Abstract
The coronavirus small envelope (E) protein plays a crucial, but poorly defined, role in the assembly of virions. To investigate E protein function, we previously generated E gene point mutants of mouse hepatitis virus (MHV) that were defective in growth and assembled virions with anomalous morphologies. We subsequently constructed an E gene deletion (ΔE) mutant that was only minimally viable. The ΔE virus formed tiny plaques and reached optimal infectious titers many orders of magnitude below those of wild-type virus. We have now characterized highly aberrant viral transcription patterns that developed in some stocks of the ΔE mutant. Extensive analysis of three independent stocks revealed that, in each, a faster-growing virus harboring a genomic duplication had been selected. Remarkably, the net result of each duplication was the creation of a variant version of the membrane protein (M) gene that was situated upstream of the native copy of the M gene. Each different variant M gene encoded an expressed protein (M*) containing a truncated endodomain. Reconstruction of one variant M gene in a ΔE background showed that expression of the M* protein markedly enhanced the growth of the ΔE mutant and that the M* protein was incorporated into assembled virions. These findings suggest that M* proteins were repeatedly selected as surrogates for the E protein and that one role of E is to mediate interactions between transmembrane domains of M protein monomers. Our results provide a demonstration of the capability of coronaviruses to evolve new gene functions through recombination.
The assembly of coronavirus virions entails concerted actions of three structural proteins: the membrane protein (M), the small envelope protein (E), and the nucleocapsid protein (N) (15, 35). Other viral structural proteins, most notably the spike glycoprotein (S), are gathered into virions, but they are not specifically required for, nor are they known to enhance, the assembly process. M protein, the major constituent of the viral envelope, is a triple-spanning integral membrane protein. M possesses a small amino-terminal ectodomain and a large carboxy-terminal endodomain, the latter making up roughly half of the molecule. Budding of virions occurs in the endoplasmic reticulum-Golgi intermediate compartment and is driven, in part, by formation of homotypic interactions between M proteins. These intermolecular M-M contacts remain to be mapped precisely, but connections between the transmembrane regions make a major contribution (16). Additionally, there are budding interactions between M endodomains and the N protein (17, 23, 30, 49, 50). N, in turn, forms a helically symmetric nucleocapsid with the ∼30-kb positive-strand viral genomic RNA (35).
Despite its dominant presence in virion structure, the M protein is not competent to form the viral envelope on its own. When M is expressed in the absence of other viral proteins, it localizes beyond the budding compartment in the Golgi, where it forms largely insoluble complexes (25, 27, 32, 33, 44). In contrast, early studies found that coexpression of the E protein with the M protein was sufficient to yield the formation of virus-like particles (VLPs) that were released from cells and appeared morphologically identical to coronavirus virions (6, 48). More recently, it has been shown that the additional coexpression of N protein substantially increases the efficiency of VLP formation (7, 47) and that coexpression of N can rescue partially defective M mutants into VLPs (3).
The critical role of E protein in coronavirus assembly is also reflected in the properties of engineered viral mutants. Particular mutants of mouse hepatitis virus (MHV) harboring E protein clustered charged-to-alanine mutations were found to be thermolabile and exhibited striking morphological anomalies (18). Complete abrogation of E protein expression has a range of effects in different coronavirus species. For transmissible gastroenteritis virus (TGEV), deletion of the E gene is lethal (11, 41). For MHV, ΔE or E knockout mutants are viable, but they are drastically impaired, replicating to 10,000-fold-lower titers than wild-type virus (29, 31). Among the coronaviruses examined to date, the severe acute respiratory syndrome coronavirus (SARS-CoV) is the most tolerant of loss of the E gene. The SARS-CoV ΔE mutant suffers only a 20- to 200-fold loss of infectious titer relative to that of its wild-type counterpart (12).
It is not yet well defined why M protein is intrinsically unable to carry out efficient budding without the assistance of E; also, the exact roles of the E protein in VLP systems or in viral infections are still unclear. Like M, E is an integral membrane protein, but E is only one-third the size of M, it has only a single transmembrane domain, and it is present in virions in only minor amounts. One role proposed for E is that it alters the budding compartment in a way that promotes assembly (48), possibly through induction of membrane curvature (42). The putative ion channel property of E (51-53) could provide a mechanism for this mode of action. Another possible role for E is to directly contact M and thereby facilitate productive M-M interactions (7, 10). Other evidence points to a need for E protein to assist the release of assembled virions from infected cells (34, 40). These roles are not mutually exclusive, and studies by the Machamer laboratory represent major steps in the assignment of individual functions to various regions of the E molecule. The carboxy-terminal endodomain of the infectious bronchitis virus (IBV) E protein has been shown to govern Golgi apparatus localization of that protein (8, 9). Additionally, a chimeric protein, composed of a heterologous transmembrane domain linked to the IBV E protein endodomain, is competent to promote VLP formation and is partially functional in a recombinant virus (34).
In the present study, we have characterized multiple viruses bearing rearranged genomes that had been independently selected in stocks of the MHV ΔE mutant. Our analysis revealed a surprising mechanism by which MHV can partially circumvent the constraints imposed by the absence of the E protein through the acquisition of a duplicated, variant form of M protein. Our findings provide insights into E protein function and have implications for coronavirus evolution.
MATERIALS AND METHODS
Cells and viruses.
Wild-type MHV-A59 and MHV mutants were propagated in mouse 17 clone 1 (17Cl1) or L2 cells. A line of 17Cl1 cells that stably expresses the MHV E protein (29) was maintained in the presence of 400 μg/ml G418 (Sigma). The wild-type virus used throughout this work was Alb240 (31); the ΔE and E knockout mutants have been described previously (29, 31). Plaque assays and plaque purifications were performed with mouse L2 cells. The interspecies chimeric virus designated fMHV.v2 (20), used for mutant construction, was grown in FCWF cells.
Analysis of viral RNA.
Metabolic labeling of virus-specific RNA with [33P]orthophosphate in the presence of actinomycin D was carried out exactly as described previously (31). In brief, 17Cl1 cells were infected at a multiplicity of 0.125 PFU per cell and incubated for 12 h at 33°C. Phosphate starvation and labeling of cells, each for 2-h periods, were carried out at 37°C. Total cytoplasmic RNA was purified from infected cells by an NP-40 gentle lysis procedure, analyzed by electrophoresis through 1% agarose containing formaldehyde, and visualized by fluorography.
For analysis of viral RNA by Northern blotting, purified total cytoplasmic RNA was fractionated in 1% agarose gels containing formaldehyde. RNA was blotted and then UV cross-linked onto Nytran Supercharge membranes (Whatman; GE Healthcare). DNA probes were generated by PCR and labeled with an AlkPhos Direct kit (Amersham; GE Healthcare). The probes used were S(mid), corresponding to nucleotides 2104 to 2845 in the middle of the S open reading frame (ORF); S(3′), corresponding to nucleotides 3440 to 3994 at the 3′ end of the S ORF; M, corresponding to nucleotides 1 to 441 of the M ORF; and 3′-end, corresponding to the 3′-most 241 nucleotides of the N ORF and the entire 3′ untranslated region. Hybridization was carried out at 55°C for 16 to 20 h, and the blots were washed and developed according to the manufacturer's instructions.
