Abstract
A major function of the hepatitis C virus (HCV) core protein is the interaction with genomic RNA to form the nucleocapsid, an essential component of the virus particle. Analyses to identify basic amino acid residues of HCV core protein, important for capsid assembly, were initially performed with a cell-free system, which did not indicate the importance of these residues for HCV infectivity. The development of a cell culture system for HCV (HCVcc) allows a more precise analysis of these core protein amino acids during the HCV life cycle. In the present study, we used a mutational analysis in the context of the HCVcc system to determine the role of the basic amino acid residues of the core protein in HCV infectivity. We focused our analysis on basic residues located in two clusters (cluster 1, amino acids [aa]6 to 23; cluster 2, aa 39 to 62) within the N-terminal 62 amino acids of the HCV core protein. Our data indicate that basic residues of the first cluster have little impact on replication and are dispensable for infectivity. Furthermore, only four basic amino acids residues of the second cluster (R50, K51, R59, and R62) were essential for the production of infectious viral particles. Mutation of these residues did not interfere with core protein subcellular localization, core protein-RNA interaction, or core protein oligomerization. Moreover, these mutations had no effect on core protein envelopment by intracellular membranes. Together, these data indicate that R50, K51, R59, and R62 residues play a major role in the formation of infectious viral particles at a post-nucleocapsid assembly step.
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (30) as well as several extrahepatic diseases (21). It has been reported that an estimated 170 million people are chronically infected with HCV worldwide (43). The currently approved treatment is a combination of pegylated alpha interferon (pegIFN-α) and ribavirin, which is effective in approximately 54% of infected patients (31).
HCV is the only member of the Hepacivirus genus in the Flaviviridae family. It is an enveloped, single-stranded positive-sense RNA virus, and its genome encodes a unique open reading frame, which is flanked by two structured nontranslated regions at its 5′ and 3′ ends (5′ NTR and 3′ NTR, respectively). The introduction of viral genome into cells leads to its translation, mediated by an internal ribosome entry site (IRES) (53). The resulting polyprotein is then processed by cellular and viral proteases (18, 20) to produce the core protein and the envelope glycoproteins E1 and E2, required for infectious virus particles production. The remaining proteins, NS2 through NS5B, are necessary for the intracellular processes of the virus life cycle. Furthermore, a small integral membrane protein, p7, has been reported to function as an ion channel (19, 42).
The HCV core protein is a basic protein, located in the N-terminal portion of the HCV precursor polyprotein. The release of core protein from the downstream E1 glycoprotein sequence requires the action of two coordinated cleavage events. The first cleavage, involving a signal peptidase (SP) (20, 49), is responsible for the anchorage of the core protein in the endoplasmic reticulum (ER) membrane through a C-terminal hydrophobic region (18). The second cleavage, mediated by a signal peptide peptidase (22, 34), is crucial for the release of the core protein from ER membrane and its targeting to lipid droplets (LDs) (6, 34). Targeting of core protein to lipid droplets was initially described by Barba and coworkers (3) and Moradpour and coworkers (37). The mature core protein that forms the viral nucleocapsid is a dimeric, α-helical protein constituted of D1 and D2 domains (8, 34). The D1 domain, composed of the N-terminal 117 amino acids (aa), contains a high proportion of basic residues involved in RNA binding and oligomerization of core protein (8, 13, 50, 52). The hydrophobic D2 domain, beginning at aa 118, has a length of about 55 aa and is involved in the targeting of HCV core protein to lipid droplets (6). Although the C-terminal end of D2 has not been mapped precisely, it likely ends between aa 177 and 182 (22, 27, 41). Furthermore, it has been shown that the folding of the D2 domain occurs in a membrane environment and is critical for the folding of the D1 domain (8). In infected cells, the lipid droplets and their associated membranes are the only subcellular compartments in which the HCV core protein accumulates and are probably the site of particle assembly (7, 36, 47). Beyond its role in the viral nucleocapsid assembly, the core protein has been shown to be involved in multiple interactions with cellular proteins, suggesting its implication in some cellular functions (44).
The major breakthrough since the discovery of HCV was the production of infectious virus in cell culture (HCVcc) (29, 55, 58). The advent of this infectious cell culture system has allowed many fundamental and applied studies to be achieved. However, many details of viral morphogenesis and infectivity are still needed to complete our knowledge. The role of the core protein in HCV infectivity has been previously investigated by alanine scanning mutagenesis or by genetic analysis (27, 40). These reports highlighted the involvement of numerous amino acids in the production of infectious virus. Other studies, using cell-free and protein expression systems, reported that the N-terminal region of the core protein is important for HCV nucleocapsid assembly (15, 25, 26) and core protein-RNA interaction (49, 57). However, this region of the core protein has not been fully investigated in the context of the HCVcc system. In this study we examined the involvement of two clusters of basic residues at the N terminus of the core protein in the replication and infectivity of HCV. We identified four basic residues essential for the production of infectious HCV particles.
