMSD of 0.94 Å. AAV6 and AAV2 (83% identical) superimpose with an RMSD of 0.67 Å, and the C-α positions in VRIV differ between 1.3 and 4.9 Å (Table 3).
TABLE 3.
RMSD in C-α position between AAV6 and the available AAV serotype crystal structures overall and for VRI and VRIV
| AAV serotype | % identity | VR C-α distance ranges (Å) |
||
|---|---|---|---|---|
| Overall | VRI | VRIV | ||
| 1 | 99 | 0.33 | 1.3-1.8 | |
| 2 | 83 | 0.67 | 1.2-5.7 | 1.3-4.9 |
| 3 | 86 | 0.57 | 1.7-5.8 | 1.3-6.4 |
| 4 | 59 | 0.94 | 0.9-6.8 | 1.0-14.6 |
| 8 | 80 | 0.57 | 1.3-7.4 | 1.4-2.4 |
The AAV VRs cluster on the capsid surface in the raised regions between the icosahedral 2- and 5-fold axes (VRs I, III, and IX) and on the wall (VRs VI and VII) and top (VRs IV, V, and VIII) of the protrusions surrounding the icosahedral 3-fold axes (23). Significantly, VRI and VRIV (Fig. 3B and C) have been shown to play a role in AAV tissue transduction and antigenic recognition (41, 59). These reports suggest that structural heterogeneity, in addition to sequence variation, confers these capsid-associated functions. The observation that VRIV adopts slightly different conformations in AAV1 and AAV6 may be related to this loop being the least-ordered region in the AAV6 structure. Atoms in the amino acids at the top of the loop exhibit high-temperature factors, consistent with high thermal motion and minor conformation variation compared to those for AAV1.
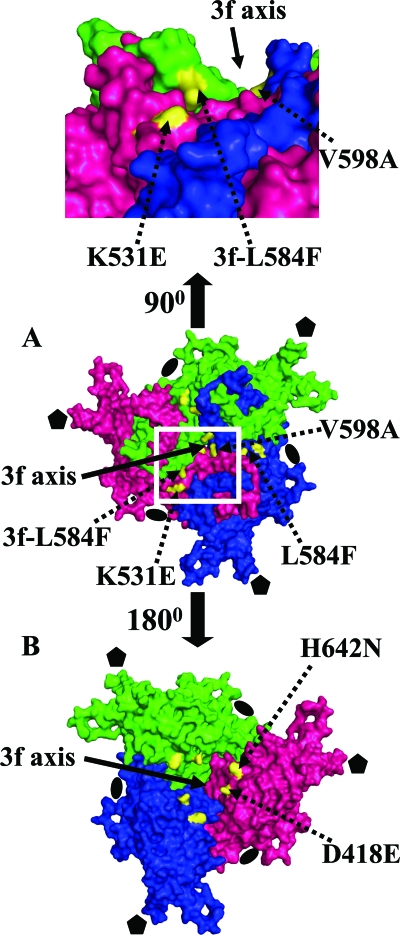
To obtain a more detailed analysis of the structural determinants that dictate differences in receptor attachment and tissue transduction in the highly homologous AAV1 and AAV6, we superimposed the AAV1 crystal structure (Govindasamy and Agbandje-McKenna, unpublished) onto the refined AAV6 structure in the electron-density map. This enabled the visualization of the positions of five of six amino acids (AAV1 and AAV6 amino acids E418D, E531K, F584L, A598V, and N642H) that differ between the AAV1 and AAV6 VPs (F584L and N642H are shown in Fig. 2A and B, respectively). Amino acid 129 (VP1 numbering) in the VP1 unique region was not present in the VP2/VP3 VLP construct used for this structure determination. Three of the ordered residues (531, 584, and 598) are located on the capsid surface, at (V598) or close to (K531 and L584) the icosahedral 3-fold-symmetry axes (Fig. 4A), whereas D418 and H642 are located on the interior surface of the capsid, below the same capsid region (Fig. 4B). Residues D418 and H642 are located in structurally conserved VP regions, whereas surface residue 531 is located in VRVI, residue 584 is located in VRVIII, and residue 598 is close to VRVIII. The localization of these five amino acids at or surrounding the icosahedral 3-fold axes, with K531 being near the icosahedral 2-fold axes in VRVI, highlights the importance of these capsid regions in AAV biology with respect to receptor attachment and transduction efficiency. The surface-exposed amino acid differences indicate potential sites for conferring the differential receptor recognition and transduction properties of AAV1 and AAV6 that are associated with entry or preuncoating interactions. The residues on the inside, with 418 in the vicinity of the DNA binding region (Fig. 4D) are unlikely to play a role in receptor attachment but could be involved in postentry/uncoating events that affect transduction efficiency.
