Abstract
RNA silencing is a potent mechanism of antiviral defense response in plants and other organisms. For counterdefense, viruses have evolved a variety of suppressors of RNA silencing (VSRs) that can inhibit distinct steps of a silencing pathway. We previously identified Pns10 encoded by Rice dwarf phytoreovirus (RDV) as a VSR, the first of its kind from double-stranded RNA (dsRNA) viruses. In this study we investigated the mechanisms of Pns10 function in suppressing systemic RNA silencing in the widely used Nicotiana benthamiana model plant. We report that Pns10 suppresses local and systemic RNA silencing triggered by sense mRNA, enhances viral replication and/or viral RNA stability in inoculated leaves, accelerates the systemic spread of viral infection, and enables viral invasion of shoot apices. Mechanistically, Pns10 interferes with the perception of silencing signals in recipient tissues, binds double-stranded small interfering RNA (siRNAs) with two-nucleotide 3′ overhangs, and causes the downregulated expression of RDR6. These results significantly deepen our mechanistic understanding of the VSR functions encoded by a dsRNA virus and contribute additional evidence that binding siRNAs and interfering with RDR6 expression are broad mechanisms of VSR functions encoded by diverse groups of viruses.
RNA silencing is an important defense mechanism against viral infection, overexpressed transgenes, and transposon activities in multiple organisms (23, 34, 36). Silencing is triggered by double-stranded RNAs (dsRNAs) generated from replicating viral RNAs, transgenes, or endogenous transposons. The dsRNAs are cleaved into duplexes of small interfering RNAs (siRNAs) of 20 to 24 nucleotides (nt) by specific DICERS (1, 7, 10, 11, 13, 18, 41). One strand of a duplex is subsequently incorporated into an Argonaute (AGO)-containing RNA-induced silencing complex to guide cleavage of homologous RNA molecules or inhibition of translation (1, 8, 14).
DICERs, AGOs, and cellular RNA-dependent RNA polymerases (RDRs) all play critical roles in RNA silencing against viruses (7, 8, 20, 28, 37). The roles of some RDRs in antiviral defense have been extensively studied. In Nicotiana benthamiana, NbRDR1 is associated with increased susceptibility to viruses (44). NbRDR6 has a role in plant defense against a broad spectrum of viruses including prevention of shoot apex invasion (27, 30). Recent work demonstrated that secondary siRNAs generated through the activities of NbRDR6 are most important for antiviral responses (32). RDRs also influence the extent to which silencing moves from cell to cell through plasmodesmata and over long distances through the phloem (34). NbRDR6 is required for a cell to respond to, but not to produce, systemic silencing signals (30).
Many plant viruses encode viral suppressors of RNA silencing (VSRs) to counteract host RNA silencing (8). The VSRs identified thus far can interfere with silencing at different steps. For instance, the helper component-proteinase (HC-Pro) of potyviruses inhibits silencing at a step upstream of the production of siRNAs (19, 24, 25, 46). In particular, the HC-Pro of Sugarcane mosaic potyvirus and the Tav2b of Tomato aspermy cucumovirus downregulate the accumulation of RDR6 mRNA, leading to lower accumulations of 3′ or 5′ secondary siRNAs (46). The 2b of Cucumber mosaic virus can prevent spread of RNA silencing signals by blocking their long-distance translocation (12). p21 of Beet yellows virus, p14 of Pothos latent virus, and p19 of tombusviruses bind and presumably inactivate siRNAs (4, 6, 24, 33). The p25 of Potato virus X (PVX) interrupts transmission of RNA silencing signals by preventing their formation (35). The p69 of Turnip yellow mosaic virus targets a step upstream of dsRNA formation by RDRs (5).
We have been using Rice dwarf virus (RDV) as a model to investigate how a virus with double-stranded genomic RNAs counteracts host silencing. Given that dsRNA genomes could be direct targets of host RNA silencing, an intriguing question is whether RDR6 is still important for generating dsRNAs for producing secondary siRNAs, as it is for single-stranded RNA viruses, and whether the dsRNA nature of the viral genomes would render its VSR function differently from those encoded by single-stranded RNA viruses. Thus, determining the full extent of VSR functions encoded by a dsRNA virus will help uncover unique and/or common mechanisms among different virus groups.
RDV is a member of the genus Phytoreovirus in the family Reoviridae (2). It is transmitted by insect vectors (Nephotettix cincticeps or Resilia dorsalis) in a circulative manner and causes severe dwarf diseases of rice (31). The RDV genome comprises 12 segmented dsRNAs, which encode seven structural proteins and at least six nonstructural proteins (48). The seven structural proteins are products of segments S1, S2, S3, S5, S7, S8, and S9 (48, 50). The six nonstructural proteins are products of S4, S6, S10, S11, and S12, respectively (3, 21). The functions of some nonstructural proteins are already known. Pns6 is a cell-to-cell movement protein that can increase RDV accumulation and symptom severity (21). Pns11 binds nucleic acids (42). Pns6, Pns11, and Pns12 together form viroplasm (38). Pns10 functions as a VSR (3, 49).
Although RDV causes systemic infection in rice, functional studies of RDV proteins have been hampered by (i) the lack of infectious RDV cDNA clones and (ii) the intractability of rice as an experimental system. Therefore, expressing individual RDV proteins in heterologous plant models remains a major approach of characterizing the functions of many RDV proteins, including Pns10. We investigated the mechanisms of Pns10 function in suppressing systemic RNA silencing in the widely used N. benthamiana model plant. We report that Pns10 suppresses local and systemic RNA silencing triggered by sense mRNA, enhances viral replication and/or viral RNA stability in inoculated leaves, accelerates systemic spread of viral infection, and enables viral invasion of shoot apices. Mechanistically, Pns10 interferes with the perception of silencing signals in recipient tissues, binds ds-siRNAs with 2-nt 3′ overhangs and causes downregulated expression of RDR6. These results significantly deepen our mechanistic understanding of the VSR functions encoded by a dsRNA virus and contribute additional evidence that binding siRNAs and interfering with RDR6 expression are broad mechanisms of VSR functions encoded by diverse groups of viruses.
MATERIALS AND METHODS
Plasmid construction.
Plasmids pE3-S10, pE3-GFP, pJawohl8-RNAi-GFP and Pns10 and its mutant proteins expressed from the chimeric PVX vector were constructed as described previously (3). pPVX-GFP, which carries mGFP5 under the control of a duplicate coat protein promoter, was as described previously (30).
Transgenic plants and grafting procedures.
