Abstract
Canine influenza virus (CIV) emerged around 2000 when an equine influenza virus (EIV) was transmitted to dogs in Florida. After 2003, the canine virus was carried by infected greyhounds to various parts of the United States and then became established in several large animal shelters, where it has continued to circulate. To better understand the evolution of CIV since its emergence, and particularly its microevolution in spatially restricted populations, we examined multiple gene segments of CIV from dogs resident in two large animal shelters in New York City during the period 2006 to 2009. In particular, we focused on viruses circulating in the two shelters in 2008 and 2009, which we found shared a common ancestor. While viruses in each shelter were generally monophyletic, we observed some gene flow between them. These shelter sequences were compared to earlier CIV isolates. The shelter viruses differed in 1 to 6 amino acids in each gene segment compared to viruses isolated in Florida between 2003 and 2005 and in Colorado in 2006 and 2008. A comparison of the sequences of equine and canine viruses revealed amino acid replacements that distinguished the viruses from the two hosts, but no clear evidence of positive selection indicative of host adaptation was detected, suggesting that any host range adaptation in CIV occurred early in the emergence of this virus or even before it transferred to dogs.
Influenza A viruses are naturally maintained in aquatic birds, occasionally transfer to mammals to cause individual infections or outbreaks of disease, and sometimes go on to cause epidemics and pandemics in their new hosts (25, 37). Influenza viruses can also transfer between different mammalian host species, as has recently been observed in the case of swine-origin pandemic H1N1 influenza A virus that emerged in 2009 in humans (16, 35). However, the determinants of host range and of interhost transmission of influenza viruses are often poorly understood. Key questions include the nature of any host barriers to viral transfer; whether host-adaptive mutants are required for initial infection of new hosts; and the role, if any, of posttransfer adaptation in determining continued transmission in the new host species. There are also considerable uncertainties about the epidemiological processes involved in host switching, including the role played by unusually dense populations of susceptible hosts, where viruses with lower transmission efficiencies may be maintained after initial transfers.
The A/H3N8 canine influenza virus (CIV) is a new pathogen of dogs (Canis familiaris), which resulted from the transfer of an intact A/H3N8 equine influenza virus (EIV) (7, 17, 38). CIV was initially recognized in greyhounds in a Florida training facility in 2004 and was then spread around the United States by infected greyhounds in 2004 and 2005, being recognized in 11 states during that period (7). Serological evidence has shown that the virus was infecting dogs in Florida in 2000 (7). Although CIV clearly originated from EIV, several differences between previously circulating equine viruses and the emerged canine viruses have been documented, including 8 amino acid replacements in the hemagglutinin (HA) segment (7, 38).
Equine influenza virus A/H3N8 was first recognized in horses (Equus caballus) in the early 1960s, with the prototype virus assigned as A/equine/Miami/1/1963 (51). EIV subsequently spread to most regions of the globe, with exceptions including New Zealand (22) and Iceland (49), and with recent outbreaks in otherwise EIV-free Australia in 2007 (2) and in South Africa in 2003 that were controlled. EIV has experienced significant evolution during its spread in horses in the >47 years since it emerged, including the emergence of two antigenically distinct sublineages that were first recognized in Florida (Florida 1 and 2) but which have subsequently spread to other regions of the United States and the world (4, 10, 26, 31). Dogs are clearly susceptible to recent strains of A/H3N8 and other influenza A viruses. Sporadic infections were reported for the A/H3N8 EIV during the Australian outbreak (24), and small outbreaks have been reported in foxhounds in the United Kingdom (9). In addition, dogs in South Korea have recently experienced widespread infections by an A/H3N2 avian influenza virus (28, 46), and canine infections by the new A/H1N1/09 swine-origin influenza virus have also been described (13). The A/H3N8 CIV that emerged in the United States was closely related to EIV strains circulating in horses around the same period and was clearly derived from a Florida 1 lineage EIV (7). Despite being an important recent example of cross-species transmission, little is known about the current spread of CIV among dogs in the United States or its evolution in its new host. The limited data available suggest that CIV is currently found mostly in the northeastern United States and in Colorado; that it is primarily being maintained in animal facilities with large numbers of incoming susceptible dogs, such as animal shelters and kennels; and that it has not spread widely within the household dog population (J. M. Scarlett and E. J. Dubovi, unpublished data).
The emergence of CIV from EIV provides an unusual opportunity to investigate the properties and dynamics of viral evolution during and shortly after a host transfer event. To this end, we examined the evolution of the CIV circulating during 2008 and 2009 in two large dog shelters in New York City and compared the viruses to those collected in some other regions of the United States since 2003. By determining the genetic diversity of the viruses within dog shelters, we were also able to explore the microevolution of CIV in defined populations, utilizing sampling that was restricted in both time and space.
MATERIALS AND METHODS
Viral samples and endemically transmitted viruses: shelters/epidemiology.
Samples were collected from naturally infected dogs from various regions of the United States between 2005 and 2009 (Table 1). Six samples were collected from dogs in New York between 2005 and 2007, two from dogs in Colorado in 2006 and 2008, and one each from Philadelphia in 2008 and Virginia in 2009. Naturally transmitted CIV samples obtained from two animal shelters that were ∼8 km apart in New York City that were controlled by the same organization (shelter A and shelter B) were examined in more detail. Each shelter accepted 300 to 500 dogs per month. Serological testing of 100 incoming dogs in 2008 indicated that the great majority of dogs entering the shelter were seronegative for CIV on arrival, but more than 20% became infected within 5 days of introduction into the shelter (Scarlett and Dubovi, unpublished).
TABLE 1.
