Abstract
Many viruses, including retroviruses, undergo frequent recombination, a process which can increase their rate of adaptive evolution. In the case of HIV, recombination has been responsible for the generation of numerous intersubtype recombinant variants with epidemiological importance in the AIDS pandemic. Although it is known that fragments of genetic material do not combine randomly during the generation of recombinant viruses, the mechanisms that lead to preferential recombination at specific sites are not fully understood. Here we reanalyze recent independent data defining (i) the structure of a complete HIV-1 RNA genome and (ii) favorable sites for recombination. We show that in the absence of selection acting on recombinant genomes, regions harboring RNA structures in the NL4-3 model strain are strongly predictive of recombination breakpoints in the HIV-1 env genes of primary isolates. In addition, we found that breakpoints within recombinant HIV-1 genomes sampled from human populations, which have been acted upon extensively by natural selection, also colocalize with RNA structures. Critically, junctions between genes are enriched in structured RNA elements and are also preferred sites for generating functional recombinant forms. These data suggest that RNA structure-mediated recombination allows the virus to exchange intact genes rather than arbitrary subgene fragments, which is likely to increase the overall viability and replication success of the recombinant HIV progeny.
Recombination is a vital source of genetic diversity for many RNA viruses (15, 18, 37, 38, 48, 49). By combining polymorphisms present in distinct genomes into a new genome in a single round of replication, recombination enables viruses to more rapidly access greater sequence space than is possible by the stepwise accumulation of point mutations. The net effect is to facilitate both the combination of advantageous mutations within individual highly fit genomes and the removal of deleterious mutations from viral populations. In the case of human immunodeficiency virus (HIV), these processes contribute to the dynamic evasion of immune responses and to the evolution of drug resistance (20, 27, 34, 45).
In addition to encoding information necessary for protein production, the genomes of RNA viruses, including HIV, convey functional information through their secondary and tertiary structures. These structures regulate many stages of the viral replication cycle, including genome replication, genome packaging into new viral particles, and intracellular trafficking (5, 6, 30, 36, 42). Studies on recombination in RNA viruses in general and in retroviruses in particular have indicated that RNA secondary structures play a potentially important role in genetic recombination (8, 9, 12, 13, 16, 21, 23, 24, 41).
In retroviruses, recombination results primarily from template switching during reverse transcription between the two RNA genomes that are present in the same viral particle (22, 50). If these two copies are genetically different, as in a heterozygous virus, template switching results in genetic recombination. RNA structures influence recombination in at least two ways. First, RNA structures at the 5′ end of the genomic RNA form base-pairing and other interactions that allow genomes to dimerize. These RNA structures thus indirectly affect recombination by regulating the efficiency with which heterodimers of genomic RNA form and are packaged into viral particles (4, 10, 11, 35, 39, 44, 52). Second, RNA structures appear to directly promote template switching via two different proposed mechanisms. Studies performed with reconstituted in vitro systems have shown that secondary structures can induce stalling of reverse transcriptase at the base of an RNA hairpin and thereby increase the probability of template switching in the ascending strand of this structure (40). In parallel, studies with both tissue culture and reconstituted in vitro systems have indicated that RNA structure-mediated template switching may occur, via a branch migration process, in the descending strand of an RNA hairpin (13, 33).
To date, experimental work aimed at dissecting the mechanism of recombination has primarily employed simplified models involving template switching between closely related model sequences. Recombination between closely related strains, including viruses belonging to the same quasispecies present in an infected individual, is certainly frequent and important for the generation and emergence of antiviral resistance and immune escape variants (17, 27, 34). However, a critical feature of the recombination process as an evolutionary force is that of allowing reshuffling of genetic information carried by distantly related viral strains. For recombination between more-divergent viruses, the degree of local sequence similarity becomes an additional crucial factor influencing the frequency and patterns of recombination (1, 31, 51).
Together, these factors regulate the production of a recombinant population on which selection acts, favoring certain progeny and purging others. A pair of recent studies focusing on intersubtype HIV-1 group M recombination emphasized that breakpoints falling within certain regions of env or pol yield chimeric proteins that have a higher probability of being dysfunctional than when breakpoints occur in other parts of these genes (14, 43). In the case of env, two factors—the mechanistic tendency for recombination to occur more frequently at certain genome sites and selection against dysfunctional recombinants—were shown to account almost entirely for recombination patterns observed in naturally sampled sequences (43).
The recent determination of the secondary structure of a full HIV-1 genome, corresponding to the NL4-3 isolate (47), provides the opportunity to test, with hitherto unachievable precision, the influence of RNA secondary structure on HIV recombination breakpoint distributions. Here we examine both experimentally determined recombination frequencies in env and the recombination breakpoint distribution detectable within natural HIV recombinants (43) and attempt to disentangle the relative contributions of RNA secondary structure, nucleotide sequence conservation, and natural selection to observed recombination patterns.
