Abstract
Peptides derived from the “stem” of dengue virus (DV) type 2 (DV2) envelope (E) protein inhibit DV2 infectivity, targeting a late-stage fusion intermediate. We show here that stem peptides from all DV serotypes cross-inhibit DV1 to DV4 but that corresponding peptides derived from related flaviviruses do not. This failure to inhibit infection is not due to poor interaction with the E protein but rather to loss of association with the virion membrane. Residues 442 to 444 of the stem are determinants of inhibition; increasing hydrophobicity in this region increases inhibitory strength. These results support a two-step model of how stem-derived peptides inhibit viral entry.
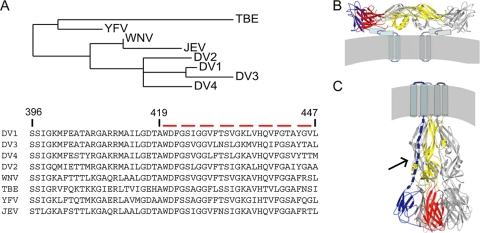
Infectious cell entry by enveloped viruses requires membrane fusion, facilitated by a viral fusion protein (5). The flavivirus envelope fusion protein (E) is the principal virion surface protein. It forms a well-ordered lattice of 90 dimers on the surface of the compact, 500-Å-diameter particle (22, 23). Crystal structures have been determined for several flavivirus envelope proteins in both prefusion (generally dimeric) and postfusion (trimeric) conformations (Fig. 1B and C) (3, 8, 11, 12, 15). These soluble forms of E (sE) include the first 395 residues of the approximately 445 ectodomain residues; they lack a membrane-proximal region called the “stem,” which is substantially conserved among all flaviviruses (Fig. 1A).
FIG. 1.
Sequences, structures, and conformational states of flavivirus E proteins. (A) Sequence alignment and distance tree of stem segments from several flavivirus envelope proteins. Sequences of residues 419 to 447 (DV2 numbering) were aligned using the program T-Coffee, and a phylogeny tree was constructed (13). JEV, Japanese encephalitis virus. (B) Prefusion conformation of DV2 E, shown as the dimer present on the virion surface. Residues 1 to 395 are in ribbon representation, derived from the sE dimer crystal structure (11). The stem (residues 396 to 447) and transmembrane (residues 448 to 491) regions are shown as cylinders and “worms,” with their approximate locations derived from subnanometer cryo-electron microscopy maps (23). For one of the subunits, domain I is in red, domain II is in yellow, and domain III is in blue. (C) E trimer after the low-pH transition. As in panel B, residues 1 to 395 are in ribbon representation, derived from the crystal structure of the sE trimer (12). One subunit is colored as described for panel B. The dashed blue line represents the stem (solid black arrow), for which the precise conformation and location are yet to be determined, and the cylinders represent the transmembrane anchor (location and clustering are merely schematic). The last stages of the fusion-promoting conformational change probably involve zipping up of the stem along the edge of domain II, so that the transmembrane anchor at the end of the stem and the fusion loop at the tip of domain II come together. We expect stem-derived peptides to interfere with this process (5).
Fusion is triggered in response to cues from the cellular compartment in which penetration occurs. Dengue virus (DV) and other flaviviruses penetrate from endosomes, following uptake by clathrin-mediated endocytosis (17, 18), and proton binding is the immediate fusion trigger. When the pH drops below about 6.2, E undergoes a large-scale conformational rearrangement that includes dissociation of the dimer and reconfiguration of the subunits into trimers (Fig. 1B and C) (2). At an intermediate stage in this molecular reorganization, a hydrophobic fusion loop at one end of the extended E subunit inserts into the outer leaflet of the target bilayer (3). Further rearrangement then draws together the fusion loop and the transmembrane segment that anchors E in the viral membrane, bringing the two membranes close enough to each other that fusion can ensue.
The sE subunit folds into three domains (domains I to III) that reorient with respect to each other during the conformational transition. The driving force for pinching the two membranes together appears to come from contacts made by domain III, as it folds back against domain I, and by the stem, as it “zips up” along domain II (Fig. 1C). Thus, interfering with either of these interfaces can block viral fusion, for example, by a soluble form of domain III or by a peptide derived from the stem (10, 16). A well-known precedent of the latter type of entry inhibitor is T-20/enfuvirtide, a peptide inhibitor of HIV-1 (4, 9, 20, 21).