Origins of CIV samples and isolates examined
| Sample | Sampling datea | Sample typeb | Location |
|---|---|---|---|
| Dog 1 | 3/20/09 | Nasal swab | Shelter A |
| Dog 2 | 3/20/09 | Trachea/nasal | Shelter A |
| Dog 3 | 3/20/09 | Tracheal wash | Shelter A |
| Dog 4 | 3/20/09 | Tracheal wash | Shelter A |
| Dog 5 | 3/20/09 | Tracheal wash | Shelter A |
| Dog 6 | 3/18/09 | Tracheal wash | Shelter A |
| Dog 7 | 11/18/08 | No further details | Shelter B |
| Dog 8 | 11/20/08 | No further details | Shelter B |
| Dog 9 | 11/20/08 | No further details | Shelter B |
| Dog 21 | 12/18/09 | Swab | Shelter B |
| Dog 22 | 9/22/09 | Lung | Shelter A |
| Dog 23 | 9/22/09 | Lung | Shelter A |
| Long Is/2005 | September 2005 | Virus (AF) | Long Island, NY |
| NewYork/Jan2006 | 1/16/06 | Virus (AF) | New York |
| A/2006 | 8/16/06 | Virus (TC) | Shelter A |
| NewYork/Dec2006 | 12/18/06 | Virus (TC) | New York |
| B/2006 | 8/16/06 | Virus (AF) | Shelter B |
| Colo/2006 | January 2006 | Virus (AF) | Colorado |
| Staten Is/2007 | 6/9/07 | Virus (TC) | Staten Island, NY |
| Colo/2008 | January 2008 | Virus (AF) | Colorado |
| Phil/2008 | November 2008 | No further details | Philadelphia |
| Vir/2009 | August 2009 | Virus (AF) | Virginia |
Unless otherwise noted, sampling dates are shown as month/day/year.
AF, allantoic fluid; Tc, tissue culture.
Sample collection.
Many samples examined were submitted as diagnostic specimens, tested for the presence of CIV by reverse transcription-PCR (RT-PCR), and isolated into embryonated eggs or MDCK tissue culture cells. Other viruses were obtained from shelters A and B in New York City between November 2008 and December 2009 (Table 1). These samples were obtained as nasal or pharyngeal swabs from dogs with respiratory disease or as lung samples from dogs that died.
RNA extraction and reverse transcriptase PCR.
RNA was extracted from the samples by the spin protocol of the QIAamp viral RNA minikit (Qiagen, Valencia, CA). A two-step RT-PCR was used to amplify the gene segments, using H3N8-specific primers (Table 2). The four largest gene segments (PB1, PB2, PA, HA) were each amplified as two overlapping fragments. Because not all segments were sequenced for viruses from every dog, we necessarily focused on the HA1, M, NS, and NA segments. The HA1 RT-PCR protocol used has been described previously (20). For the other 11 fragments, the protocol was as follows. Briefly, cDNA was made with universal primer Uni12a (AGC AAA AGC AGG), deoxynucleoside triphosphates (dNTPs), Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA), and RNase inhibitor (New England Biolabs, Ipswich, MA), with ∼5 μg total RNA, and incubated at 55°C for 60 min followed by 70°C for 15 min. A 5-μl aliquot of the cDNA was included in the final PCR step, with 1 μM each primer, 2.5 mM MgCl2, 0.2 mM dNTPs, and 0.1 U AmpliTaq Gold (Applied Biosystems, Carlsbad, CA). The reaction cycle consisted of an initial denaturation at 94°C for 10 min, followed by 45 cycles of 94°C for 20 s, 53°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min. PCR products were purified using the spin protocol of the Qiagen PCR purification kit.
TABLE 2.
Locations of H3N8-specific primers used for RT-PCR
| Gene segment | Position ina: |
|||
|---|---|---|---|---|
| Fragment 1 |
Fragment 2 |
|||
| Forward primer | Reverse primer | Forward primer | Reverse primer | |
| PB2 | 1 | 1250 | 1150 | 2342 |
| PB1 | 1 | 1253 | 1148 | 2341 |
| PA | 1 | 1252 | 1150 | 2233 |
| HA | 1 | 1008 | 927 | 1773 |
| NP | 1 | 1567 | ||
| NA | 1 | 1465 | ||
| M | 1 | 1027 | ||
| NS | 1 | 892 | ||
The numbering shown is from A/equine/Miami/1963 (GenBank accession no. CY028836 to CY028843).
Cloning and analysis of single sequences.
PCR products were cloned into the pCR4Blunt-TOPO vector in the Zero Blunt TOPO PCR cloning kit (Invitrogen) and electrotransformed into supplied TOP10 electrocompetent Escherichia coli cells. Plasmid DNAs prepared by the Miniprep method (Promega, Madison, WI) were sequenced with the T3 and T7 plasmid primers. Forward and reverse sequences were assembled with SeqMan (DNASTAR, Madison, WI) and visually checked. Primer regions were trimmed.
Evolutionary analysis.
All EIV sequences (between 77 and 205 per gene segment) and CIV sequences (between 4 and 10 per gene segment) available up to 1 March 2010 were downloaded from the Influenza Virus Resource, National Center for Biotechnology Information (NCBI). A multiple alignment of the sequences for each gene region was done in ClustalX v1.83.1 (6, 27). The output alignment was visually checked and edited in Se-Al v2.0a11 (kindly provided by Andrew Rambaut, University of Edinburgh). Maximum likelihood (ML) trees were then inferred in PAUP* (48), using substitution model parameters as determined by Modeltest3.7 (40). Specifically, for the trees containing both EIV and CIV sequences, these were the K81uf+I+G, TIM+I+G, and TVM+I substitution models for the HA1, M, and NS segments, respectively. For the trees with the shelter CIV sequences, the models used were TIM, HKY+I, and TVM for HA1, M, and NS, respectively. In all cases, a heuristic search was used, with 10 replicates, random sequence addition, and tree bisection-reconnection branch-swapping. Bootstrap analysis was also performed in PAUP* with 1,000 replicates of neighbor-joining trees under the ML substitution model.
We used the Bayesian Markov Chain Monte Carlo (MCMC) approach available in the BEAST package (11) to estimate both rates of nucleotide substitution and the time to the most recent common ancestor (TMRCA) of the HA1, M, NA, and NS gene segments from CIV and EIV using information on the month and year of sampling. The sequences in question were very closely related, so there will be little multiple substitution at single-nucleotide sites. Because of this, we used the relatively simple HKY85 model of nucleotide substitution in each case (which accords with the models described above) and assumed a relaxed log-normal clock and a Bayesian skyline coalescent tree prior (12). The MCMC chain was run for 200 or 300 million generations with a 10% burn-in. Statistical uncertainty is provided by values of the 95% highest-probability density (HPD). To avoid being biased by the large number of intrahost CIV sequences available for the HA1, M, and NS gene regions, we also used an alignment of only the consensus sequences from each dog to estimate the rates of nucleotide substitution in these cases.
