Abstract
Using two-photon-induced fluorescence lifetime imaging microscopy, we corroborate an interaction (previously demonstrated by yeast two-hybrid domain analysis) of full-length vaccinia virus (VACV; an orthopoxvirus) A36 protein with the cellular microtubule motor protein kinesin. Quenching of enhanced green fluorescent protein (EGFP), fused to the C terminus of VACV A36, by monomeric red fluorescent protein (mDsRed), fused to the tetratricopeptide repeat (TPR) domain of kinesin, was observed in live chicken embryo fibroblasts infected with either modified vaccinia virus Ankara (MVA) or wild-type fowlpox virus (FWPV; an avipoxvirus), and the excited-state fluorescence lifetime of EGFP was reduced from 2.5 ± 0.1 ns to 2.1 ± 0.1 ns due to resonance energy transfer to mDsRed. FWPV does not encode an equivalent of intracellular enveloped virion surface protein A36, yet it is likely that this virus too must interact with kinesin to facilitate intracellular virion transport. To investigate possible interactions between innate FWPV proteins and kinesin, recombinant FWPVs expressing EGFP fused to the N termini of FWPV structural proteins Fpv140, Fpv168, Fpv191, and Fpv198 (equivalent to VACV H3, A4, p4c, and A34, respectively) were generated. EGFP fusions of intracellular mature virion (IMV) surface protein Fpv140 and type II membrane protein Fpv198 were quenched by mDsRed-TPR in recombinant FWPV-infected cells, indicating that these virion proteins are found within 10 nm of mDsRed-TPR. In contrast, and as expected, EGFP fusions of the IMV core protein Fpv168 did not show any quenching. Interestingly, the p4c-like protein Fpv191, which demonstrates late association with preassembled IMV, also did not show any quenching.
Viruses, including the best-studied and prototypic poxvirus, vaccinia virus (VACV), lack intrinsic motility. During the early stages of infection, VACV exploits the microtubule cytoskeleton and the dynein-dynactin complex to permit delivery of infecting virions to the site of future replication (34). Later in the infection, it disrupts microtubule organization, centrosome function, and actin organization and polymerizes actin beneath membrane-bound, enveloped virus particles to assist dispersal (11). During morphogenesis, various different types of VACV particles are observed within the infected cell: immature virions (IV), intracellular mature virus (IMV; also known as mature virions), intracellular enveloped virus (IEV; also known as wrapped virions), cell-associated enveloped virus (CEV), and extracellular enveloped virus (EEV); the last two particle types are also known collectively as extracellular virions (EV) (9, 32, 37, 46). These particles display different VACV proteins on their surfaces, dependent primarily on the nature and origins of the particular outer membrane that surrounds the nucleic acid-containing core.
VACV surface proteins, both IMV specific (A27 [38]) and IEV specific (F12 [27, 56] and A36 [57]), have been implicated in microtubule-based intracellular transport of VACV (56, 60). It was reported that A27 is required for transport of IMV to the sites of membrane wrapping where IEV are formed (40) although this was not substantiated in a later report (59). In contrast, F12 (56) and A36 (60) are involved in transport of the IEV to the cell surface and eventual egress of the virus (27, 31).
Conventional kinesin is classified as a heterotetramer that contains two heavy and two light chains (55). The light chain associates with the heavy chain through its N-terminal domain while its C-terminal domain, which contains six tetratricopeptide repeat (TPR) motifs, binds to cellular cargoes (18). The motor domain present in the N terminus of each heavy chain binds to microtubules (54). A study using a yeast two-hybrid system tested the ability of the short cytoplasmic domains of five VACV proteins (F12 [56], A36 [57], A33 [39], A34 [16], and B5 [17, 25]) to bind to the TPR region of the kinesin light chain (KLC-TPR) (60). It revealed that, of the five domains tested, only the fragment from the N terminus of A36 showed KLC-TPR-binding capability and suggested that it was a likely candidate to interact with kinesin during VACV intracellular microtubule-based transport. In other studies, it has been shown that deletion of A36 from VACV causes partial inhibition of IEV transport to the plasma membrane in infected cells (21, 22). Deletion of F12 from VACV impaired >99% of CEV formation (21). A recent study has shown that F12 has structural similarity to KLC and, as well as possessing TPR motifs, has a conserved tryptophan and aspartic acid (WD) motif essential to the binding of KLC and IEV transport (31). A36 and E2, which have been shown to interact with F12 and to be involved in IEV transport (14, 15), were also shown to contain TPR motifs (31).
Fowlpox virus (FWPV), type species of the Avipoxvirus genus, offers a safe model for investigating infection and is currently undergoing clinical trials as a nonreplicating recombinant vector for vaccination of mammals, including humans, in the fight against, malaria, HIV, avian influenza, cancer, and other diseases (45). A recombinant FWPV (rFWPV) expressing the avian influenza virus H5 hemagglutinin is probably the most extensively deployed live recombinant virus vector vaccine used in any sector, with some 2 billion doses having been used in Mexico against highly pathogenic H5N2 avian influenza virus in poultry (8). The molecular and cell biology of FWPV has been much less extensively studied than that of VACV. In contrast to VACV, however, it has been demonstrated that production of EV of FWPV and of another avipoxvirus, pigeonpox virus, occurs primarily by budding of “naked” IMV rather than by wrapping of IMV to produce IEV, which would in VACV, in turn, form EV by fusion of their outer membranes with the cell membrane (5, 19). Whether as a consequence or a cause of this strategy, it is notable that the FWPV genome (and that of the distantly related avipoxvirus, canarypox virus, better known as the ALVAC live recombinant vaccine vector used for many commercial veterinary vaccines and for the recent Thai HIV vaccine trial [35]) clearly lacks equivalents of a number of genes encoding proteins that play key roles in the egress of VACV and in the function of EV (2, 28, 52); specifically, the following proteins are lacking: A27L, A33R, A36R, A56R, and B5R. FWPV retains equivalents of A34R (fpv198), E2L (fpv101), F12L (fpv109), and F13L (fpv108). Lest this might be considered a peculiarity of the avipoxviruses, it is worth noting that this same gene spectrum is shared with the only reptilian poxvirus sequenced thus far, crocodilepox virus (1). Furthermore, among mammalian poxviruses, parapoxviruses lack equivalents of A36R and B5R (13, 20) and molluscum contagiosum virus lacks equivalents of A27L, B5R, and E2L (43).
Interaction of proteins in cells can be followed by utilizing fluorescence resonance energy transfer (FRET) between protein pairs tagged with appropriate fluorophores, such as enhanced green fluorescent protein (EGFP), acting as a donor, and monomeric red fluorescent protein (mDsRed), acting as the acceptor. The fluorescent lifetime imaging microscopy (FLIM) approach, whereby quenching of the excited-state lifetime is evidence for a direct physical interaction, is an improvement over steady-state FRET. This FRET-FLIM technique is highly sensitive, and the change in the excited-state lifetime of the donor fluorophore is concentration independent (58) and does not suffer from fluorophore bleed-through. So far, it has been employed in only a few studies of the interaction of viral proteins in live cells (42, 47, 48). Two-photon-induced FRET-FLIM (2P-FRET-FLIM) provides several advantages over the single-photon method, including reduced phototoxicity (by use of near-infrared excitation light that is not absorbed by cellular components) and reduced bleaching of the fluorophore (10, 50, 51).