FIG. 4.
Locations of amino acid differences in AAV1/AAV6 capsids. (A) Surface representation of an AAV6 VP3 trimer viewed from outside approximately along a 3-fold axis (middle) with the white boxed section rotated by 90° (top). The monomers are colored pink (reference), green (3-fold), and blue (3-fold) (as in Fig. 1B), with the differing AAV1/AAV6 amino acids colored in yellow. Residues K531E, L584F (3f-L584 is from a 3-fold related monomer), and V598A (first letter, AAV6; second letter, AAV1) are located on the capsid surface. The panel on the top shows the close proximity of residues 531 and 584 at the base of the 3-fold protrusions facing the icosahedral 2-fold axis. (B) Same as panel A, but rotated 180° to show the location of residues D418E and H642N on the interior surface of the capsid. The approximate position of the 3-fold axis (3f axis) is indicated with a solid arrow in all three panels. Approximate positions of icosahedral and 2- and 5-fold symmetry axes on the capsid are depicted as described in the legend to Fig. 1. This figure was generated using the PyMol program (14).
With respect to receptor attachment, residue K531 in AAV6 (E531 in AAV1) has been reported to be important for its HS binding properties, and an E531K mutant of AAV1 shows strong affinity for HS and also confers liver cell transduction (69), identifying a position at the base of the protrusions and close to the depression at the icosahedral 2-fold axes (Fig. 4A) that confers this phenotype. None of the mutations in AAV1 converting the remaining five amino acids which differ from those in AAV6 to the type found in the latter virus conferred an HS binding phenotype (70). A novel primate AAV variant, AAV(VR-942), which also uses HS as a primary receptor, contains a K528 residue that is predicted to be structurally equivalent to the AAV6 K531 residue (57) (Table 4). AAV2 also binds HS (60) but lacks this basic residue and, instead, utilizes two critical residues, R585 and R588, along with R484, R487, K527, and K532 (minor contributors) (AAV2 VP1 numbering) (Table 4) for this interaction (32, 38, 49, 50, 60). Except for R487, these residues form a basic footprint on the surface (Fig. 5B) of the AAV2 capsid on the inner face of the protrusions surrounding the 3-fold axes. Interestingly, AAV1 and AAV6 contain basic residues R485, R488, K528, and K533 (equivalent to R484, R487, K527, and K532, respectively, in AAV2) in the equivalent region of the mapped AAV2 HS binding site, plus R576 and H597 (Fig. 5A, B, and D). A basic region is created on the AAV1 and AAV6 capsid surface by residues R485, R576, and H597 close to the 3-fold axis and is missing in AAV2, which contains R484, Q575, and N596 at the structurally equivalent positions (Fig. 5A, B, and D). However, a role in HS binding has not been reported for R576 and H597. On the other hand, in AAV6, K531 is located adjacent to R488 (equivalent to AAV2 R487, but is now surface exposed), K528, and K533, which creates a second continuous basic patch on the capsid surface, whereas in AAV1, E531 creates a gap in this patch (Fig. 5A and D). Hence, this continuous, basic region is likely sufficient for and the determinant of HS binding by AAV6. AAV5, which binds sialic acid, is missing all these basic residues, except for R471, which is equivalent to AAV6 R485 (Table 4).
TABLE 4.
Comparison of residues reported to be involved in heparan sulfate and sialic acid binding for AAV6, AAV1, AAV2, AAV5, and AAV-VR942 and the amino acids at equivalent positions in each virus
| Glycan target | AAV serotype | Residue at position (AAV6 numbering): |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 485 | 488 | 528 | 531 | 533 | 586 | 589 | 592 | ||
| HS/Sa | 6 | R | R | K | K | K | S | T | A |
| S | 1 | R (485)b | R (488) | K (528) | E (531) | K (533) | S (586) | T (589) | A (592) |
| HS | 2 | R (484) | R (487) | K (527) | E (530) | K (532) | R (585) | R (588) | A (591) |
| S | 5 | R (471) | G (474) | L (515) | S (518) | N (519) | S (575) | T (578) | A (581) |
| HS | VR942 | R (482) | R (485) | K (525) | K (528) | K (530) | N (583) | A (586) | T (589) |
HS, heparan sulfate; S, sialic acid.
Numbers in parentheses are based on VP1 numbering for the respective serotypes.
FIG. 5.