To determine the strength of geographical structure in these data, and specifically whether sequences were clustered according to (i) host dog, (ii) dog shelter, and (iii) sampling date, we employed a Bayesian MCMC approach that accounts for phylogenetic uncertainty by considering a large set of plausible trees (36). Hence, using the Bayesian Tip-Associated Analysis (BaTS) program, we computed the parsimony score (PS) and association index (AI) statistics of phylogeny-trait association, making use of the posterior sample of trees previously output from the BEAST analysis and employing 1,000 randomizations.
Finally, to determine the nature of the selection pressures acting on CIV, we estimated the relative numbers of nonsynonymous (dN) and synonymous (dS) nucleotide substitutions per site (dN/dS) in the protein-coding gene regions of HA1, M, NS, and NA, using HyPhy (39). For each gene segment, a neighbor-joining tree was used and the single likelihood ancestor counting (SLAC), two-rate fixed-effects likelihood (FEL), and random-effects likelihood (REL) methods were all employed. In addition, we used the TestBranchDNDS method in HyPhy to look for differences in the dN/dS ratio on the EIV-to-CIV main internal branches compared to the rest of the phylogeny. Finally, we used the two-ratio model implemented in the CODEML program from the PAML package (54) to determine the dN/dS ratio on external branches and internal branches of an ML tree constructed with only a single representative sequence from each dog. If those mutations fixed on internal branches were primarily due to positive selection, we would expect a higher dN/dS ratio on internal branches than on external branches of the phylogeny. Conversely, an elevated dN/dS ratio on external branches is suggestive of the presence of transient deleterious mutations that have yet to be removed by purifying selection.
Nucleotide sequence accession numbers.
New sequences generated here have been submitted to GenBank and assigned accession numbers HQ237502 to HQ238128.
RESULTS
Evolution of CIV in dogs since 2003.
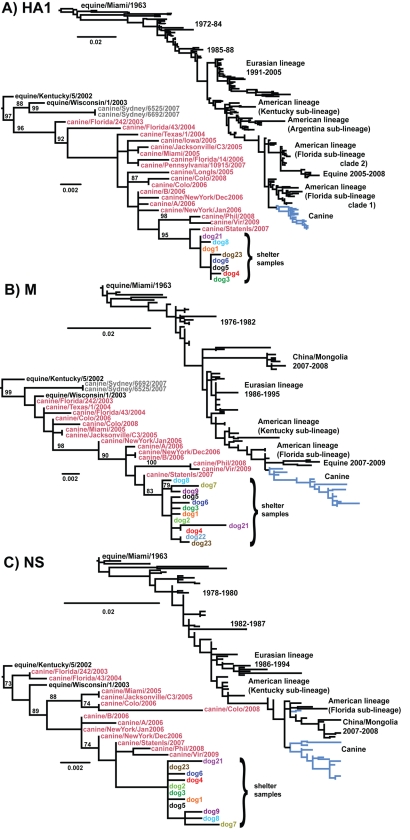
We examined the evolution of CIV since its initial recognition in Florida, with a particular focus on those viruses circulating in the northeastern United States, including those transmitted within two large shelters in New York City during 2008 and 2009. We examined sequences from all gene segments, but since there were many more sequences for the HA1, NA, M, and NS segments, we focused our studies of the dynamic aspects of the viral evolution on these segments. Our phylogenetic analysis confirmed that the CIV lineage was derived from a single EIV strain circulating around 2000 (Fig. 1). More notably, our analysis of multiple gene segments from viruses collected at different times and places since it emerged suggested a marked degree of sequence variation among the CIV lineages (Fig. 1 and 2; see Fig. S1 in the supplemental material). One lineage contained the viruses collected from New York City between 2006 and 2009, and as expected for a rapidly evolving RNA virus such as influenza virus, additional mutations were observed in the later samples compared to those collected in earlier years (Fig. 1 and see Table 4). Samples taken from dogs in Virginia in 2009 and Philadelphia in 2008 were related to the other viruses from the northeast United States, although they were sufficiently distinct to indicate that the virus was not derived directly from the New York virus population (Fig. 1).
FIG. 1.
Phylogenetic trees of EIV (from 1963 to the present), CIV, and representative shelter sequences. CIV sequences are shown in blue; their detailed phylogenetic relationships are shown in the insets. Virus samples in black are equine sequences, and those in pink are canine sequences. Sequences of viruses from the dogs in the New York shelters are color coded and indicate the consensus sequence of the virus from an individual dog. All horizontal branches are drawn to a scale of nucleotide substitutions per site, and bootstrap values of >70% are shown. (A) HA1; (B) M; (C) NS.
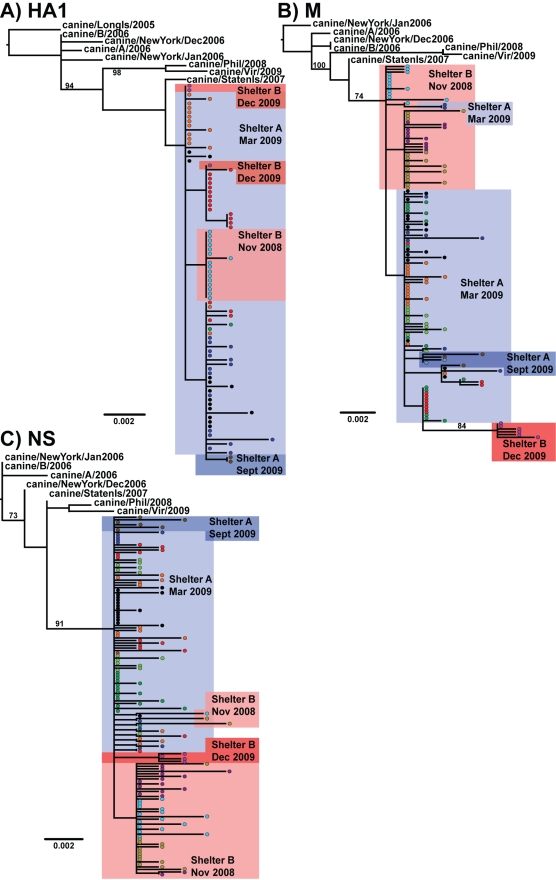
FIG. 2.