The transgenic line Nb/Pns10 was generated by transformation of N. benthamiana with a construct pE3-S10 (3) expressing Pns10. N. benthamiana line GFP16c was described as line 16c previously in which the green fluorescent protein (GFP) transgene was constitutively expressed in the N. benthamiana plant (29). Pollen grains from line 16c were used to pollinate Nb/Pns10 plants (49) to generate 16c/Pns10. A wedge-grafting method (12, 26) was used to generate graft unions. N. benthamiana plants that were 5 to 6 weeks old were used for grating. A scion stem was cut into a wedge shape that was then inserted into a vertical slit cut into the rootstock stem ∼4 cm above the soil level. The graft junction was wrapped with Parafilm. All grafted plants were kept humid under a plastic cover for 1 week (30). All plants were grown in a greenhouse with 16-h light and 8-h dark periods at a temperature regime of 24/22°C (day/night).
Agroinfiltration and GFP imaging.
Agrobacterium infiltration operation was carried out according to the method described previously (13, 16). For coinfiltration, equal volumes of individual Agrobacterium tumefaciens cultures (optical density at 600 nm of 1) were mixed prior to infiltration. GFP expression was visualized with a 100-W handheld long-wave UV lamp (Black Ray model B 100A; UV Products) and photographed with a Nikon D70 digital camera (46).
Northern blots.
Total RNA from leaves was isolated by using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. For gel blot analysis of high-molecular-weight RNA, 10 μg of total RNA was extracted from leaves and separated on 1% agarose-formaldehyde gels, transferred to Hybond-N+ membranes, and hybridized with digoxigenin (DIG)-labeled probes according to the instructions of the DIG system user's guide (Roche). For the detection of small RNA, 20 μg of total RNA sample were separated on 15% polyacrylamide gels containing 0.5× Tris-borate-EDTA (TBE) and transferred onto Hybond-N+ membranes. The membranes were hybridized with DIG-labeled DNA probes corresponding to the full-length open reading frames (ORFs) of Pns10, PVX coat protein (CP), or GFP. Hybridization and detection of siRNAs were performed as described previously (3). Western blotting analysis of Pns10 expression was performed as described previously (47).
Real-time RT-PCR analysis.
Transcript levels of RDR and Argonaute genes were analyzed by quantitative real-time reverse transcription-PCR (RT-PCR) using a Chromo4 detector in combination with a PTC-200 thermal cycler (MJ Research/Bio-Rad). For each plant line, total RNA was extracted from three plants at the six-leaf stage using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Total cDNA was first synthesized by using the oligo(dT) 15 primer (Promega) and Superscript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Reactions without reverse transcriptase or without template were included as negative controls. For quantitative real-time PCR, cDNA corresponding to 100 ng of total RNA was used in 20-μl reactions using DyNAmo HS SYBR green qPCR kit (Finnzymes, Finland) according to the manufacturer's instructions. Primers were designed to amplify similar-sized regions of GAPDH (glyceraldehyde-3-phosphate dehydrogenase), serving as an internal standard, and the N. benthamiana orthologs of RDR6, RDR1, AGO1, and AGO4. Primer sequences were as follows: GAPDH, 5′-GGAGGAGGGAACAACAAGAGG-3′ and 5′-AGATGCCGTCAGTGCCGA-3′; RDR6, 5′-CTCAGCTTGGGGACCTCA-3′ and 5′-CAGC CTCCAGAATCCTCAC-3′; RDR1, 5′-GCATTGAACACGCCTTGGA-3′ and 5′-GCAGAACCCGATTGGATACG-3′; AGO1, 5′-CATACCCAGTGGCCTTGTCT-3′ and 5′-CTCCTTCGCATATGGGTCAT-3′; and AGO4, 5′-CCAGGCTGGATCTCCTGATA-3′ and 5′-AACTTACCGGAGCCACAATG-3′. The primer sequences in rice were as follows: Os-actin, 5′-CCCTTAGCACCTTCCAACAG-3′ and 5′-TAGAAGCACTTCCGGTGGAC-3′; Os-RDR1, 5′-TGTCGTCTTTCCACAGCAAG-3′ and 5′-GAGATGGGTCCCAAGAAACA-3′; Os-RDR6, 5′-CCTGACTTCATGGGAAAGGA-3′ and 5′-TGGAGTGCAAACCTCTTGTG-3′; Os-AGO1, 5′-AAGAGGGCAACTGGACAGAA-3′ and 5′-CGCTGGTCCTTGTGGTTATT-3′; and Os-AG4, 5′-GTGGGCACATTCCTCAAGTT-3′ and 5′-GTATGACCTCCTTGGCTGGA-3′. Amplicons were also analyzed on agarose gels to verify single PCR products.
Gel mobility shift assay.
siRNA binding experiments were carried out based on the procedure of Duan et al. (9) with modifications. Synthetic 21- and 24-nt RNA duplexes with 2-nt 3′ overhangs were used as probes for RNA binding assays. The sequences of the 21- and 24-nt RNA duplexes were as follows: 21-nt sequence, 5′-pCGUACGCGGAAUACUUCGAUU-3′ and 3′-UUGCAUGCGCCUUAUGAAGCUp-5′; and 24-nt sequence, 5′-pCGUACGCGGAAUACUUCGAAAGUU-3′ and 3′-UUGCAUGCGCCUUAUGAAGCUUUCp-5′. The Pns10 and Pns10 mutants of RDV were cloned into pMAL-p2x vector containing a maltose-binding protein (MBP) at the N terminus and expressed in Escherichia coli TB1. The recombinant proteins were purified by using an amylose prepacked column according to the manufacturer's protocols (New England Biolabs). The typical binding reaction (20 μl) contained 50 pmol of dsRNA, 10 mM HEPES/KOH (pH 7.6), 10 mM KCl, 1 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM MgCl2, and 10 μg of purified wild-type Pns10 protein or mutant proteins. His-P19 was used as a positive control. The binding reaction mixture was incubated at room temperature for 30 min, and the complexes was separated from the free duplex in a 5% nondenaturing polyacrylamide 0.5× TBE gel. The gel was stained with ethidium bromide. Images were captured by using the fluorescence imaging system (Alpha Innotech Corp.).
In situ hybridization.
Procedures for the preparation of plant shoot meristem sections and in situ hybridization using a DIG-labeled RNA probe complementary to the CP sequence of PVX were carried out according to a previously described method (43). A specific sequence of PVX CP was amplified as a probe using the primers 5′-ACAGGCTGCTTGGGACTTAG-3′ and 5′-TCTAGGCTGGCAAAGTCGTT-3′ in PCR. The fragment was cloned into the T-Easy vector for labeling. The sense and antisense probes were synthesized using linear plasmid templates according to the DIG RNA labeling kit manual (Roche, Mannheim, Germany). The stained sections were examined under a BX-50 light microscope (Leica DMRE).