MATERIALS AND METHODS
All sequences used in this work, including that of NL4-3, were aligned to the HXB2 reference sequence (GenBank accession no. K03455), as outlined by Korber et al. (25). Position numbers in the text and figures correspond to HXB2 numbering.
Experimental data set.
Recombination frequencies across overlapping fragments of the entire env gene for a range of HIV-1 primary isolates (6 isolates of HIV-1 group M, subtype A, 1 isolate of subtype B, 1 isolate of subtype C, 2 isolates of subtype D, and 3 isolates of subtype G) have been described previously (43).
Sequences sampled from nature.
A previously described HIV-1 group M envelope sequence alignment (27) was analyzed. Briefly, this alignment included 30 “pure” HIV-1 subtype sequences (3 for each subtype), 106 circulating recombinant form (CRF) sequences (2 for each CRF), and 197 apparently unique recombinant sequences retrieved from the Los Alamos National Laboratory (LANL) HIV Sequence Database (http://hiv-web.lanl.gov/).
Extent of conserved RNA secondary structure in env in primary HIV-1 isolates.
The degree of RNA secondary structure across the env gene, expressed as the percentage of paired nucleotides over a 50-nucleotide (nt) sliding window, was first determined using the NL4-3 secondary structure model (47). We then estimated whether these RNA structures were present in the individual primary isolates used in the recombination experiments (43). The sequence of each primary isolate was aligned to that of NL4-3. For each paired nucleotide in the NL4-3 structure model, we determined whether the corresponding nucleotide in each primary isolate could pair with the nucleotide at the complementary base-paired position in the NL4-3 structure. The rationale behind this calculation was to include biologically relevant base pairings that have apparently been maintained through coevolution. Nucleotides within the primary isolate env sequences that could pair (A-U, G-C, or G-U) were assigned a score of 1; those that could not form a canonical pair were given a score of 0. The extent of RNA secondary structure was calculated for each position in the NL4-3 model as the mean pairing score for the primary isolates, averaged over a 50-nt sliding window along env.
Median SHAPE reactivity and recombinant breakpoint distributions.
Median SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) reactivity values were calculated over a sliding 75-nt window, based on the single-nucleotide resolution analysis of a complete HIV-1 NL4-3 genome (47). The recombination breakpoint distribution along the whole genome analyzed here is essentially identical to that published previously (43), except that a 75-nt instead of 200-nt sliding window was used to calculate breakpoint clustering probabilities.
Distribution of experimental and simulated recombinant breakpoints relative to RNA hairpin motifs in env.
We defined an RNA hairpin as a structure containing a stem of three or more paired nucleotides with no additional embedded hairpins. By this definition, there are 37 hairpin elements in the env coding region. Simulated recombination breakpoints were generated as described previously (3). Briefly, 15 sets of breakpoints (one for each of the 15 pairs studied in reference 43, for a total of 371 breakpoints) were generated 1,000 times. For both experimental and simulated data sets, the central nucleotide of each breakpoint window was numbered according to the position of its homologue within the HXB2 reference genome. The position of this central nucleotide relative to RNA hairpin motifs present in the current secondary structure model for the NL4-3 genome (see Fig. 2A) was computed. If this central nucleotide fell outside a hairpin, the distance to the closest hairpin was calculated.
Permutation test of associations between recombination breakpoint clustering and RNA secondary structure (permutation test A).
A permutation test of recombination breakpoint clustering based on that described previously (29) was used to determine associations between RNA secondary structure and breakpoint clustering. This test is a modification of one described previously (19) and accounts for uncertainties in recombination breakpoint site identification due to the underlying degree of sequence conservation (recombination is easier to detect in more divergent regions). The summed total of all median SHAPE reactivity estimates for all mapped recombination breakpoint sites (371 in the experimental env analysis and 691 in the full-genome analysis) was compared with those determined for 10,000 simulated data sets. The proportion of simulated data sets with summed SHAPE reactivity scores that were lower than or equal to those of the real data sets was taken as the probability that there was not a significant tendency for recombination breakpoints to fall at sites with low SHAPE reactivity scores (corresponding to sites that had a high probability of having base-paired secondary structures).
Permutation test of associations between recombination breakpoint clustering, genome location, and RNA secondary structure (permutation test B).
The same permutation test as that described previously (29) was used to determine whether recombination breakpoints were significantly more or less clustered within specified pairs of genome regions. In all cases, observed breakpoint distributions were compared with breakpoint distributions determined for 10,000 simulated data sets, each displaying precisely the same number and character of recombination signals (spacing between breakpoint positions, degrees of parental sequence relatedness, and numbers of sequences carrying evidence of recombination), but with randomized breakpoint positions. In comparing, for example, breakpoint densities within region A (for example, low-SHAPE gene border sites) with those in region B (for example, high-SHAPE gene border sites), the breakpoint numbers observed within regions A and B in the real data set were randomly distributed between the regions in each of the 10,000 simulated data sets. Simulated data sets in which the number of breakpoints in region A was equal to or greater than the number observed in region A of the real data set were counted. This count was then divided by 10,000 to yield the probability that breakpoints were not significantly more clustered in region A than in region B. The inverse test (whether breakpoints were not significantly more clustered in region B than in region A) was also performed.