We reported recently the sequence-specific binding of stem-derived peptides to the postfusion form of dengue virus type 2 (DV2) sE trimer. These peptides, which blocked in vitro fusion, as anticipated, also inhibited DV2 infectivity (16). The latter observations, although consistent with an earlier result (6a), were puzzling, because we expected a site for stem-peptide binding to be present only after induction of a conformational transition by the low pH of the endosome and an externally applied peptide to be inactive unless carried into the endosome by some nonspecific process. A series of experiments supported a two-step mechanism, in which an initial, nonspecific interaction of the hydrophobic C terminus of the peptide inhibitor with the viral membrane carries it with the virion into the endosome, allowing a specific interaction of the peptide with domain II as the protein reconfigures, inhibiting the final zipping of the stem.
The high degree of stem sequence conservation among DV serotypes and among flaviviruses more generally (Fig. 1A) prompted us to examine cross inhibition by these peptides. We find that stem peptides derived from each of the dengue virus serotypes inhibit the other serotypes to various degrees. None of the related flavivirus stem peptides inhibit dengue entry, but mutation of their C-terminal residues to the DV2 consensus sequence or to sequences with enhanced hydrophobicity confers inhibitory capacity. These data support the two-step mechanism proposed previously, in which a reversible, nonspecific interaction with the viral membrane brings virion-associated peptides into the low-pH endosome, where full exposure of the peptide site on the E conformational intermediate leads to the tight, specific binding that blocks membrane fusion. This mechanism suggests strategies for inhibiting entry of viruses that fuse in internal compartments.
MATERIALS AND METHODS
Peptide synthesis.
Peptides were synthesized as previously described (16). They were purified by reverse-phase high-pressure liquid chromatography and analyzed by mass spectrometry. Fluorescein isothiocyanate (FITC) was conjugated to the N terminus using a beta-alanine linker.
Preparation of DV2 sE trimers.
The postfusion sE trimer was produced as described previously (12). Briefly, purified sE (Hawai'i Biotech) was incubated in the presence of liposomes, acidified with sodium acetate to pH 5.5. Liposomes were solubilized with n-octyl-β-d-glucoside (β-OG) and n-undecyl-maltopyranoside (UDM). sE trimers were purified by cation exchange using a MonoS column (GE Healthcare) and further purified by size-exclusion chromatography on a Superdex 200 chromatograph (GE Healthcare). Protein was dialyzed extensively using a 50-kDa-molecular-mass cutoff membrane (Spectrapor).
Fluorescence polarization.
Binding experiments were carried out in 384-well microplates and analyzed in a PerkinElmer EnVisions instrument. FITC-conjugated peptide stocks were made in dimethyl sulfoxide (DMSO) and dissolved in assay buffer immediately before use. Increasing concentrations of protein were used with a constant 20 nM concentration of peptide. Data were collected at 3 and 24 h. Kd (equilibrium dissociation constant) was estimated as the concentration of protein with a half-maximal increase in fluorescence polarization.
Cells and viruses.
C6/36 cells were maintained in L-15 medium (Mediatech) supplemented with 10% fetal bovine serum. For plaque-forming assays (PFAs), BHK-21 cells were seeded (5 × 104 cells/well) in 24-well plates in minimum essential medium (α-MEM) supplemented with the antibiotics penicillin and streptomycin and 5% fetal bovine serum (FBS).
Dengue virus serotypes 1 to 4 were propagated by infection of confluent monolayers of C6/36 cells for 1 h at 25°C with rocking every 15 min. L-15 medium (Mediatech) was added, and the cells were incubated at 25°C until syncytium formation was observed. The supernatant was clarified by centrifugation, separated into aliquots, and stored at −80°C.
Plaque reduction inhibition assays.