In this study, as a positive control we have demonstrated that an interaction between full-length VACV A36, fused at its N terminus to EGFP, and KLC-TPR, attached to mDsRed, can be observed in cells infected with modified vaccinia virus Ankara (MVA) using a highly sensitive FRET-FLIM technique. Infection of cells with FWPV, which does not express it own A36 ortholog, does not abrogate the interaction between the ectopically expressed VACV A36 and KLC-TPR. Furthermore, N-terminal EGFP fusions of Fpv140 (equivalent to VACV H3L) and Fpv198 (equivalent to VACV A34), could also be observed to interact with KLC-TPR-mDsRed in live, FWPV-infected cells even though no such interactions have been reported between the equivalent VACV proteins and KLC-TPR.
MATERIALS AND METHODS
All reagents were purchased from Sigma, United Kingdom, or Invitrogen and used without further treatment unless otherwise stated.
Virus and cell culture.
The origins and propagation of FWPV FP9 (FP9), as well as the isolation and purification or rFWPV, were as described previously (4, 26, 28).
Antibody.
For immunofluorescence staining, the following mouse monoclonal antibodies (MAbs) were used: GB9 (anti-Fpv168), DH6 (anti-Fpv191), and DF6 (anti-Fpv140), as described previously (3). Anti-β-tubulin antibody to stain microtubules was from Sigma (United Kingdom). Alexa Fluor 568 goat anti-mouse immunoglobulin G (IgG) secondary antibody was from Invitrogen (United Kingdom).
Construction of plasmids.
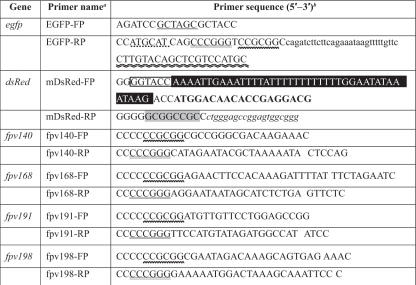
A plasmid that encodes enhanced green fluorescent protein was constructed in two steps. Briefly, the DNA spanning the coding sequence of EGFP was amplified from pEGFP-C1 vector (Molecular Probes) using primers EGFP-FP (forward) and EGFP-RP (reverse) (Table 1). The amplified PCR product was gel purified (Qiagen) for digestion with NheI and NsiI enzymes and ligated into the XbaI and PstI sites of an FWPV expression/transfer vector, pEFGPT12S (7), under the control of an early/late synthetic poxvirus promoter. Following bacterial transformation with this intermediate plasmid, positive clones were identified by PCR and restriction enzyme analysis. DNA spanning fpv140, fpv168, fpv191, or fpv198 was amplified by PCR from FWPV FP9 genomic DNA as a template using the following primer pairs, fpv140-FP and fpv140-RP, fpv168-FP and fpv168-RP, fpv191-FP and fpv191-RP, and fpv198-FP and fpv198-RP, respectively. The resultant PCR product was gel purified (Qiagen) and digested then ligated into SacII and XmaI sites of the generated intermediate plasmid. Similarly, transformation and subsequent identification of inserts of positive clones were performed as described above. The final recombinant plasmids were named pFPVEGFP140, pFPVEGFP168, pFPVEGFP191, and pFPVEGFP198, and they express N-terminal EGFP fusions of Fpv140, Fpv168, Fpv191, and Fpv198, respectively.
TABLE 1.
Gene-specific primers used for constructing recombinant plasmids
FWPV, egfp, and dsRed gene-specific forward (FP) and reverse (RP) primers were designed based on sequences of the FWPV genome, pEGFP-C1 (Clontech), and pDsRed-Monomer-C1 (Clontech), respectively.
Restriction sites are as follows: NheI (single underlining), NsiI (single dotted line), XmaI (double underlining), SacII (wavy line), KpnI (boxed), and NotI (light shading). Also indicated are a Myc tag coding sequence (lowercase roman), the synthetic vaccinia virus early/late promoter (dark shading), the complement of sequence encoding the C terminus of the egfp gene (double wavy line) plus the N terminus (bold), and the C terminus (lowercase italics) of the dsRed.
A vector (pEL-KLC-TPR-mDsRed) was constructed to express kinesin light chain tetratricopeptide repeat (KLC-TPR) motifs containing monomeric red fluorescent protein (mDsRed) from Discosoma sp. at its N terminus, under the control of an early/late poxvirus promoter (pEL) from pEL-GFP-TPR plasmid (36). Briefly, the DNA corresponding to the coding sequence of mDsRed was amplified from pDsRed-Monomer-C1 vector (Molecular Probes) using primers mDsRed-FP (forward) and mDsRed-RP (reverse) (Table 1). To generate pEL-KLC-TPR-mDsRed vector, the KpnI-NotI fragment in the pEL-EGFP-TPR was replaced with the resultant PCR product, which was cut with KpnI-NotI. The pEL-GFP-TPR (36), pEL-A36-EGFP (encoding vaccinia virus A36 with a C-terminal EGFP), and p-Actin-EGFP (encoding actin with an N-terminal EGFP) used for plasmid construction and protein-protein interaction studies were a kind gift from Michael Way, Cancer Research UK, London, United Kingdom.
Confocal immunofluorescence imaging.
Infected or mock-infected chicken embryo fibroblasts (CEFs) on coverslips were prepared for immunofluorescence microscopy as described previously (3). Cells were transfected as required using Lipofectamine according to the manufacturer's recommendations (Invitrogen). Confocal fluorescence images were obtained using either a Leica TCS NT confocal microscope or a Nikon eC1 attached to a TE2000 inverted microscope with a 60× objective (water immersion; numerical aperture [NA], 1.2; Nikon). Images were processed and annotated using Photoshop, version CS4 (Adobe).
FRET-FLIM analysis.
Protein-protein interaction analysis in live, infected/cotransfected CEFs was carried out at the Central Laser Facility of the Rutherford Appleton Laboratory using a FRET-FLIM setup, as previously described (33). Briefly, multiphoton excitation was delivered from a titanium sapphire laser (Mira; Coherent, United Kingdom) and a frequency-doubled vanadate laser (18 W) as a pump laser (Coherent Lasers; United Kingdom), optimized for delivering laser light at a wavelength of 910 ± 5 nm with 180-fs pulses at a 75-MHz repetition rate. The laser was focused to generate a diffraction-limited spot through a 60× objective (water immersion; NA, 1.2; Nikon) to excite specimens on the microscope stage (Nikon TE2000U). Fluorescence emission from the specimen was obtained through a band-pass filter (BG39; Comar) using a non-descanned port of the confocal microscope. Single photon pulses were detected through an external high-speed microchannel plate photomultiplier tube (R3809U; Hamamatsu), and reference laser, frame sync, line sync, and pixel clock signals of the scanning system were collected using a time-correlated single photon counting (TCSPC) PC module (SPC-830; Becker and Hickl, Germany).