Comparison of AAV surface residues. Schematic Roadmap projections (72) of surface residues in a portion of the icosahedral asymmetric unit for crystal structures of AAV1 (A), AAV2 (B), AAV5 (C), and AAV6 (D) are shown. The area occupied by each amino acid residue correlates to surface exposure when the capsid is viewed down an icosahedral 2-fold axis. The boundary for each residue is shown in black, and the colors correspond to acidic (red), basic (blue), polar (yellow), and hydrophobic (green) residues. Dashed outlines highlight regions proposed to play a role in glycan binding by the respective serotypes. Residues are labeled by type and number. The icosahedral 3-fold axis is depicted by the filled triangle.
In addition to these basic residues, the juxtaposition of acidic residues on the capsid surface and their interactions with amino acids in the vicinity of the mapped basic HS binding residues appears to be important for the binding of this glycan by AAV serotypes. Mutation of an acidic residue, D532 to N532, adjacent to K533 on the AAV6 capsid surface (Fig. 5D and 6A), in an AAV variant derived by directed evolution from AAV libraries, shH10, was reported to confer HS binding dependence and sialic acid binding independence (Table 1) for cellular transduction by the variant (33). In contrast, though wild-type AAV6 binds HS, it can transduce cells in the absence of HS but not in the absence of sialic acid (33, 58, 70). Residue D532 is predicted to stabilize the surface loop containing K531 by means of electrostatic interactions with H527 and D562 (Fig. 6A), which are likely to be disrupted if the acid group at position 532 is lost. This loop also contains K528 and K533, which flank K531 on the capsid surface, as discussed above (Fig. 5D and 6A). The D532N mutation reduces the negative charge on the capsid and likely disrupts the D532-H527-D562 interaction; consequently, it could alter the conformation of this loop and thus the cellular interactions of constituent residues. The predicted loop-stabilizing interaction is conserved in HS binding AAV2 through the interactions of E531-H526-D561 and also E563 (Fig. 6B); thus, the stabilization is expected to be tighter in this virus. The side chain of AAV6 residue E564, with a C-α position equivalent to AAV2 E563, adopts a different orientation and does not participate in the stabilizing interactions (Fig. 6C). The shH10 mutant has improved HS binding compared to that of wild-type AAV6 and exhibits an improved transduction phenotype. AAV6, on the other hand, binds HS more weakly than AAV2 but also exhibits better transduction properties. Thus, binding affinity alone does not control transduction efficiency (33). Also, improved HS binding of the AAV6 D532N mutant nullified the virus's need for sialic acid, although it still transduced cells that contain sialic acid. These observations highlight the complex nature of cellular interactions that control cellular transduction mechanisms.
FIG. 6.
Stabilizing interactions for the K531 loop. (A) Residues D532, H527, and D562 that form electrostatic interactions at the base of the loop containing basic residues K528, K531, and K533 are shown along with neighboring residues R485 and R488, equivalent to R484 and R487, respectively, involved in HS binding by AAV2. 3f-L584 is contributed from a 3-fold (3f) related VP3 monomer. The residues (in stick form) are colored according to atom type: carbon, yellow; nitrogen, blue; and oxygen, red. Dashed lines indicate the distance between interacting residues. Disruption of the D532-H527-D562 interaction by a D532N mutation is predicted to alter the conformation of the basic loop and in turn could alter AAV6 HS binding properties. (B) AAV2 residues E531, H526, D561, and E563 that form electrostatic interactions which stabilize an equivalent surface loop in this serotype containing residues K527, E530, and K532 and the neighboring amino acids, R484 and R487, involved in HS binding. Mutation of D561 and E563 to alanine disrupts HS binding in AAV2. 3f-L583 is contributed from a 3f VP3 monomer. Residues are colored as in panel A. (C) A superimposition of the residues shown in panels A and B with the AAV6 amino acids (labeled in black) shown according to atom type, as described for panels A and B, and those for AAV2 are colored blue (labeled in blue). This figure was generated using the PyMol program (14).
Further support for the role of acidic residues in HS binding was reported by Wu et al., who found that alanine scanning mutagenesis of acidic AAV2 residues 561-DEEE-564 to 561-AAAA-564 resulted in a noninfectious HS-negative (HS−) mutant (69). AAV6 residue D562 is not on the capsid surface but is structurally equivalent to AAV2 D561, which along with E563 and E531 participates in interactions which stabilize the basic residues involved in AAV HS binding (Fig. 6C), as discussed above. A disruption of the D561 and E563 interactions with neighboring residues, which is predicted to occur when these acidic residues are mutated to alanine in AAV2, is likely involved in the HS− phenotype of the 561-AAAA-564 mutant. Interestingly, AAV2 E531, D561, and E563 are contained in two highly conserved acidic stretches of amino acids in the representative AAV clade members, with the exception of clonal isolates AAV4 and AAV5 (52). The above observations suggest that their interactions are important for stabilizing the configuration of HS binding regions on the AAV2 capsid as well as the AAV6 capsid.