Phylogenetic trees of cloned CIV sequences prepared after direct RT-PCR of virus in swabs of infected dogs sampled from the New York City shelters, as well as some other isolates. Samples were taken from shelter B in November 2008 (light pink boxes) and December 2009 (dark pink boxes) and from shelter A in March 2009 (light blue boxes) and September 2009 (dark blue boxes). Each colored circle represents a sequence, and sequences from the same dog are colored the same across all trees. All horizontal branches are drawn to a scale of nucleotide substitutions per site, and bootstrap values of >70% are shown. (A) HA1; (B) M; (C) NS.
Viruses from dogs in Colorado in 2006 and 2008 were distinct from those seen in New York City, and in many segments were very similar to the viruses from the original Florida outbreaks, suggesting that they represent a separate lineage (Fig. 1). Our analysis of rates of nucleotide substitution and times to common ancestry of the HA1, NS, and M segment sequences (see below) suggests that the Colorado lineage emerged between December 2004 and December 2005 (range of 95% HPD values), likely during the original spread of CIV with racing greyhounds.
Finally, it is important to note that there appeared to be little variation deriving from the virus isolation in eggs, as comparison of two New York 2006 egg isolation virus M sequences with two New York 2006 tissue culture-derived virus M sequences showed there is only one mutation, in one of the latter sequences, not seen in earlier canine or equine reference sequences.
Two canine sequences isolated from a dog in Sydney, Australia, during the 2007 EIV outbreak (24) are included in the HA1 and M phylogenies (Fig. 1) and NA phylogeny (see Fig. S1 in the supplemental material). These Sydney CIV sequences are identical to an EIV isolated from a horse in the same stable as the dog, and there have been no reports of interdog CIV transmission in Australia (24). As a result, the Sydney CIV sequences are considered equine viruses isolated from dogs and were not included in further CIV analyses in this study.
Rates of molecular evolution.
To explore the evolutionary dynamics of CIV in its new host species, we estimated rates of nucleotide substitution for the HA, NA, M, and NS gene segments and compared those to the rates estimated from publicly available EIV isolates (Table 3). The highest EIV substitution rates were observed in the HA1 and NA gene segments (mean rates of 1.5 × 10−3 substitutions/site/year; 95% HPD = 1.1 × 10−3 to 1.8 × 10−3), while rates estimated for other gene segments were between 0.7 × 10−3 and 0.8 × 10−3 substitutions/site/year (combined 95% HPD = 0.6 × 10−3 to 1.1 × 10−3). These rates are broadly similar to the ∼0.5 × 10−3 substitutions/site/year documented for M and NS in a previous EIV study (29), although a rather different methodology has been employed here. The highest CIV substitution rates were observed in the NA (6.2 × 10−3 substitutions/site/year; 95% HPD = 3.8 × 10−3 to 9.0 × 10−3) and M (4.0 × 10−3 substitutions/site/year; 95% HPD = 2.9 × 10−3 to 5.2 × 10−3) gene segments and hence were higher than those observed in EIV. As expected, the HA1, M, and NS substitution rates obtained using only the consensus sequences were lower than those calculated with all the sequence data included (Table 3), suggesting that the intrahost data sets contain an abundance of deleterious mutational polymorphisms. Indeed, it is important to note that the CIV sequences were sampled over a far shorter time frame than the EIV sequences, which will generally result in an inflation of substitution rates due to the presence of transient deleterious mutations yet to be purged by purifying selection, as has also been suggested for the swine-origin H1N1/09 influenza virus (21, 45).
TABLE 3.
Rates of nucleotide substitutions per site for CIV and EIV sequences
| Gene segment | CIV |
EIV |
||||||
|---|---|---|---|---|---|---|---|---|
| CIV no. | Mean no. of nta substitutions/site/yr (10−3) | 95% HPD |
EIV no. | Mean no. of nta substitutions/site/yr (10−3) | 95% HPD |
|||
| Lower | Upper | Lower | Upper | |||||
| HA1 | 104 | 2.1 | 1.1 | 2.9 | 205 | 1.5 | 1.2 | 1.7 |
| Consensus | 26 | 1.8 | 1.1 | 2.6 | ||||
| M | 149 | 4.0 | 2.9 | 5.2 | 104 | 0.7 | 0.6 | 0.9 |
| Consensus | 26 | 2.7 | 1.9 | 3.7 | ||||
| NS | 156 | 3.6 | 2.8 | 4.6 | 96 | 0.8 | 0.6 | 1.1 |
| Consensus | 24 | 3.3 | 1.2 | 5.1 | ||||
| NA | 36 | 6.2 | 3.8 | 9.0 | 91 | 1.5 | 1.1 | 1.8 |
nt, nucleotide.
Our analysis of selection pressures in CIV using dN/dS revealed no evidence for positive selection in the HA1, HA2, M1, NS1, NA, or NP gene regions, with global dN/dS ratios ranging from 0.27 to 0.66. Similarly, we found no significant difference in the dN/dS ratios on main EIV-to-CIV trunk branches compared to the rest of the phylogeny. Finally, in all segments analyzed, the external branches of the ML trees were characterized by higher dN/dS ratios than the internal branches (differences in dN/dS ratios for external and internal branches ranged from 0.06 to 0.51), strongly suggesting the presence of transient deleterious mutations in this viral population.
Microevolution of CIV in dog shelters.