Definition of local sequence identity in env.
To define regions of high and low similarity within env, pairwise alignments between two given isolates were made and the level of sequence identity was computed over a 30-nt sliding window. Based on previous observations (3), the presence of more than 15% nonidentical residues between the two parental sequences in the 30-nt region 3′ of the potential breakpoint strongly decreases the probability of recombination. Regions presenting ≤4 nonidentical residues in a pairwise alignment in the 30-nt window were therefore taken to be highly similar; conversely, sequences with ≥5 discordant residues were considered not highly similar.
RESULTS
Strong correlation between recombination and RNA structure in HIV-1.
Mechanisms of recombination vary among RNA viruses. For retroviruses, including HIV, recombination occurs primarily during reverse transcription, when the viral reverse transcriptase switches, midreplication, from one to the other of the two genomic RNA copies that are copackaged within individual viral particles (21). This mechanism of recombination, termed copy choice, is strongly influenced by two factors. Recombination is enhanced by local sequence similarity between the two genomic RNAs (3, 12, 31, 51), and specific stem-loop structures efficiently promote template switching, generating defined recombination hot spots (12, 13).
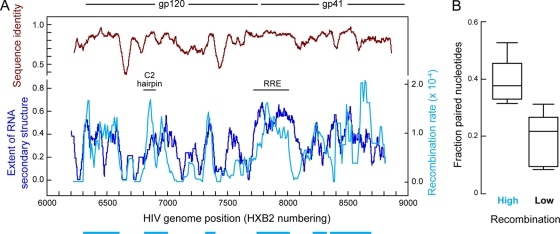
We recently investigated recombination frequencies within the envelope genes (env) of primary HIV-1 isolates by using a tissue culture system that generates recombinant forms in the absence of any selective pressure (43). This work showed that recombination rates varied significantly across env (Fig. 1A, light blue line), with recombination hot spots corresponding to the most conserved parts of the gene and cold spots generally corresponding to the least conserved regions. However, sequence similarity is clearly not the only determinant of the recombination breakpoint distribution, because recombination rates varied widely even within gene regions with high degrees of sequence identity (Fig. 1A, compare brown and light blue traces).
FIG. 1.
Recombination rates, sequence similarity, and RNA secondary structure in the env gene of HIV-1. (A) Comparison of recombination rates (light blue) and degrees of sequence identity (brown) in the analyzed HIV-1 group M isolates (43) with the distribution of conserved RNA structures (dark blue). The distribution of conserved RNA secondary structures was calculated based on the SHAPE-constrained structure determined for the NL4-3 HIV-1 genome (47). Recombination hot spots, defined as regions containing at least one significant hot spot (P < 0.01) for breakpoint clustering and bounded by statistically significant (P < 0.05) cold spots for breakpoint clustering (43), are indicated by light blue bars. (B) Comparison of the predicted extents of RNA secondary structure within and outside recombination hot spots. The large box encompasses the central one-half of the data, the central horizontal line gives the mean, and the upper and lower whiskers indicate standard deviations. The regions with the highest recombination rates have significantly higher degrees of RNA structure (P = 0.007) than the remainder of the gene. For this analysis, regions 5 and 6 were taken as a single region.
Two recombination-prone regions in env correspond to previously described RNA structures: the Rev-responsive element (RRE) (32) and an RNA hairpin located in the C2 portion of the gp120 coding region (33) (Fig. 1A, bars). This correlation raised the possibility that recombination efficiencies in env might be modulated by previously undetected RNA secondary structures. A recently developed RNA secondary structure model for an entire HIV-1 genome (47) now makes it possible to test this hypothesis.
Based on the secondary structure model for the HIV-1 NL4-3 genome, we calculated the probability that secondary structures identified in NL4-3 (47) are also present in the RNA genomes of the isolates used in the selection-free recombination assays (43). Since recombination was measured using multiple HIV-1 strains from different group M subtypes, we normalized the extent of conserved base pairing (and covariant residues) to account for the effects of sequence variation with respect to NL4-3. To estimate the probability that a structure identified in NL4-3 is also present in primary isolates, we calculated the number of NL4-3 pairings maintained in the primary isolates over a sliding 50-nt window across env.
Strikingly, the local extent of conserved RNA secondary structure was highly correlated with the sites of recombination across env (Fig. 1A, compare light and dark blue profiles). The correlation seen by visual comparison was readily shown to be statistically significant (P < 0.0001; permutation test A [see Materials and Methods]).
Recombination breakpoints within env were previously shown to be clustered at six hot spots (43) (light blue bars at bottom of Fig. 1A). Therefore, in addition to the whole-gene analysis, we evaluated the relationship between recombination breakpoint frequency and RNA secondary structure by comparing the extents of conserved RNA secondary structure within and outside these hot spots. The recombination hot spots had a significantly higher degree of predicted base pairing than the remainder of the gene (P = 0.007; unpaired t test) (Fig. 1B).