The region of the AAV capsid required for interaction with sialic acid has not been structurally mapped, but mutagenesis studies with AAV5 suggest the involvement of A581 at the icosahedral 3-fold axes (19) (Fig. 5C). An alanine is conserved at the equivalent positions in AAV1, AAV2, and AAV6 (A592, A591, and A592, respectively) (Table 4; Fig. 5A to D). Of note, the region at or immediately adjacent to the icosahedral 3-fold axis is hydrophobic in AAV1, AAV5, and AAV6, which bind sialic acid, and polar in AAV2, which does not (Fig. 5). Thus, if this is a conserved, sialic acid recognition site among the AAVs, the V598A difference between AAV6 and AAV1, which, along with V582 and A592, forms a continuous hydrophobic surface at the 3-fold axes (Fig. 5A and D), may be involved in the sialic acid binding interaction of both viruses. Both viruses are reported to have common sialic acid linkage recognition (71), and thus, if residue 598 exhibits a serotype-specific phenotype in transduction, it is unlikely to be due to this interaction.
Efforts to engineer AAV variants with improved/tissue-specific transduction properties have led to chimeric AAV1 and AAV6 vectors that show differential lung epithelial cell transduction efficiency dependent on which residues in the two serotypes are located at VP1 unique position 129 and within the common VP3 sequence (39, 40). The AAV6 F129L mutation (AAV6.2 in Table 1) confers 2-fold better transduction in airway epithelium (40) compared to the parental serotype and AAV6 K531E (AAV6.1 in Table 1), which eliminates HS binding, confers an AAV1 transduction phenotype that is reduced compared to that of the parental AAV6. This observation suggests that amino acids at both the 129 and 531 positions affect cellular transduction. Residue 129, located in the VP1 unique region, is part of a PLA2 domain in the parvoviruses that is predicted to be located inside the assembled capsid but that later becomes externalized through the 5-fold channel during capsid trafficking through the endocytic pathway. This is purported to aid endosomal escape for nuclear localization and subsequent genome replication (35). Residue 531, as discussed above, facilitates HS binding in AAV6. Thus, residues 129 and 531 are likely involved in postentry events and receptor recognition, respectively. Consistent with this suggestion, a mutant containing F129L and K531E (Table 1, AAV6R2) had the reduced transduction phenotype of the virus with the single K531E mutation (40), indicating that the K531E mutation functions early in infection, prior to the step affected by amino acid F129L.
Li et al. used directed evolution from an AAV library to identify chimeric human airway epithelia (HAE) transducing vectors, HAE-1 and HAE-2 (Table 1), which contain mostly AAV1 and AAV6 sequences and which have improved transduction efficiency relative to that of the parental serotypes (39). HAE-1 contains AAV1 residues 1 to 583/641 to 736 and AAV6 residues 584 to 640, and HAE-2 contains AAV9 residues 1 to 30/104 to 193, AAV1/AAV6 residues 31 to 103, AAV6 residues 194 to 641, and AAV1 residues 642 to 736. The reported transduction efficiencies for these viruses compared to the those of the parental serotypes were in the order AAV1/AAV9 < AAV6 < HAE-1 < HAE-2, with HAE-1 and HAE-2 showing ∼3- to 4-fold and ∼2-fold improved transduction compared to that of AAV1 and AAV6, respectively. HAE-1 contains AAV1 residue L129 (not observed in the crystal structures), capsid surface residue E531, and interior residues E418 and N642, with AAV6 contributing surface residues L584 and V598 (Table 1). Thus, L584 and V598 confer the ∼3-fold improvement in HAE-1 transduction compared to that of AAV1. The improvement in transduction relative to that in AAV6 could be due to the L129 from AAV1, as discussed above. HAE-2 contains the equivalent of AAV1 L129 (contributed from AAV9) and interior residue N642 from AAV1 and AAV6 capsid surface residues, K531, L584, and V598, as well as interior residue D418 from AAV6 (Table 1). For this vector, the AAV6 K531 residue likely combines with the L129, L584, and V598 residues of HAE-1 to achieve the further improvement in transduction compared to that of the parental viruses and HAE-1. Both chimeras were observed to bind equally to the apical surface of HAE, suggesting that their difference in transduction was postentry, consistent with an intracellular step in the viral life cycle at which L129 is critical. A functional role for the interior residues, 418 and 642, is yet to be defined.