MATERIALS AND METHODS
Cell culture.
Huh-7 and Huh-7w7 (CD81−) cells (46) were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) and supplemented with 100 nM nonessential amino acids (NEAA; Invitrogen). Huh-7w7 (CD81−) cells support viral replication and virus production but cannot be reinfected with the virus. These cells were used in this study for viral titration and replication experiments to avoid any effect of reinfection, which allows the mutants to be tested in a single-cycle production assay (48).
Antibodies.
Mouse anti-core protein monoclonal antibodies (MAbs) (ACAP-27/IgG2a and 2H9/IgG1) were kindly provided by J. F. Delagneau and T. Wakita (National Institute of Infectious, Tokyo, Japan), respectively. Rat MAb anti-E2 (3/11) (14), kindly provided by J. A. Mckeating, and mouse MAb anti-E1 (A4) (12) were produced in vitro by using a MiniPerm apparatus (Heraeus), according to the manufacturer's instructions. Mouse MAb anti-NS5A (9E10/IgG2a) was kindly provided by C. M. Rice (The Rockefeller University). Goat anti-β-actin polyclonal antibody was from Santa Cruz. All conjugated secondary antibodies for indirect immunofluorescence (IF) were from Invitrogen. Peroxidase-conjugated donkey anti-goat IgG was from Santa Cruz, peroxidase-conjugated goat anti-rat IgG was from Jackson ImmunoResearch, and peroxidase-conjugated goat anti-mouse IgG was from Sigma.
Plasmid construction.
Mutants used in this study were based on the genotype 2a plasmid pJFH1/CS-N6-A4 (11, 17). In this work, this virus is referred to as JFH1 or wild type (wt). All core protein mutations and pJFH1/CS-N6-A4 with a deletion of residues 62 to 160 in the core protein [pJFH1/CS-N6-A4/Δcore (62-160)] were introduced into the plasmid by standard PCR methods using AgeI, FspAI, and BsiWI sites in pJFH1/CS-N6-A4 plasmid. pJFH1/CS-N6-A4/ΔE1E2 and pJFH1/CS-N6-A4/GND mutants were constructed as described previously (55). The pJFH1/CS-N6-A4/NS5A-Gluc plasmid was constructed by introducing a small nucleotide sequence to create two unique sites, NruI and BglII, and leading to the insertion of four amino acids, RSSR, between aa 2394 and 2395 of the NS5A protein (38). The coding sequence of the nonsecreted form of Gaussia luciferase (Gluc) was cloned between these two sites. All mutations were also introduced into this reporter construction for replication study. Mutants with Gluc were not infectious or only very poorly infectious.
To construct the wt and mutant core protein-expressing pcDNA3.1 plasmids, DNA fragments encoding the core protein gene from wt and mutant genomes were generated by PCR using primers containing EcoRI and XbaI restriction sites. The resulting sequence was then cloned into pcDNA3.1 to obtain pcDNA/core-wt, pcDNA/core-R50, pcDNA/core-K51, pcDNA/core-R59, and pcDNA/core-R62 plasmids.
In vitro transcription.
Viral RNA was prepared as described previously (11, 17). Briefly, plasmids linearized by XbaI and treated with Mung bean nuclease were transcribed using a MEGAscript kit (Ambion) according to the manufacturer's instructions. After 4 h of incubation at 37°C, the DNA template was degraded by DNase treatment for 15 min at 37°C. Synthesized RNA was precipitated by LiCl and then purified and quantified by absorbance at 260 nm.
RNA transfection.
Cells were electroporated using in vitro transcribed viral RNA as described previously (23). Briefly, 4 × 106 cells in OptiMEM (Invitrogen) were electroporated with 10 μg of viral RNA. For replication analysis, electroporated Huh-7w7 cells with Gluc-harboring mutants were plated in 24-well plates and lysed with 100 μl of 1× Renilla lysis buffer (Promega) at 4 h and 72 h postelectroporation. Twenty microliters of cell lysate was used to determine the luciferase activity. Values at 72 h were normalized relative to those at 4 h.
Virus titration.
Clarified Huh-7w7 cell culture supernatants of T25 flasks harvested at 72 h postelectroporation were used, and virus titers were determined by endpoint dilution assays on Huh-7 cells as described previously (58). Briefly, cell supernatants were serially diluted 10-fold in complete DMEM, and 50 μl was used to infect 7 × 103 Huh-7 cells in 96-well plates. Infected cells were fixed at 48 h postinoculation and stained with anti-E1 antibody (A4). The viral titer is expressed as the number of focus-forming units (FFU) per milliliter of supernatant.
Intracellular infectivity.