In sum, both the whole-gene and recombination hot spot analyses indicated that recombination has a strong tendency to occur more frequently in genomic regions rich in experimentally detected RNA structures.
Topological mapping of recombination breakpoints on HIV-1 env RNA structures.
The close association between recombination and the local level of RNA structure, inferred statistically, was further investigated by mapping recombination rates onto the experimentally constrained HIV-1 env secondary structure for the NL4-3 genome (47). Regions with the highest recombination rates (Fig. 2A, red areas) were highly overrepresented in regions of the RNA that form strong secondary structures. Conversely, recombination was the least frequent in large loops and in regions connecting hairpin structures (Fig. 2A, black areas). The secondary structure model for env contains many hairpins, including structures in which a hairpin is nested within the structure of a larger hairpin element. This organization complicates the statistical analysis of whether local structures influence recombination. We therefore focused on a conservative hairpin definition (see Materials and Methods). We then evaluated whether this set of hairpins (37 total hairpins, covering 1,149 of 2,895 nt, or 40% of env) contained more recombination breakpoints than would be expected by chance.
FIG. 2.
Locations of recombination breakpoints on HIV-1 env RNA structures. (A) Recombination rates per nucleotide (43) plotted on the structure of the HIV-1 NL4-3 env gene (47). Nucleotides are numbered relative to the HXB2 proviral DNA sequence. Two previously characterized RNA structures, the RRE and the previously identified hairpin in C2, are labeled. (B) Distribution of 371 experimental recombination breakpoints obtained in the absence of selection relative to hairpin RNA structural elements in env (black bars) versus randomly generated breakpoints (white bars) (error bars indicate standard deviations). Breakpoints falling outside hairpin structures are shown both binned by distance from the nearest hairpin and summed.
The bias for recombination to occur experimentally more frequently inside hairpins was statistically highly significant (Wilcoxon test; P < 0.0001). The converse observation was also clear, such that breakpoints fell less frequently than expected by chance in regions outside hairpin structures (Fig. 2B).
Relationship between recombination, protein domains, and HIV-1 gene structure.
It was recently shown, by replication experiments performed in cell culture, that recombination in env and in pol does not yield uniformly functional variants. Instead, many recombination events yield dysfunctional proteins that are presumably strongly selected against under natural infection conditions. In addition, the probability that a recombinant is functional varies significantly with the position at which recombination occurs within a gene (14, 43). The clustering of breakpoints, as observed in natural group M HIV-1 recombinants, appears to be linked to the structure of the encoded protein. For example, breakpoints tend to cluster at discrete sites within the part of env that encodes gp120, a protein which has a complex tertiary structure organization. In the region encoding the gp41 protein, which does not have the same complex organization, breakpoints are instead distributed more homogeneously (26).
The analyses described above emphasize that highly structured RNA elements promote recombination and that highly structured regions within the RNA genome tend to occur at the junctions between domains in HIV-1 proteins. In addition, the RNA structure at protein domain junctions is expected to facilitate recombination in a way that is less likely to create dysfunctional protein chimeras. These three ideas—the correlation between RNA structure and recombination, the enrichment of RNA structure at protein domain junctions, and the higher probability of generating functional products when recombination occurs at protein domain junctions—suggest that there might be large-scale relationships between RNA structure and recombination in HIV-1.
To determine how gene arrangement and the distribution of RNA secondary structure elements might influence natural recombination patterns, we reanalyzed the locations of recombination breakpoints within an alignment of near-full-length genome sequences from 274 HIV-1 group M isolates (described previously in reference 43) (Fig. 3A). Specifically, we used a permutation test (permutation test B [see Materials and Methods]) to evaluate recombination breakpoint densities at four distinct sets of nucleotide sites: (i) those within 30 nucleotides of gene junctions (called “gene border” sites) (Fig. 3A); (ii) those more than 30 nucleotides distant from gene junctions (called “internal gene” sites); (iii) those with median SHAPE reactivity of <0.25, which correspond to the most highly structured genome regions (called “low-SHAPE” sites [blue shading in Fig. 3B]); and (iv) genome sites with SHAPE reactivity of >0.25, which are therefore in relatively unstructured regions of the genome (called “high-SHAPE” sites).
FIG. 3.
Association between RNA structures predicted across the HIV-1 genome and recombination breakpoint distributions detectable in 247 near-full-length sequences sampled from nature (691 breakpoints). (A) HIV-1 gene organization, border regions between individual peptide coding regions (horizontal black bars), and positions of breakpoints inferred from circulating natural sequences. (B) Breakpoint density map inferred from an alignment of natural circulating HIV-1 sequences (red line) (43) compared to the distribution of RNA structures inferred from the median SHAPE reactivity profile (blue line) (47). SHAPE data (blue) are plotted on an inverted scale such that peaks indicate high levels of RNA structure. The height of the breakpoint density map (red) indicates the probability that recombination breakpoint distributions are not more clustered than would be expected by chance within a 75-nucleotide window centered on a given position. SHAPE reactivity values for sites where recombination breakpoints cluster are significantly lower than can be accounted for by chance (P = 0.0008).