In summary, this comparative analysis of AAV1 and AAV6 highlights key AAV residues that control host interactions, including receptor recognition and attachment as well as postentry events, which enable successful infection and improved cellular transduction. These results should facilitate further molecular characterization and manipulation of AAV vectors for improved tissue-specific targeting.
Supplementary Material
Acknowledgments
We thank the staff at CHESS, especially Katherine Dedrick, for assistance in obtaining beam time; and Bill Miller, Chae Un Kim, David J. Schuller, Irina Kriksunov, Marian D. Szebenyi, Mike Cook, Scott Smith, and Ulrich Englich for help during X-ray diffraction data collection. We also thank Kim Van Vliet for careful reading of the manuscript and the Department of Cell Biology and Anatomy, University of Florida, for providing a JEOL JEM-100CX II electron microscope for sample visualization.
CHESS is supported by NSF grant DMR-0225180 and NIH/NCRR grant RR-01646. This project was funded by NIH projects R01 GM082946 (parent project and ARRA funding; to M.A.-M. and R.M.), R01 AI072176-01 (to M.A.-M. and R.J.S.), R37 GM-33050 (to T.S.B.), and 1S10 RR020016 (to T.S.B.); by NIH NRSA fellowship F32A1078624 (to K.N.P.); and by support from the University of California, San Diego (UCSD), and the Agouron Foundation (to T.S.B.) to establish cryo-EM facilities at UCSD.
Footnotes
Published ahead of print on 22 September 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adams, P. D., R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter, and T. C. Terwilliger. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948-1954. [DOI] [PubMed] [Google Scholar]
- 2.Adrian, M., J. Dubochet, J. Lepault, and A. W. McDowall. 1984. Cryo-electron microscopy of viruses. Nature 308:32-36. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 4.Atchison, R. W., B. C. Casto, and W. M. Hammon. 1965. Adenovirus-associated defective virus particles. Science 149:754-756. [DOI] [PubMed] [Google Scholar]
- 5.Bankiewicz, K. S., J. Forsayeth, J. L. Eberling, R. Sanchez-Pernaute, P. Pivirotto, J. Bringas, P. Herscovitch, R. E. Carson, W. Eckelman, B. Reutter, and J. Cunningham. 2006. Long-term clinical improvement in MPTP-lesioned primates after gene therapy with AAV-hAADC. Mol. Ther. 14:564-570. [DOI] [PubMed] [Google Scholar]
- 6.Brunger, A. T. 2007. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2:2728-2733. [DOI] [PubMed] [Google Scholar]
- 7.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 8.Buller, R. M., J. E. Janik, E. D. Sebring, and J. A. Rose. 1981. Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. J. Virol. 40:241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter, B. J. 2005. Adeno-associated virus vectors in clinical trials. Hum. Gene Ther. 16:541-550. [DOI] [PubMed] [Google Scholar]
- 10.Chapman, M. S., and M. Agbandje-McKenna. 2006. Atomic structure of viral particles, p 109-123. In M. E. Bloom, S. F. Cotmore, R. M. Linden, C. R. Parrish, and J. R. Kerr (ed.), Parvoviruses. Edward Arnold, Ltd., London, United Kingdom.
- 11.Chirmule, N., K. Propert, S. Magosin, Y. Qian, R. Qian, and J. Wilson. 1999. Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther. 6:1574-1583. [DOI] [PubMed] [Google Scholar]
- 12.Cideciyan, A. V. 2010. Leber congenital amaurosis due to RPE65 mutations and its treatment with gene therapy. Prog. Retin. Eye Res. 29:398-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coura Rdos, S., and N. B. Nardi. 2007. The state of the art of adeno-associated virus-based vectors in gene therapy. Virol. J. 4:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- 15.DiMattia, M., L. Govindasamy, H. C. Levy, B. Gurda-Whitaker, A. Kalina, E. Kohlbrenner, J. A. Chiorini, R. McKenna, N. Muzyczka, S. Zolotukhin, and M. Agbandje-McKenna. 2005. Production, purification, crystallization and preliminary X-ray structural studies of adeno-associated virus serotype 5. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 61:917-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiPrimio, N., A. Asokan, L. Govindasamy, M. Agbandje-McKenna, and R. J. Samulski. 2008. Surface loop dynamics in adeno-associated virus capsid assembly. J. Virol. 82:5178-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubochet, J., M. Adrian, J. J. Chang, J. C. Homo, J. Lepault, A. W. McDowall, and P. Schultz. 1988. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21:129-228. [DOI] [PubMed] [Google Scholar]