Cells expressing both EGFP (488-nm excitation) and mDsRed (543-nm excitation) fusion proteins were selected following confocal microscopy. FRET-FLIM data were collected as optical Z-stacks of infected/cotransfected cells due to the diffraction-limited size of the viral particles (∼250 nm). Cells expressing EGFP alone were selected as an FLIM control for a noninteracting EGFP excited-state lifetime. The changes in the lifetime of the EGFP in the region of interest (the contour of individual cells) were analyzed using SPCImage analysis software (Becker and Hickl, Germany), and the determined average lifetime of EGFP data was analyzed by an independent-sample t test using statistical analysis software (SPSS, version 17).
RESULTS
Use of 2P-FRET-FLIM to demonstrate interaction between VACV A36 and KLC-TPR in live cells infected with VACV MVA.
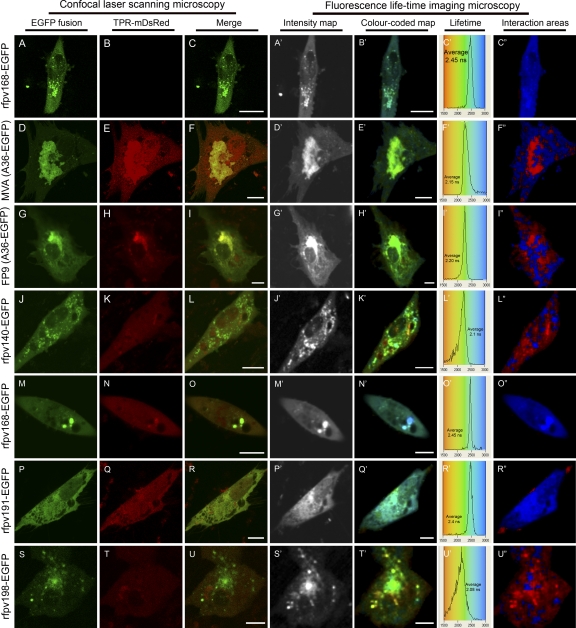
The 2P-FRET-FLIM technique was employed to demonstrate the expected in vivo interaction of the EGFP-VACV A36 (A36-EGFP) full-length fusion with KLC-TPR-mDsRed. Live CEF cells infected with MVA and transfected with plasmid expression constructs for either A36-EGFP or actin-EGFP alone, without KLC-TPR-mDsRed, were used to provide controls to establish the unquenched lifetime of EGFP in the context of the fusion proteins. At 24 h postinfection (hpi), the average excited-state fluorescence lifetime of EGFP in the A36, actin, or Fpv168 fusion, following two-photon excitation (910 nm) in the absence of KLC-TPR-mDsRed acceptor, was determined to be 2.45 ± 0.1 ns (Table 2). Images of a representative control cell expressing Fpv168-EGFP, including a pseudo-color-coded image displaying a diffraction-limited resolution image of the lifetime of the Fpv168-EGFP fluorophore, are shown in Fig. 1A to C". The excited-state lifetime of A36-EGFP in CEFs at 24 h after infection with MVA and transiently expressing KLC-TPR-mDsRed decreased to an average of 2.15 ± 0.14 ns (Fig. 1F′ and Table 2), indicating that the EGFP and mDsRed moieties are located within 10 nm of each other and that A36 therefore interacts with KLC-TPR in MVA-infected CEFs, consistent with the previous yeast two-hybrid and glutathione-S-transferase (GST) pulldown data (60). At higher magnification, faint puncta indicative of intracellular virions are present (panels F to E′), labeled by the A36-EGFP. The puncta are less bright than those seen in FWPV-infected cells labeled with Fpv140-EGFP (panel J′) or Fpv198-EGFP (panel S′). This may be due to low efficiency of incorporation of A36 into the MVA strain, which has not been well studied in this regard, or to a kinetic effect as MVA infection of CEFs progresses faster than FWPV infection. The locations of the puncta are compatible with areas of interaction in the cytoplasm (panels E′ and F").
TABLE 2.
Analysis of FRET-FLIM data of fowlpox virus protein interactions with kinesina
| Virus | Donor | Acceptor | Lifetime (ns [mean] ± SD) | Significance (P) |
|---|---|---|---|---|
| FWPV or MVA | Actin-EGFP or A36-EGFP or Fpv168-EGFP | None | 2.45 ± 0.1 | |
| MVA | A36-EGFP | KLC-TPR-mDsRed | 2.15 ± 0.14 | 0.0 |
| FWPV | A36-EGFP | KLC-TPR-mDsRed | 2.20 ± 0.1 | 1.58 × 10−7 |
| rFWPV | Fpv140-EGFP (VV H3) | KLC-TPR-mDsRed | 2.1 ± 0.15 | 2.97 × 10−7 |
| rFWPV | Fpv168-EGFP (VV A4) | KLC-TPR-mDsRed | 2.45 ± 0.1 | 0.964 |
| rFWPV | Fpv191-EGFP (VV p4c) | KLC-TPR-mDsRed | 2.4 ± 0.1 | 0.327 |
| rFWPV | Fpv198-EGFP (VV A34) | KLC-TPR-mDsRed | 2.08 ± 0.12 | 1.37 × 10−7 |
Chicken embryo fibroblasts-infected with MVA (positive control) or parental FWPV (FP9 strain) or recombinant FWPV FP9 expressing FWPV EGFP fusion proteins (vaccinia virus [VV] orthologs in brackets) were either transfected with plasmids expressing actin-EGFP, A36-EGFP, or KLC-TPR-mDsRed or cotransfected with two plasmids, one expressing vaccinia virus A36-EGFP and the other expressing KLC-TPR-mDsRed. FRET-FLIM data were collected at 24 h postinfection/transfection and analyzed statistically using a two-tailed Student's t test. Acceptor refers to cotransfection, or not, of plasmid expressing KLC-TPR-mDsRed. n = 5 infected cells.
FIG. 1.