Virus supernatant produced from C6/36 cells was diluted in Earle's balanced salt solution (EBSS) to a stock concentration that would allow infection at a multiplicity of infection of 1, on the basis of 50,000 seeded cells. Virus inoculum with peptide or vehicle (DMSO) was preincubated for 15 min at 37°C. BHK-21 cells were infected for 1 h at 37°C. Virus (or virus-peptide mixtures) was washed from cells with 1 ml of phosphate-buffered saline (PBS); and α-MEM supplemented with HEPES, the antibiotics penicillin and streptomycin, and 2% FBS was added. Supernatants were harvested 24 h later and stored at −80°C for plaque assay as described below.
Plaque assay.
BHK-21 cells were seeded as described above. Aliquots from infections were thawed, and 10-fold dilutions in EBSS were prepared in 96-well plates. One hundred microliters of each dilution was added to the cells. The plates were incubated for 1 h at 37°C. Unadsorbed virus was removed by two washes with PBS, after which 1 ml of α-MEM supplemented with carboxymethyl cellulose (CMC), the antibiotics penicillin and streptomycin, and 2% FBS was added to each well and the plates were incubated for 4 days. The CMC overlay was removed, and the cells were washed with PBS and stained with crystal violet. The plates were washed with water to remove excess crystal violet and dried overnight.
Focus-forming assay.
The conditions for the focus-forming assay (FFA) were identical to those described above for the plaque assays. After the CMC was aspirated, the plates were washed 2 times with PBS, blocked with 5% milk in PBS-Tween 20 (PBS-T) for 1 h at room temperature, washed once with PBS, and incubated with the monoclonal antibody 4G2 for 1 h at 37°C at a 1:1,000 dilution. The plates were washed with 1 ml PBS-T, incubated with a secondary antibody, goat-anti-mouse horseradish peroxidase (Bio-Rad) at a 1:10,000 dilution for 30 min at 37°C, washed twice, and developed using a 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) kit (Vector Labs).
Direct plaque assay.
BHK-21 cells were seeded at 50,000 cells/ml. A total of 50,000 PFU of virus was preincubated with peptide or carrier (DMSO) at 37°C for 15 min, diluted 10-, 100-, and 1,000-fold, and adsorbed to cells for 1 h at 37°C. The cells were washed 2 times with PBS and overlaid with 1 ml of α-MEM supplemented with CMC, the antibiotics penicillin and streptomycin, and 2% FBS and incubated for 4 days. The CMC overlay was removed, and the cells were washed with PBS and stained with crystal violet or as described above for the focus-forming assay.
RESULTS
Serotype cross inhibition by stem-derived peptides.
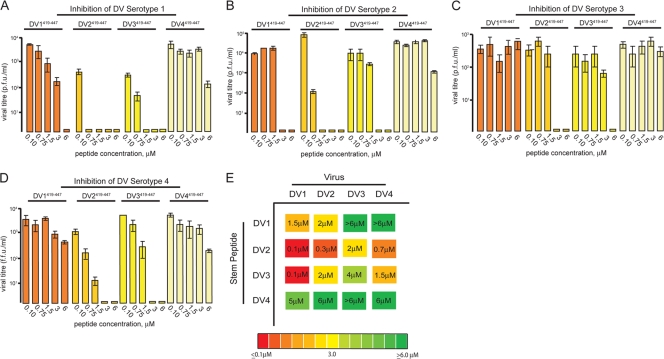
We synthesized peptides corresponding to residues 419 to 447 of each of the four DV serotypes (DV419-447 peptides), adding a C-terminal solubility tag, RGKGR, shown previously to be compatible with inhibitory activity, and tested these peptides against all serotypes, as described (16). Briefly, DV419-447 peptides were preincubated with virus inoculum at 37°C for 30 min prior to adsorption to cells. Viral supernatants were harvested 24 h later and assayed for infectivity with a standard PFA for DV1 WP74, DV2 NGC, and DV3 THD 3 or an FFA for DV4 TVP360 (6). We further validated the results for DV2 inhibition by the DV2-derived peptide with a direct plaque assay (see Methods and Materials and Fig. S1 in the supplemental material).
Titrations of the peptide of residues 419 to 447 against the dengue serotypes show that DV2419-447 is a strong inhibitor of all four serotypes, followed in strength by DV1419-447, DV3419-447, and DV4419-447 (Fig. 2A to D). The peptide derived from the DV4 E was a poor inhibitor, inactive even against its own serotype. While all serotypes could be inhibited to some degree, DV3 was particularly insensitive to inhibition by stem-derived peptides from DV1 and DV4 (Fig. 2C).