CD spectroscopic assays.
All circular dichroism (CD) spectra of the wild-type and mutant Pns10 proteins were recorded on a Jobin Yvon CD 6 spectrometer (Longjumeau, France) at 298K according to a previously described method (9). The CD spectra of all proteins were recorded in phosphate-buffered saline (pH 7.5). For the near-UV/CD spectrum, a cell with a path length of 1 mm was used. Each spectrum was the average of four scans corrected by subtracting a spectrum of the buffer solution in the absence of proteins recorded under identical conditions. Each scan in the range of 195 to 260 nm was obtained by taking data points every 0.5 nm with a 2-nm bandwidth and an integration time of 1 s.
RESULTS
Transgenically expressed Pns10 in N. benthamiana suppresses local and systemic RNA silencing triggered by sense mRNA.
We previously showed that transiently expressed Pns10 suppressed local and systemic silencing induced by a sense mRNA but did not interfere with local and systemic silencing induced by a dsRNA in N. benthamiana 16c (3). To facilitate further investigation into the role of Pn10 in systemic RNA silencing suppression, it will be convenient to use transgenic plant lines that stably express this protein. To this end, we generated transgenic N. benthamiana 16c/Pns10 plants in which Pns10 was constitutively expressed by crossing a transgenic line Nb/Pns10 (46) with line 16c (see Materials and Methods). Expression of Pns10 in 16c/Pns10 lines was confirmed by Western blots. One example is shown in Fig. 1A.
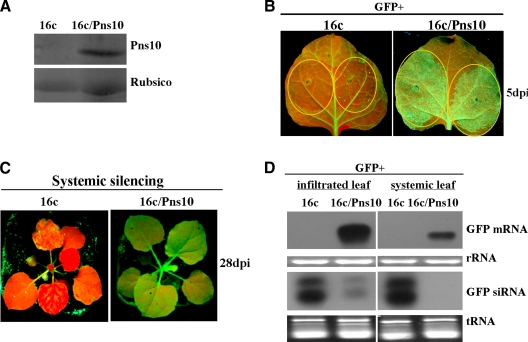
FIG. 1.
RDV Pns10 suppressor activity in transgenic N. benthamiana 16c/Pns10, in which GFP and Pns10 were stably expressed. (A) Western blot of total proteins extracted from 16c and 16c/Pns10 plant leaves, probed with polyclonal antisera specific for Pns10. Coomassie blue-stained Rubisco proteins were used as the loading control. (B) N. benthamiana 16c and 16c/Pns10 plants were infiltrated with Agrobacterium carrying 35S-GFP. GFP fluorescence was imaged under long-wavelength UV light at 5 dpi. (C) GFP fluorescence was monitored for the onset of systemic silencing at 28 dpi. Of 12 infiltrated 16c/Pns10 plants, 11 showed GFP fluorescence indicative of suppression of silencing by Pns10. All 13 infiltrated 16c plants, as controls, lacked GFP fluorescence, a finding indicative of GFP silencing. (D) Northern blots showing the steady-state levels of GFP mRNAs and siRNAs extracted from the different infiltrated patches shown in panels B and C. The bottom panel shows rRNA or tRNA stained with ethidium bromide as a loading control.
To test the VSR function of Pns10 in 16c/Pns10 transgenic plants, we infiltrated the leaves with an Agrobacterium strain carrying 35S-GFP. Infiltration of 16c plant leaves served as a control. GFP fluorescence intensity in the infiltrated leaves of 16c plants started to decline at 3 days postinfiltration (dpi) (data not shown) and became hardly visible at 5 dpi (Fig. 1B, left panel). In contrast, GFP fluorescence in the infiltrated 16c/Pns10 leaves reached the highest level at 3 dpi (data not shown) and remained strong at 5 dpi (Fig. 1B, right panel). At 28 dpi, GFP fluorescence was lost in the systemic leaves (including newly emerged leaves) of infiltrated 16c plants (Fig. 1C, left panel) but remained strong in the systemic leaves of infiltrated 16c/Pns10 plants (Fig. 1C, right panel). Northern blots showed no accumulation of GFP mRNA in infiltrated patches and systemic leaves of 16c plants but an abundant presence in the 16c/Pns10 plants (Fig. 1D). GFP siRNAs showed reverse accumulation patterns, with relatively high levels in the infiltrated 16c leaves and low levels in the infiltrated 16c/Pns10 leaves (Fig. 1D). Furthermore, they were abundantly present in the systemic leaves of 16c plants but were not detectable in the systemic leaves of 16c/Pns10 plants (Fig. 1D).
These data indicate that the stably expressed Pns10 in the transgenic 16c/Pns10 line suppresses RNA silencing induced by a sense mRNA as it does when transiently expressed (3). Therefore, the transgenic 16c/Pns10 plants were suitable for further studies on the mechanisms of Pns10 function.
The presence of Pns10 enhanced viral infection and invasion of shoot apices in transgenic N. benthamiana plants.
Our previous work showed that expression of Pns10 from a chimeric PVX (PVX-S10) enhanced viral systemic infection and symptom expression in N. benthamiana (3). To better understand the influence of Pns10 on the temporal and spatial infection patterns of viral infection, we used transgenic N. benthamiana plants expressing Pns10 (Nb/Pns10) (49) and then monitored the infection of these plants by chimeric PVX-GFP.
As shown in Fig. 2A, the GFP fluorescence intensity in inoculated leaves of Nb/Pns10 plants was much stronger than that in nontransgenic plants at 5 to 7 dpi, reflecting more active replication of the virus in the Pns10 transgenic plants. This was further confirmed by Northern blots showing a higher accumulation of viral RNAs in infected Nb/Pns10 plants than in infected nontransgenic plants (Fig. 2B).
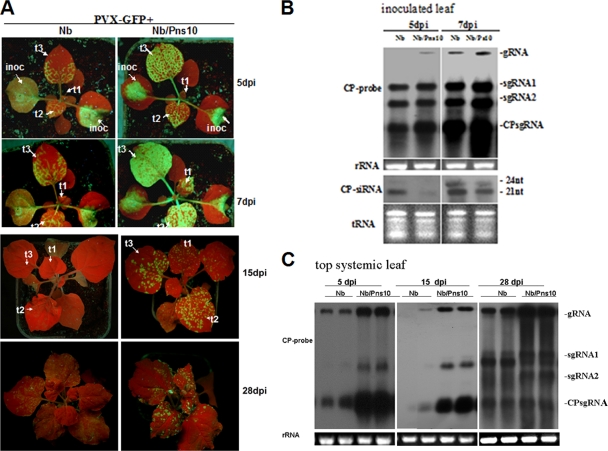
FIG. 2.