Confocal laser scanning microscopy (CLSM) and FRET-FLIM analysis of transfected and/or poxvirus-infected live chicken embryo fibroblasts at 24-h postinfection. Panels A to C" represent infections with recombinant FWPV (rfpv168-EGFP) showing expression of fusion protein Fpv168-EGFP alone as a representative, unquenched, negative control. In panels D to I" cells were transfected with plasmids expressing A36-EGFP and KLC-TPR-mDsRed and infected with MVA (D to F") or parental FWPV (strain FP9; G to I"). MVA- and FP9-infected cells show colocalization (F and I, respectively) and interaction (E′ to F" and H′ to I", respectively) of VACV A36 with kinesin-TPR. In panels J to U" cells were transfected with plasmid expressing KLC-TPR-mDsRed and infected with recombinant FWPV expressing Fpv140-EGFP (rfpv140-EGFP; J to L"), Fpv168-EGFP (rfpv168-EGFP; M to O"), Fpv191-EGFP (rfpv191-EGFP; P to R"), or Fpv198-EGFP (rfpv198-EGFP; S to U"). Color-coded excited lifetimes of EGFP are shown in panels C′, F′, I′, L′, O′, R′, and U′. Panels C", F", I", L", O", R", and U" show areas of strong interaction (red shades) and noninteraction (blue shades). The distribution curves indicate the relative occurrence frequency of the lifetimes within the lifetime image. The distribution curves in panels L′ and U′ describe a distribution for interaction spread from 1.6 to 2.2 ns. Yellow in CLSM images indicates colocalization. To highlight interacting and noninteracting areas in the color-coded image, the green channel was switched off, and the intensity level was adjusted using Adobe Photoshop (CS4 version). Scale bar, 10 μm.
2P-FRET-FLIM demonstrates that the interaction between ectopically expressed VACV A36 and KLC-TPR is not abrogated in live cells infected with fowlpox virus.
Since interaction of VACV A36 with kinesin was detected in MVA-infected cells by FRET-FLIM, we investigated whether it could also interact with KLC-TPR when expressed ectopically in cells infected with FWPV, a virus which lacks an ortholog of A36 (2, 28). At 24 hpi, the average fluorescence lifetime of A36-EGFP in the presence of KLC-TPR-mDsRed in FWPV-infected CEFs was 2.2 ± 0.1 ns (Fig. 1 and Table 2), comparable to the level in MVA-infected cells (2.15 ± 0.14 ns) and significantly lower than the control value of 2.45 ± 0.1 ns. This indicates that A36 is able to bind to KLC-TPR when expressed in cells infected with either VACV or FWPV. No puncta indicative of FWPV virions labeled with A36-EGFP were visible though there was significant interaction outside the viral factory (Fig. 1I"). It may be that in FWPV-infected cells, the interaction occurs only between KLC-TPR and A36 embedded in cellular, rather than viral, membranes.
Expression and localization of EGFP fusions of FWPV structural proteins in live, FWPV-infected cells.
Because FWPV does not encode an equivalent of VACV IEV surface protein A36 and because the IMV of FWPV are not generally wrapped to produce IEV, it is obvious that FWPV proteins other than an A36 equivalent must be responsible for recruiting kinesin so that the progeny virions, which are predominantly IMV, might be transported from the viral factory directly to the cell surface for budding. Equivalents of VACV IMV surface protein A27, as well as IEV surface proteins A33, A56, and B5, are also not encoded. The most likely remaining candidates are the known FWPV IMV surface proteins, namely, Fpv140, equivalent to VACV IMV surface protein H3, and Fpv191, equivalent to the “virion occlusion protein” encoded by the p4c gene (A26L) found in VACV Western Reserve (Joklik strain) (30). The FWPV equivalents of proteins found on the surface of VACV IEV (such as VACV A34 and F12) were considered less likely as KLC-TPR-interacting partners for IMV motility, but their distribution, location, and topology, which have not been described in FWPV-infected cells, might conceivably be different from those observed for their VACV equivalents.
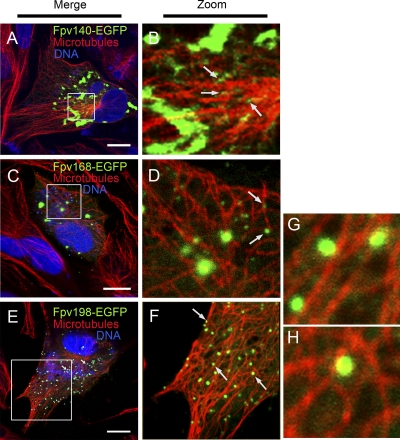
EGFP fusions of FWPV equivalents of H3, p4c, A34, and, as a likely clear negative control, IMV core protein A4, were expressed in cells infected with rFWPV carrying the appropriate EGFP fusion gene, as well as the parental gene expressing the respective native protein (Fpv140, Fpv191, Fpv198, and Fpv168, respectively). The locations of the expressed EGFP fusion proteins were confirmed by confocal microscopy using specific FWPV monoclonal antibodies for colocalization studies. The intracellular locations of EGFP fusions of three of these FWPV proteins (Fpv140, Fpv168, and Fpv191) in the cytoplasm at 24 hpi (Fig. 2, 3, and 4) were comparable to locations of the corresponding native proteins determined by immunofluorescence microscopy using MAbs, as reported previously (3); no antibodies are available for Fpv198. In cells infected with rFWPV expressing Fpv140-EGFP, Fpv168-EGFP, or Fpv198-EGFP and stained with anti-tubulin, the EGFP fusion proteins were seen to localize to virions, some of which were found to be in close proximity to microtubules (Fig. 2). At 24 hpi, in addition to diffuse distribution throughout the cytoplasm, Fpv191-EGFP showed labeling of cytoplasmic virions, which was partially coincident with labeling of virions by monoclonal antibody to Fpv191 (Fig. 2).
FIG. 2.
Localization of three of the EGFP-tagged FWPV proteins to FWPV virions in close proximity to microtubules in recombinant FWPV FP9-infected chicken embryo fibroblast cells. Cells infected with individual recombinant FWPVs expressing one of the EGFP fusions (green) of FWPV protein Fpv140-EGFP (A and B), Fpv168-EGFP (C and D), or Fpv198-EGFP (E to H) were stained at 24 hpi for microtubules (using mouse anti-tubulin antibody/Alexa Fluor 568 goat anti-mouse IgG; red) and DNA (with ToPRO-3; blue). Merged channels are shown in panels A, C, and E; zoomed sections of merged green and red channels are shown in panels B, D, and F to H. Solid arrows indicate virus particles. Scale bar, 10 μm.
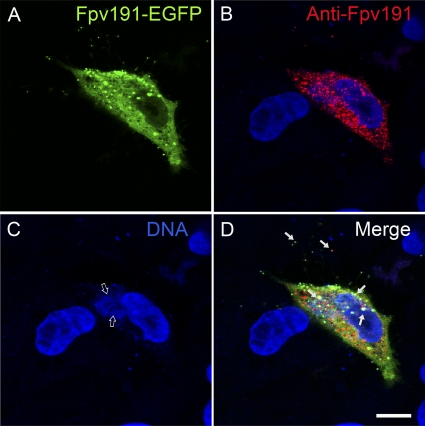
FIG. 3.
Recognition of EGFP fusion of FWPV protein Fpv191-EGFP by monoclonal antibody raised against native protein Fpv191. A cell infected with recombinant FWPV (A to D) expressing both parental Fpv191 and Fpv191-EGFP (green), stained (red) using anti-Fpv191 monoclonal antibody (mDH6) and Alexa Fluor 568 goat anti-mouse IgG, and analyzed by immunofluorescence microscopy, at 24 h postinfection. Colocalization is incomplete; solid arrows indicate incidences of colocalization compatible with virions. DNA staining (blue; labeled with ToPRO-3) reveals the location of the viral factories (indicated by open arrows). Scale bar, 10 μm.