Reverse transcription (RT) of RNA from infected cells was carried out with a random hexanucleotide primer and avian myeloblastosis virus reverse transcriptase (Life Sciences). Amplicons of cDNA were generated with the Expand High Fidelity PCR System (Roche) under standard conditions with various pairs of the primers listed in Table 1. The PCR products were analyzed by agarose gel electrophoresis or were purified with QIAquick spin columns (Qiagen) prior to DNA sequencing. In some cases, PCR products were cloned with a Topo-TA kit (Invitrogen), according to the manufacturer's instructions, using the vector pCR4-TOPO with kanamycin selection.
TABLE 1.
Oligonucleotide primers used in RT-PCR analysis
| Primer | Genea | Sense | Sequence (5′ to 3′) |
|---|---|---|---|
| CK3 | S (2104-2121) | − | AACGCATAAGCCAGCACC |
| CK4 | S (3662-3679) | + | TCGAGAAGTTAAATGTTA |
| CK20 | S (3908-3925) | + | GTTGTGATGAGTATGGAG |
| CM49 | S (1451-1468) | + | CGGACATAGTTAGCCCTT |
| CM56 | S (1995-2012) | − | TTCTCCTCACGGGAAATA |
| CM58 | S (2226-2243) | − | TCATATAATCCATCAACG |
| CM73 | S (3567-3584) | + | GAACACTTCAATACCTAA |
| CM76 | S (3817-3834) | − | AGCTACACCAGCTAATCC |
| CM89 | M (545-562) | − | GCTTATAAGTGCAAAGGT |
| LK59 | S (3863-3880) | + | GCTGCACAGGTTGTGGCT |
| LK98 | S (3911-3934) | − | CCTGGTGTCCTCCATACTCATCAC |
| LK99 | Leader (16-39) | + | GCGTCCGTACGTACCCTCTCAACT |
| PM145 | N (169-186) | − | CACACTCCCGGAGTTGGG |
| PM148 | M (175-192) | + | ATCTTGTGGTTAATGTGG |
| PM204 | M (424-442) | − | TGTTAGTGTATGGTAATC |
| PM210 | N (142-159) | − | TGCAGTCTGCTTTGGCTG |
Nucleotide coordinates are given in parentheses.
MHV mutant construction.
MHV gene 2 substitutions and mutant M protein reconstructions were carried out through targeted RNA recombination and host-range-based selection, as described in detail previously (28, 36). In brief, feline FCWF cells that had been infected with fMHV.v2 were transfected with in vitro-synthesized donor RNAs (mMessage mMachine T7; Ambion) containing the mutations of interest. Progeny viruses were harvested and purified by plaque titration on mouse L2 cells. The incorporation of genes, gene deletions, or point mutations in candidate recombinants was confirmed by the sequencing of RT-PCR products that were amplified from viral RNA.
Transcription vectors for the synthesis of donor RNAs originated with pMH54 (28), which encodes 5′ elements of the MHV-A59 genome linked to the 3′-most 8.6 kb of the MHV-A59 genome. Vector pLK70 is a derivative of pMH54 containing the original deletion of genes 4, 5a, and E (31). Vector pPM9 is a derivative of pMH54 containing the 3′-most 2.9 kb of gene 1b, downstream of which there is a deletion of transcription-regulating sequence (TRS) 2, gene 2a, and almost all of the hemagglutinin-esterase (HE) gene; in place of the deletion, unique SalI and AscI sites were created (26). The vector for the construction of the Δ(2a-HE)/ΔE mutant, pPM9ΔE, was created from pPM9 by replacement of the XhoI-EagI fragment, running from the S gene to the start of the M gene, with its counterpart from pLK70. The vector for the construction of the M*/ΔE mutant, pLK150, was generated by replacement of the SalI-AscI fragment of pPM9ΔE with a PCR product containing the M*-290 ORF and its associated TRS, which had been amplified from a cDNA subclone of Alb290 RNA. The vector for the construction of the M*-KO/ΔE mutant, pLK151, was generated in the same manner as pLK150, except that mutations were incorporated into the upstream PCR primer. The knockout mutations in pLK151 resulted in (i) alteration of the fourth base of the canonical TRS (AAUCUAAAC) from C to U, a change that is known to abolish transcription (39, 54); disruption of the start codon by its replacement with CUA; and (iii) replacement of the fourth codon with the stop codon UAA. Vectors for the incorporation of M gene mutations into a ΔE background were constructed by RT-PCR amplification of the relevant regions of individual mutant viral genomes, followed by transfer into pLK70 of the SbfI-BssHII fragment, running from immediately upstream of the M gene to the 3′ end of the M gene.
Western blotting.
Virion purification and preparation of lysates from infected 17Cl1 cell monolayers were carried out exactly as described previously (22). Samples of infected cell lysates or purified virions were separated by SDS-PAGE through 12 or 15% polyacrylamide gels, with prestained protein standards (Invitrogen or NEB) in flanking lanes. Proteins were transferred to polyvinylidene difluoride membranes, blots were probed with the indicated antibodies, and bound primary antibodies were visualized by enhanced-chemiluminescence detection (GE Healthcare or Pierce). The primary antibodies used were anti-N monoclonal antibody J.3.3 and anti-M monoclonal antibody J.1.3 (both provided by John Fleming, University of Wisconsin, Madison, WI) and polyclonal rabbit antiserum raised against the bacterially expressed carboxy-terminal tail of the MHV E protein (29).
RESULTS
Unusual transcription patterns of particular ΔE mutant isolates.
In a previous study of E protein function, we constructed an MHV ΔE mutant, which we obtained as eight separate isolates resulting from targeted RNA recombination (31). These isolates, designated Alb289 through Alb296, consecutively represented four independent sibling pairs. The ΔE viruses were only minimally viable, forming very small plaques and growing to maximal infectious titers that were some 3 to 4 orders of magnitude below those of the wild type. We had designed the ΔE mutation so that, in addition to deletion of the entire E gene, the adjacent accessory genes 4 and 5a were also removed (Fig. 1). However, we subsequently showed, through knockout of the E gene by point mutations, that the severely impaired phenotype of the ΔE mutant was entirely due to deletion of the E gene (29).
FIG. 1.
Production of unexpected viral RNA species by certain ΔE isolates of MHV. At the top is a schematic of the MHV genome, with expanded regions comparing the downstream ends of the wild-type (wt) and ΔE mutant genomes. The arrowheads denote TRSs. At the bottom is shown RNA synthesis in 17Cl1 cells that were mock infected or infected at a multiplicity of 0.125 PFU per cell. RNA was metabolically labeled with [33P]orthophosphate, purified, and electrophoretically separated, as described in Materials and Methods. gRNA, genomic RNA. The stars indicate additional RNA species from cells infected with ΔE isolate Alb290 or Alb289.
In our characterization of RNA synthesis by ΔE viruses, we found that some of the isolates, specifically Alb291, Alb292, Alb293, and Alb296, displayed exactly the pattern of transcription that was expected to result from the constructed genomic deletion (31). MHV infection entails synthesis of a 3′ nested set of six subgenomic (sg) RNAs, each consisting of a short 5′ leader segment joined at a TRS to all genomic sequence downstream of that TRS (46). Consequently, those ΔE isolates exemplified by Alb291 exhibited sgRNAs 6 and 7 that were identical to those of the wild type, but sgRNAs 4 and 5 were absent, and sgRNAs 2 and 3 were displaced to higher mobilities owing to the 1,023-nucleotide (nt) deletion of genes 4, 5a, and E (Fig. 1). It should be noted that the minor mobility variations seen for sgRNAs 6 and 7 of the viruses in Fig. 1 were artifacts of electrophoresis, as was evident from differences among replicate samples in other lanes of the same agarose gel (not shown).