Pns10-enhanced PVX-GFP infection and spread in N. benthamiana Nb/Pns10 plant lines. (A) PVX-GFP-infected nontransgenic N. benthamiana (Nb) and Nb/Pns10 plants under UV light at 5, 7, 15, and 28 dpi. The inoculated leaves (inoc) and successive systemic leaves (t1 to t3) are indicated. (B) Northern blots of PVX-GFP RNA accumulations in inoculated leaves of N. benthamiana (Nb) and Nb/Pns10 plants at 3 and 7 dpi. The membranes were hybridized with DIG-labeled probes corresponding to the full-length ORF of PVX CP. (C) Northern blot of PVX-GFP RNA in top systemic leaves of N. benthamiana (Nb) and Nb/Pns10 plants at 5, 15, and 28 dpi.
The systemically infected leaves of Nb/Pns10-transgenic plants also exhibited stronger GFP fluorescence than those from nontransgenic plants as early as 5 dpi (Fig. 2A). At 7 dpi, the GFP fluorescence spread throughout the leaf blades in Nb/Pns10 plants compared to the appearance of a few patches of fluorescent areas in the nontransgenic plants (Fig. 2A, leaves t2 and t3 at 5 and 7 dpi). By 15 dpi, the GFP fluorescence decreased significantly in the older leaves of Nb/Pns10 plants, but the newly emerged leaves of these plants exhibited GFP fluorescence, indicating continuous viral replication (Fig. 2A, t1 at 15 dpi). In contrast, GFP fluorescence almost completely disappeared in all leaves of nontransgenic plants, indicating suppressed viral replication throughout the plant (Fig. 2A, t1 at 15 dpi). At 28 dpi, some level of GFP fluorescence recovered in the nontransgenic plants, but it was still weaker than that in the Nb/Pns10 plants. This conclusion was further supported by Northern blot data (Fig. 2C) and RT-PCR analysis of GFP-CP derived from PVX-GFP (data not shown).
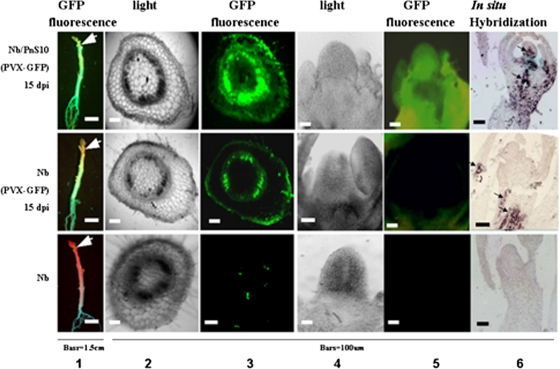
The presence of Pns10 not only enhanced the temporal spread of PVX-GFP but also the spatial invasion of tissues. Most significantly, strong GFP fluorescence was observed in the shoot apices of infected Nb/Pns10 plants (Fig. 3, top row), compared to its absence from the shoot apices of infected nontransgenic plants (Fig. 3, middle row). The GFP fluorescence in the stems was also much stronger in the infected transgenic plants than in the infected nontransgenic plants. To understand the cellular basis of these differences, we examined transverse sections of stems and longitudinal sections of shoot apices with fluorescence microscopy. As shown in panel 3 of Fig. 3, strong GFP fluorescence was present in the stem cortex of an infected Nb/Pns10 plant (top image) but barely visible in the cortex of an infected nontransgenic plant (middle image), indicating effective viral invasion of stem cortex facilitated by Pns10. Furthermore, panel 5 shows bright GFP fluorescence in the shoot apex of an infected Nb/Pns10 plant (top) and its absence from the apex of an infected nontransgenic plant (middle image). Pns10-enabled viral invasion of shoot apices was further confirmed by in situ localization of PVX CP mRNA (Fig. 3, panel 6).
FIG. 3.
Distribution of PVX-GFP in infected stems and shoot apices of N. benthamiana (Nb) and Nb/Pns10 plants at 15 dpi. Column 1 shows GFP fluorescence under long-wavelength UV light at 15 dpi, and columns 3 and 5 show fluorescent images of transverse sections of stems and longitudinal sections of shoot apices. Columns 2 and 4 show the corresponding bright-field images. Column 6 shows PVX CP signals in shoot apices detected by in situ hybridization.
Collectively, these data indicate that Pns10 enhanced (i) viral replication and/or viral RNA stability in inoculated leaves and (ii) temporal and spatial spread of viral infection. In the latter case, the most important finding is that presence of Pns10 enabled effective invasion of the stem cortex and shoot apices.
The presence of Pns10 interfered with the perception in recipient tissues, but not the production in source tissues, of mobile RNA silencing signals triggered by GFP dsRNA.
To gain insights into the mechanisms of Pns10 function in interfering with systemic RNA silencing, we first sought to determine whether Pns10 blocks the production of mobile RNA silencing signals. To this end, we analyzed systemic RNA silencing triggered by GFP dsRNA produced by agroinfiltration in 16c/Pns10, as well as 16c control plants.
In both lines, GFP fluorescence (Fig. 4A) and GFP mRNA (data not shown) were hardly detectable at 3 dpi. At 5 dpi, RNA silencing emerged in systemic leaves of both lines (Fig. 4B, bottom left panel). Detailed analyses, however, revealed some clear differences between the two lines. In 100% (30/30) of the 16c plants GFP silencing occurred throughout an entire leaf blade, whereas in more than 70% (21/29) of the 16c/Pns10 plants GFP silencing was restricted to the veins. At 28 dpi, the newly emerged leaves from these plants exhibited the same differences of silencing patterns (Fig. 4B, top panel). Northern blots with RNA samples taken from the GFP-silenced vein tissues of systemic leaves showed a much higher accumulation of GFP siRNAs in 16c plants than in 16c/Pns10 plants (Fig. 4C). These data indicate that stably expressed Pns10 in transgenic N. benthamiana did not block initiation of local RNA silencing triggered by dsGFP RNA.