FIG. 4.
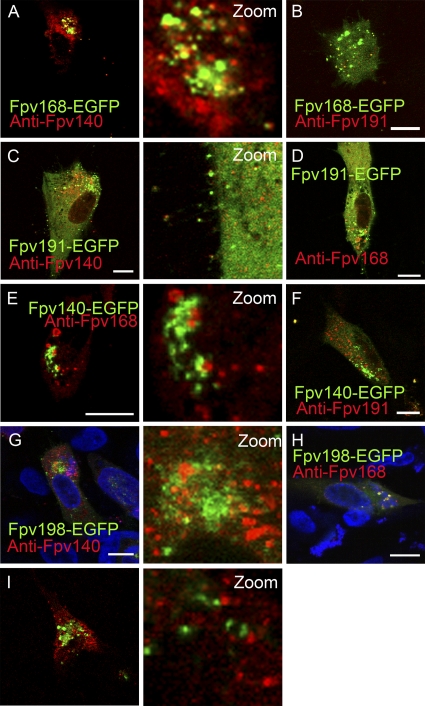
Colocalization of FWPV EGFP fusion proteins with FWPV structural proteins recognized by MAbs. Chicken embryo fibroblasts infected with recombinant FWPV FP9 (rFWPV) expressing the native structural proteins and one of the EGFP fusion protein (green; indicated on the figure) were immunolabeled at 24 hpi with anti-Fpv140 (MAb DF6; A, C, and G), anti-Fpv168 (MAb GB9; D, E, and H) or anti-Fpv191 (MAb DH6; B, F, and I) antibody. DNA was labeled with ToPRO-3 (blue; G and H). Merged images are shown, as are zoomed green and red merged channels. Yellow indicates colocalization of EGFP fusion proteins with immunolabeled proteins. Scale bar, 10 μm.
Staining of cells infected with rFWPV expressing each of the EGFP fusion proteins using monoclonal antibodies against the various structural proteins revealed considerable colocalization, especially on virions, indicating that the EGFP fusion proteins functioned as expected. Fpv168-EGFP showed good, incomplete colocalization on cytoplasmic virions immunolabeled with anti-Fpv140 (Fig. 4A) or anti-Fpv191 (Fig. 4B). Fpv191, whether as an EGFP fusion protein or immunolabeled, displayed good, incomplete colocalization with Fpv140, immunolabeled or as an EGFP-fusion protein, on extracellular virions (Fig. 4C and F, respectively). Fpv140 and Fpv168 did not colocalize on cytoplasmic pools, regardless of the route of labeling (Fig. 4A and E), nor did Fpv191 colocalize on cytoplasmic pools with Fpv168 or Fpv140-EGFP as an EGFP fusion protein (Fig. 4 D) or immunolabeled (Fig. 4F), respectively. Fpv198-EGFP showed little sign of colocalization with immunolabeled Fpv140 and Fpv191 (Fig. 4G and I, respectively), but there was evidence of colocalization with immunolabeled Fpv168 in cytoplasmic pools (Fig. 4 H).
Use of 2P-FRET-FLIM to probe interactions between EGFP fusions of FWPV proteins and KLC-TPR-mDsRed in live, FWPV-infected cells.
Having verified that the EGFP fusions of the FWPV structural proteins behaved as expected in vivo, cells infected with rFWPV expressing each EGFP fusion protein were transfected with the KLC-TPR-mDsRed expression plasmid to permit 2P-FRET-FLIM analysis (Fig. 1, panels J to U"). By confocal microscopy, the infected cells expressing EGFP fusions of Fpv140 (panel L), Fpv168 (panel O), Fpv191 (panel R), and Fpv198 (panel U) did not show overt colocalization with mDsRed kinesin-TPR. Subsequent 2P-FRET-FLIM analysis showed that the average fluorescence lifetime of Fpv168-EGFP in the presence of KLC-TPR-mDsRed in FWPV-infected CEFs was 2.45 ± 0.1 ns (Fig. 1N′ to O" and Table 2), indicating the expected lack of interaction between the internal IMV core protein (equivalent to VACV A4) and KLC-TPR-mDsRed. However, the average fluorescence lifetime of Fpv140-EGFP was 2.1 ± 0.15 ns, comparable to the value observed for A36-EGFP in MVA-infected cells expressing KLC-TPR-mDsRed (Fig. 1K′ to L" and Table 2), indicating an interaction between IMV surface protein Fpv140 (equivalent to VACV H3) and KLC-TPR. In contrast, Fpv191-EGFP, which, like its VACV equivalent p4c, is also found on the surface of IMV, had an average fluorescence lifetime of 2.4 ± 0.1 ns (Fig. 1Q′ to R" and Table 2), indicating that it does not interact with KLC-TPR-mDsRed. Somewhat surprisingly, the average fluorescence lifetime for Fpv-198-EGFP was 2.08 ± 0.12 ns (Fig. 1T′ to U" and Table 2), indicating interaction between the FWPV equivalent of VACV A34 and KLC-TPR.
DISCUSSION
It is likely that the absence from the FWPV genome (and from genomes of other avipoxviruses) of a gene encoding a homologue of A36 (as well as homologues of A33, A56, and B5) is linked, either causally or consequentially, to the formation of FWPV EV by a budding rather than by a wrapping pathway. Nevertheless, FWPV virions, in the form of IMV, still need to be transported, presumably directly, from the factory to the cell membrane. Whether the mechanism for this transport, with the viral proteins required, is the same as that used to transport VACV IMV from the factory to the site of IEV formation or is a different mechanism is not clear. Indeed, the mechanism of transport of VACV IMV has received limited attention (40, 59).
The sensitivity of EGFP fused to the C terminus of A36 (a type I glycoprotein) to quenching by mDsRed fused to KLC-TPR demonstrates that the VACV protein can be expressed in a suitable location and in a suitable topology for its C terminus to interact with cytoplasmic kinesin, not only in cells infected by MVA but also in cells infected by FWPV. In VACV, the C terminus of A36 is displayed on the cytoplasmic face of the IEV (as well as on the cytoplasmic side of the plasma membrane, under CEV). The localization of A36-EGFP when transiently expressed in FWPV-infected cells has not been carefully investigated though confocal imaging revealed that FWPV, like VACV, is able to recruit VACV A36-EGFP to a perinuclear region. It has been shown that in VACV-infected cells, A36 is unable to recruit kinesin by itself and that it requires the activity of VACV F12 (31). FWPV does express an equivalent of F12, namely, Fpv109, which is presumably competent at recruiting VACV A36 to kinesin though the nature of the structure at which the recruitment occurs is not known.
The use of the highly sensitive FRET-FLIM technique was then extended to detect interactions in rFWPV-infected live CEFs between KLC-TPR and EGFP-tagged FWPV structural proteins. That all of these proteins are present in or on virions is indicated by colocalization of the EGFP fusions for Fpv140, Fpv168, Fpv191, and, rarely, Fpv198 with small puncta labeled with monoclonal antibody to Fpv191 (Fig. 3 and 4).