Very surprisingly, not all of the ΔE isolates conformed to the expected pattern. Passage 3 (and higher-passage) stocks of Alb289, Alb290, Alb294, and Alb295 produced a second nested set of sgRNAs, which were superimposed upon the expected ΔE transcripts. The second set of transcripts appeared to result from an extra transcription unit that ranged in size from 0.5 to 2.6 kb. RNAs synthesized by Alb289 and Alb290, examples of this class of ΔE isolate, are shown in Fig. 1. In some cases, the presence of the second set of transcripts correlated with a noticeable increase in the rate of growth of the virus compared to the growth rate of the original ΔE mutants, suggesting that each of these stocks had accumulated substantial amounts of a faster-growing variant. The presence of the unexpected transcripts initially made us concerned that a subset of the ΔE isolates had somehow regained the E gene. However, this possibility was ruled out both by RT-PCR analysis, using primers internal to the E and 5a genes, and by Northern dot blots with a probe specific for the E gene, as described previously (31).
Genomic analysis of ΔE isolate Alb290.
The transcription profiles of the four aberrant ΔE isolates suggested that each of these stocks contained two populations of virus: (i) the original ΔE mutant and (ii) the ΔE mutant harboring an insertion of variable size upstream of the M gene. In some cases, e.g., Alb290 (Fig. 1), comparison of sgRNAs 2 and 3 to their larger counterparts indicated that the insert-containing variant comprised the predominant subpopulation. To attempt to separate and characterize the variant viruses, we purified multiple plaques from each stock, and following a single passage of each plaque, we examined intracellular RNA by Northern blotting and RT-PCR analysis. For Alb289, RNA derived from all 12 purified plaques had the same pattern as the original ΔE mutant; therefore, this isolate was not pursued further.
For Alb290, extra bands were seen by Northern blotting of infected cellular RNA from all 11 purified plaques, a subset of which are shown in Fig. 2A. Hybridization with a probe specific for the MHV 3′ genomic end revealed that the primary (i.e., smallest) extra RNA species for Alb290 migrated slightly faster than wild-type sgRNA5. Hybridization with a probe specific for the M gene showed that the primary extra band contained M sequence (Fig. 2A). Although this M hybridization confirmed that the extra band fit into the typical nested set structure of coronavirus RNAs, it did not necessarily indicate that the inserted material was from the M gene. Proceeding further upstream, probes specific for either the 3′ end or the middle of the S gene failed to hybridize with the extra band (data not shown), which showed that the inserted material in Alb290 RNA did not contain sequence from the S gene.
FIG. 2.
Genomic composition of ΔE mutant Alb290. (A) Northern blots of RNA from cells infected with stocks started from individual plaques of ΔE isolate Alb290 and detected with probes specific for the MHV 3′ genomic end (left) or the M gene (right). Control RNA samples were from mock-infected cells or from cells infected with the nonrearranged ΔE mutant Alb293 or the wild-type Alb240 (wt). The stars indicate the primary extra RNA band in Alb290 stocks. (B) RT-PCR analysis of the duplicated region of the Alb290 genome. Random-primed RT products obtained with RNA isolated from infected cells were amplified with primer pairs neighboring the region of the E gene deletion. PCR products were analyzed by agarose gel electrophoresis; the sizes (bp) of DNA markers are indicated on the left of each gel. (C) Schematic of the Alb290 genome compared with the ΔE genome. Beneath each genome are shown the loci of individual primers (arrows) or of RT-PCR products (bars); above each genome are shown the hybridizing regions of the probes used in the Northern blots. The M gene fragment in Alb290 is indicated by brackets; l denotes the embedded leader RNA.
To more precisely define the nature of the insertion in Alb290, we carried out RT-PCR analysis of infected cellular RNA from purified plaques. Since the Northern blot results situated the insertion between the S and the M genes, we found, by trial and error, primers that fell outside what turned out to be a region of duplication. The primer pair LK59 and PM204, spanning the 3′ end of the S gene to the downstream portion of the M gene, amplified two products from Alb290 RNA (Fig. 2B and C). The first PCR product appeared identical to the 595-bp fragment obtained with RNA from the basic ΔE mutant Alb291, and this identity was confirmed by sequencing. This result indicated that, even following plaque purification, the Alb290 variants contained a substantial subpopulation of the original ΔE mutant. The second PCR product was larger, roughly 1.1 kb, showing that the insertion in A290 was on the order of 0.5 kb. Similar results were obtained with the primer pair CK20 and CM89 (Fig. 2B and C), i.e., both a band corresponding to that of the ΔE mutant and a second band some 0.5 kb larger were produced. The larger PCR fragments obtained with each of these primer pairs often appeared as the major band in a ladder of products. In retrospect, we believe that these ladders were likely created by iterative self-priming of duplicated regions of the amplified genomic segments. In contrast, no PCR products were obtained by the pairing of LK98, a negative-sense primer at the 3′ end of the S gene, with PM148, a positive-sense primer in the middle of the M gene (Fig. 2C). The latter negative result verified that the insert in Alb290 did not contain a duplication of the 3′ end of the S gene, as was found for Alb295 and Alb294 (see below).
The larger PCR product obtained with LK59 and PM204 was sequenced directly. Additionally, the larger PCR products obtained with CK20 and CM89, as well as with another primer pair, CK20 and PM145, were sequenced following TA cloning. The bulk PCR product sequence and the sequences of 14 of 18 TA clones were identical, establishing that Alb290 contained a 484-nt insert in which nucleotide 413 of a partially duplicated M ORF was fused to nucleotide 2 of the MHV leader RNA (Fig. 2C). However, the sequences of the remaining four TA clones revealed that there was a minor subpopulation within Alb290 containing a 485-nt insert in which nucleotide 423 of a partially duplicated M ORF was fused to nucleotide 11 of the MHV leader RNA. Curiously, in both the major and the minor forms of the Alb290 insert, there was an exact deletion of codon 90 of the duplicated M gene segment; this same codon was intact in the wild-type copy of the M gene in Alb290.
Genomic analysis of ΔE isolates Alb295 and Alb294.