In order to test whether low-SHAPE sites (corresponding to regions with high degrees of RNA structure) had higher breakpoint densities than high-SHAPE sites (regions with lower degrees of RNA structure), we first considered gene border and internal gene sites separately. For both the gene border and internal gene sites, breakpoints were significantly more clustered at low-SHAPE sites than at high-SHAPE sites (P = 0.0004 for gene border sites and P = 0.0261 for internal gene sites; permutation test B). This indicates that RNA structure has a large and readily detectable influence on recombination breakpoint positions, independent of where these structures occur in relation to gene boundaries (Fig. 3).
We next tested the influence of gene junctions on recombination breakpoint densities by comparing gene border and internal gene sites either within low-SHAPE sites or within high-SHAPE sites (again equivalent to high versus low levels of RNA structure, respectively). Gene border sites had significantly higher breakpoint densities than internal gene sites, irrespective of whether low-SHAPE or high-SHAPE sites were considered (P = 0.0002 for both low-SHAPE and high-SHAPE sites; permutation test B). Thus, in HIV-1, recombination breakpoints identified in intersubtype recombinants found in nature cluster at gene boundaries.
To determine if one of these two factors—RNA structure or gene position—constitutes the predominant determinant of the HIV-1 breakpoint patterns, we compared high-SHAPE regions located at gene border sites (for which a high density of breakpoints is expected due to their location, but not due to RNA structure) to low-SHAPE regions found in internal gene sites (for which RNA structure is predictive of a large number of breakpoints, but the location in the genome is not). Although we observed a slightly higher density of breakpoints at high-SHAPE/gene border sites than at low-SHAPE/internal gene sites, the difference was not significant (P = 0.382; permutation test B).
In sum, these results indicate that both high levels of RNA secondary structure and the locations of gene boundaries contribute to the distribution of natural recombination breakpoints in HIV-1 and that neither of these factors prevails significantly over the other.
Recombination, local sequence identity, and RNA structures.
Recombination between two retroviral genomes tends to be enhanced in regions of high sequence similarity (1-3, 31, 51). Similarly, base-paired sites within the RNA secondary structure model of the NL4-3 genome occur more frequently in regions with high sequence similarity (47). The latter correlation likely reflects, in part, conservation driven by both protein coding and RNA structure formation constraints (47). We therefore tested whether breakpoint clustering within highly structured genome regions was attributable simply to sequence similarity rather than to the direct influence of RNA structure.
Paradoxically, in the full genome sequences analyzed in this work, the most highly structured genome regions (defined as those with a median SHAPE reactivity of <0.25 over a 75-nt window) had an average similarity of 0.849 (standard deviation [SD] = 0.176), which is essentially identical to that for less-structured genome regions (with a median SHAPE reactivity of >0.25), with an average similarity of 0.889 (SD = 0.152). The higher density of breakpoints found in the highly structured regions of HIV-1 genomes is thus not attributable simply to these genome regions having a higher degree of sequence similarity than less-structured regions.
The env gene appears to be a special case in HIV-1, because the association between sequence similarity and secondary structure is different from that seen in the remainder of the genome. Highly structured RNA regions within env have an average similarity of 0.923, whereas sequence similarity in unstructured regions is only 0.859. Therefore, focusing on env, we sought to evaluate the impact of RNA structure on recombination breakpoint distributions by comparing regions with similar degrees of sequence identity but different degrees of structure. We divided env into highly similar sequences, defined as regions with sequence identities of ≥0.85; the remaining regions were defined as not highly similar. This similarity cutoff reflects prior work indicating that a sequence divergence of >0.15 is associated with a significant reduction in recombination probability (3). There was a significantly higher frequency of breakpoints within the highly structured regions than in the less-structured regions (0.3196 versus 0.1502 per nt; P < 0.0001).
Thus, RNA structure makes a readily detected contribution to the recombination breakpoint pattern, even within env, where highly structured regions do colocalize with regions of high sequence similarity.
DISCUSSION
The RNA genomes of HIV and other RNA viruses contain secondary and tertiary structures that convey critical functional information. The locations and stabilities of these RNA structures within HIV genomes evolve in response to multiple selective pressures. For example, highly stable RNA secondary structures are found in the linker segments that separate individual proteins and that define domains within the HIV-1 Gag, Gag-Pol, and Env proteins. These structures appear to modulate ribosome processivity, consistent with a role in facilitating stepwise folding of individual protein domains (47). Here we identified a second, independent role of genome-wide RNA secondary structure: the promotion of genetic recombination at specific locations within the genome. There are two broad implications of our analysis.