CIV transmission is sustained by the high numbers of susceptible dogs entering animal shelters and kennels, where the animals are housed in close proximity. We examined the viruses circulating in 2006, 2008, and 2009 in two large shelters (denoted A and B) located in New York City. Phylogenies of multiple cloned CIV sequences from individual dogs in the shelters are shown for three gene regions: HA1, M, and NS (Fig. 2). These trees showed that all of the 2008/2009 viruses in the two shelters derive from a single common ancestor that likely existed between August 2007 and September 2008 (range of 95% HPD values). The 2006 isolates from each of these shelters (A/2006 and B/2006) were interspersed with the earlier northeast virus samples and do not appear to be the direct ancestors of the 2008 and 2009 viruses. The patterns obtained suggest either that the viruses had evolved in situ or had been replaced after 2006 by a different viral lineage, or possibly the viruses in these shelters were seeding other areas. For the viruses collected during November 2008 for shelter B and March 2009 for shelter A, there was a separation of sequences, suggesting that each shelter was maintaining a distinct viral population. Indeed, an analysis of the extent of phylogeographic structure revealed a highly significant geographical clustering by shelter (P < 0.001 for both the PS and AI tests), indicative of relatively little viral gene flow even among shelters that were only ∼8 km apart. However, some limited mixing of the viruses between the two shelters during 2008 and 2009 was observed. For example, two M sequences from a shelter A dog grouped within the shelter B sequences (Fig. 2B). Additionally, HA1 and M sequences from a dog sampled in December 2009 from shelter B were located within the shelter A clade (Fig. 2).
By comparing sequences of up to 20 clones of three gene segments from the same dog sample, we also sought to explore some aspects of dog infection bottleneck size and intrahost evolution. Contrasting with the variation among samples from different dogs, the multiple sequences recovered from each dog were mostly identical in sequence or very closely related, indicative of a single founding virus within each animal. This was supported by our BaTS analysis, which showed significant clustering by host dog (for the PS and AI tests, P < 0.001). However, as the sequences found within each shelter were generally very similar, there may not be sufficient phylogenetic resolution to determine whether dogs were multiply infected.
Sequence and amino acid changes associated with canine infection.
Our analysis of viruses associated with the dog shelters in New York City along with the database CIV and EIV sequences also enabled us to identify, on a genome-wide basis, mutations that are unique or enriched in the CIV sequences compared to those in EIV (Table 4; see Table S1 in the supplemental material). In each gene segment, between one and nine nonsynonymous mutations were identified as exclusively present in CIV isolates. HA has been the particular focus for studies of genetic variation in influenza virus strains, and several changes in HA that have been previously reported as CIV specific were seen in all our CIV sequences (7, 38). Interestingly, two A/H3N8 viruses recently isolated from swine in China differ from early European equine A/H3N8 sequences at six HA sites (50), three of which (N159S, W222L, and I328T) were also found in the CIV sequences, suggesting that these mutations may play an important role in the infection of new host species.
TABLE 4.
Amino acids in the different gene regions showing differences in each gene segment found only in CIV and not in any EIV sequence examined
| Gene segment | Amino acid site (mature) | Amino acid found in: |
|||||
|---|---|---|---|---|---|---|---|
| Human H3N2 New York | Equine isolates |
Canine isolates |
Canine sequences (shelter) | ||||
| Miami 1963 | Florida lineage | 2002-2005 | 2006-2009 | ||||
| HA | 29a | I | I | I | I/M | M | M |
| 54a | S | N | N | K | K | K | |
| 75 (site E) | Q | H | H | H | H | Q | |
| 92a | K | S | S | S/N | N | N | |
| 118a | L | L | L | L/V | V | V | |
| 216 (site D) | N | N | N | N | N/H | H | |
| 222a | R | W | W/L | L | L | L | |
| 261a | R | R | K | K/N | N | N | |
| 262 | S | T | T | T | T/P | P | |
| 328a (cleavage site) | T | I | I | T | T | T | |
| 483a | N | N | N | T | T | T | |
| NA | 62 | n/ab | I | I | I | L | L |
| 250 | n/a | Q | K | K | N | N | |
| NS | 21 | Q | R | R | R | R/Q | Q |
| 193 | R | R | R | R | R | K | |
| 214 | L | F | F | F | F | L | |
| M | 138 | V | V | V | V | V/I | I |
| NP | 27 | A | A | A | A | n/a | T |
| 375a | G | D | D | D/N | n/a | N | |
| PA | 327 | E | E | E | E/K | n/a | K |
| 444 | N | N | N | N | N/D | D | |
| 675a | N | N | N | D | N/D | D | |
| PB1 | 200 | V | V | V | V/I | I | I |
| 338 | S | S | S | S | S/N | N | |
| 529 | V | V | V | V | V/I | I | |
| 591 | V | V | V | V | V/I | I | |
| 687 | Q | Q | Q | Q | Q/H | H | |
| 754 | R | R | R | R | R/K | K | |
| PB2 | 389 | R | R | R | R | n/a | K |
| 559 | A | I | I | I | n/a | N | |
In Table S1 in the supplemental material, we identify all of the nucleotide substitutions that are CIV specific or that showed increased occurrence in CIV compared to EIV (some of which were identified in previous studies [7, 38]). Some changes found in only a small proportion of the EIV sequences appeared to become fixed in the CIV isolates, including eight additional sites in HA, six of which were nonsynonymous. Furthermore, each gene segment also had between 3 and 15 synonymous substitutions that appeared to be fixed in the CIV sequences, with the exception of NS, in which only nonsynonymous CIV-fixed substitutions were seen (see Table S1 in the supplemental material). Several CIV-specific or CIV-dominant sites may influence viral function. HA1 sites 75 and 216 possibly occur at antigenic sites E and D, respectively (52). HA sites 222 (Trp to Leu) and 223 (Val to Ile) are adjacent to HA structures that influence binding to modified sialic acids or those in different (α2,3 or α2,6) linkages (44). The M1 substitution V15I results in a high-pathogenicity H5N1 influenza virus in mice (23). A truncated M1 protein due to a termination codon at site 195 was seen in the consensus sequences B/2006 and NewYork/Dec2006, which were sampled from dogs 4 months apart. The presence of this clearly deleterious mutation in the consensus sequences suggests the transmission, for at least 4 months, of a defective virus possibly through complementation, as has previously been suggested for dengue virus (1). None of the other CIV sequences had the site 220 stop codon mutation seen in 17 equine sequences and the 2003 and 2004 Florida canine NS sequences.
DISCUSSION
The emergence of CIV provides a unique opportunity to examine the evolution of an influenza virus before and after it transferred to a different mammalian host, in which it created a new self-sustaining epidemic of infections and disease. By examining the evolutionary history of CIV in the United States since its emergence from EIV, as well as the smaller-scale “microevolution” of CIV in two confined areas where the virus has been circulating continuously for 3 years or more, we can begin to describe the evolutionary forces that occur during the circulation of this influenza virus in its new canine host compared to that seen in the donor equine host.