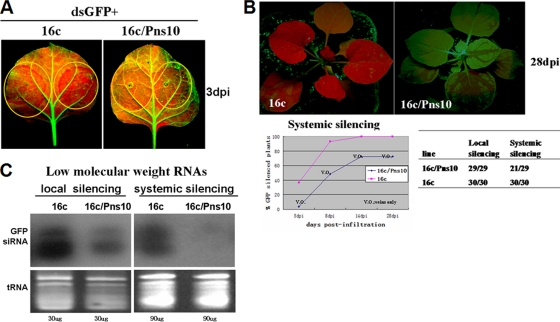
FIG. 4.
Effect of Pns10 on systemic RNA silencing induced by 35S-dsGFP. (A) N. benthamiana 16c and 16c/Pns10 plants were infiltrated with Agrobacterium carrying 35S-dsGFP. GFP fluorescence was imaged under long-wavelength UV light at 3 dpi. (B) GFP fluorescence was monitored for the onset of systemic silencing at 5, 8, 14, and 28 dpi. (C) Northern blots of siRNAs extracted from the veins of the different infiltrated plants shown in panels A and B. The bottom panel shows tRNA stained with ethidium bromide as a loading control.
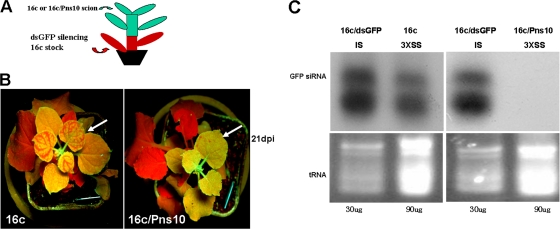
We then asked the question of whether Pns10 inhibits perception of silencing signals in recipient tissues by grafting experiments. We carried out one-way grafting assays with a 16c/Pns10 or 16c plant as scion and a silenced 16c as rootstock (Fig. 5A). We produced silenced 16c rootstocks by agroinfiltrating these plants with 35S-dsGFP at the four-leaf stage. When silenced expression of GFP was confirmed by red fluorescence under UV illumination at the eight-leaf stage (28 dpi), the16c/Pns10 or 16c scions were grafted onto rootstocks of these silenced plants. At 21 days after grafting, GFP silencing spread into more than 80% (21/26) of the 16c scions (Fig. 5B, left). In contrast, GFP silencing was restricted to a few spotty veins of the lower leaves that emerged in more than 85% (23/27) of the 16c/Pns10 scions (Fig. 5B, right). Consistent with these results, GFP-derived siRNAs were detected in strongly silenced 16c scions but not in weakly silenced 16c/Pns10 scions (Fig. 5C). These data indicate that the presence of Pns10 in the 16c/Pns10 scions interfered with the perception of systemic silencing signals.
FIG. 5.
Grafting of N. benthamiana 16c or 16c/Pns10 scions onto 35S-dsGFP silenced N. benthamiana 16c stocks. (A) Schematic representation of the grafting experiments. (B) Young 16c rootstocks were infiltrated with Agrobacterium carrying 35S-dsGFP. At 14 dpi, 16c or 16c/Pns10 scions were wedge grafted onto the rootstocks. GFP silencing in the scions was monitored by UV illumination. In the 16c scions GFP silencing spread into the entire leaves of more than 80% (21/26) of the plants. In the 16c/Pns10 scion plants, more than 85% (23/27) showed GFP fluorescence in the new leaves, and GFP silencing was restricted to a few veins of lower leaves. The images were taken 3 weeks after grafting. The arrows indicate the graft scions. (C) Northern blots of small RNAs in the grafts. The samples in lanes IS (infiltration silenced) were from rootstock leaves of N. benthamiana 16c exhibiting local silencing after infiltration with Agrobacterium carrying 35S-dsGFP. Lanes SS (systemic silencing) were from systemic, upper scion leaves. “3X” indicates the amounts of RNAs loaded in the SS lanes relative to those loaded into the IS lanes.
The ability of Pns10 to bind ds-siRNAs with 2-nt 3′ overhangs was required for silencing suppressor activity.
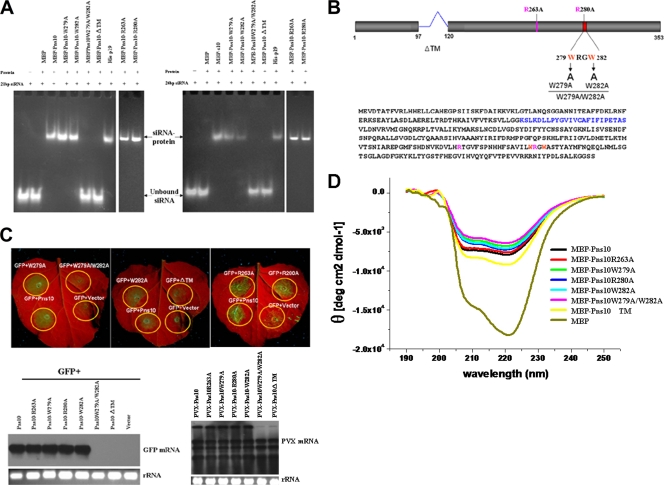
Many viral RNA silencing suppressors bind siRNAs as a means of suppressing RNA silencing (8). Therefore, one potential mechanism of Pns10 function is to bind siRNAs to interfere with the spread of RNA silencing. To test this possibility, we first investigated whether Pns10 binds siRNAs. We synthesized 21- and 24-nt RNA duplexes with 2-nt 3′ overhangs as probes for RNA binding assays. As shown in Fig. 6A, gel mobility shift assays demonstrated retarded mobility of siRNAs in the presence of MBP-Pns10, but not in the presence of MBP, indicating the binding of Pns10 to the siRNAs.
FIG. 6.
Gel mobility shift assay of recombinant Pns10 binding to ds-siRNA. (A) EMSA showing binding of Pns10 and its mutants to 21- and 24-bp siRNAs. A total of 10 μg of protein was used in each lane. His-P19 was used as a positive control. Expressed MBP or buffer (−) were used as negative controls. (B) Schematic representation of Pns10 amino acid sequence and mutations. (C) Suppressor activities of Pns10 and its mutants. N. benthamiana leaves either were infiltrated with mixtures of agrobacteria harboring a GFP construct, an empty vector as control, and a GFP construct with Pns10 or Pns10 mutants, or PVX containing Pns10 or its mutants. The upper fluorescent images were taken 7 days after infiltration. The lower left Northern blot image showed the steady-state levels of GFP mRNAs extracted from the different infiltrated patches as shown in the upper panels. The lower right Northern blot image showed the steady-state levels of PVX-Pns10 and related Pns10 mutant mRNAs from the chimeric PVX vector in extracted from the different infiltrated patches. rRNA stained with ethidium bromide was used as a loading control. (D) CD analysis of MBP-Pns10 and MBP-Pns10 mutants. Different proteins are distinguished from each other by colors. The protein concentration was 0.16 mg/ml in 20 mM Tris-HCl buffer (pH 7.4). The spectrum is background corrected, and the observed optical activity is expressed as the mean residue molar ellipticity [θ] (degrees cm2 dmol−1).