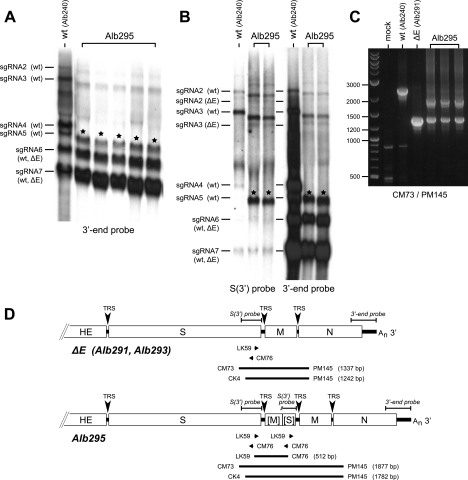
The genomic compositions of the other two aberrant ΔE isolates were determined in a manner similar to that described for Alb290. For Alb295, infected cellular RNA from all 12 purified plaques exhibited extra bands, as detected by Northern blot hybridization with a probe specific for the 3′ genomic end. A subset of these samples are shown in Fig. 3A. As was seen for Alb290, the primary extra RNA species for Alb295 was comparable in size to wild-type sgRNA5. In the case of Alb295, however, the primary (and larger) extra bands also hybridized to a probe specific for the 3′ end of the S gene (Fig. 3B), indicating that the Alb295 insert must include a duplication of some downstream segment of the S gene. This conclusion was confirmed by RT-PCR analysis with the primers LK59 and CM76, which are closely spaced primers that fall within the 3′ end of the S gene but have diverging polarities (Fig. 3D). As expected, this primer pair could not amplify a product from RNA of either the wild type or the basic ΔE mutant Alb291. However, a 0.5-kb PCR fragment was obtained with Alb295 RNA (data not shown), supporting the notion that Alb295 contained an insert on the order of 0.5 kb that included a partial duplication of the S gene. More conclusively, primers CM73 and PM145, which spanned the insertion, were found to produce two PCR products (Fig. 3C). The smaller product was identical to that obtained with the basic ΔE mutant Alb291, while the larger product indicated the presence of a 0.5-kb insert. A similar pair of products was obtained with the primer pair CK4 and PM145 (Fig. 3D). Direct sequences of the LK59-CM76 and CK4-PM145 PCR products, as well as all six sequences obtained from TA clones of the CM73-PM145 product, were in complete accord. They showed that Alb295 contained a 540-nt insert in which the 5′-most 303 nucleotides of the M ORF were fused to the 3′-most 196 nucleotides of the S ORF (Fig. 3D).
FIG. 3.
Genomic composition of ΔE mutant Alb295. (A and B) Northern blots of RNA from cells infected with stocks started from individual plaques of ΔE isolate Alb295 and detected with probes specific for the MHV 3′ genomic end or the 3′ end of the S gene. Control RNA samples were from cells infected with the wild-type Alb240 (wt). The stars indicate the primary extra RNA band in Alb295 stocks. In panel A, the wild-type control lane is a lower exposure than that shown for the other lanes. (C) RT-PCR analysis of the duplicated region of the Alb295 genome. Random-primed RT products obtained with RNA isolated from infected cells were amplified with a primer pair neighboring the region of the E gene deletion. The PCR products were analyzed by agarose gel electrophoresis; the sizes (bp) of DNA markers are indicated on the left of each gel. (D) Schematic of the Alb295 genome compared with the ΔE genome. Beneath each genome are shown the loci of individual primers (arrows) and of RT-PCR products (bars); above each genome are shown the hybridizing regions of the probes used in the Northern blots. The M and S gene fragments in Alb295 are indicated by brackets.
For Alb294, extra RNA species were detected in infected cellular RNA from 9 out of 12 purified plaques. Northern blots for a subset of these nine, hybridized with a probe specific for the 3′ genomic end, are shown in Fig. 4A. In this case, the primary extra band was quite large, migrating approximately midway between the bands for wild-type sgRNA3 and sgRNA4. This mobility showed that the insert in Alb294 was some 2.5 kb in length. Moreover, the primary extra band also hybridized with probes specific for both the middle and the 3′ end of the S gene (Fig. 4A), indicating that the Alb294 insert contained a duplication of at least half of the S gene. The extensive magnitude of the duplication in Alb294 confounded our ability to obtain unique RT-PCR products for much of the region of the insertion. Despite many attempts, we could not amplify a predicted 6.1-kb PCR product with primers CM49 and PM210, which were expected to flank the genomic duplication (Fig. 4C). However, we were able to obtain a 0.7-kb PCR product (data not shown) with PM148, a positive-sense primer in the M gene, and CM58, a negative-sense primer in the S gene (Fig. 4C). These products established that the Alb294 insert contained a fragment of the M gene upstream of the partial duplication of the S gene, and sequencing of the PM148-CM58 PCR fragment defined the aberrant M-S junction. We also obtained a 0.7-kb PCR product with the leader RNA-specific primer LK99 and primer CM58, which falls in the middle of the S gene (Fig. 4B and C). The sequence of that product, as well as the sequence of an overlapping amplicon from LK99 and CK3, clearly delineated the extent of the duplicated M gene fragment and confirmed the composition of the novel genomic junction. Taken together, the results from the Northern blotting and RT-PCR analysis were consistent with Alb294 containing a 2,647-nt insert in which the 5′-most 242 nucleotides of the M ORF were fused to the 3′-most 2,364 nucleotides of the S ORF (Fig. 4C).
FIG. 4.
Genomic composition of ΔE mutant Alb294. (A) Northern blots of RNA from cells infected with stocks started from individual plaques of ΔE isolate Alb294 and detected with probes specific for the MHV 3′ genomic end (left), the middle of the S gene (middle), or the 3′ end of the S gene (right). Control RNA samples were from mock-infected cells or from cells infected with the nonrearranged ΔE mutant Alb293 or the wild type Alb240 (wt). The stars indicate the primary extra RNA band in Alb294 stocks. (B) RT-PCR analysis of the duplicated region of the Alb294 genome. Random-primed RT products obtained with RNA isolated from infected cells were amplified with a primer pair neighboring the region of the E gene deletion. The PCR products were analyzed by agarose gel electrophoresis; the sizes (bp) of DNA markers are indicated on the left of each gel. (C) Schematic of the Alb294 genome compared with the ΔE genome. Beneath each genome are shown the loci of individual primers (arrows) or of RT-PCR products (bars); the dashed lines indicate expected PCR products that we were unable to obtain. Note that PCR products obtained with primer LK99, which falls in the MHV leader sequence, originated from subgenomic templates. Above each genome are shown the hybridizing regions of the probes used in the Northern blots. The M and S gene fragments in Alb294 are indicated by brackets.
All three aberrant ΔE isolates encode a second, truncated form of the M protein.
Notwithstanding the clear differences among the inserts in the aberrant ΔE isolates, examination of the genomic structures of these viruses revealed three common characteristics. First, in each case, a substantial duplication in the ΔE genome had been created, apparently by a nonhomologous crossover event, either between a ΔE genome and sgRNA6 (for Alb290) or between two ΔE genomes (for Alb295 and Alb294) (Fig. 5A). For each variant genome, then, the insert size could be defined as the distance between any two exactly duplicated points. Second, as demonstrated by our detection of persistent ΔE RNA by Northern blotting and RT-PCR, each variant genome existed in a dynamic steady state with the original ΔE genome, through constant regeneration of the latter by a single intramolecular homologous crossover event between duplicated regions. Third, and most striking, in all three aberrant ΔE genomes, the net result of the duplication was the creation of a new version of the M gene situated upstream of the native copy of the M gene. In each case, the new M gene encoded a protein (which we termed M*) that was identical to a portion of wild-type M at its amino terminus but was fused at its carboxy terminus to a short stretch of heterologous polypeptide determined by the juxtaposed sequence at the crossover junction (Fig. 5A and B). The heterologous tails of the M*-290 and M*-290m proteins, encoded by the major and minor populations of Alb290, were derived from different ORFs contained within the MHV leader RNA. Similarly, the M*-295 protein tail was in the −1 reading frame with respect to the S ORF and the M*-294 protein tail was in the +1 reading frame with respect to the S ORF. Additionally, M*-290 (and M*-290m) contained an exact deletion of the codon for F90 in the third transmembrane domain of M (Fig. 5B).
FIG. 5.