T25 flasks of electroporated cells were harvested at 72 h postelectroporation, washed with phosphate-buffered saline (PBS), trypsinized, and pelleted by centrifugation. Cell pellets were resuspended in 1 ml of DMEM containing 10% FBS and supplemented with nonessential amino acids and subjected to four freeze-thaw cycles (16). Cell debris was then removed by centrifugation, and supernatants were used to determine intracellular infectivity as described above.
Quantification of HCV RNA.
Huh-7 cells, in 35-mm wells of six-well cell culture plates, were passaged at 72 h postelectroporation. Cells and supernatants were harvested at 72 h postpassage (i.e., 6 days postelectroporation [P1/6d]), and intra- and extracellular HCV RNA was titrated by quantitative real-time reverse transcription-PCR (qRT-PCR) assay as described previously (10). One microgram of total RNA was used for intracellular HCV RNA quantification.
Quantification of HCV core protein.
HCV core antigen (Ag) expressed within cells or secreted into the supernatant was quantified at P1/6d with an immunoassay using chemiluminescent microparticle technology (Architect HCV Ag Test; Abbott, France) (39). The concentration of HCV core Ag in the specimen was determined using a previously generated calibration curve. The cutoff value was set at 3.00 fmol/liter, equivalent to 0.06 pg/ml of recombinant c11 Ag (residues 1 to 160 of the HCV genotype 2a isolate [GenBank accession number I49748]). Specimens with concentration values of <3.00 fmol/liter were considered nonreactive for HCV Ag. For intracellular core protein quantification, cells were lysed in 450 μl of PBS supplemented with 1% Triton-X, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 50 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK), and protease inhibitor cocktail (Complete; Roche) at 4°C for 20 to 30 min. Cleared lysates were then measured at a dilution of 1:500 in PBS. Supernatants were clarified by centrifugation prior to core protein quantification.
Velocity sedimentation and sucrose gradients.
Electroporated cells of wt and mutant genomes were lysed at 72 h postelectroporation in lysis buffer containing 50 mM tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.3% NP-40, 1 mM PMSF, and protease inhibitor cocktail (Roche) for 15 min at room temperature and then incubated on ice. Cell lysates were precleared by centrifugation at 14,000 rpm for 5 min at 4°C. Each sample was then layered on top of 12 ml of a 10% to 60% sucrose gradient prepared in the same buffer without detergent and centrifuged in a Beckman SW41 Ti rotor (Beckman) at 29,000 rpm for 16 h at 4°C. Fractions of 1 ml were collected from the top of each tube. Proteins were precipitated by the methanol-chloroform method, resuspended in sample buffer, and then analyzed by SDS-PAGE and immunoblotting.
Encapsidated RNA analysis.
Cells were lysed at 72 h postelectroporation using the lysis buffer used above. The nucleocapsids were then pelleted over a 30% sucrose cushion in a Beckman TLA 55 rotor at 48,000 rpm for 5 h at 4°C. Pellets were resuspended in 50 μl of Dulbecco's PBS (DPBS) overnight. Ten percent of each pellet was used for SDS-PAGE. The rest was divided into two parts: one part was treated with RNase and DNase to eliminate any nonincorporated and nonprotected nucleic acids by the nucleocapsid, and the other part remained untreated. RNAs were extracted by an RNeasy Mini kit (Qiagen) according to the manufacturer's protocol and eluted in 30 μl of RNase-free water. Five microliters was used for RNA quantification as described above.
Core trans-complementation.
Huh-7w7 cells were electroporated with mutant or truncated (deletion of residues 62 to 160) core protein RNAs. At 72 h postelectroporation, cells were seeded onto 35-mm wells of six-well cell culture plates and cultured overnight before transfection. Mutants and wt pcDNA/core protein plasmids or empty pcDNA3.1 (2 μg) were transfected into cells using TransIT-T1 reagent (Mirus). Supernatants of transfected cells were collected after a further 48-h period and titrated as described above.
Membrane protection assay.
A membrane protection assay was performed as described previously by Ai et al. (1) to assess the core protein envelopment by cellular membranes. Briefly, electroporated cells were harvested at 72 h postelectroporation and resuspended in buffer containing 50 mM Tris-HCl, pH 8, 10 mM CaCl2, and 1 mM dithiothreitol (DTT). Cells were passed 25 to 30 times through a 27-gauge syringe. After a centrifugation at 1,000 × g for 10 min at 4°C, the postnuclear supernatant was used in a protease protection assay with proteinase K (10 μg/ml) in the presence or absence of 1% Triton X-100 on ice for 1 h. The reaction was quenched by the addition of 5 mM PMSF (final concentration), followed by SDS-PAGE and immunoblotting.
Western blot analysis.