High rates of viral transmission in dog shelters.
A feature of influenza viruses and other pathogens spread by respiratory routes (such as severe acute respiratory syndrome [SARS] coronavirus and measles) is that even low-efficiency viruses are able to spread in dense populations with large numbers of incoming susceptible hosts (3, 8, 30). This appears to be the situation for the canine shelter populations that we studied here, which had sufficient seronegative dogs entering into relatively confined areas to allow continuous endemic CIV transmission. Observations of clinical signs and analysis of viral sequences indicated that CIV entered the shelters around 2005. We show here that by late 2008 and early 2009, the virus circulating in both shelters was derived from a single common ancestor, and there appeared to be only limited mixing of viruses between the shelters over a period of 2 to 3 years. Because they likely allow continuing circulation of even inefficiently transmitting viruses, dog shelters are important in facilitating virus persistence, and they also may represent important source populations for CIV evolution, generating viral lineages with a range of phenotypic properties.
Evolution of CIV in dogs over a 9-year period.
Overall, CIV in the United States has experienced rapid evolutionary change, with substitution rates of 1.8 × 10−3 to 6.2 × 10−3 substitutions/site/year in the HA1, NA, M, and NS gene segments (for which we had sufficient samples to undertake an analysis). These rates overlap with those seen in human A/H3N2 (3.8 × 10−3 to 5.7 × 10−3 substitutions/site/year) and A/H1N1 (5 × 10−3 substitutions/site/year) viruses (14, 41, 42), although the short time scale of sampling of CIV means that the rates determined for that virus have likely been artificially inflated due to the presence of transient deleterious mutations. Indeed, despite the emergence of mutations that are found only in the CIV isolates, there was no evidence of strong positive selection acting on these viruses, although this may be due to inherent limitations in detecting adaptation on single nucleotide sites on individual lineages. We have previously shown that CIV can experience measurable sequence evolution even within single dogs, including some tentative evidence for immune selection in the viral HA in partially immune dogs (20). In the present study, we also saw a concentration of changes in the internal gene segments, such as M, NS, and NP, in addition to the genes that encode proteins involved in receptor or antibody recognition (HA and NA). In the case of HA, the relative lack of selection at antigenic sites over the 10 years since its emergence may be due to the specific circumstances of the transmission of the CIV, with continual introduction of naïve animals into the shelters with little or no reintroduction of immune animals, so that no evasion of the host immune response is required.
Evolution of influenza virus in its new host and potential canine adaptation.
One of the most important questions in the study of disease emergence is the degree to which pre- or posttransfer adaptation is required for the successful establishment in the new host species. As the emergence of CIV is well characterized and the canine viruses clearly derive from the transfer of a single EIV ancestor, it is now possible to directly examine the evolution of that virus over 10 years of circulation in dogs.
The host barrier presented by dogs to the recent A/H3N8 EIV viruses may be relatively low, as a number of independent transfers of the virus into dogs have been reported, including the current outbreak in the United States and a less extensive outbreak involving foxhounds in the United Kingdom (9). As such, it is unclear whether significant amounts of adaptative evolution are required or would be expected in EIV when it is transferred to dogs. A number of nucleotide substitutions were shown to uniquely characterize CIV, and the CIV sequences in the northeastern United States showed a more rapid rate of substitution than that reported for EIV over a period of almost 50 years. However, we were unable to conclusively show that any of the substitutions we document in CIV have been subject to positive selection, using the bioinformatics methods currently available. Indeed, any nucleotide changes strongly required for CIV transmission in dogs likely occurred early in the evolutionary history of the virus, prior to the first isolates being collected in 2003, and so would have been fixed in the population before the sampling took place. However, even if some canine adaptation has occurred, the virus appears to be insufficiently transmissible to be able to spread into the general population of household dogs. High-density dog populations, such as those that characterize dog shelters, may therefore be of critical importance for the maintenance of this host-transferred virus even after a period of 10 years. This raises the question of whether a further increase in transmissibility would allow CIV to circulate widely in household dogs in New York City or in other regions, potentially exposing large numbers of people to the virus. The current apparent low transmissibility also suggests that it may be possible to control CIV by using currently available methods such as quarantine, vaccination, or antiviral drugs.
Host range switching in influenza viruses often involves changes of many different viral genome segments, and a number of signature changes have been identified in the genomes of such host-transferred viruses (reviewed in references 5 and 32-34). However, despite the finding of CIV-associated changes in most gene segments (Table 4; see Table S1 in the supplemental material), only a few were in sites that have been identified as influencing host ranges of other influenza viruses. Alteration in sialic acid specificity of the HA has been associated with alterations within, or close to, the sialic acid binding pocket, including residues 222, 226, and 228 (5, 15, 44). Horses and most breeds of domestic dogs differ in the sialic acids expressed, since horses express the sialic acid N-glycolyl neuraminic acid, while dogs primarily express the N-acetyl neuraminic acid. Those sialic acids in the respiratory tissues of horses and dogs may also differ in the linkages of the terminal sialic acids and their acetylation (18, 19, 43, 46, 47, 53, 55). The differences in residues 222 and 223 from primarily Trp and Val to Leu and Ile, respectively, are likely associated with changes in the binding of sialic acids. However, it is clear that the roles of these or other changes in host adaptation of CIV to dogs need to be identified by experimental testing.
Supplementary Material
Acknowledgments
We thank Wendy S. Weichert, Virginia Scarpino, and Changhao (Bobby) Yu for technical support and Elodie Ghedin (University of Pittsburgh), Pablo Murcia, and Karin Hoelzer for assistance with methods.
This work was supported by NIH grant GM080533-03 to E.C.H. and C.R.P.