MHV variants evolved from ΔE mutants. (A) Summary of the 3′-end genomic structures of the five ΔE isolates that were analyzed in detail. Alb291 and Alb293 (top) were previously shown to contain only the predicted deletion between the S and M genes (31). Alb290, Alb295, and Alb294 each contain a duplication resulting from nonhomologous recombination, either between the ΔE genome and sgRNA6 (for Alb290) or between two ΔE genomes (for Alb295 and Alb294). In each case, the second component of the nonhomologous recombination event is shaded gray, and the crossover junction is indicated by an X. Fragments of genes are indicated by brackets; l denotes the leader sequence embedded in the Alb290 genome. The arrowheads denote TRSs. Schematics of the encoded wild-type M and variant (M*) proteins are shown below each genome. (B) Alignment of wild-type and M* protein sequences; for the latter, only those residues that differ from the wild type are shown. M*-290m is the M* protein encoded by a minor form of the Alb290 insert. The circles above the wild-type M sequence denote the three transmembrane domains, as defined by Rottier and coworkers (45); Δ, deletion of the F90 residue in M*-290. Following the crossover junction in M*-295, the extension of the M* ORF is in the −1 reading frame with respect to the S ORF; for M*-294, the extension of the M* ORF is in the +1 reading frame with respect to the S ORF. (C) Western blots of lysates from 17Cl1 cells infected at a multiplicity of 0.1 PFU per cell with wild-type MHV (Alb240) or the ΔE mutant Alb290, Alb294, or Alb295 and harvested at 18 h postinfection. The blot was probed with monoclonal antibody J.1.3, which is specific for the M ectodomain (13). mock, control lysate from mock-infected cells. Molecular mass standards (kDa) are indicated on the left of the panel.
We expected that the different M* proteins would be expressed by their respective viruses because, as shown above, their corresponding sgRNAs were strongly transcribed. To determine whether the inferred M* ORFs were indeed functional, we carried out Western blot analysis of lysates from cells infected with the each of the three aberrant ΔE isolates. The blots were probed with monoclonal antibody J1.3 (19), which recognizes an epitope in the amino-terminal ectodomain of M (13). We found that, in addition to wild-type M protein, each variant ΔE isolate produced a smaller immunoreactive protein of a size consistent with that predicted for its M* protein (Fig. 5C), although we observed that the electrophoretic resolution of M and M* proteins varied, with respect to marker proteins, in polyacrylamide gels of differing densities. (The calculated molecular masses of unglycosylated M, M*-290, M*-295, and M*-294 are 26.0, 18.0, 16.2, and 11.4 kDa, respectively.) We were not able to grow sufficient quantities of Alb290, Alb295, or Alb294 virus to determine whether their respective M* proteins were incorporated into purified virions.
Expression of M* protein partially compensates for the absence of E protein.
Our analyses of the aberrant ΔE isolates showed that, remarkably, three independent crossover events at different loci in ΔE genomes had selected for the same type of partial M gene duplication. Moreover, each of the partially duplicated M genes encoded a version of the M protein possessing an effectively truncated endodomain. This outcome strongly implied that the M* proteins conferred some significant advantage on MHV in the absence of the E protein. However, we could not rule out the possibility that the genomic duplications themselves were necessary to remedy some global genomic RNA structural requirement that had been disrupted by the deletion in the ΔE mutant.
To determine whether the M* proteins per se were important, rather than the genomic rearrangements in which they were situated, we repositioned an M* ORF in a ΔE background that was free of any gross duplication. This was accomplished through construction of a mutant, designated M*/ΔE, a ΔE virus in which the M* ORF and its associated TRS from Alb290 replaced the nonessential 2a and HE genes, immediately upstream of the S gene (Fig. 6A). The 2a and HE genes are not required for MHV assembly, and their deletion has no effect on viral replication in tissue culture (14, 26). M*-290 was chosen for this test, because it showed the strongest level of expression (Fig. 5C). Further, Alb290 was the ΔE variant that exhibited the most enhanced growth relative to the original ΔE mutant, producing discernibly larger plaques and growing to 10-fold-higher titers. In contrast, we were unable to clearly quantitate a growth difference between Alb294 or Alb 295 and the basic ΔE mutant Alb291.
FIG. 6.
Expression of M*-290 from the gene 2 region of MHV. (A) Comparison of the downstream ends of the genomes of wild-type MHV and the mutants Δ(2a-HE), Δ(2a-HE)/ΔE, M*/ΔE, and M*-KO/ΔE. Beneath the schematics are shown details of the 1b-S or 1b-M* intergenic junction; SalI and AscI sites that were introduced into the parent transcription vectors are marked by dashed underlines; TRSs are boxed; indicates a nonfunctional TRS. Point mutations created to disrupt the TRS and the M* ORF in the M*-KO mutant are indicated in boldface with solid underlines. Deletion and substitution mutants were constructed by targeted RNA recombination, as described in Materials and Methods. (B) Plaques of mutants Δ(2a-HE)/ΔE, M*/ΔE, and M*-KO/ΔE compared with those of the wild type (Alb240). Plaque titrations were carried out on L2 cells at 37°C. Monolayers were stained with neutral red at 96 h postinfection and were photographed 18 h later. For the Δ(2a-HE)/ΔE and M*-KO/ΔE mutants, one quadrant of the photograph is shown with altered contrast to allow better visualization of the tiny plaques of these mutants. (C) Growth kinetics of the M*/ΔE mutant relative to those of its Δ(2a-HE) and Δ(2a-HE)/ΔE counterparts. Confluent monolayers of 17Cl1 cells were infected at a multiplicity of 0.1 PFU per cell. At the indicated times postinfection, aliquots of medium were removed, and infectious titers were determined by plaque assay on L2 cells. The open and shaded symbols represent results from two independent experiments.
The relocation of the M* ORF also precluded the persistent regeneration of the original ΔE mutant through recombination. Although homologous recombination between the M* and M ORFs was still possible in the M*/ΔE genome, such an event would be lethal, owing to deletion of the intervening S gene. As a control, we also isolated a parallel construct of the original ΔE mutant, Δ(2a-HE)/ΔE. Additionally, we created M*-KO/ΔE, a version of the M*/ΔE mutant in which the relocated M*-290 ORF, and also its associated TRS, were knocked out with point mutations (Fig. 6A). Each of these viruses was then compared either to the wild type or to a previously constructed isogenic Δ(2a-HE) control (26), which was phenotypically identical to the wild type.
The M*/ΔE mutant was found to produce plaques intermediate in size between those of the wild type and those of the Δ(2a-HE)/ΔE mutant (Fig. 6B). As expected, the properties of the Δ(2a-HE)/ΔE mutant were indistinguishable from those previously reported for the ΔE and E knockout viruses (29, 31). Significantly, we observed that the M*-KO/ΔE mutant exhibited the same tiny plaques as did the Δ(2a-HE)/ΔE mutant, demonstrating that the enlarged plaques of the otherwise isogenic M*/ΔE mutant must result from expression of the M* protein by the latter virus. A second set of independently isolated mutants behaved identically to those shown in Fig. 6B.
The benefit conferred by the M* protein in the absence of E protein was explored in more detail through measurement of growth kinetics. The growth of the Δ(2a-HE)/ΔE mutant displayed a protracted lag and peaked at infectious titers that were some 4 orders of magnitude below those of the wild type (Fig. 6C). In contrast, the growth of the M*/ΔE mutant closely followed that of the wild type for the first 12 h of infection. The M*/ΔE mutant subsequently reached peak infectious titers at 24 h postinfection that were at least 100-fold higher than those of the Δ(2a-HE)/ΔE mutant, although they were still 100-fold lower than those of the wild type. The M*-KO/ΔE mutant behaved exactly like its Δ(2a-HE)/ΔE counterpart (data not shown), again confirming that the enhanced growth of the M*/ΔE virus could be attributed to expression of the M* protein. It should be noted that, in order to obtain a sufficient inoculum for the growth experiment shown in Fig. 6C, it was necessary for us to propagate a stock of the Δ(2a-HE)/ΔE virus in a line of 17Cl1 cells that expresses the MHV E protein (29). Consequently, the observed growth kinetics for this mutant, although very poor, may actually be artificially robust.