Western blot analysis was performed as described previously (11) with some modifications. Briefly, cells were lysed in a buffer containing 10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.5% Triton-X, 0.5 mM PMSF, 50 μg/ml TPCK, and protease inhibitor cocktail (Complete; Roche) at 4°C for 20 to 30 min. After a precleaning by centrifugation, protein concentration was determined in the postnuclear supernatants by the Bradford method (Bio-Rad). The proteins were then resolved by SDS-12.5% PAGE and transferred onto nitrocellulose membranes (Hybond-ECL; Amersham) using a Trans-Blot apparatus (Bio-Rad). Proteins of interest were revealed with specific primary antibodies, followed by species-specific secondary antibodies conjugated to peroxidase.
Indirect immunofluorescence microscopy.
Immunofluorescent detection of viral proteins was performed as previously described (17, 47). Briefly, cells were fixed in 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in PBS. After being blocked with PBS-10% goat serum, cells were incubated with primary antibodies for 1 h at room temperature. Cells were washed with PBS and then incubated with secondary antibodies for 30 to 45 min at room temperature. In a last step, lipid droplets were stained with BODIPY (4,4-difluoro-4-bora-3a,4a-diaza-s-indacene) 493/503 (0.5 μg/ml; Invitrogen) for 10 min at room temperature. Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI). Image acquisition was carried out using an Axiophot 2 microscope (Zeiss) equipped with a Coolsnap ES camera (Photometrix). For colocalization experiments, confocal microscopy was performed with an LSM710 confocal microscope (Zeiss) using a 63× oil immersion objective with a 1.4 numerical aperture. Signals were sequentially collected using single fluorescence excitation and acquisition settings to avoid crossover. Images were assembled using Adobe Photoshop software.
RESULTS
Two clusters rich in basic residues are located at the N terminus of HCV core protein.
In recent reports (27, 40), the role of amino acids 57 to 191 of the HCV core protein has been investigated. In these two studies, many residues were shown to be essential during the viral life cycle. Nevertheless, the role of the N terminus of core protein in viral infectivity has not yet been studied. When analyzed in a cell-free system, this part of core protein seemed important for HCV nucleocapsid assembly (25, 26). This region contains many basic residues located between aa 1 to 62 of the core; however, their role in HCV infectivity has not yet been investigated. We therefore analyzed the basic residues located between aa 1 to 62 for their role in HCV infectivity. The N terminus of the core protein contains 18 basic residues (5 lysines and 13 arginines), and a RNA binding activity has also been described as being associated with this region (49). The basic residues can be grouped into two clusters (cluster 1, aa 6 to 23; cluster 2, aa 39 to 62). As shown in Fig. 1, five basic residues (K6, R9, K10, K12, and R13) are located in the first 14 aa of the core protein. In addition, 10 basic residues (R17, R18, K23, R39, R40, R43, R47, R50, K51, and R55) are found in a region of the core protein for which the RNA coding sequence is well structured in stem-loops V and VI (SLV and SLVI, respectively) (51). The last three basic residues (R59, R61, and R62) are located outside these structures. The strategy used in this study was to replace basic residues with alanines to create mutants with double or triple mutations and to determine whether they affect the virus life cycle. The residues were then modified separately in mutants of interest to point out the most essential amino acids for HCV infectivity.
FIG. 1.
Proposed model of secondary RNA structures of JFH1 within the 5′ NTR and the 5′ end of the capsid-coding sequence. The major structural domains are indicated by SL (stem-loop) followed by roman numerals. The authentic initiator AUG codon is highlighted in SLIV. The presence of SLV and SLVI has been suggested by Smith and Simmonds (51) and adapted to JFH1 based on the structure also presented by Vassilaki and collaborators (54). Mutated residues are indicated with arrows that point out the two nucleotides of each codon that were replaced with GC to obtain an alanine. The position of each basic residue and the different amino acid mutants that were initially modified are indicated. Basic residues that are conserved with more than 99% of the listed sequences in the European HCV database (http://euhcvdb.ibcp.fr) are indicated by asterisks. The other residues are conserved in 95 to 99% of these sequences.
Substitution of the basic residues with alanines was performed by site-directed mutagenesis. Specifically, the first two nucleotides of each codon were modified, and the third nucleotide was unchanged. Mutations were created in the context of a modified JFH1 virus (JFH1/CS-N6-A4, considered here as the wt) (11, 17) to investigate viral production. To quantify RNA replication, the luciferase reporter mutant JFH1/NS5A-Gluc was generated from the JFH1 virus. In this mutant, the Gluc-coding sequence was introduced into the NS5A-coding sequence of JFH1 at a position where viral replication is still favored (38).
Modification of basic residues located in cluster 1 (aa 6 to 23).