- 18.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 19.Excoffon, K. J., J. T. Koerber, D. D. Dickey, M. Murtha, S. Keshavjee, B. K. Kaspar, J. Zabner, and D. V. Schaffer. 2009. Directed evolution of adeno-associated virus to an infectious respiratory virus. Proc. Natl. Acad. Sci. U. S. A. 106:3865-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher, A. J., B. R. McKinney, A. Schneemann, R. R. Rueckert, and J. E. Johnson. 1993. Crystallization of viruslike particle assembled from flock house virus coat protein expressed in a baculovirus system. J. Virol. 67:2950-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, G., L. H. Vandenberghe, M. R. Alvira, Y. Lu, R. Calcedo, X. Zhou, and J. M. Wilson. 2004. Clades of adeno-associated viruses are widely disseminated in human tissues. J. Virol. 78:6381-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geoffroy, M. C., and A. Salvetti. 2005. Helper functions required for wild type and recombinant adeno-associated virus growth. Curr. Gene Ther. 5:265-271. [DOI] [PubMed] [Google Scholar]
- 23.Govindasamy, L., E. Padron, R. McKenna, N. Muzyczka, N. Kaludov, J. A. Chiorini, and M. Agbandje-McKenna. 2006. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J. Virol. 80:11556-11570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimm, D., and M. A. Kay. 2003. From virus evolution to vector revolution: use of naturally occurring serotypes of adeno-associated virus (AAV) as novel vectors for human gene therapy. Curr. Gene Ther. 3:281-304. [DOI] [PubMed] [Google Scholar]
- 25.Grimm, D., S. Zhou, H. Nakai, C. E. Thomas, T. A. Storm, S. Fuess, T. Matsushita, J. Allen, R. Surosky, M. Lochrie, L. Meuse, A. McClelland, P. Colosi, and M. A. Kay. 2003. Preclinical in vivo evaluation of pseudotyped adeno-associated virus vectors for liver gene therapy. Blood 102:2412-2419. [DOI] [PubMed] [Google Scholar]
- 26.Gurda, B. L., K. N. Parent, H. Bladek, R. S. Sinkovits, M. A. Dimattia, C. Rence, A. Castro, R. McKenna, N. Olson, K. Brown, T. S. Baker, and M. Agbandje-McKenna. Human bocavirus capsid structure: insights into the structural repertoire of the parvoviridae. J. Virol. 84:5880-5889. [DOI] [PMC free article] [PubMed]
- 27.Halbert, C. L., J. M. Allen, and A. D. Miller. 2001. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J. Virol. 75:6615-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hauck, B., and W. Xiao. 2003. Characterization of tissue tropism determinants of adeno-associated virus type 1. J. Virol. 77:2768-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernando, E., A. L. Llamas-Saiz, C. Foces-Foces, R. McKenna, I. Portman, M. Agbandje-McKenna, and J. M. Almendral. 2000. Biochemical and physical characterization of parvovirus minute virus of mice virus-like particles. Virology 267:299-309. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, H., D. Lillicrap, S. Patarroyo-White, T. Liu, X. Qian, C. D. Scallan, S. Powell, T. Keller, M. McMurray, A. Labelle, D. Nagy, J. A. Vargas, S. Zhou, L. B. Couto, and G. F. Pierce. 2006. Multiyear therapeutic benefit of AAV serotypes 2, 6, and 8 delivering factor VIII to hemophilia A mice and dogs. Blood 108:107-115. [DOI] [PubMed] [Google Scholar]
- 31.Jones, T. A., and S. Thirup. 1986. Using known substructures in protein model building and crystallography. EMBO J. 5:819-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kern, A., K. Schmidt, C. Leder, O. J. Muller, C. E. Wobus, K. Bettinger, C. W. Von der Lieth, J. A. King, and J. A. Kleinschmidt. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 77:11072-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klimczak, R. R., J. T. Koerber, D. Dalkara, J. G. Flannery, and D. V. Schaffer. 2009. A novel adeno-associated viral variant for efficient and selective intravitreal transduction of rat Muller cells. PLoS One 4:e7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kontou, M., L. Govindasamy, H.-J. Nam, N. Bryant, A. L. Llamas-Saiz, C. Foces-Foces, E. Hernando, M.-P. Rubio, R. McKenna, J. M. Almendral, and M. Agbandje-McKenna. 2005. Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J. Virol. 79:10931-10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kronenberg, S., B. Bottcher, C. W. von der Lieth, S. Bleker, and J. A. Kleinschmidt. 2005. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 79:5296-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kronenberg, S., J. A. Kleinschmidt, and B. Böttcher. 2001. Electron cryo-microscopy and image reconstruction of adeno-associated virus type 2 empty capsids. EMBO Rep. 2:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lerch, T. F., Q. Xie, and M. S. Chapman. The structure of adeno-associated virus serotype 3B (AAV-3B): insights into receptor binding and immune evasion. Virology 403:26-36. [DOI] [PMC free article] [PubMed]