As expected, the 39-kDa internal core protein Fpv168 (homologous to VACV A4) showed no evidence by FRET-FLIM of interacting with KLC-TPR. However, the observed interaction between Fpv140 (homologous to the VACV IMV surface protein H3) and KLC-TPR-mDsRed represents evidence of a novel interaction among poxviruses. Evidence that this interaction is not merely a serendipitous interaction between any protein on the IMV surface and KLC-TPR is provided by the lack of interaction observed between Fpv191 (homologous to VACV p4c) and KLC-TPR.
Fpv140, like its VACV homolog H3 (30 and 37 kDa), has been shown to have multiple nonglycosylated forms (30 and 35 kDa) and has been identified as a peripheral membrane protein present on the external surface of IMV particles (3). Fpv140 contains a C-terminal hydrophobic sequence but lacks a signal sequence, and the protein is expected to be incorporated posttranslationally into the IMV membrane via the C-terminal hydrophobic domain, as is the VACV H3 protein (12). Like VACV H3 (29), Fpv140 contains potential heparan sulfate binding sites (3), which in VACV allow IMV entry. However, no interaction between H3 and kinesin has been reported for VACV.
The detection of an interaction between Fpv198 (homologous to VACV A34) and KLC-TPR raises the question of how such an interaction might be mediated, given the localization of VACV A34 to the IEV surface and the general lack of production of FWPV IEV. In some ways, though, that interaction might be viewed as similar to the interaction observed when VACV A36-EGFP is expressed ectopically in the FWPV-infected cell, as the tagged N terminus of A34 and the tagged C terminus of A36 are displayed on the cytoplasmic surface of VACV IEV. However, the C-terminal domain of A36 is much longer than the N-terminal domain of A34. In VACV, A34 is a type II integral membrane glycoprotein found in the Golgi compartment, on EV, and on the surface of IEV. Interaction between VACV A34 and kinesin was excluded based on the evidence that the N-terminal (cytoplasmic) domain did not bind to KLC-TPR in a yeast two-hybrid system (60). It has recently been shown that migration of A34 from the Golgi compartment is dependent on the presence of the “stalk” region of B5 (6), but FWPV encodes no homolog of B5. The location of the FWPV equivalent of VACV A34, Fpv198 (which shares only 31% amino acid identity with A34), has not been characterized, but the morphogenesis of FWPV primarily by IMV budding means that it might be considered surprising for Fpv198-EGFP to be found on cytoplasmic IEV unless they represent a minority population of intracellular virions. It would be less surprising for Fpv198 to traffic on cellular vesicles, displaying the same topology as in VACV IEV, to the cell surface, where it might be acquired as an EV protein by budding FWPV IMV, though the EV surface proteins of FWPV have not yet been defined. Confocal imaging shows rare virions (it is not clear whether these are intra- or extracellular), which are labeled for DNA and immunolabeled for Fpv191 (p4c equivalent), also displaying Fpv198-EGFP. It is even remotely conceivable that in FWPV Fpv198 might represent an IMV surface protein, transferred from the endoplasmic reticulum to IMV with retention of topology, as has been observed for VACV A9 (24).
Fpv191 (63 kDa) is a member of the A-type inclusion (ATI)-like protein family. However, the protein has been shown to be present in IMV and EV, but, unlike Fpv140, Fpv168, and Fpv198, the protein does not colocalize with viral factories in FWPV-infected cells (3). The origins, nature, and relationships of this protein and its orthologs have been considered in considerable detail elsewhere (3, 30). The manner of association of Fpv191 with FWPV IMV is unclear but is probably equivalent to the IMV association of VACV proteins p4c and A27 (as they do not colocalize with virus factories but bind after formation of IMV [41, 44, 49], involving protein-protein interactions with other VACV IMV membrane proteins, such as A17 [23]). It was suggested that VACV A26 (p4c) might act as a regulator to suppress formation of EV in favor of IMV production (53). It is possible, therefore, that Fpv191 (and possibly p4c) might be involved in disrupting the interaction of membrane-bound IMV with KLC-TPR (though ATI-bound IMV might require an alternative transport mechanism).
The observation of interactions between Fpv140 and KLC-TPR and between Fpv198 and KLC-TPR raises the question of the nature of the molecular determinants for those interactions. Recent analysis of VACV F12 revealed the presence of TPRs and structural similarity between the TPR domain of F12 and that of KLC (31). The same authors also report the presence in VACV A36 and E2 of TPRs. Fpv140 also shows the presence of repeats with some similarity to TPRs (Gareth Morgan, personal communication), which could account for the observed interaction with KLC-TPR in FWPV-infected cells.
It is much more difficult to account for the observed interaction between Fpv198 and KLC-TPR, particularly if Fpv198 displays the same topology and similar localization in FWPV-infected cells as A34 in VACV-infected cells. In that case, Fpv198 would display only a short N-terminal cytoplasmic sequence (the location of the EGFP tag) of 13 amino acids for interaction with KLC-TPR. It is possible that localization of the TPR to this domain is mediated by another FWPV protein with a longer cytoplasmic domain, with this protein interacting with the longer domain of Fpv198 in the lumenal compartment. Resolution of this issue will, no doubt, require further investigation into the localization and topology of Fpv198 in FWPV-infected cells.
In conclusion, using the highly sensitive time-resolved FRET-FLIM imaging technique, this study has demonstrated that Fpv140 and Fpv198 interact with KLC-TPR in FWPV-infected cells. VACV A36 and Fpv168 served as positive and negative controls, respectively. The Fpv198 interaction may represent interaction of a rare population of FWPV IEV with kinesin though it might represent interaction of host cell transport vesicles with kinesin. The Fpv140 interaction, a novel interaction among poxviruses, probably represents an interaction between FWPV IMV and kinesin, allowing transport directly from the viral factory to the cell membrane for budding. Although it is an intriguing possibility that the VACV ortholog H3 might similarly interact with KLC-TPR, the divergence in sequence (FWPV and VACV orthologs share only 31% amino acid identity) and in morphogenesis (budding versus wrapping) indicates that this is by no means a foregone conclusion. The lack of interaction for Fpv191 demonstrates that not all IMV proteins interact with KLC-TPR and might even indicate a regulatory role for Fpv191, as has been suggested for VACV p4c.
Acknowledgments
We thank Amanda Lewis and Alasdair Mackenzie for assistance with the FLIM data collection. We are grateful to Gareth Morgan and Adrien Breiman for discussions and access to prepublished data.
The Biotechnology and Biological Sciences Research Council is acknowledged for CSG funding (Avipox Viromics Project, reference BBSEI00001019) to M.A.S. for construction of recombinant fowlpox viruses conducted while he was at the Institute for Animal Health, Compton, Berkshire RG20 7NN, United Kingdom. Oxford Brookes University is acknowledged for a research fellowship fund awarded to A.J. Access to the Central Laser Facility and the multiphoton laboratory was funded by the Science and Technology Facilities Council.