Footnotes
Published ahead of print on 13 October 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aaskov, J., K. Buzacott, H. M. Thu, K. Lowry, and E. C. Holmes. 2006. Long-term transmission of defective RNA viruses in humans and Aedes mosquitoes. Science 311:236-238. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 2008. Summary of the Australian equine influenza outbreak. Vet. Rec. 163:378. [DOI] [PubMed] [Google Scholar]
- 3.Bauch, C. T., J. O. Lloyd-Smith, M. P. Coffee, and A. P. Galvani. 2005. Dynamically modeling SARS and other newly emerging respiratory illnesses: past, present, and future. Epidemiology 16:791-801. [DOI] [PubMed] [Google Scholar]
- 4.Bryant, N. A., A. S. Rash, C. A. Russell, J. Ross, A. Cooke, S. Bowman, S. MacRae, N. S. Lewis, R. Paillot, R. Zanoni, H. Meier, L. A. Griffiths, J. M. Daly, A. Tiwari, T. M. Chambers, J. R. Newton, and D. M. Elton. 2009. Antigenic and genetic variations in European and North American equine influenza virus strains (H3N8) isolated from 2006 to 2007. Vet. Microbiol. 138:41-52. [DOI] [PubMed] [Google Scholar]
- 5.Chen, G. W., S. C. Chang, C. K. Mok, Y. L. Lo, Y. N. Kung, J. H. Huang, Y. H. Shih, J. Y. Wang, C. Chiang, C. J. Chen, and S. R. Shih. 2006. Genomic signatures of human versus avian influenza A viruses. Emerg. Infect. Dis. 12:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford, P. C., E. J. Dubovi, W. L. Castleman, I. Stephenson, E. P. Gibbs, L. Chen, C. Smith, R. C. Hill, P. Ferro, J. Pompey, R. A. Bright, M. J. Medina, C. M. Johnson, C. W. Olsen, N. J. Cox, A. I. Klimov, J. M. Katz, and R. O. Donis. 2005. Transmission of equine influenza virus to dogs. Science 310:482-485. [DOI] [PubMed] [Google Scholar]
- 8.Cross, P. C., P. L. Johnson, J. O. Lloyd-Smith, and W. M. Getz. 2007. Utility of R0 as a predictor of disease invasion in structured populations. J. R. Soc. Interface 4:315-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daly, J. M., A. S. Blunden, S. Macrae, J. Miller, S. J. Bowman, J. Kolodziejek, N. Nowotny, and K. C. Smith. 2008. Transmission of equine influenza virus to English foxhounds. Emerg. Infect. Dis. 14:461-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daly, J. M., A. C. Lai, M. M. Binns, T. M. Chambers, M. Barrandeguy, and J. A. Mumford. 1996. Antigenic and genetic evolution of equine H3N8 influenza A viruses. J. Gen. Virol. 77:661-671. [DOI] [PubMed] [Google Scholar]
- 11.Drummond, A. J., and A. Rambaut. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drummond, A. J., A. Rambaut, B. Shapiro, and O. G. Pybus. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22:1185-1192. [DOI] [PubMed] [Google Scholar]
- 13.Dubovi, E. J. 2010. Canine influenza. Vet. Clin. Small Anim. 40:1063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitch, W. M., R. M. Bush, C. A. Bender, and N. J. Cox. 1997. Long term trends in the evolution of H(3) HA1 human influenza type A. Proc. Natl. Acad. Sci. U. S. A. 94:7712-7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamblin, S. J., L. F. Haire, R. J. Russell, D. J. Stevens, B. Xiao, Y. Ha, N. Vasisht, D. A. Steinhauer, R. S. Daniels, A. Elliot, D. C. Wiley, and J. J. Skehel. 2004. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 303:1838-1842. [DOI] [PubMed] [Google Scholar]
- 16.Gatherer, D. 2009. The 2009 H1N1 influenza outbreak in its historical context. J. Clin. Virol. 45:174-178. [DOI] [PubMed] [Google Scholar]
- 17.Gibbs, E. P., and T. C. Anderson. 2010. Equine and canine influenza: a review of current events. Anim. Health Res. Rev. 11:43-51. [DOI] [PubMed] [Google Scholar]
- 18.Hanaoka, K., T. J. Pritchett, S. Takasaki, N. Kochibe, S. Sabesan, J. C. Paulson, and A. Kobata. 1989. 4-O-Acetyl-N-acetylneuraminic acid in the N-linked carbohydrate structures of equine and guinea pig alpha 2-macroglobulins, potent inhibitors of influenza virus infection. J. Biol. Chem. 264:9842-9849. [PubMed] [Google Scholar]
- 19.Hashimoto, Y., T. Yamakawa, and Y. Tanabe. 1984. Further studies on the red cell glycolipids of various breeds of dogs. A possible assumption about the origin of Japanese dogs. J. Biochem. 96:1777-1782. [DOI] [PubMed] [Google Scholar]
- 20.Hoelzer, K., P. R. Murcia, G. J. Baillie, J. L. Wood, S. M. Metzger, N. Osterrieder, E. J. Dubovi, E. C. Holmes, and C. R. Parrish. 2010. Intrahost evolutionary dynamics of canine influenza virus in naive and partially immune dogs. J. Virol. 84:5329-5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes, E. C. 2010. Evolution in health and medicine Sackler Colloquium: the comparative genomics of viral emergence. Proc. Natl. Acad. Sci. U. S. A. 107(Suppl. 1):1742-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horner, G. W., and A. M. Ledgard. 1988. A serological survey for equine influenza in New Zealand horses. N. Z. Vet. J. 36:205-206. [DOI] [PubMed] [Google Scholar]
- 23.Katz, J. M., X. Lu, T. M. Tumpey, C. B. Smith, M. W. Shaw, and K. Subbarao. 2000. Molecular correlates of influenza A H5N1 virus pathogenesis in mice. J. Virol. 74:10807-10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirkland, P. D., D. S. Finlaison, E. Crispe, and A. C. Hurt. 2010. Influenza virus transmission from horses to dogs, Australia. Emerg. Infect. Dis. 16:699-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobasa, D., and Y. Kawaoka. 2005. Emerging influenza viruses: past and present. Curr. Mol. Med. 5:791-803. [DOI] [PubMed] [Google Scholar]
- 26.Lai, A. C., K. M. Rogers, A. Glaser, L. Tudor, and T. Chambers. 2004. Alternate circulation of recent equine-2 influenza viruses (H3N8) from two distinct lineages in the United States. Virus Res. 100:159-164. [DOI] [PubMed] [Google Scholar]