We then addressed the question of whether the siRNA-binding capacity of Pns10 is required for its silencing suppression activity by analyzing several types of Pns10 mutants. As shown in Fig. 6B, several mutations were designed to target (i) the basic amino acids (263R and 280R) that are conserved in homologous proteins from other phytoreoviruses; (ii) the WXXW motif, which in P19 is crucial for siRNA binding and silencing suppression activity (6, 33, 40); and (iii) the putative transmembrane motif (TM; 98 to 119 amino acids), which is essential for the formation of tubular structure (mutation with the deletion of this motif disrupts the tubular structures, which resembled the tubules observed in RDV-infected insect vector monolayers) (39). In addition, the Pns10 mutation with the deletion of this motif lost the ability to display Pns10 out the of the cell membrane (39). Gel mobility shift assays showed that, among all mutants, Pns10W279A/W282A and Pns10ΔTM failed to bind 21- and 24-nt siRNAs (Fig. 6A). All of these results are summarized in Table 1.
TABLE 1.
VSR and binding activities of Pns10 mutants
| Pns10 mutant | VSR activity | Binding activity |
|---|---|---|
| R263A | + | + |
| W279A | + | + |
| R280A | + | + |
| W282A | + | + |
| W279A/282A | - | - |
| ΔTM | - | - |
The silencing suppression activity of each mutant was then directly tested by agroinfiltrating a mixture of 35S-GFP and a 35S-Pns10 mutant in N. benthamiana leaves. Both GFP fluorescence and mRNA analyses showed that, among all mutants, Pns10W279A/W282A and Pns10ΔTM lost their silencing suppression activities (Fig. 6C). The promotion of virus infection was demonstrated by agroinfiltrating PVX-Pns10 or mutant in N. benthamiana leaves. The mRNA analyses showed, that among all mutants, only PVX-Pns10W279A/W282A and PVX-Pns10ΔTM lost their promotion of virus infection activities (Fig. 6C). Thus, there is a direct correlation between the ability of a mutant to bind siRNAs and its ability to suppress RNA silencing and the promotion of virus infection.
Because mutants Pns10W279A/W282A and Pns10ΔTM lost both siRNA-binding and silencing suppression functions, it was important to test whether this was attributed to altered conformations of the mutant proteins. We analyzed the conformations of all of these mutants, as well as the wild-type Pns10, by using CD. The CD spectra of these mutants were similar to that of the wild-type Pns10, indicating that they were still folded correctly (Fig. 6D). Therefore, protein conformation change was unlikely the cause of dysfunction for these two mutants. All data thus indicate that the siRNA-binding function of Pns10 is necessary for its silencing suppression activity.
The presence of Pns10 led to decreased expression of RDR6.
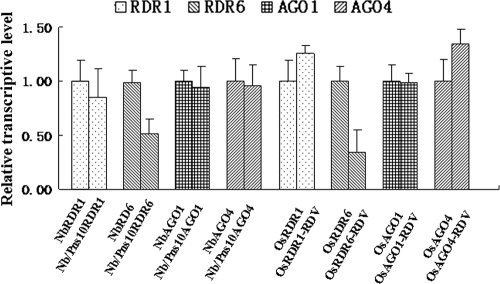
To gain further insights into the mechanisms of Pns10 function, we sought to determine whether the presence of Pns10 would affect the expression of any components of the RNA silencing machinery, besides binding siRNAs. We used real-time RT-PCR to examine the mRNA levels for NbRDR1, NbRDR6, NbAGO1, and NbAGO4. As shown in Fig. 7, the accumulation of NbRDR6 was reduced to 47% in Nb/Pns10 transgenic plants compared to that in nontransgenic plants. The mRNAs of NbRDR1, NbAGO1, and NbAGO4 did not show significant differences between the transgenic and nontransgenic plants (Fig. 7). Therefore, the presence of Pns10 appeared to specifically contribute to the downregulated expression of NbRDR6.
FIG. 7.
Quantitative real-time RT-PCR analysis of RDR1, RDR6, AGO1, and AGO4 transcript levels in N. benthamiana (Nb), Nb/Pns10, RDV-infected rice and mock-inoculated healthy rice. Each mean value is derived from three pools of plants, with each pool comprising three plants. Error bars represent SEs of the mean. Relative transcript levels were calculated by using the DDC(t) method (22) with N. benthamiana GAPDH or Oryza sativa actin transcripts serving as an internal standard.
To determine whether downregulated expression of RDR6 in the presence of Pns10 has relevance to RDV infection in the natural host of rice, we examined the rice OsRDR6 mRNA levels in RDV-infected rice plants. As shown in Fig. 7, the OsRDR6 mRNA level in RDV-infected plants decreased to nearly 34% of that in noninfected plants, whereas the OsRDR1, OsAGO1, and OsAGO4 levels did not differ significantly between the infected and noninfected plants.
These data suggest that presence of Pns10 can cause downregulated expression of RDR6 in N. benthamiana and RDV-infected rice plants. This function may be an important counterdefense mechanism for the virus, allowing efficient systemic infection of a host.
DISCUSSION
We previously identified the RDV Pns10 as a VSR by using a transient agroinfiltration expression system (3). In the present study, we confirmed this finding with Pns10 stable-expressing transgenic N. benthamiana plants. More importantly, this new experimental system, in combination with multiple approaches, has enabled us to further investigate the mechanisms of Pns10 function. The findings described above significantly extend our understanding of Pns10 function and broaden our knowledge of VSR functions in general.