To further characterize the set of mutants shown in Fig. 6, we performed Western blot analyses. We probed blots of lysates prepared from infected cells with anti-M monoclonal antibody and ascertained that the M*/ΔE mutant indeed expressed the M* protein and—equally importantly—that the M*-KO/ΔE mutant failed to express the M* protein (Fig. 7A). Additionally, probing of blots with anti-E antiserum verified that the Δ(2a-HE)/ΔE, M*/ΔE, and M*-KO/ΔE mutants did not synthesize E protein. Analysis of purified virions of the wild type and the M*/ΔE mutant revealed that the M* protein was incorporated as a constituent of assembled viruses (Fig. 7B). This finding showed that the M* protein was appropriately localized to be able to participate in the viral budding process and that M* must have been closely associated with the native M protein.
FIG. 7.
Expression and virion incorporation of M* protein. (A) Western blots of lysates from 17Cl1 cells infected at a multiplicity of 0.1 PFU per cell with wild-type MHV or with each of the mutants shown in Fig. 6A and harvested at 16 h postinfection. The blots were probed with anti-M monoclonal antibody J.1.3 (α-M) (top) or anti-E polyclonal antibody (α-E) (bottom). mock, control lysate from mock-infected cells. (B) Western blots of purified wild-type or M*/ΔE mutant virions probed with anti-E polyclonal antibody (left), anti-M monoclonal antibody J.1.3 (middle), or anti-N monoclonal antibody J.3.3 (α-N) (right). Molecular mass standards (kDa) are indicated on the left of each panel.
Despite the positive effects of M* expression on viral replication in the absence of E protein, it was evident that the support provided by the M* protein was incomplete. After reaching a peak at 24 h postinfection, the infectious titers of the M*/ΔE mutant dropped more than 10-fold by 48 h postinfection (Fig. 6C). By comparison, the infectious titers of both the wild type and the Δ(2a-HE)/ΔE mutant remained stable over the same period. This discrepancy suggested that virions that were assembled with the assistance of M* protein were less stable than wild-type virions. Further support for such a notion came from viral purifications, during which we noted that M*/ΔE virions sedimented to a different position and formed bands that were more diffuse than those seen for wild-type virions on glycerol-tartrate gradients. Another indication of the limitations of M* protein arose when we propagated the M*/ΔE virus in E-expressing 17Cl1 cells. Subsequent titration on L2 cells yielded roughly equal numbers of tiny plaques among the intermediate-size plaques that were characteristic of the M*/ΔE mutant. We analyzed six of these tiny plaques and found that each contained a different deletion, ranging from 15 to 190 nucleotides, in the region of the M* gene. All of the deletions disrupted the start of the M* ORF, and all but one of them also removed the upstream TRS. The disruption of M* expression in these mutants was confirmed by Western blotting (data not shown). It is not currently clear whether this outcome demonstrated that a selection against M* expression occurred in the presence of E protein. Alternatively, this result may have merely signified that, since M* expression was superfluous in the presence of E protein, there was an accumulation of mutants that contained disruptions in the M* gene.
Other partially reverting mutations of ΔE or E knockout mutants map in the M gene.
In addition to the M* duplications, we found other, less dramatic mutations that allowed MHV to partially suppress the effects of the ΔE phenotype. Some of these mutations arose in higher-passage stocks of the E knockout mutant (29); others arose during our unsuccessful attempts to replace the E ORF with an ORF encoding the influenza virus M2 protein. In all cases, these mutations mapped to the M gene and created amino acid changes, principally in the first and third transmembrane domains, but also in the amino-terminal ectodomain and in the external loop between the second and third transmembrane domains (Fig. 8A). Some of the resulting M protein alterations, such as T15M, F28L, I92T, and S94F, were found as single point mutations. However, more commonly, T15M was paired with one additional mutation, the most frequent of which was F28L. In one case, a single residue was replaced by 5 residues, due to a 12-nt insertion within the codon for N81. The localization of these partial suppressors may, in some ways, parallel the recent finding that mutations in the first transmembrane domain of M were partially compensatory for an apparently lethal mutation that was constructed in a highly conserved region of M a short way downstream from the third transmembrane domain (3).
FIG. 8.
Mutations, other than M* duplications, that partially counteract the ΔE phenotype. (A) Summary of mutations in the M protein that were found to produce larger-plaque-size variants of E mutants. The solid boxes represent transmembrane (Tm) domains. The three M* proteins are also shown for comparison; the hatched boxes represent heterologous sequences resulting from frameshifts (Fig. 5). (B) Plaques of reconstructed mutants containing the ΔE mutation and one or more compensating M mutations compared to plaques of an original ΔE mutant (Alb291) and the wild type (Alb240). Plaque titrations were carried out on L2 cells at 37°C. The monolayers were stained with neutral red at 72 h postinfection and photographed 18 h later.
To assess whether any of the mutations that we found were capable of enhancing MHV growth in the absence of E protein, we reconstructed a subset of them in a ΔE background (Fig. 8B). The two most commonly occurring single mutations, T15M and F28L, were each found to individually provide partial compensation for the ΔE phenotype. Moreover, we noted a small additive effect when the two mutations were constructed together in a ΔE virus. Likewise, V11F and W98S, each of which we had isolated in the company of T15M, could each individually augment the effect of the T15M mutation. None of the point mutations appeared to be as strong a suppressor as was the M* partial duplication. Nevertheless, these results showed that relatively subtle modifications of the M protein transmembrane region are capable of producing compensatory effects similar to those brought about by duplication of the M protein transmembrane region.
DISCUSSION
At the time of our original construction of the MHV ΔE mutant, all prior evidence—from VLP studies of multiple coronaviruses (5, 6, 8, 48), from the defective assembly of MHV E point mutants (18), and from the demonstrated lethality of E gene deletions in TGEV (11, 41)—indicated that the E gene must be essential. Thus, our finding that the ΔE mutant was viable, albeit severely impaired, was surprising. We used a number of criteria to confirm the genotype of ΔE isolates, in order to rule out any possible extraneous source of the E gene or its presence at an ectopic location in the genome (31). It was therefore alarming to observe that passaged stocks of some isolates of the ΔE mutant exhibited grossly incongruous patterns of RNA synthesis, patterns that appeared to result from the acquisition of an extra transcription unit of unknown origin (Fig. 1).