- 38.Levy, H. C., V. D. Bowman, L. Govindasamy, R. McKenna, K. Nash, K. Warrington, W. Chen, N. Muzyczka, X. Yan, T. S. Baker, and M. Agbandje-McKenna. 2009. Heparin binding induces conformational changes in adeno-associated virus serotype 2. J. Struct. Biol. 165:146-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, W., L. Zhang, J. S. Johnson, W. Zhijian, J. C. Grieger, X. Ping-Jie, L. M. Drouin, M. Agbandje-McKenna, R. J. Pickles, and R. J. Samulski. 2009. Generation of novel AAV variants by directed evolution for improved CFTR delivery to human ciliated airway epithelium. Mol. Ther. 17:2067-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Limberis, M. P., L. H. Vandenberghe, L. Zhang, R. J. Pickles, and J. M. Wilson. 2009. Transduction efficiencies of novel AAV vectors in mouse airway epithelium in vivo and human ciliated airway epithelium in vitro. Mol. Ther. 17:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lochrie, M. A., G. P. Tatsuno, B. Christie, J. W. McDonnell, S. Zhou, R. Surosky, G. F. Pierce, and P. Colosi. 2006. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J. Virol. 80:821-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lovell, S. C., I. W. Davis, W. B. Arendall III, P. I. de Bakker, J. M. Word, M. G. Prisant, J. S. Richardson, and D. C. Richardson. 2003. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins 50:437-450. [DOI] [PubMed] [Google Scholar]
- 43.Manno, C. S., G. F. Pierce, V. R. Arruda, B. Glader, M. Ragni, J. J. Rasko, M. C. Ozelo, K. Hoots, P. Blatt, B. Konkle, M. Dake, R. Kaye, M. Razavi, A. Zajko, J. Zehnder, P. K. Rustagi, H. Nakai, A. Chew, D. Leonard, J. F. Wright, R. R. Lessard, J. M. Sommer, M. Tigges, D. Sabatino, A. Luk, H. Jiang, F. Mingozzi, L. Couto, H. C. Ertl, K. A. High, and M. A. Kay. 2006. Successful transduction of liver in hemophilia by AAV-factor IX and limitations imposed by the host immune response. Nat. Med. 12:342-347. [DOI] [PubMed] [Google Scholar]
- 44.McKenna, R., D. Xia, P. Willingmann, L. L. Ilag, S. Krishnaswamy, M. G. Rossmann, N. H. Olson, T. S. Baker, and N. L. Incardona. 1992. Atomic structure of single-stranded DNA bacteriophage phi X174 and its functional implications. Nature 355:137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller, E. B., B. Gurda-Whitaker, L. Govindasamy, R. McKenna, S. Zolotukhin, N. Muzyczka, and M. Agbandje-McKenna. 2006. Production, purification and preliminary X-ray crystallographic studies of adeno-associated virus serotype 1. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 62:1271-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitchell, M., H. J. Nam, A. Carter, A. McCall, C. Rence, A. Bennett, B. Gurda, R. McKenna, M. Porter, Y. Sakai, B. J. Byrne, N. Muzyczka, G. Aslanidi, S. Zolotukhin, and M. Agbandje-McKenna. 2009. Production, purification and preliminary X-ray crystallographic studies of adeno-associated virus serotype 9. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 65:715-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nam, H. J., M. D. Lane, E. Padron, B. Gurda, R. McKenna, E. Kohlbrenner, G. Aslanidi, B. Byrne, N. Muzyczka, S. Zolotukhin, and M. Agbandje-McKenna. 2007. Structure of adeno-associated virus serotype 8, a gene therapy vector. J. Virol. 81:12260-12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navaza, J. 2001. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D Biol. Crystallogr. 57:1367-1372. [DOI] [PubMed] [Google Scholar]
- 49.O'Donnell, J., K. A. Taylor, and M. S. Chapman. 2009. Adeno-associated virus-2 and its primary cellular receptor—cryo-EM structure of a heparin complex. Virology 385:434-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Opie, S. R., K. H. Warrington, Jr., M. Agbandje-McKenna, S. Zolotukhin, and N. Muzyczka. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 77:6995-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otwinowski, Z. W., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 52.Padron, E., V. Bowman, N. Kaludov, L. Govindasamy, H. Levy, P. Nick, R. McKenna, N. Muzyczka, J. A. Chiorini, T. S. Baker, and M. Agbandje-McKenna. 