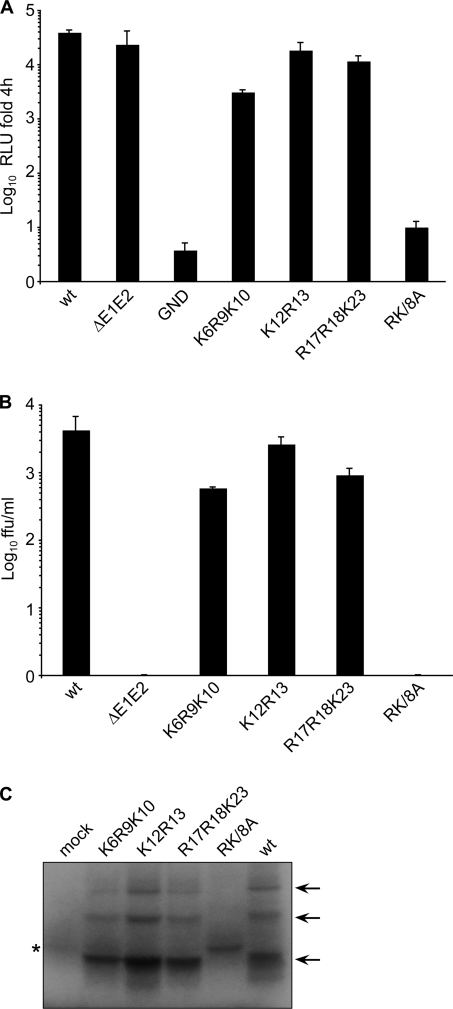
To study the function of the first cluster, basic residues were mutated to alanines, as mentioned above. In this way four mutants, K6A/R9A/K10A (K6R9K10), K12A/R13A (K12R13), R17A/R18A/K23A (R17R18K23), and a mutant with all eight mutations (RK/8A), were first generated (Fig. 1). Thus, replication fitness of the wt and mutants was determined in CD81-deficient Huh-7w7 cells (46) after electroporation of the in vitro transcribed RNAs. Viral replication and viral titers were analyzed, respectively, by quantification of luciferase activity and assay of the number of FFU in naïve Huh-7 cells. As shown in Fig. 2A, the replication levels of the K6R9K10 and R17R18K23 mutants were about 12- and 3-fold lower, respectively, than those obtained with the parental virus. The replication rate of the K12R13 mutant was comparable to that of the wt. As expected, no replication was observed with the polymerase-defective mutant (JFH1/NS5A-Gluc/GND). Furthermore, the RK/8A mutant showed a replication level comparable to that of the polymerase-defective mutant (GND). The titration of each virus produced in a single cycle of virus infection revealed that the titers of the K6R9K10 and R17R18K23 mutants were reduced 7-fold and 5-fold, respectively, while the titer of the K12R13 mutant was similar to that of the parental virus (Fig. 2B). As expected from its replication defect, the RK/8A mutant did not produce any detectable infectious viral particles, as observed for the control virus lacking envelope glycoproteins JFH1/ΔE1E2.
FIG. 2.
Replication and infectivity of core mutants generated in the first basic cluster. (A) Replication of core protein mutants was measured by Gaussia luciferase activity at 72 h postelectroporation. Values are expressed relative to the quantity of luciferase activity measured at 4 h postelectroporation. (B) Determination of infectious virus production released from transfected cells. Supernatants were harvested at 72 h postelectroporation and were assayed by endpoint dilution assays of FFU using naïve Huh-7 cells. Means and standard errors of the means of results from at least duplicate electroporations are shown. (C) T7 RNA transcripts were obtained from plasmids containing the HCV IRES and core protein-coding sequences of the first cluster mutants. Transcripts were then translated for 1 h in a rabbit reticulocyte lysate with addition of [35S]methionine in the reaction mixture. Arrows indicate monomeric, dimeric, and trimeric forms of the core protein. The nonspecific band is indicated by an asterisk. RLU, relative light units.
As the 5′ core protein coding sequence is also involved in the IRES function (45), we hypothesized that modifications in this nucleotide sequence might affect the viral genome translation. The IRES-dependent expression of mutant core proteins was analyzed by an in vitro translation assay. Compared to expression of the wt core protein, only slight differences were observed for K6R9K10 and R17R18K23 core protein expression, suggesting that nucleotide modifications of these mutants had little impact on translation, whereas core protein expression was undetectable in the RK/8A mutant (Fig. 2C). Furthermore, bands that might correspond to dimeric and trimeric forms of the core protein were detected for all mutants, as observed previously (33), except RK/8A. However, a band with reduced mobility compared to that of the wt and other mutants was observed for this mutant. This band comigrated with a faint band observed with a translation control in the absence of RNA, suggesting that the band was not specific. The decreased level of RNA replication observed in the RK/8A mutant was therefore probably due to lower translation caused by nucleotide modifications introduced in the core protein sequence close to the IRES. Consequently, the combination of mutations interferes with infectivity by affecting the translation and/or the replication of the viral genome. Taken together, our data indicate that basic residues of this cluster (aa 6 to 23) have no major role in HCV infectivity when they are modified separately.
Modification of basic residues located in cluster 2 (aa 39 to 62).