First, RNA structure is strongly predictive of recombination breakpoint sites. In the absence of selection, there is a clear predictive association between SHAPE reactivity data and the frequency of recombination in the env gene (P < 0.0001; permutation test A). Local analysis of the breakpoint distribution further strengthens this association, since breakpoints are preferentially located within hairpins predicted in the experimentally defined NL4-3 HIV-1 secondary structure (Fig. 2). These results clearly indicate a direct involvement of RNA structures in promoting recombination and are consistent with the predominant mechanisms proposed for the copy choice process, which emphasize a crucial role for double-stranded structures in the RNA (33, 40). Remarkably, for natural recombinants, which additionally experience natural selection, there remains a strong association between SHAPE-detected RNA structures and recombination breakpoint distributions (P = 0.0008; permutation test A [see Materials and Methods]). However, in natural recombinants, the distribution is additionally influenced independently by the positions of gene junctions.
Second, RNA structures near gene borders promote recombination locally, with the net effect of minimizing the apparent chance that recombination will generate genes that express dysfunctional proteins. The probability of recombination-induced protein misfolding or dysfunctionality decreases as recombination breakpoints move from the center of genes toward their borders (7, 28, 46). Genome sites within 30 nucleotides of gene borders have significantly higher breakpoint densities than internal gene sites, even after controlling for the extent of RNA folding at these sites. Since these gene border sites have a greater tendency to be base paired than regions in the rest of the HIV-1 genome (P = 0.057; one-tailed Fisher's exact test), we infer that RNA structures also function to guide recombination toward gene junctions.
Overall, this analysis supports the basic hypothesis that the global pattern of HIV-1 genomic RNA structure plays a central role in multiple facets of the biology of this virus. This work reveals that structured RNA sequences that encode interprotein linkages within viral polyproteins are favored recombination breakpoint sites. Our data imply that recombination between viruses belonging to different HIV-1 subtypes would then tend to shuffle entire genes or subgene fragments that encode autonomously folding protein domains. By promoting the modular assembly of genomes through recombination, the risks of fitness losses associated with genetic exchanges between divergent genomes would be reduced. RNA structures within the genome therefore appear to directly enhance the adaptive value of recombination during HIV-1 evolution.
Acknowledgments
This work was supported by grants from the Wellcome Trust (to D.P.M.), the U.S. National Institutes of Health (AI068462 to K.M.W.), the French National Agency for AIDS Research (2007/290), Sidaction (51005-02-00/AO16-2), and the CNRS (to M.N.).
Footnotes
Published ahead of print on 29 September 2010.
REFERENCES
- 1.An, W., and A. Telesnitsky. 2002. Effects of varying sequence similarity on the frequency of repeat deletion during reverse transcription of a human immunodeficiency virus type 1 vector. J. Virol. 76:7897-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, W., and A. Telesnitsky. 2001. Frequency of direct repeat deletion in a human immunodeficiency virus type 1 vector during reverse transcription in human cells. Virology 286:475-482. [DOI] [PubMed] [Google Scholar]
- 3.Baird, H. A., R. Galetto, Y. Gao, E. Simon-Loriere, M. Abreha, J. Archer, J. Fan, D. L. Robertson, E. J. Arts, and M. Negroni. 2006. Sequence determinants of breakpoint location during HIV-1 intersubtype recombination. Nucleic Acids Res. 34:5203-5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin, M. P., T. D. Rhodes, J. Chen, W. Fu, and W. S. Hu. 2005. Identification of a major restriction in HIV-1 intersubtype recombination. Proc. Natl. Acad. Sci. U. S. A. 102:9002-9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cochrane, A. W., M. T. McNally, and A. J. Mouland. 2006. The retrovirus RNA trafficking granule: from birth to maturity. Retrovirology 3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cros, J. F., and P. Palese. 2003. Trafficking of viral genomic RNA into and out of the nucleus: influenza, Thogoto and Borna disease viruses. Virus Res. 95:3-12. [DOI] [PubMed] [Google Scholar]
- 7.Drummond, D. A., J. J. Silberg, M. M. Meyer, C. O. Wilke, and F. H. Arnold. 2005. On the conservative nature of intragenic recombination. Proc. Natl. Acad. Sci. U. S. A. 102:5380-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duch, M., M. L. Carrasco, T. Jespersen, L. Aagaard, and F. S. Pedersen. 2004. An RNA secondary structure bias for non-homologous reverse transcriptase-mediated deletions in vivo. Nucleic Acids Res. 32:2039-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dykes, C., M. Balakrishnan, V. Planelles, Y. Zhu, R. A. Bambara, and L. M. Demeter. 2004. Identification of a preferred region for recombination and mutation in HIV-1 gag. Virology 326:262-279. [DOI] [PubMed] [Google Scholar]