The secondary siRNAs generated through the activities of NbRDR6 are most important for antiviral responses (32), and NbRDR6 has a role in preventing viral invasion of shoot apices (27, 30). Furthermore, both NbAGO1- and NbAGO4-like genes are required for full systemic silencing (17). In most studies, the roles of these host proteins were demonstrated through RNA interference-based downregulation of gene expression. To understand better the roles of these proteins, it will be useful to determine how their expression might be affected by VSRs. We presented here evidence that the presence of Pns10 in transgenic N. benthamiana led to downregulated expression of NbRDR6. This observation is significant in several aspects. First, this downregulated expression appears to be specific to NbRDR6, because the expression of NbRDR1, NbAGO1, and NbAGO4 was not affected. However, we cannot rule out the possibility that the expression of other genes in the RNA silencing machinery is also affected. Second, the expression of RDR6 is also reduced significantly and specifically in RDV-infected rice, the natural host, supporting the biological relevance of findings from N. benthamiana. Third, Pns10 did not have direct interactions with RDR6 in our yeast two-hybrid and coimmunoprecipitation experiments (data not shown). Therefore, Pns10 likely affects the expression of RDR6 gene rather than its activity. These findings set the stage for further studies on the mechanisms underlying Pns10 modulation of RDR6 expression.
Crystal structural studies on the tombusvirus p19-siRNA complex revealed several conserved basic and polar residues as binding sites for siRNAs (33, 45). Specifically, the end-capping interaction was made by two conserved tryptophan residues (WXXW) in the N terminus of p19. Interestingly, amino acid sequence alignments for Pns10 of different Phytoreoviruses also revealed a conserved WXXW motif (WRGW) (data not shown). The substitution mutants Pns10W279A/W282A lost both siRNA binding and silencing suppression activities. Furthermore, p19 binds siRNA as a dimer, with two molecules of p19 per siRNA duplex. Although the structure of Pns10 is not yet known, this protein interacts with itself and forms a tubular structure in insect host cells that plays a major role in RDV cell-to-cell movement in insect vector cells (39). The possibility that Pns10 binds siRNAs as a dimer or multimer is therefore worth further investigation. Nonetheless, our findings, together with those on p19, suggest that WXXW is an important binding site for siRNAs from diverse virus groups.
The putative TM motif of Pns10 is essential for the formation of tubular structure and cell-to-cell movement in insect vector cells (39). Our results showed that deleting this motif led to the loss of siRNA binding and silencing suppression functions. Because this motif is not reported for other VSRs, it is likely a unique function associated with Pns10. The specific mechanisms remain to be determined.
Taken together, our results show that RDV Pns10 plays important roles in enhancing viral replication, systemic movement, and invasion of new tissues and especially the shoot apices. The siRNA binding activity is required for the silencing suppression function. Significantly, Pns10 uses a unique TM and a conserved WXXW for siRNA binding, both being functionally important for silencing suppression. In addition, Pns10 can downregulate the expression of RDR6, a critical enzyme for the generation of secondary siRNAs for antiviral responses. Our data suggest that this particular activity appears to be critical for viral invasion of shoot apices. Indeed, our recent work showed that downregulated expression of RDR6 by antisense method in rice enhanced the infection and disease symptoms of RDV, as well as Rice stripe virus (D. Q. Jiang Lin, L.-Y. Meng, J. Chen, W.-J. Le, T. Zhou, J.-C. Tian, L. Liang, Y.-J. Zhou, G.-Y. Ye, C.-H. Wei, and Y. Li, unpublished data). Thus, our data, together with published studies from other labs, demonstrate that RDR6 homologues from different plants play a conserved role of inhibiting infection by viruses of different genome types.
Acknowledgments
We are indebted to David Baulcombe for providing plasmid pPVX-GFP, A. tumefaciens strain GV3101 containing pJIC SA_Rep, and A. tumefaciens strain GV3101 containing pPVX-GFP. We thank Bekir Ülker for providing plasmid pJawohl8-RNAi. We also thank Feng Qu and Biao Ding for helpful comments on the manuscript.
This study was supported by grants from the Ministry of Science and Technology 973 program (2006CB101903) and NSFC (31030005, 30910103904) to C.W. and Y.L.
Footnotes
Published ahead of print on 6 October 2010.
REFERENCES
- 1.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 2.Boccardo, G., and R. G. Milne. 1984. Plant reovirus group: descriptions of plant viruses. [Online.] http://www.dpvweb.net/dpv/showdpv.php?dpvno=294.
- 3.Cao, X., P. Zhou, X. Zhang, S. Zhu, X. Zhong, Q. Xiao, B. Ding, and Y. Li. 2005. Identification of an RNA silencing suppressor from a plant double-stranded RNA virus. J. Virol. 79:13018-13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman, E. J., A. I. Prokhnevsky, K. Gopinath, V. V. Dolja, and J. C. Carrington. 2004. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18:1179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J., W. X. Li, D. Xie, J. R. Peng, and S. W. Ding. 2004. Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microrna in host gene expression. Plant Cell 16:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu, M., B. Desvoyes, M. Turina, R. Noad, and H. B. Scholthof. 2000. Genetic dissection of tomato bushy stunt virus p19-protein-mediated host-dependent symptom induction and systemic invasion. Virology 266:79-87. [DOI] [PubMed] [Google Scholar]
- 7.Deleris, A., J. Gallego-Bartolome, J. Bao, K. D. Kasschau, J. C. Carrington, and O. Voinnet. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313:68-71. [DOI] [PubMed] [Google Scholar]
- 8.Ding, S. W., and O. Voinnet. 2007. Antiviral immunity directed by small RNAs. Cell 130:413-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan, M., J. Nan, Y. Liang, P. Mao, L. Lu, L. Li, C. Wei, L. Lai, and Y. Li. 2007. DNA binding mechanism revealed by high-resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Res. 35:1145-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunoyer, P., C. Himber, and O. Voinnet. 2005. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 37:1356-1360. [DOI] [PubMed] [Google Scholar]
- 11.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 12.Guo, H. S., and S. W. Ding. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21:398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton, A., O. Voinnet, L. Chappell, and D. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates posttranscriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 15.Reference deleted.