Four mutants were created with modifications in the second basic cluster (aa 39 to 62). Two of them are located in stem-loop VI, R39A/R40A/R43A and R47A/R50A/K51A (R39R40R43 and R47R50K51, respectively), and the other two, R55A/R59A and R61A/R62A (R55R59 and R61R62, respectively), are outside this structure (Fig. 1). As shown in Fig. 3A, all mutants were able to replicate the viral genome. R55R59 and R61R62 mutants, whose modifications were introduced outside the SLVI, were able to replicate the viral genome as efficiently as the wt at 72 h postelectroporation. However, replication levels of R39R40R43 and R47R50K51, whose mutations were introduced into the SLVI, were about 6- and 8-fold lower, respectively, than the level of the wt. Mutations introduced into SLVI were predicted to disturb the upper or the middle half of the stem-loop (Fig. 1). These data suggest that the combination of mutations in SLVI could impact viral replication. Similar effects on HCV replication induced by SLVI destabilization have been described by Vassilaki and collaborators (54). Importantly, none of our mutants was able to produce infectious viral particles when tested with the inoculation of naïve Huh-7 cells with the supernatant of the different electroporated cells (Fig. 3B). As shown in Fig. 3C, core protein was detectable in all the mutants at 72 h postelectroporation, with decreased expression levels in the R39R40R43 and R47R50K51 mutants. The decreased replication levels of these two mutants might explain these differences in core protein expression. Together, these data suggest that the inability to produce infectious virus does not result from the instability of mutant core proteins.
FIG. 3.
Replication and infectivity of core protein mutants generated in the second basic cluster. (A) Replication of core protein mutants and normalization of values were performed as described in the legend of Fig. 2. (B) Determination of intracellular and extracellular infectious virus produced from electroporated cells at 72 h postelectroporation. Means and standard errors of the means of results from at least duplicate electroporations are shown. (C) Western blot analysis of lysates harvested at 72 h postelectroporation. Results for core protein, E2, NS5A, and actin are indicated.
Since the release of infectious particles could be altered by these changes, we next investigated the intracellular production of viral particles. Electroporated cells were washed and lysed at 72 h postelectroporation by multiple freeze-thaw cycles, as described previously (16). Clarified lysates were then used to infect naïve Huh-7 cells. In contrast to the wt, none of these lysates was able to infect new cells, indicating that no intracellular infectious virus was produced by these mutants (Fig. 3B). Altogether, these data suggest that one or more of the basic residues located in each mutant of the second cluster are necessary for HCV infectivity.
Identification of basic residues necessary for HCV infectivity.
In an attempt to identify basic residues of the core protein necessary for infectious virus production, we separately mutated each basic residue to alanine in the second cluster (aa 39 to 62) as indicated in Fig. 1. For most single mutants, the replication rates were comparable to the rate of the wt. However, slightly lower levels were observed for R39 and R47 mutants (Fig. 4A). The values were comparable to those obtained with the R39R40R43 and R47R50K51 mutants, suggesting that modifications of nucleotides (nt) 455 to 456 (mutant R39) or 479 to 480 (mutant R47) in the SLVI affected RNA replication. Since these two mutations disrupted two base pairings in the SLVI structure (Fig. 1), our data further support the role of this portion of the stem-loop structure in HCV replication.
FIG. 4.
Analysis of individual core protein mutants of the second cluster. (A) Replication of core protein mutants and normalization of values were performed as described in the legend of Fig. 2. (B) Determination of infectious virus production released from electroporated cells at 72 h postelectroporation. Means and standard errors of the means of results from at least duplicate electroporations are shown. (C) Western blot analysis of lysates harvested at 72 h postelectroporation. Results for core protein, E2, NS5A, and actin are indicated.
We next investigated the effect of our single mutations on the production of infectious viral particles. As shown in Fig. 4B, only four of these mutants were unable to produce infectious viral particles. Indeed, only the R50, K51, R59, and R62 mutants were unable to infect naïve cells. The infectivity of the R39, R40, R43, and R47 mutants was about 5- to 8-fold lower than that of the wt, whereas R55 and R61 infectivity was comparable to that of the wt. Interestingly, in contrast to the R39R40R43 mutant, which was not infectious, the single mutants R39, R40, and R43 were still infectious. This might be explained by the cumulative effect of these mutations. The absence of infectivity for the R50, K51, R59, and R62 mutants was not due to the instability of mutant core proteins since the proteins were detected at 72 h postelectroporation by Western blotting (Fig. 4C). Furthermore, intracellular infectivity of these defective mutants was tested as described above. However, no infectivity was detected after naïve cells were inoculated with clarified lysates obtained at 72 h postelectroporation of cells with mutant RNAs (data not shown). Together, these data suggest that the R50, K51, R59, and R62 residues are critical for the production of infectious viral particles. Given their importance, all further experiments were performed only on these four mutants.
No release of noninfectious particles by core mutants.