- 10.Flynn, J. A., W. An, S. R. King, and A. Telesnitsky. 2004. Nonrandom dimerization of murine leukemia virus genomic RNAs. J. Virol. 78:12129-12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flynn, J. A., and A. Telesnitsky. 2006. Two distinct Moloney murine leukemia virus RNAs produced from a single locus dimerize at random. Virology 344:391-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galetto, R., V. Giacomoni, M. Veron, and M. Negroni. 2006. Dissection of a circumscribed recombination hot spot in HIV-1 after a single infectious cycle. J. Biol. Chem. 281:2711-2720. [DOI] [PubMed] [Google Scholar]
- 13.Galetto, R., A. Moumen, V. Giacomoni, M. Veron, P. Charneau, and M. Negroni. 2004. The structure of HIV-1 genomic RNA in the gp120 gene determines a recombination hot spot in vivo. J. Biol. Chem. 279:36625-36632. [DOI] [PubMed] [Google Scholar]
- 14.Galli, A., M. Kearney, O. A. Nikolaitchik, S. Yu, M. P. Chin, F. Maldarelli, J. M. Coffin, V. K. Pathak, and W. S. Hu. 2010. Patterns of human immunodeficiency virus type 1 recombination ex vivo provide evidence for coadaptation of distant sites, resulting in purifying selection for intersubtype recombinants during replication. J. Virol. 84:7651-7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Andres, S., D. M. Tomas, S. Sanchez-Campos, J. Navas-Castillo, and E. Moriones. 2007. Frequent occurrence of recombinants in mixed infections of tomato yellow leaf curl disease-associated begomoviruses. Virology 365:210-219. [DOI] [PubMed] [Google Scholar]
- 16.Hanson, M. N., M. Balakrishnan, B. P. Roques, and R. A. Bambara. 2005. Effects of donor and acceptor RNA structures on the mechanism of strand transfer by HIV-1 reverse transcriptase. J. Mol. Biol. 353:772-787. [DOI] [PubMed] [Google Scholar]
- 17.Harrington, P. R., J. A. Nelson, K. M. Kitrinos, and R. Swanstrom. 2007. Independent evolution of human immunodeficiency virus type 1 env V1/V2 and V4/V5 hypervariable regions during chronic infection. J. Virol. 81:5413-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, C. Q., Z. X. Xie, G. Z. Han, J. B. Dong, D. Wang, J. B. Liu, L. Y. Ma, X. F. Tang, X. P. Liu, Y. S. Pang, and G. R. Li. 2009. Homologous recombination as an evolutionary force in the avian influenza A virus. Mol. Biol. Evol. 26:177-187. [DOI] [PubMed] [Google Scholar]
- 19.Heath, L., E. van der Walt, A. Varsani, and D. P. Martin. 2006. Recombination patterns in aphthoviruses mirror those found in other picornaviruses. J. Virol. 80:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes, E. C. 2001. On the origin and evolution of the human immunodeficiency virus (HIV). Biol. Rev. Camb. Philos. Soc. 76:239-254. [DOI] [PubMed] [Google Scholar]
- 21.Hu, W. S., and H. M. Temin. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. U. S. A. 87:1556-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jetzt, A. E., H. Yu, G. J. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, J. K., C. Palaniappan, W. Wu, P. J. Fay, and R. A. Bambara. 1997. Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J. Biol. Chem. 272:16769-16777. [DOI] [PubMed] [Google Scholar]
- 24.Klaver, B., and B. Berkhout. 1994. Premature strand transfer by the HIV-1 reverse transcriptase during strong-stop DNA synthesis. Nucleic Acids Res. 22:137-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korber, B., B. Foley, C. Kuiken, S. Pillai, and J. Sodroski. December 1998, posting date. Numbering positions in HIV relative to HXB2CG. Los Alamos National Laboratory, Los Alamos, NM. http://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/1998/III/HXB2.pdf.
- 26.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labrosse, B., L. Morand-Joubert, A. Goubard, S. Rochas, J. L. Labernardiere, J. Pacanowski, J. L. Meynard, A. J. Hance, F. Clavel, and F. Mammano. 2006. Role of the envelope genetic context in the development of enfuvirtide resistance in human immunodeficiency virus type 1-infected patients. J. Virol. 80:8807-8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefeuvre, P., J. M. Lett, B. Reynaud, and D. P. Martin. 2007. Avoidance of protein fold disruption in natural virus recombinants. PLoS Pathog. 3:e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lefeuvre, P., J. M. Lett, A. Varsani, and D. P. Martin. 2009. Widely conserved recombination patterns among single-stranded DNA viruses. J. Virol. 83:2697-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, Y., E. Wimmer, and A. V. Paul. 2009. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim. Biophys. Acta 1789:495-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magiorkinis, G., D. Paraskevis, A.-M. Vandamme, E. L. Magiorkinis, V. Sypsa, and A. Hatzakis. 2003. In vivo characteristics of human immunodeficiency virus type 1 intersubtype recombination: determination of hot spots and correlation with sequence similarity. J. Gen. Virol. 84:2715-2722. [DOI] [PubMed] [Google Scholar]