- 27.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 28.Lee, C., D. Song, B. Kang, D. Kang, J. Yoo, K. Jung, G. Na, K. Lee, B. Park, and J. Oh. 2009. A serological survey of avian origin canine H3N2 influenza virus in dogs in Korea. Vet. Microbiol. 137:359-362. [DOI] [PubMed] [Google Scholar]
- 29.Lindstrom, S. E., Y. Hiromoto, R. Nerome, K. Omoe, S. Sugita, Y. Yamazaki, T. Takahashi, and K. Nerome. 1998. Phylogenetic analysis of the entire genome of influenza A (H3N2) viruses from Japan: evidence for genetic reassortment of the six internal genes. J. Virol. 72:8021-8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lloyd-Smith, J. O., P. C. Cross, C. J. Briggs, M. Daugherty, W. M. Getz, J. Latto, M. S. Sanchez, A. B. Smith, and A. Swei. 2005. Should we expect population thresholds for wildlife disease? Trends Ecol. Evol. 20:511-519. [DOI] [PubMed] [Google Scholar]
- 31.Martella, V., G. Elia, N. Decaro, L. Di Trani, E. Lorusso, M. Campolo, C. Desario, A. Parisi, N. Cavaliere, and C. Buonavoglia. 2007. An outbreak of equine influenza virus in vaccinated horses in Italy is due to an H3N8 strain closely related to recent North American representatives of the Florida sub-lineage. Vet. Microbiol. 121:56-63. [DOI] [PubMed] [Google Scholar]
- 32.Matrosovich, M., J. Stech, and H. D. Klenk. 2009. Influenza receptors, polymerase and host range. Rev. Sci. Tech. 28:203-217. [DOI] [PubMed] [Google Scholar]
- 33.Naffakh, N., A. Tomoiu, M. A. Rameix-Welti, and S. van der Werf. 2008. Host restriction of avian influenza viruses at the level of the ribonucleoproteins. Annu. Rev. Microbiol. 62:403-424. [DOI] [PubMed] [Google Scholar]
- 34.Neumann, G., and Y. Kawaoka. 2006. Host range restriction and pathogenicity in the context of influenza pandemic. Emerg. Infect. Dis. 12:881-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neumann, G., T. Noda, and Y. Kawaoka. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459:931-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parker, J., A. Rambaut, and O. G. Pybus. 2008. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect. Genet. Evol. 8:239-246. [DOI] [PubMed] [Google Scholar]
- 37.Parrish, C. R., and Y. Kawaoka. 2005. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu. Rev. Microbiol. 59:553-586. [DOI] [PubMed] [Google Scholar]
- 38.Payungporn, S., P. C. Crawford, T. S. Kouo, L. M. Chen, J. Pompey, W. L. Castleman, E. J. Dubovi, J. M. Katz, and R. O. Donis. 2008. Influenza A virus (H3N8) in dogs with respiratory disease, Florida. Emerg. Infect. Dis. 14:902-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pond, S. L., S. D. Frost, and S. V. Muse. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676-679. [DOI] [PubMed] [Google Scholar]
- 40.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 41.Rambaut, A., and E. Holmes. 2009. The early molecular epidemiology of the swine-origin A/H1N1 human influenza pandemic. PLoS Curr. 1:RRN1003. doi;10.1371/currents.RRN1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rambaut, A., O. G. Pybus, M. I. Nelson, C. Viboud, J. K. Taubenberger, and E. C. Holmes. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453:615-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scocco, P., and V. Pedini. 2008. Localization of influenza virus sialoreceptors in equine respiratory tract. Histol. Histopathol. 23:973-978. [DOI] [PubMed] [Google Scholar]
- 44.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 45.Smith, G. J., D. Vijaykrishna, J. Bahl, S. J. Lycett, M. Worobey, O. G. Pybus, S. K. Ma, C. L. Cheung, J. Raghwani, S. Bhatt, J. S. Peiris, Y. Guan, and A. Rambaut. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122-1125. [DOI] [PubMed] [Google Scholar]
- 46.Song, D., B. Kang, C. Lee, K. Jung, G. Ha, D. Kang, S. Park, B. Park, and J. Oh. 2008. Transmission of avian influenza virus (H3N2) to dogs. Emerg. Infect. Dis. 14:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki, Y., T. Ito, T. Suzuki, R. E. Holland, Jr., T. M. Chambers, M. Kiso, H. Ishida, and Y. Kawaoka. 2000. Sialic acid species as a determinant of the host range of influenza A viruses. J. Virol. 74:11825-11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swofford, D. L. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. Sinauer Associates. Sunderland, MA.
- 49.Timoney, P. J. 1996. Equine influenza. Comp. Immunol. Microbiol. Infect. Dis. 19:205-211. [DOI] [PubMed] [Google Scholar]
- 50.Tu, J., H. Zhou, T. Jiang, C. Li, A. Zhang, X. Guo, W. Zou, H. Chen, and M. Jin. 2009. Isolation and molecular characterization of equine H3N8 influenza viruses from pigs in China. Arch. Virol. 154:887-890. [DOI] [PubMed] [Google Scholar]
- 51.Waddell, G. H., M. B. Teigland, and M. M. Sigel. 1963. A new influenza virus associated with equine respiratory disease. J. Am. Vet. Med. Assoc. 143:587-590. [PubMed] [Google Scholar]
- 52.Wiley, D. C., I. A. Wilson, and J. J. Skehel. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289:373-376. [DOI] [PubMed] [Google Scholar]
- 53.Yachida, Y., K. Tsuchihashi, and S. Gasa. 1996. Characterization of novel mono-O-acetylated GM3s containing 9-O-acetyl sialic acid and 6-O-acetyl galactose in equine erythrocytes. Glycoconj. J. 13:225-233. [DOI] [PubMed] [Google Scholar]
- 54.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555-556. [DOI] [PubMed] [Google Scholar]
- 55.Yasue, S., S. Handa, S. Miyagawa, J. Inoue, A. Hasegawa, and T. Yamakawa. 1978. Difference in form of sialic acid in red blood cell glycolipids of different breeds of dogs. J. Biochem. 83:1101-1107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.