The lack of intra- and extracellular infectivity of our mutants might result from a defect in particle assembly or from some alteration during the disassembly process in HCV entry. To discriminate between these two hypotheses, we investigated if noninfectious particles were produced by analyzing the release of HCV RNA and core protein into the supernatants. The HCV RNA in cell culture medium was quantified after one passage to avoid any contamination by the RNA used for electroporation. RNA levels released by noninfectious R50, K51, R59, and R62 mutants were about 2 to 3 logs lower than the level of the wt and were similar to the RNA level released from the assembly-defective JFH1/ΔE1E2 control (Fig. 5A). In addition, none of these mutants released larger amounts of core protein than JFH1/ΔE1E2, and these amounts were about 20- to 150-fold lower than the amount released by the wt (Fig. 5C). The reduced levels of secreted RNA and core protein were not due to diminished accumulation in electroporated cells since the intracellular levels of RNA and core protein of mutants were comparable to those of the wt (Fig. 5B and D). Taken together, these data indicate that these mutants do not release noninfectious particles and that the defect in viral particle production probably occurs at the assembly step.
FIG. 5.
Defective core mutants do not release viral particles. (A and B) Cells were electroporated with the wt and core protein mutant RNAs. At 72 h postelectroporation, cells were passaged, and extra- and intracellular viral RNA was quantified after a further 72 h (i.e., 6 days postelectroporation [P1/6d]) as described in Materials and Methods. (C and D) Quantification of extra- and intracellular HCV core protein of electroporated cells at P1/6d. Dashed horizontal lines indicate background levels based on those of negative controls. Means and standard errors of the means of results from at least duplicate electroporations are shown.
Intracellular localization of core protein.
As previously described, core protein localizes to the lipid droplets (LDs) (6, 34), which have been defined as important organelles for HCV assembly (36). We therefore analyzed the subcellular localization of the mutant core proteins to determine the effect of these mutations on the association with the LDs. As shown in Fig. 6A, all core mutants colocalized with the LDs in a manner indistinguishable from that of the wt. Moreover, as recent data indicated that NS5A also plays a key role in the HCV particle formation by interacting with the core protein (32), colocalization of NS5A with our mutant core proteins was also investigated. A partial colocalization of core protein and NS5A was observed in the perinuclear area of electroporated cells (Fig. 6B). However, no alteration in the subcellular localization of these two proteins was observed in the mutants (Fig. 6B). Together, our data suggest that these mutations do not disturb the subcellular localization of the core protein with LDs and NS5A protein.
FIG. 6.
Subcellular localization of core protein. (A) Huh-7 cells were electroporated with in vitro transcribed HCV genomes containing the indicated mutations. Cells were fixed at 72 h postelectroporation, and core protein was detected with MAb anti-core 2H9 (red). Lipid droplets were stained with BODIPY 493/503 (green). (B) Fixed cells at 72 h postelectroporation were double labeled with MAb anti-core (2H9/IgG1) (green) and MAb anti-NS5A (9E10/IgG2a) (red). For both panels, merged images and zoomed views of the indicated areas are shown. Nuclei were stained with DAPI (blue). Representative confocal images are shown. Bar, 10 μm.
Core protein trans-complementation.
Since mutations introduced in the core protein-coding sequence also lead to modification of RNA nucleotide sequence, we wanted to determine whether the effects observed with the mutants were due to amino acid or to RNA nucleotide changes. For this purpose, a trans-complementation assay was developed. A JFH1/Δcore (62-160) RNA, expressing a truncated HCV core protein, was introduced by electroporation into Huh-7w7 cells, and the cells were passaged 72 h later. Then, these cells were transfected with plasmids expressing the wt or mutant core proteins. Supernatants harvested at 48 h posttransfection were used to infect naïve Huh-7 cells for 48 h. As shown in Fig. 7A, other than the wt core protein-expressing plasmid, no mutant core protein could rescue the core protein-truncated mutant, in spite of successful expression of core protein (Fig. 7B). In contrast, when cells were electroporated with defective mutant genomes followed by transfection with the wt core protein-expressing plasmid, all supernatants were infectious (Fig. 7C). These data indicate that these mutations were rescued by the wt core protein. As shown in Fig. 7D, infected cells with trans-complemented JFH1/Δcore (62-160) supernatant were positive for NS5A protein but negative for core protein, as observed in immunofluorescence studies. Moreover, the supernatants of trans-complemented mutants were able to initiate only a single round of infection (data not shown). These results indicate that the observed infectivity was due to the trans-complementation, and not to the recombination, between mutant RNAs and core protein transcripts. Together, our results indicate that the loss of infectivity observed for R50, K51, R59, and R62 mutants is due to the mutation of basic residues rather than to RNA sequence changes.