The current study stemmed from our analysis of those aberrant ΔE isolates. In retrospect, we now see that a number of factors confounded our initial efforts to determine the nature of the unexpected RNA species. The combined effects of partial genomic duplications and the nested set structure of coronavirus RNAs created unpredicted target sites or orientations for PCR primers. Additionally, a second layer of target sites in each stock was generated from the background population of the original ΔE mutant, which was formed via recombination between identical regions of the partial duplications. Nevertheless, we were able to confirm the genomic compositions of Alb290 and Alb295 by means of the sequences of PCR products that were anchored in unique regions and spanned the duplicated segments of each of these genomes (Fig. 2 and 3). For Alb294, the genomic composition was pieced together from the combined evidence from Northern blots and PCR analysis of the unique junction of the duplication (Fig. 4). Together, our results showed that the genomes of the aberrant ΔE isolates had arisen from nonhomologous recombination, either between the ΔE genome and sgRNA6 or between two ΔE genomes, producing respective net duplications of 487, 540, and 2,647 nucleotides for Alb290, Alb295, and Alb294. Although such a situation could have suggested that removal of the E gene somehow promoted overall genomic instability, the salient feature of each duplication was that it created a second version of the M gene that encoded a shortened protein, designated M* (Fig. 5). All three types of M* protein were found to be expressed by their respective viruses. To directly assess the role of this second form of M protein, we repositioned one M* ORF in a ΔE background, replacing the 2a and HE genes. This placement of the M* ORF so that it could not be lost through recombination revealed that expression of M* substantially enhanced the growth of the ΔE mutant (Fig. 6). Moreover, we showed that the M* protein became incorporated into assembled virions (Fig. 7).
The severity of the growth phenotype of the ΔE virus creates a tremendous selective pressure on this mutant to offset its deficiency. Our results strongly suggest that, in three separate instances, different M* proteins were selected because they were capable of assisting wild-type M protein in virion assembly and budding in the absence of the E protein. A framework for an understanding of how M* proteins could act in this capacity is provided by recent cryo-electron microscopic (37, 38) and cryo-electron tomographic (4) reconstructions of MHV, SARS-CoV, and other coronaviruses. These structural studies supply remarkably detailed glimpses of molecular organization that can be correlated with previous and current genetic and biochemical results. The main findings of the structural studies with respect to M protein are shown schematically in Fig. 9. First, the M protein has a small ectodomain and transmembrane stalk. Most of the mass of M is found in its compact globular endodomain, consistent with the results of very early analyses of M protein that showed that only ∼15 carboxy-terminal endodomain residues were susceptible to protease digestion (2, 43, 45). Second, based on estimated domain volumes, each observed M density is most likely a dimer (38). Third, the only detectable M(dimer)-M(dimer) contacts occur between endodomains, and not between transmembrane domains (4, 37, 38).
FIG. 9.
Model of M-M interactions based on cryo-electron microscopic (37, 38) and cryo-electron tomographic (4) reconstructions of coronaviruses. M multimers are depicted as dimers, but the same relationships would pertain for higher-order M oligomers. At the bottom is shown the domain structure of M protein in comparison to that of M* protein. Tm, transmembrane.
One crucial inference to be drawn from the developing structural model is that two distinct sets of M-M interactions exist: those between monomers and those between dimers. M(monomer)-M(monomer) contacts seem to be determined principally by transmembrane domain interactions. In contrast, M(dimer)-M(dimer) contacts are seen to occur exclusively between endodomains. In keeping with elegant work presented by the Gallagher laboratory (7), we believe that a major function for E protein is to act as an antiaggregant for M protein, keeping M in an assembly-competent state for both monomer and dimer interactions. Such a chaperone-like role, which does not require highly sequence-specific contacts (1, 55), would be consistent with our demonstration of the diverse range of heterologous sequences that can support E protein function in MHV (29). We hypothesize that M*, as a surrogate for E, also facilitates the establishment of proper transmembrane domain interactions between M monomers before M(dimer)-M(dimer) endodomain interactions can take place. The fact that M* protein is found in purified virions of the M*/ΔE mutant shows that M* interacts with native M protein. Such a conclusion is supported by findings from VLP studies that included M endodomain deletion mutants (16). Since the endodomains of M* proteins are almost entirely truncated (Fig. 5 and 9), the contacts between M* and M must occur via their respective transmembrane domains; accordingly, such contacts affect M(monomer)-M(monomer) interactions. In this manner, M* protein would be able to partially solve one problem that is normally addressed by E protein, although perhaps not via an identical mechanism. Similarly, particular M transmembrane domain mutations that are compensatory in the absence of E (Fig. 8) could aid the removal of some barrier to successful M(monomer)-M(monomer) interactions. On the other hand, the inability of M* to mediate M(dimer)-M(dimer) interactions may explain why growth of the M*/ΔE mutant is not restored to wild-type levels and why virions produced by the M*/ΔE mutant appear to be less stable than those of the wild type.
The propensity of ΔE mutants to generate M* ORFs dramatically highlights the inherent plasticity of coronavirus genomes and how the capacity of these viruses for recombination helps to drive their evolution. We speculate that a partial gene duplication, such as we have seen, could be how the ancestral E gene was originally acquired. It could subsequently have become fixed in the genome because it greatly potentiated the efficiency of viral assembly. In this regard, we note that the coronavirus E gene is always found upstream of, and adjacent to, the M gene. Coronaviruses, and other members of the nidovirus order, harbor evidence of a number of similar duplication and divergence events. The replicase genes of coronaviruses and arteriviruses contain as many as four copies of active and vestigial papain-like proteinase domains (21). Likewise, the reported homologies between SARS-CoV accessory gene products and coronavirus structural proteins (24) would be consistent with an origin for the former in a process of gene duplication and divergence. In particular, the SARS-CoV accessory protein 3a, which is encoded by a gene upstream of the M gene, is (like M) a triple-spanning membrane protein. The gene encoding another triple-spanning membrane protein, the GP5 protein, is located immediately upstream of the M gene in arterivirus genomes. There thus appears to be ample precedent for nidoviruses having improvised new functions from old genes. Conceivably, then, the M* protein mimics the precursor of the present-day E protein, and with sufficient selective pressure, M* could further evolve toward a more E-like functionality.
Acknowledgments
We are grateful to John Fleming (University of Wisconsin, Madison, WI) for generously providing monoclonal antibodies. We thank the Applied Genomics Technology Core Facility of the Wadsworth Center for DNA sequencing.
This work was supported by National Institutes of Health (National Institute of Allergy and Infectious Diseases) grants R01 AI064603 and R56 AI064603.
Footnotes
Published ahead of print on 6 October 2010.