FIG. 2.
Cross inhibition of dengue viruses by stem peptides from heterologous serotypes. (A to D) Inhibition of DV1 to DV4 by stem-derived peptides from DV1 to DV4, residues 419 to 447. Peptides were preincubated with virus inoculum and assayed by PFA or FFA as described in the text. (E) Schematic summary of panels A to D. The inhibitory activity is color coded from green (IC90 > 6 μM) to red (IC90 < 0.1 μM). Approximate IC90s are displayed within each colored box. Viral infectivity for each concentration point was determined in duplicate, with plaque assays being carried out in triplicate.
Stem peptides from related flaviviruses do not inhibit dengue serotypes.
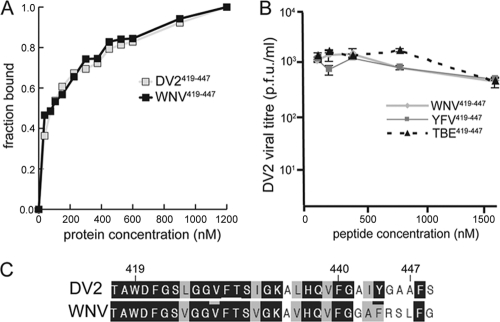
The stem sequence is reasonably well conserved in other flaviviruses, including tick-borne encephalitis virus (TBE), yellow fever virus (YFV) and West Nile virus (WNV) (Fig. 1A). We therefore asked whether stem peptides 419 to 447 derived from related flaviviruses could inhibit DV infectivity. None of these stem peptides inhibited DV2 infectivity to any measurable extent (Fig. 3B), nor did they inhibit any of the other dengue virus serotypes (data not shown).
FIG. 3.
DV2 envelope interation with stem peptides from heterologous flaviviruses. (A) Binding of DV2419-447 and WNV419-447 stem peptides to trimeric DV2 sE. The fraction bound is plotted as a function of the concentration of sE (in nM). Each curve is an average of three independent experiments. (B) Effects of WNV, TBE, and YFV stem peptides from residues 419 to 447 on DV2 infectivity. Peptides were preincubated with the virus inoculum and assayed by PFA as described in the text. Viral infectivity for each concentration point was determined in duplicate, with plaque assays being carried out in triplicate. (C) Sequence alignment showing conservation and variation of the stem peptides from WNV and DV2 used in these experiments.
WNV stem peptide binds to the postfusion DV2 sE trimer.
Is the failure of stem peptides derived from the related flaviviruses to inhibit DV2 due to low affinity for the DV2 envelope protein or to poor interaction with the viral membrane? Using the WNV stem peptide of residues 419 to 447, we determined whether a fluoresceinated derivative could bind trimeric DV2 sE. Both the preparation of trimeric DV2 sE and the determination by fluorescence anisotropy of the Kds for WNV419-447 and DV2419-447 were carried out as described previously (16).
As shown in Fig. 3A, WNV419-447 and DV2419-447 have nearly identical affinities for trimeric DV2 sE. Indeed, the sequences of these two peptides are extremely similar, particularly between residues 419 and 440, the minimal sequence required for high-affinity sE binding (16). This region has only three conservative differences between the two viruses: L425V, I432V, and L436V. There is, however, significant variation in the C-terminal 7 residues. Our earlier work demonstrated that these residues greatly enhance the inhibitory potency of DV2419-447 (with respect to that of the shorter and essentially inactive peptide DV2419-440) but do not contribute to affinity for trimeric sE. We concluded that these residues interact with the viral membrane, concentrating peptide on the virion surface and allowing it to be carried with the virion into endosomes. The comparison of the properties of WNV419-447 and DV2419-447 suggests that the failure of WNV419-447 to inhibit DV2 infection is due to poor interaction with the viral membrane (i.e., loss of the first step in the inhibitory mechanism) rather than poor affinity for the rearranging protein.
Effects of mutations at the C terminus of stem peptides on inhibition of DV2.