- 16.Johansen, L. K., and J. C. Carrington. 2001. Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 126:930-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, L., T. Keining, A. Eamens, and F. E. Vaistij. 2006. Virus-induced gene silencing of Argonaute genes in Nicotiana benthamiana demonstrates that extensive systemic silencing requires Argonaute1-like and Argonaute4-like genes. Plant Physiol. 141:598-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. Plasterk. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in Caenorhabditis elegans. Genes Dev. 15:2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lakatos, L., T. Csorba, V. Pantaleo, E. J. Chapman, J. C. Carrington, Y. P. Liu, V. V. Dolja, L. F. Calvino, J. J. Lopez-Moya, and J. Burgyan. 2006. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25:2768-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, F., and S. W. Ding. 2006. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60:503-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, Y., Y. M. Bao, C. H. Wei, Z. S. Kang, Y. W. Zhong, P. Mao, G. Wu, Z. L. Chen, J. Schiemann, and R. S. Nelson. 2004. Rice dwarf phytoreovirus segment S6-encoded nonstructural protein has a cell-to-cell movement function. J. Virol. 78:5382-5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 23.Matzke, M., and A. J. Matzke. 2003. RNAi extends its reach. Science 301:1060-1061. [DOI] [PubMed] [Google Scholar]
- 24.Merai, Z., Z. Kerenyi, A. Molnar, E. Barta, A. Valoczi, G. Bisztray, Z. Havelda, J. Burgyan, and D. Silhavy. 2005. Aureusvirus P14 is an efficient RNA silencing suppressor that binds double-stranded RNAs without size specificity. J. Virol. 79:7217-7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mlotshwa, S., S. E. Schauer, T. H. Smith, A. C. Mallory, J. M. Herr, Jr., B. Roth, D. S. Merchant, A. Ray, L. H. Bowman, and V. B. Vance. 2005. Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. Plant Cell 17:2873-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palauqui, J. C., T. Elmayan, J. M. Pollien, and H. Vaucheret. 1997. Systemic acquired silencing: transgene-specific posttranscriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16:4738-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu, F., and T. J. Morris. 2005. Suppressors of RNA silencing encoded by plant viruses and their role in viral infections. FEBS Lett. 579:5958-5964. [DOI] [PubMed] [Google Scholar]
- 28.Qu, F., X. Ye, and T. J. Morris. 2008. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. U. S. A. 105:14732-14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratcliff, F., A. M. Martin-Hernandez, and D. C. Baulcombe. 2001. Technical advance: tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25:237-245. [DOI] [PubMed] [Google Scholar]
- 30.Schwach, F., F. E. Vaistij, L. Jones, and D. C. Baulcombe. 2005. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138:1842-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki, N., M. Sugawara, T. Kusano, H. Mori, and Y. Matsuura. 1994. Immunodetection of rice dwarf phytoreoviral proteins in both insect and plant hosts. Virology 202:41-48. [DOI] [PubMed] [Google Scholar]
- 32.Vaistij, F. E., and L. Jones. 2009. Compromised virus-induced gene silencing in RDR6-deficient plants. Plant Physiol. 149:1399-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vargason, J. M., G. Szittya, J. Burgyan, and T. M. Hall. 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115:799-811. [DOI] [PubMed] [Google Scholar]
- 34.Voinnet, O. 2005. Non-cell autonomous RNA silencing. FEBS Lett. 579:5858-5871. [DOI] [PubMed] [Google Scholar]
- 35.Voinnet, O., C. Lederer, and D. C. Baulcombe. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157-167. [DOI] [PubMed] [Google Scholar]
- 36.Wang, M. B., and M. Metzlaff. 2005. RNA silencing and antiviral defense in plants. Curr. Opin. Plant Biol. 8:216-222. [DOI] [PubMed] [Google Scholar]
- 37.Wassenegger, M., and G. Krczal. 2006. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 11:142-151. [DOI] [PubMed] [Google Scholar]
- 38.Wei, T., A. Kikuchi, Y. Moriyasu, N. Suzuki, T. Shimizu, K. Hagiwara, H. Chen, M. Takahashi, T. Ichiki-Uehara, and T. Omura. 2006. The spread of Rice dwarf virus among cells of its insect vector exploits virus-induced tubular structures. J. Virol. 80:8593-8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei, T., T. Shimizu, and T. Omura. 2008. Endomembranes and myosin mediate assembly into tubules of Pns10 of rice dwarf virus and intercellular spreading of the virus in cultured insect vector cells. Virology 372:349-356. [DOI] [PubMed] [Google Scholar]
- 40.Xia, Z., Z. Zhu, J. Zhu, and R. Zhou. 2009. Recognition mechanism of siRNA by viral p19 suppressor of RNA silencing: a molecular dynamics study. Biophys. J. 96:1761-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie, Z., L. K. Johansen, A. M. Gustafson, K. D. Kasschau, A. D. Lellis, D. Zilberman, S. E. Jacobsen, and J. C. Carrington. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2:E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu, H., Y. Li, Z. Mao, Y. Li, Z. Wu, L. Qu, C. An, X. Ming, J. Schiemann, R. Casper, and Z. Chen. 1998. Rice dwarf phytoreovirus segment S11 encodes a nucleic acid binding protein. Virology 240:267-272. [DOI] [PubMed] [Google Scholar]
- 43.Xu, Y. Y., K. Chong, Z. H. Xu, and K. H. Tan. 2001. Expression patterns of a vernalization-related genes responding to jasmonate. Acta Botanica Sinica 43:871-873. [Google Scholar]
- 44.Yang, S. J., S. A. Carter, A. B. Cole, N. H. Cheng, and R. S. Nelson. 2004. A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. U. S. A. 101:6297-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye, K., L. Malinina, and D. J. Patel. 2003. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 426:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, X., P. Du, L. Lu, Q. Xiao, W. Wang, X. Cao, B. Ren, C. Wei, and Y. Li. 2008. Contrasting effects of HC-Pro and 2b viral suppressors from Sugarcane mosaic virus and Tomato aspermy cucumovirus on the accumulation of siRNAs. Virology 374:351-360. [DOI] [PubMed] [Google Scholar]
- 47.Zheng, H., L. Yu, C. Wei, D. Hu, Y. Shen, Z. Chen, and Y. Li. 2000. Assembly of double-shelled, virus-like particles in transgenic rice plants expressing two major structural proteins of rice dwarf virus. J. Virol. 74:9808-9810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, F., Y. Pu, T. Wei, H. Liu, W. Deng, C. Wei, B. Ding, T. Omura, and Y. Li. 2007. The P2 capsid protein of the nonenveloped rice dwarf phytoreovirus induces membrane fusion in insect host cells. Proc. Natl. Acad. Sci. U. S. A. 104:19547-19552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, P., B. Ren, X. M. Zhang, Y. Wang, C. H. Wei, and Y. Li. 2010. Stable expression of Rice dwarf virus Pns10 suppresses the posttranscriptional gene silencing in transgenic Nicotiana benthamiana plants. Acta Virol. 54:99-104. [DOI] [PubMed] [Google Scholar]
- 50.Zhu, S., F. Gao, X. Cao, M. Chen, G. Ye, C. Wei, and Y. Li. 2005. The rice dwarf virus P2 protein interacts with ent-kaurene oxidases in vivo, leading to reduced biosynthesis of gibberellins and rice dwarf symptoms. Plant Physiol. 139:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]