Footnotes
Published ahead of print on 13 October 2010.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, G. Delhon, Z. Lu, G. J. Viljoen, D. B. Wallace, G. F. Kutish, and D. L. Rock. 2006. Genome of crocodilepox virus. J. Virol. 80:4978-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2000. The genome of fowlpox virus. J. Virol. 74:3815-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boulanger, D., P. Green, B. Jones, G. Henriquet, L. G. Hunt, S. M. Laidlaw, P. Monaghan, and M. A. Skinner. 2002. Identification and characterization of three immunodominant structural proteins of fowlpox virus. J. Virol. 76:9844-9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulanger, D., P. Green, T. Smith, C. P. Czerny, and M. A. Skinner. 1998. The 131-amino-acid repeat region of the essential 39-kilodalton core protein of fowlpox virus FP9, equivalent to vaccinia virus A4L protein, is nonessential and highly immunogenic. J. Virol. 72:170-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulanger, D., T. Smith, and M. A. Skinner. 2000. Morphogenesis and release of fowlpox virus. J. Gen. Virol. 81:675-687. [DOI] [PubMed] [Google Scholar]
- 6.Breiman, A., and G. L. Smith. 2010. Vaccinia virus B5 protein affects the glycosylation, localization and stability of the A34 protein. J. Gen. Virol. 91:1823-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, M., Y. Zhang, S. Dermine, E. A. de Wynter, C. Hart, H. Kitchener, P. L. Stern, M. A. Skinner, and S. N. Stacey. 2000. Dendritic cells infected with recombinant fowlpox virus vectors are potent and long-acting stimulators of transgene-specific class I restricted T lymphocyte activity. Gene Ther. 7:1680-1689. [DOI] [PubMed] [Google Scholar]
- 8.Bublot, M., N. Pritchard, J. S. Cruz, T. R. Mickle, P. Selleck, and D. E. Swayne. 2007. Efficacy of a fowlpox-vectored avian influenza H5 vaccine against Asian H5N1 highly pathogenic avian influenza virus challenge. Avian Dis. 51:498-500. [DOI] [PubMed] [Google Scholar]
- 9.Chan, W. M., and B. M. Ward. 2010. There is an A33-dependent mechanism for the incorporation of B5-GFP into vaccinia virus extracellular enveloped virions. Virology 402:83-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Y., and A. Periasamy. 2004. Characterization of two-photon excitation fluorescence lifetime imaging microscopy for protein localization. Microsc. Res. Tech. 63:72-80. [DOI] [PubMed] [Google Scholar]
- 11.Cudmore, S., P. Cossart, G. Griffiths, and M. Way. 1995. Actin-based motility of vaccinia virus. Nature 378:636-638. [DOI] [PubMed] [Google Scholar]
- 12.da Fonseca, F. G., E. J. Wolffe, A. Weisberg, and B. Moss. 2000. Characterization of the vaccinia virus H3L envelope protein: topology and posttranslational membrane insertion via the C-terminal hydrophobic tail. J. Virol. 74:7508-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delhon, G., E. R. Tulman, C. L. Afonso, Z. Lu, A. de la Concha-Bermejillo, H. D. Lehmkuhl, M. E. Piccone, G. F. Kutish, and D. L. Rock. 2004. Genomes of the parapoxviruses ORF virus and bovine papular stomatitis virus. J. Virol. 78:168-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dodding, M. P., T. P. Newsome, L. M. Collinson, C. Edwards, and M. Way. 2009. An E2-F12 complex is required for IEV morphogenesis during vaccinia infection. Cell Microbiol. 11:808-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domi, A., A. S. Weisberg, and B. Moss. 2008. Vaccinia virus E2L null mutants exhibit a major reduction in extracellular virion formation and virus spread. J. Virol. 82:4215-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan, S. A., and G. L. Smith. 1992. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J. Virol. 66:1610-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelstad, M., S. T. Howard, and G. L. Smith. 1992. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology 188:801-810. [DOI] [PubMed] [Google Scholar]
- 18.Gindhart, J. G., Jr., and L. S. Goldstein. 1996. Tetratrico peptide repeats are present in the kinesin light chain. Trends Biochem. Sci. 21:52-53. [PubMed] [Google Scholar]
- 19.Hatano, Y., M. Yoshida, F. Uno, S. Yoshida, N. Osafune, K. Ono, M. Yamada, and S. Nii. 2001. Budding of fowlpox and pigeonpox viruses at the surface of infected cells. J. Electron Microsc. (Tokyo) 50:113-124. [DOI] [PubMed] [Google Scholar]
- 20.Hautaniemi, M., N. Ueda, J. Tuimala, A. A. Mercer, J. Lahdenpera, and C. J. McInnes. 2010. The genome of Pseudocowpoxvirus: comparison of a reindeer isolate and a reference strain. J. Gen. Virol. 91:1560-1576. [DOI] [PubMed] [Google Scholar]
- 21.Herrero-Martinez, E., K. L. Roberts, M. Hollinshead, and G. L. Smith. 2005. Vaccinia virus intracellular enveloped virions move to the cell periphery on microtubules in the absence of the A36R protein. J. Gen. Virol. 86:2961-2968. [DOI] [PubMed] [Google Scholar]
- 22.Hollinshead, M., G. Rodger, H. Van Eijl, M. Law, R. Hollinshead, D. J. Vaux, and G. L. Smith. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howard, A. R., T. G. Senkevich, and B. Moss. 2008. Vaccinia virus A26 and A27 proteins form a stable complex tethered to mature virions by association with the A17 transmembrane protein. J. Virol. 82:12384-12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Husain, M., A. S. Weisberg, and B. Moss. 2006. Existence of an operative pathway from the endoplasmic reticulum to the immature poxvirus membrane. Proc. Natl. Acad. Sci. U. S. A. 103:19506-19511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isaacs, S. N., E. J. Wolffe, L. G. Payne, and B. Moss. 1992. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J. Virol. 66:7217-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeshtadi, A., G. Henriquet, S. M. Laidlaw, D. Hot, Y. Zhang, and M. A. Skinner. 2005. In vitro expression and analysis of secreted fowlpox virus CC chemokine-like proteins Fpv060, Fpv061, Fpv116 and Fpv121. Arch. Virol. 150:1745-1762. [DOI] [PubMed] [Google Scholar]
- 27.Johnston, S. C., and B. M. Ward. 2009. Vaccinia virus protein F12 associates with intracellular enveloped virions through an interaction with A36. J. Virol. 83:1708-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laidlaw, S. M., and M. A. Skinner. 2004. Comparison of the genome sequence of FP9, an attenuated, tissue culture-adapted European strain of Fowlpox virus, with those of virulent American and European viruses. J. Gen. Virol. 85:305-322. [DOI] [PubMed] [Google Scholar]