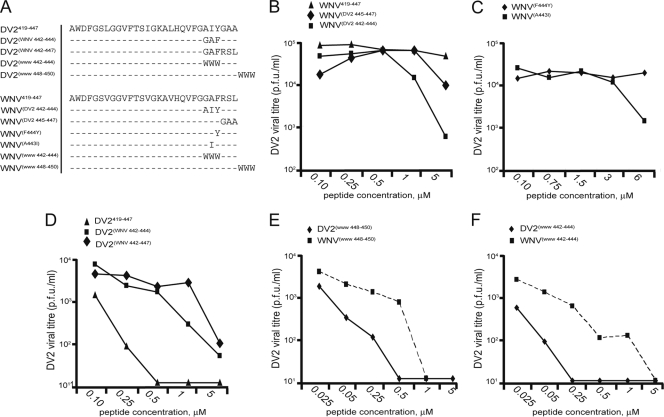
Because WNV419-447 binds strongly to the sE DV2 envelope protein, we sought to identify changes in residues between 441 and 447 that could confer on the peptide a capacity to inhibit DV2 infection. We synthesized a series of WNV-derived stem peptides, maintaining the consensus sequence from positions 419 to 440 while varying the identities of the residues at positions 441 to 447. Sequences were designed to alter specific residues to those of their DV2 counterparts. Substituting residues 442 to 444 from DV2 into the WNV sequence markedly enhanced DV2 inhibition, while substituting residues 445 to 447 did not [compare peptides WNV(DV2 445-447) and WNV(DV2 442-444) in Fig. 4B]. We further found that a single substitution, alanine to isoleucine at position 443 (WNVA443I), was sufficient to achieve some degree of inhibition, while introduction of the conserved tyrosine at position 444 from the DV serotypes (WNVF444Y) had no effect (Fig. 4C).
FIG. 4.
Effects of modifications in the C-terminal residues of stem-derived peptides on inhibition of DV2 infectivity. (A) Sequences of DV and WNV residue 419 to 447 variants used in this study; (B) comparison of inhibition of DV2 infectivity by WNV-derived stem peptides and the changes to the DV2 consensus sequence; (C) single-residue changes made in WNV419-447 to match the DV2 sequence and their effects on viral infectivity; (D) comparison of inhibition of DV2 infectivity by DV2-derived stem peptides, including changes to match the WNV sequence; (E) inhibition of DV2 infectivity by addition of WWW to the C terminus of DV2419-447 and WNV419-447; (F) inhibition by WWW variants of DV2419-447 and WNV419-447 at residues 442 to 444. Each experiment was performed in duplicate; plaque assays were carried out in triplicate.
If WNV419-447 can be made into an inhibitor of DV2 by mutating positions 441 to 447 to the DV2 sequence, is it possible to mutate DV2419-447 at the similar positions and reduce its inhibitory strength? We synthesized a peptide that replaced positions 442 to 444 of DV2 with the WNV sequence. As seen in Fig. 4D, DV2(WNV 442-444) has a nearly 10-fold higher 90% inhibitory concentration (IC90; ∼2 μM) than unaltered DV2419-447 (∼250 nM), confirming the importance of these residues and their likely role in the inhibitory mechanism. None of the WNV-derived peptides inhibit DV2 infectivity as strongly as does native DV2419-447. We therefore cannot exclude a contribution from the three residues between positions 419 and 440 that differ between the two viruses.
Increasing the hydrophobicity of the C terminus enhances inhibition.
If the C-terminal residues 441 to 447 contribute little to specific protein-protein interactions but are necessary for membrane targeting, could the inclusion of an unrelated hydrophobic sequence in this segment enhance inhibitory strength? We synthesized two different sets of DV2 and WNV residue 419 to 447 stem peptides: one set in which three consecutive tryptophans replace native residues 442 to 444 and another in which three consecutive tryptophans extend the peptide at its C terminus. We tested these peptides against DV2 in a standard viral infectivity assay. As shown in Fig. 4F, replacing residues 442 to 444 with tryptophans reduced the IC90 of DV2419-447 to less than 25 nM and conferred inhibitory activity on the otherwise inactive WNV419-447 (IC90, ∼250 nM). Addition of three tryptophans to the C terminus maintained DV2419-447 activity and made WNV419-447 into a modest inhibitor, but in neither case was the effect as strong as that obtained by replacement of residues 442 to 444 with tryptophans (compare Fig. 4E and F).