2005. Structure of adeno-associated virus type 4. J. Virol. 79:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pang, J., S. E. Boye, B. Lei, S. L. Boye, D. Everhart, R. Ryals, Y. Umino, B. Rohrer, J. Alexander, J. Li, X. Dai, Q. Li, B. Chang, R. Barlow, and W. W. Hauswirth. 2010. Self-complementary AAV-mediated gene therapy restores cone function and prevents cone degeneration in two models of Rpe65 deficiency. Gene Ther. 17:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 55.Quesada, O., B. Gurda, L. Govindasamy, R. McKenna, E. Kohlbrenner, G. Aslanidi, S. Zolotukhin, N. Muzyczka, and M. Agbandje-McKenna. 2007. Production, purification and preliminary X-ray crystallographic studies of adeno-associated virus serotype 7. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 63:1073-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rutledge, E. A., C. L. Halbert, and D. W. Russell. 1998. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 72:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidt, M., L. Govindasamy, S. Afione, N. Kaludov, M. Agbandje-McKenna, and J. A. Chiorini. 2008. Molecular characterization of the heparin-dependent transduction domain on the capsid of a novel adeno-associated virus isolate, AAV(VR-942). J. Virol. 82:8911-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seiler, M. P., A. D. Miller, J. Zabner, and C. L. Halbert. 2006. Adeno-associated virus types 5 and 6 use distinct receptors for cell entry. Hum. Gene Ther. 17:10-19. [DOI] [PubMed] [Google Scholar]
- 59.Shen, X., T. Storm, and M. A. Kay. 2007. Characterization of the relationship of AAV capsid domain swapping to liver transduction efficiency. Mol. Ther. 15:1955-1962. [DOI] [PubMed] [Google Scholar]
- 60.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang, Y., J. Cummins, J. Huard, and B. Wang. 2010. AAV-directed muscular dystrophy gene therapy. Expert Opin. Biol. Ther. 10:395-408. [DOI] [PubMed] [Google Scholar]
- 62.Tickle, I. J. 2007. Experimental determination of optimal root mean square deviations of macromolecular bond lengths and angles from their restrained ideal values Acta Crystallogr. D Biol. Crystallogr. 63:1274-1281. [DOI] [PubMed] [Google Scholar]
- 63.Tong, L. A., and M. G. Rossmann. 1990. The locked rotation function. Acta Crystallogr. A 46(Pt 10):783-792. [DOI] [PubMed] [Google Scholar]
- 64.Urzhumtseva, L., P. V. Afonine, P. D. Adams, and A. Urzhumtsev. 2009. Crystallographic model quality at a glance. Acta Crystallogr. D Biol. Crystallogr. 65:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Heel, M., and M. Schatz. 2005. Fourier shell correlation threshold criteria. J. Struct. Biol. 151:250-262. [DOI] [PubMed] [Google Scholar]
- 66.Walters, R. W., M. Agbandje-McKenna, V. D. Bowman, T. O. Moninger, N. H. Olson, M. Seiler, J. A. Chiorini, T. S. Baker, and J. Zabner. 2004. Structure of adeno-associated virus serotype 5. J. Virol. 78:3361-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walz, C., A. Deprez, T. Dupressoir, M. Durst, M. Rabreau, and J. R. Schlehofer. 1997. Interaction of human papillomavirus type 16 and adeno-associated virus type 2 co-infecting human cervical epithelium. J. Gen. Virol. 78(Pt 6):1441-1452. [DOI] [PubMed] [Google Scholar]
- 68.Wriggers, W., R. A. Milligan, and J. A. McCammon. 1999. Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 125:185-195. [DOI] [PubMed] [Google Scholar]
- 69.Wu, P., W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74:8635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu, Z., A. Asokan, J. C. Grieger, L. Govindasamy, M. Agbandje-McKenna, and R. J. Samulski. 2006. Single amino acid changes can influence titer, heparin binding, and tissue tropism in different adeno-associated virus serotypes. J. Virol. 80:11393-11397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu, Z., E. Miller, M. Agbandje-McKenna, and R. J. Samulski. 2006. Alpha2,3 and alpha2,6 N-linked sialic acids facilitate efficient binding and transduction by adeno-associated virus types 1 and 6. J. Virol. 80:9093-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiao, C., and M. G. Rossmann. 2007. Interpretation of electron density with stereographic roadmap projections. J. Struct. Biol. 158:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. U. S. A. 99:10405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yan, X., R. S. Sinkovits, and T. S. Baker. 2007. AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 157:73-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.