- 29.Lin, C. L., C. S. Chung, H. G. Heine, and W. Chang. 2000. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 74:3353-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKelvey, T. A., S. C. Andrews, S. E. Miller, C. A. Ray, and D. J. Pickup. 2002. Identification of the orthopoxvirus p4c gene, which encodes a structural protein that directs intracellular mature virus particles into A-type inclusions. J. Virol. 76:11216-11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan, G. W., M. Hollinshead, B. J. Ferguson, B. J. Murphy, D. C. Carpentier, and G. L. Smith. 2010. Vaccinia protein F12 has structural similarity to kinesin light chain and contains a motor binding motif required for virion export. PLoS Pathog. 6:e1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moss, B. 2006. Poxvirus entry and membrane fusion. Virology 344:48-54. [DOI] [PubMed] [Google Scholar]
- 33.Osterrieder, A., C. M. Carvalho, M. Latijnhouwers, J. N. Johansen, C. Stubbs, S. Botchway, and C. Hawes. 2009. Fluorescence lifetime imaging of interactions between Golgi tethering factors and small GTPases in plants. Traffic 10:1034-1046. [DOI] [PubMed] [Google Scholar]
- 34.Ploubidou, A., V. Moreau, K. Ashman, I. Reckmann, C. Gonzalez, and M. Way. 2000. Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO J. 19:3932-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rerks-Ngarm, S., P. Pitisuttithum, S. Nitayaphan, J. Kaewkungwal, J. Chiu, R. Paris, N. Premsri, C. Namwat, M. de Souza, E. Adams, M. Benenson, S. Gurunathan, J. Tartaglia, J. G. McNeil, D. P. Francis, D. Stablein, D. L. Birx, S. Chunsuttiwat, C. Khamboonruang, P. Thongcharoen, M. L. Robb, N. L. Michael, P. Kunasol, and J. H. Kim. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209-2220. [DOI] [PubMed] [Google Scholar]
- 36.Rietdorf, J., A. Ploubidou, I. Reckmann, A. Holmstrom, F. Frischknecht, M. Zettl, T. Zimmermann, and M. Way. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 3:992-1000. [DOI] [PubMed] [Google Scholar]
- 37.Roberts, K. L., and G. L. Smith. 2008. Vaccinia virus morphogenesis and dissemination. Trends Microbiol. 16:472-479. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez, J. F., and M. Esteban. 1987. Mapping and nucleotide sequence of the vaccinia virus gene that encodes a 14-kilodalton fusion protein. J. Virol. 61:3550-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roper, R. L., L. G. Payne, and B. Moss. 1996. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J. Virol. 70:3753-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanderson, C. M., M. Hollinshead, and G. L. Smith. 2000. The vaccinia virus A27L protein is needed for the microtubule-dependent transport of intracellular mature virus particles. J. Gen. Virol. 81:47-58. [DOI] [PubMed] [Google Scholar]
- 41.Sarov, I., and W. K. Joklik. 1973. Isolation and characterization of intermediates in vaccinia virus morphogenesis. Virology 52:223-233. [DOI] [PubMed] [Google Scholar]
- 42.Scolari, S., S. Engel, N. Krebs, A. P. Plazzo, R. F. De Almeida, M. Prieto, M. Veit, and A. Herrmann. 2009. Lateral distribution of the transmembrane domain of influenza virus hemagglutinin revealed by time-resolved fluorescence imaging. J. Biol. Chem. 284:15708-15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Senkevich, T. G., E. V. Koonin, J. J. Bugert, G. Darai, and B. Moss. 1997. The genome of molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology 233:19-42. [DOI] [PubMed] [Google Scholar]
- 44.Shida, H., K. Tanabe, and S. Matsumoto. 1977. Mechanism of virus occlusion into A-type inclusion during poxvirus infection. Virology 76:217-233. [DOI] [PubMed] [Google Scholar]
- 45.Skinner, M. A., S. M. Laidlaw, I. Eldaghayes, P. Kaiser, and M. G. Cottingham. 2005. Fowlpox virus as a recombinant vaccine vector for use in mammals and poultry. Expert Rev. Vaccines 4:63-76. [DOI] [PubMed] [Google Scholar]
- 46.Smith, G. L., A. Vanderplasschen, and M. Law. 2002. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 83:2915-2931. [DOI] [PubMed] [Google Scholar]
- 47.Snippe, M., J. W. Borst, R. Goldbach, and R. Kormelink. 2005. The use of fluorescence microscopy to visualise homotypic interactions of tomato spotted wilt virus nucleocapsid protein in living cells. J. Virol. Methods 125:15-22. [DOI] [PubMed] [Google Scholar]
- 48.Snippe, M., J. Willem Borst, R. Goldbach, and R. Kormelink. 2007. Tomato spotted wilt virus Gc and N proteins interact in vivo. Virology 357:115-123. [DOI] [PubMed] [Google Scholar]
- 49.Sodeik, B., S. Cudmore, M. Ericsson, M. Esteban, E. G. Niles, and G. Griffiths. 1995. Assembly of vaccinia virus: incorporation of p14 and p32 into the membrane of the intracellular mature virus. J. Virol. 69:3560-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stubbs, C. D., S. W. Botchway, S. J. Slater, and A. W. Parker. 2005. The use of time-resolved fluorescence imaging in the study of protein kinase C localisation in cells. BMC Cell Biol. 6:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suhling, K., P. M. French, and D. Phillips. 2005. Time-resolved fluorescence microscopy. Photochem. Photobiol. Sci. 4:13-22. [DOI] [PubMed] [Google Scholar]
- 52.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2004. The genome of canarypox virus. J. Virol. 78:353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ulaeto, D., D. Grosenbach, and D. E. Hruby. 1996. The vaccinia virus 4c and A-type inclusion proteins are specific markers for the intracellular mature virus particle. J. Virol. 70:3372-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vale, R. D. 2003. The molecular motor toolbox for intracellular transport. Cell 112:467-480. [DOI] [PubMed] [Google Scholar]
- 55.Vale, R. D., T. S. Reese, and M. P. Sheetz. 1985. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 42:39-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Eijl, H., M. Hollinshead, G. Rodger, W. H. Zhang, and G. L. Smith. 2002. The vaccinia virus F12L protein is associated with intracellular enveloped virus particles and is required for their egress to the cell surface. J. Gen. Virol. 83:195-207. [DOI] [PubMed] [Google Scholar]
- 57.van Eijl, H., M. Hollinshead, and G. L. Smith. 2000. The vaccinia virus A36R protein is a type Ib membrane protein present on intracellular but not extracellular enveloped virus particles. Virology 271:26-36. [DOI] [PubMed] [Google Scholar]
- 58.Wallrabe, H., and A. Periasamy. 2005. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotechnol. 16:19-27. [DOI] [PubMed] [Google Scholar]
- 59.Ward, B. M. 2005. Visualization and characterization of the intracellular movement of vaccinia virus intracellular mature virions. J. Virol. 79:4755-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ward, B. M., and B. Moss. 2004. Vaccinia virus A36R membrane protein provides a direct link between intracellular enveloped virions and the microtubule motor kinesin. J. Virol. 78:2486-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]