DISCUSSION
Stem-derived peptides from the DV serotypes inhibit other serotypes to various extents. The proposed two-step mechanism for inhibition of infectivity by these peptides includes both a relatively nonspecific, membrane-binding step, followed by a specific interaction with a trimeric intermediate of E during its low-pH-triggered, fusion-promoting conformational transition. Thus, variation of cross-inhibitory strength may derive either from differences in the specific interaction of these stem peptides with the envelope proteins or from differences in the initial, nonspecific recruitment of the stem peptide to the virion membrane, or both. The principal contribution to the membrane interaction comes from residues 441 to 447, which have little or no influence on binding to trimeric DV2 sE in vitro; the principal contribution to affinity for trimeric sE comes from residues 419 to 440 (16). Thus, the two contributions to inhibition of infectivity appear to be separable. The level of conservation in the sequence between residues 419 and 440 is so high that it is very difficult to discern any pattern relating cross inhibition to the identities of the residues at the eight positions of extremely conservative substitution. Indeed, the DV3 peptide, which is the most distant of the three other peptides from DV2, is the most similar in its inhibitory strength and specificity. Additional confounding factors could be the extent (for different serotypes) of E conformational fluctuations, which we have shown previously to be important for access of peptides to the viral membrane, and the relative rates of specific peptide binding and E-stem zippering during the fusion-inducing conformational transition (16). The latter relationship could also influence the number of peptides required for inhibition.
The one clear correlate of inhibition is hydrophobicity of the segment between residues 441 and 447. Residues 442 to 444 appear to be the most important. We can estimate the membrane-targeting properties of these residues using the Wimley-White scale for transfer of a peptide from an aqueous environment to a palmitoyl-oleoyl-phosphatidyl choline interface (19). The rank order for the segment centered on position 443 of peptides from the four DV serotypes and WNV is DV2 > DV4 ≈ DV1 > DV3 > WNV, using either of two different weighting schemes (see Fig. S2 in the supplemental material). Changes at residues 442 to 444 that enhance hydrophobicity generate antiviral activity when they are introduced into the stem peptide from WNV. In particular, altering residues 442 to 444 to WWW has a very pronounced enhancing effect. Tryptophan residues in membrane proteins are often located at the interface between hydrophilic head groups and hydrophobic fatty acyl chains. Residues 442 to 444, removed by no more than 2 to 3 residues from the E transmembrane anchor, will approach this hydrophilic-hydrophobic interface, and binding of a WWW-containing, stem-derived peptide through association of the tryptophans with the viral membrane should place the peptide in an optimal position to interfere with the endogenous stem as it draws domain II toward the viral membrane. The observed effect of the WWW substitutions is therefore fully consistent with the two-step mechanism described in our previous paper. We suggest that suitable modification of any flavivirus-derived stem peptide at positions between residues 441 and 447 should generate an inhibitor of the corresponding virus and potentially also of heterologous flaviviruses.
Preconcentration of a fusion inhibitor in the membrane of an enveloped virus is likely to be a general mechanism for enhancing inhibitory activity. Certain broadly neutralizing antibodies that bind the membrane-proximal external region (MPER) of HIV-1 target the extended, prehairpin intermediate of gp41 and block infection by preventing completion of its fusion-promoting conformational transition (1). They appear to neutralize by a two-step mechanism similar to one that we have characterized for flavivirus stem-derived peptides. The activity of the gp41 MPER-derived peptide T-20/enfuvirtide may also be enhanced by preassociation with the viral membrane of its hydrophobic C-terminal residues (including at least one tryptophan). Membrane binding of these residues, like that of residues 441 to 447 from flavivirus stem-derived peptides, should position T-20/enfuvirtide optimally to interfere with the final stages of the gp41 conformational change. The addition of membrane-targeting sequences to peptides derived from membrane-proximal regions may be a useful strategy for creating reagents to block enveloped viral entry (7, 14).
Supplementary Material
Acknowledgments
We thank the ICCB Screening Facility at Harvard Medical School for use of instrumentation, staff at Hawai'i Biotech for ongoing interactions, and members of the S. C. Harrison and P. L. Yang laboratories for helpful discussions.