REFERENCES
- 1.Anelli, T., and R. Sitia. 2008. Protein quality control in the early secretory pathway. EMBO J. 27:315-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, J., H. Niemann, S. Smeekens, P. Rottier, and G. Warren. 1984. Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature 308:751-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arndt, A. L., B. J. Larson, and B. G. Hogue. 2010. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. J. Virol. 84:11418-11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bárcena, M., G. T. Oostergetel, W. Bartelink, F. G. A. Faas, A. Verkleij, P. J. M. Rottier, A. J. Koster, and B. J. Bosch. 2009. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc. Natl. Acad. Sci. U. S. A. 106:582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudoux, P., C. Carrat, L. Besnardeau, B. Charley, and H. Laude. 1998. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 72:8636-8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bos, E. C. W., W. Luytjes, H. van der Meulen, H. K. Koerten, and W. J. M. Spaan. 1996. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 218:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boscarino, J. A., H. L. Logan, J. J. Lacny, and T. M. Gallagher. 2008. Envelope protein palmitoylations are crucial for murine coronavirus assembly. J. Virol. 82:2989-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corse, E., and C. E. Machamer. 2000. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 74:4319-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corse, E., and C. E. Machamer. 2002. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J. Virol. 76:1273-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corse, E., and C. E. Machamer. 2003. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology 312:25-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtis, K. M., B. Yount, and R. S. Baric. 2002. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 76:1422-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeDiego, M. L., E. Alvarez, F. Almazán, M. T. Rejas, E. Lamirande, A. Roberts, W. J. Shieh, S. R. Zaki, K. Subbarao, and L. Enjuanes. 2007. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 81:1701-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Haan, C. A. M., L. Kuo, P. S. Masters, H. Vennema, and P. J. M. Rottier. 1998. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 72:6838-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Haan, C. A. M., P. S. Masters, X. Shen, S. Weiss, and P. J. M. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Haan, C. A. M., and P. J. M. Rottier. 2005. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 64:165-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Haan, C. A. M., H. Vennema, and P. J. M. Rottier. 2000. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 74:4967-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escors, D., J. Ortego, H. Laude, and L. Enjuanes. 2001. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 75:1312-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72:7885-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleming, J. O., S. A. Stohlman, R. C. Harmon, M. M. C. Lai, J. A. Frelinger, and L. P. Weiner. 1983. Antigenic relationships of murine coronaviruses: analysis using monoclonal antibodies to JHM (MHV-4) virus. Virology 131:296-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goebel, S. J., B. Hsue, T. F. Dombrowski, and P. S. Masters. 2004. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 78:669-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurst, K. R., C. A. Koetzner, and P. S. Masters. 2009. Identification of in vivo interacting domains of the murine coronavirus nucleocapsid protein. J. Virol. 83:7221-7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurst, K. R., L. Kuo, C. A. Koetzner, R. Ye, B. Hsue, and P. S. Masters. 2005. A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein. J. Virol. 79:13285-13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inberg, A., and M. Linial. 2004. Evolutional insights on uncharacterized SARS coronavirus genes. FEBS Lett. 577:159-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klumperman, J., J. Krijnse Locker, A. Meijer, M. C. Horzinek, H. J. Geuze, and P. J. M. Rottier. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koetzner, C. A., L. Kuo, S. J. Goebel, A. B. Dean, M. M. Parker, and P. S. Masters. 2010. Accessory protein 5a is a major antagonist of the antiviral action of interferon against murine coronavirus. J. Virol. 84:8262-8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krijnse Locker, J., D.-J. E. Opstelten, M. Ericsson, M. C. Horzinek, and P. J. M. Rottier. 1995. Oligomerization of a trans-Golgi/trans-Golgi network retained protein occurs in the Golgi complex and may be part of its recognition. J. Biol. Chem. 270:8815-8821. [DOI] [PubMed] [Google Scholar]
- 28.Kuo, L., G.-J. Godeke, M. J. B. Raamsman, P. S. Masters, and P. J. M. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuo, L., K. R. Hurst, and P. S. Masters. 2007. Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. J. Virol. 81:2249-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo, L., and P. S. Masters. 2002. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 76:4987-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo, L., and P. S. Masters. 2003. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 77:4597-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machamer, C. E., S. A. Mentone, J. K. Rose, and M. G. Farquhar. 1990. The E1 glycoprotein of an avian coronavirus is targeted to the cis Golgi complex. Proc. Natl. Acad. Sci. U. S. A. 87:6944-6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machamer, C. E., and J. K. Rose. 1987. A specific transmembrane domain of a coronavirus E1 glycoprotein is required for its retention in the Golgi region. J. Cell Biol. 105:1205-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Machamer, C. E., and S. Youn. 2006. The transmembrane domain of the infectious bronchitis virus E protein is required for efficient virus release. Adv. Exp. Med. Biol. 581:193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masters, P. S. 2006. The molecular biology of coronaviruses. Adv. Virus Res. 66:193-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masters, P. S., and P. J. M. Rottier. 2005. Coronavirus reverse genetics by targeted RNA recombination. Curr. Top. Microbiol. Immunol. 287:133-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neuman, B. W., B. D. Adair, C. Yoshioka, J. D. Quispe, G. Orca, P. Kuhn, R. A. Milligan, M. Yeager, and M. J. Buchmeier. 2006. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 80:7918-7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neuman, B. W., G. Kiss, J. Klaus, F. Baksh, P. Kuhn, S. Makino, and M. J. Buchmeier. 2008. Structural analysis of M protein interactions in virions and virus-like particles. Scientific program, p. 47-48. XIth International Nidovirus Symposium. St. Catherine's College, Oxford, United Kingdom.
- 39.Ontiveros, E., L. Kuo, P. S. Masters, and S. Perlman. 2001. Inactivation of expression of gene 4 of mouse hepatitis virus strain JHM does not affect virulence in the murine CNS. Virology 289:230-238. [DOI] [PubMed] [Google Scholar]
- 40.Ortego, J., J. E. Ceriani, C. Patiño, J. Plana, and L. Enjuanes. 2007. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology 368:296-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ortego, J., D. Escors, H. Laude, and L. Enjuanes. 2002. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 76:11518-11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raamsman, M. J. B., J. Krijnse Locker, A. de Hooge, A. A. F. de Vries, G. Griffiths, H. Vennema, and P. J. M. Rottier. 2000. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 74:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rottier, P., D. Brandenburg, J. Armstrong, B. van der Zeijst, and G. Warren. 1984. Assembly in vitro of a spanning membrane protein of the endoplasmic reticulum: the E1 glycoprotein of coronavirus mouse hepatitis virus A59. Proc. Natl. Acad. Sci. U. S. A. 81:1421-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rottier, P. J. M., and J. K. Rose. 1987. Coronavirus E1 protein expressed from cloned cDNA localizes in the Golgi region. J. Virol. 61:2042-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rottier, P. J. M., G. W. Welling, S. Welling-Wester, H. G. M. Niesters, J. A. Lenstra, and B. A. M. Van der Zeijst. 1986. Predicted membrane topology of the coronavirus protein E1. Biochemistry 25:1335-1339. [DOI] [PubMed] [Google Scholar]
- 46.Sawicki, S. G., D. L. Sawicki, and S. G. Siddell. 2007. A contemporary view of coronavirus transcription. J. Virol. 81:20-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siu, Y. L., K. T. Teoh, J. Lo, C. M. Chan, F. Kien, N. Escriou, S. W. Tsao, J. M. Nicholls, R. Altmeyer, J. S. Peiris, R. Bruzzone, and B. Nal. 2008. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 82:11318-11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vennema, H., G.-J. Godeke, J. W. A. Rossen, W. F. Voorhout, M. C. Horzinek, D.-J. E. Opstelten, and P. J. M. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verma, S., V. Bednar, A. Blount, and B. G. Hogue. 2006. Identification of functionally important negatively charged residues in the carboxy end of mouse hepatitis coronavirus A59 nucleocapsid protein. J. Virol. 80:4344-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verma, S., L. A. Lopez, V. Bednar, and B. G. Hogue. 2007. Importance of the penultimate positive charge in mouse hepatitis coronavirus A59 membrane protein. J. Virol. 81:5339-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson, L., P. Gage, and G. Ewart. 2006. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology 353:294-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, L., C. McKinlay, P. Gage, and G. Ewart. 2004. SARS coronavirus E protein forms cation-selective ion channels. Virology 330:322-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye, Y., and B. G. Hogue. 2007. Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J. Virol. 81:3597-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yokomori, K., and M. M. C. Lai. 1991. Mouse hepatitis virus S RNA sequence reveals that nonstructural proteins ns4 and ns5a are not essential for murine coronavirus replication. J. Virol. 65:5605-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young, J. C., V. R. Agashe, K. Siegers, and F. U. Hartl. 2004. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5:781-791. [DOI] [PubMed] [Google Scholar]