- 32.Malim, M. H., J. Hauber, S. Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338:254-257. [DOI] [PubMed] [Google Scholar]
- 33.Moumen, A., L. Polomack, T. Unge, M. Veron, H. Buc, and M. Negroni. 2003. Evidence for a mechanism of recombination during reverse transcription dependent on the structure of the acceptor RNA. J. Biol. Chem. 278:15973-15982. [DOI] [PubMed] [Google Scholar]
- 34.Nora, T., C. Charpentier, O. Tenaillon, C. Hoede, F. Clavel, and A. J. Hance. 2007. Contribution of recombination to the evolution of human immunodeficiency viruses expressing resistance to antiretroviral treatment. J. Virol. 81:7620-7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onafuwa, A., W. An, N. D. Robson, and A. Telesnitsky. 2003. Human immunodeficiency virus type 1 genetic recombination is more frequent than that of Moloney murine leukemia virus despite similar template switching rates. J. Virol. 77:4577-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paillart, J. C., M. Shehu-Xhilaga, R. Marquet, and J. Mak. 2004. Dimerization of retroviral RNA genomes: an inseparable pair. Nat. Rev. Microbiol. 2:461-472. [DOI] [PubMed] [Google Scholar]
- 37.Rakoto-Andrianarivelo, M., N. Gumede, S. Jegouic, J. Balanant, S. N. Andriamamonjy, S. Rabemanantsoa, M. Birmingham, B. Randriamanalina, L. Nkolomoni, M. Venter, B. D. Schoub, F. Delpeyroux, and J. M. Reynes. 2008. Reemergence of recombinant vaccine-derived poliovirus outbreak in Madagascar. J. Infect. Dis. 197:1427-1435. [DOI] [PubMed] [Google Scholar]
- 38.Ramirez, B. C., E. Simon-Loriere, R. Galetto, and M. Negroni. 2008. Implications of recombination for HIV diversity. Virus Res. 134:64-73. [DOI] [PubMed] [Google Scholar]
- 39.Rasmussen, S. V., and F. S. Pedersen. 2006. Co-localization of gammaretroviral RNAs at their transcription site favours co-packaging. J. Gen. Virol. 87:2279-2289. [DOI] [PubMed] [Google Scholar]
- 40.Roda, R. H., M. Balakrishnan, J. K. Kim, B. P. Roques, P. J. Fay, and R. A. Bambara. 2002. Strand transfer occurs in retroviruses by a pause-initiated two-step mechanism. J. Biol. Chem. 277:46900-46911. [DOI] [PubMed] [Google Scholar]
- 41.Shen, W., L. Gao, M. Balakrishnan, and R. A. Bambara. 2009. A recombination hot spot in HIV-1 contains guanosine runs that can form a G-quartet structure and promote strand transfer in vitro. J. Biol. Chem. 284:33883-33893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon, A. E., and L. Gehrke. 2009. RNA conformational changes in the life cycles of RNA viruses, viroids, and virus-associated RNAs. Biochim. Biophys. Acta 1789:571-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon-Loriere, E., R. Galetto, M. Hamoudi, J. Archer, P. Lefeuvre, D. P. Martin, D. L. Robertson, and M. Negroni. 2009. Molecular mechanisms of recombination restriction in the envelope gene of the human immunodeficiency virus. PLoS Pathog. 5:e1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song, R., J. Kafaie, L. Yang, and M. Laughrea. 2007. HIV-1 viral RNA is selected in the form of monomers that dimerize in a three-step protease-dependent process; the DIS of stem-loop 1 initiates viral RNA dimerization. J. Mol. Biol. 371:1084-1098. [DOI] [PubMed] [Google Scholar]
- 45.Streeck, H., B. Li, A. F. Poon, A. Schneidewind, A. D. Gladden, K. A. Power, D. Daskalakis, S. Bazner, R. Zuniga, C. Brander, E. S. Rosenberg, S. D. Frost, M. Altfeld, and T. M. Allen. 2008. Immune-driven recombination and loss of control after HIV superinfection. J. Exp. Med. 205:1789-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voigt, C. A., C. Martinez, Z. G. Wang, S. L. Mayo, and F. H. Arnold. 2002. Protein building blocks preserved by recombination. Nat. Struct. Biol. 9:553-558. [DOI] [PubMed] [Google Scholar]
- 47.Watts, J. M., K. K. Dang, R. J. Gorelick, C. W. Leonard, J. W. Bess, Jr., R. Swanstrom, C. L. Burch, and K. M. Weeks. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460:711-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White, K. A., and P. D. Nagy. 2004. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucleic Acid Res. Mol. Biol. 78:187-226. [DOI] [PubMed] [Google Scholar]
- 49.Worobey, M., and E. C. Holmes. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 80:2535-2543. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, J., L. Y. Tang, T. Li, Y. Ma, and C. M. Sapp. 2000. Most retroviral recombinations occur during minus-strand DNA synthesis. J. Virol. 74:2313-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, J., and H. M. Temin. 1994. Retrovirus recombination depends on the length of sequence identity and is not error prone. J. Virol. 68:2409-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhuang, J., S. Mukherjee, Y. Ron, and J. P. Dougherty. 2006. High rate of genetic recombination in murine leukemia virus: implications for influencing proviral ploidy. J. Virol. 80:6706-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]