The work was supported by NIH grants U54AI057159 (the SCH project of the New England Regional Center of Excellence for Biodefense and Emerging Infectious Diseases, D. Kasper, principal investigator), an award from the Giovanni Armenise-Harvard Foundation (to P.L.Y.), and an Albert J. Ryan Fellowship (to A.G.S.). S.C.H. is an investigator in the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 29 September 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alam, S. M., M. Morelli, S. M. Dennison, H. X. Liao, R. Zhang, S. M. Xia, S. Rits-Volloch, L. Sun, S. C. Harrison, B. F. Haynes, and B. Chen. 2009. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc. Natl. Acad. Sci. U. S. A. 106:20234-20239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 69:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 5.Harrison, S. C. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henchal, E. A., M. K. Gentry, J. M. McCown, and W. E. Brandt. 1982. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am. J. Trop. Med. Hyg. 31:830-836. [DOI] [PubMed] [Google Scholar]
- 6a.Hrobowski, Y. M., R. F. Garry, and S. F. Michael. 2005. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol. J. 2:49 doi: 10.1186/1743-422X-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingallinella, P., E. Bianchi, N. A. Ladwa, Y. J. Wang, R. Hrin, M. Veneziano, F. Bonelli, T. J. Ketas, J. P. Moore, M. D. Miller, and A. Pessi. 2009. Addition of a cholesterol group to an HIV-1 peptide fusion inhibitor dramatically increases its antiviral potency. Proc. Natl. Acad. Sci. U. S. A. 106:5801-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanai, R., K. Kar, K. Anthony, L. H. Gould, M. Ledizet, E. Fikrig, W. A. Marasco, R. A. Koski, and Y. Modis. 2006. Crystal structure of West Nile virus envelope glycoprotein reveals viral surface epitopes. J. Virol. 80:11000-11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 10.Liao, M., and M. Kielian. 2005. Domain III from class II fusion proteins functions as a dominant-negative inhibitor of virus membrane fusion. J. Cell Biol. 171:111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313-319. [DOI] [PubMed] [Google Scholar]
- 13.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 14.Porotto, M., C. C. Yokoyama, L. M. Palermo, B. Mungall, M. Aljofan, R. Cortese, A. Pessi, and A. Moscona. Viral entry inhibitors targeted to the membrane site of action. J. Virol. 84:6760-6768. [DOI] [PMC free article] [PubMed]
- 15.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt, A. G., P. L. Yang, and S. C. Harrison. Peptide inhibitors of dengue-virus entry target a late-stage fusion intermediate. PLoS Pathog. 6:e1000851. [DOI] [PMC free article] [PubMed]
- 17.van der Schaar, H. M., M. J. Rust, C. Chen, H. van der Ende-Metselaar, J. Wilschut, X. Zhuang, and J. M. Smit. 2008. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 4:e1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Schaar, H. M., M. J. Rust, B. L. Waarts, H. van der Ende-Metselaar, R. J. Kuhn, J. Wilschut, X. Zhuang, and J. M. Smit. 2007. Characterization of the early events in dengue virus cell entry by biochemical assays and single-virus tracking. J. Virol. 81:12019-12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White, S. H., and W. C. Wimley. 1994. Peptides in lipid bilayers: structural and thermodynamic basis for partitioning and folding. Curr. Opin. Struct. Biol. 4:79-86. [Google Scholar]
- 20.Wild, C., J. W. Dubay, T. Greenwell, T. Baird, Jr., T. G. Oas, C. McDanal, E. Hunter, and T. Matthews. 1994. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc. Natl. Acad. Sci. U. S. A. 91:12676-12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. U. S. A. 89:10537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu, I. M., W. Zhang, H. A. Holdaway, L. Li, V. A. Kostyuchenko, P. R. Chipman, R. J. Kuhn, M. G. Rossmann, and J. Chen. 2008. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319:1834-1837. [DOI] [PubMed] [Google Scholar]
- 23.Zhang, Y., J. Corver, P. R. Chipman, W. Zhang, S. V. Pletnev, D. Sedlak, T. S. Baker, J. H. Strauss, R. J. Kuhn, and M. G. Rossmann. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.