FIG. 7.
trans-complementation assays. (A) Huh-7w7 cells were electroporated with a core protein deletion mutant, JFH1/Δcore (62-160). At 72 h postelectroporation, cells were seeded onto 35-mm wells of a six-well cell culture plate and cultured overnight. Cells were transfected with empty pcDNA3.1 or with plasmids expressing the wt or mutant core proteins. Infectious virus production was assayed at 48 h posttransfection as described in Materials and Methods. (B) Western blot analysis of electroporated cells at 48 h posttransfection. Results for core protein and actin are shown as controls of transfection. (C) Electroporated Huh-7w7 cells with mutant genomes were passaged at 72 h postelectroporation as in panel A. Cells were then transfected with the pcDNA/core-wt plasmid, and infectivity was titrated at 48 h posttransfection as described in Materials and Methods. An empty pcDNA plasmid was used as a negative control. Means and standard errors of the means of results from at least duplicate electroporations are shown. (D) Supernatant of JFH1/Δcore (62-160)-electroporated Huh-7w7 cells that were trans-complemented with wt core protein was used to infect naïve Huh-7 cells. Cells were fixed at 48 h postinoculation, and immunostaining was performed with anti-core protein and anti-NS5A antibodies (upper panel). Supernatant of wt-electroporated Huh-7w7 cells was used as a control (lower panel).
Analysis of core protein oligomerization and RNA encapsidation.
To further understand the molecular basis of the absence of intracellular and extracellular infectious virus particles, we investigated whether the core protein was able to oligomerize into nucleocapsid structures. Oligomerization of core proteins expressed by these mutants was analyzed on a sucrose gradient by ultracentrifugation, as described in Materials and Methods. The core protein complexes of the wt were detected in all the fractions (Fig. 8A), with a peak of intensity in fractions 6 to 8, as described previously (1). The first fractions likely correspond to monomeric forms of core proteins, whereas the other fractions must correspond to highly ordered multimeric complexes. A similar profile of sedimentation was observed with the assembly-defective control JFH1/ΔE1E2 (Fig. 8B). In contrast, when the wt core protein was treated with 1% SDS before ultracentrifugation, only the monomeric form of the core protein was detected in fractions 1 and 2 at the top of the gradient. This profile is due to the disruption of core protein complexes by SDS, as observed by Ai and collaborators (1) (Fig. 8B). Although the core protein of the mutants also showed a peak of sedimentation in fractions 6 to 8, some slight differences could be observed (Fig. 8A). In the case of R50, a sharp peak was observed in fraction 7 only, whereas the core protein peaked more toward the bottom of the gradient for the K51 and R59 mutants. Despite these differences, it appeared that the core proteins of all noninfectious mutants were able to assemble into nucleocapsid-like structures, just like the wt core protein (Fig. 8A). These results therefore suggest that the mutations do not affect core protein multimerization. However, the differences observed in the sedimentation profiles of the mutants suggest that there might be differences in the compactness of the nucleocapsid-like structures of the mutants.
FIG. 8.
Characterization of mutant core protein oligomerization by velocity sedimentation. (A) Cells electroporated with mutant and wt RNA genomes were lysed at 72 h postelectroporation as described in Materials and Methods. Lysates were subjected to 10 to 60% sucrose density gradient centrifugation, followed by Western blot analysis of core protein. (B) Sucrose density gradient (10 to 60%) centrifugation from cells electroporated with JFH1/ΔE1E2 genome (upper panel) and lysate of wt electroporated cells pretreated with 1% SDS (lower panel), followed by Western blot analysis of core protein. The input represents 10% of lysates. (C) Nucleocapsids were isolated as described in Materials and Methods, and 10% was resolved on 12.5% SDS-PAGE followed by Western blot analysis of core protein.
To determine whether the viral genome could be encapsidated by the mutants, we quantified the viral RNA associated with the isolated nucleocapsid by quantitative RT-PCR. For this, nucleocapsids were isolated on a cushion of 30% sucrose, as reported for virus-like particles (4) and hepatitis B virus (HBV) nucleocapsid (5, 28). Ten percent of the resulting pellets were analyzed by Western blotting, and the rest were divided into two parts. One part remained untreated, whereas the other part was treated by RNase A and DNase to eliminate any nucleic acids and RNA that were not incorporated and protected by the nucleocapsid. As shown in Fig. 8C, core protein was detectable in the pellet for all mutants, indicating the presence of core protein. As a control, no detection of core was observed with a core deletion mutant, Δcore (62-160), and wt lysate pretreated with 1% SDS. As shown in Table 1, a 182-fold decrease was observed for wt RNA after treatment with RNase, whereas it was 16,129-fold lower when the lysate was treated with 1% SDS before ultracentrifugation and RNase treatment. These results indicate that the difference in RNA detection between treated and untreated samples was due to RNA protection by an SDS-sensitive factor, most likely the capsid. As shown in Table 1, all core mutants were able to protect viral RNA from RNase treatment as efficiently as the wt core protein. Altogether, these data suggest that core mutations do not inhibit core protein-RNA interaction and RNA encapsidation.
TABLE 1.
RNA encapsidation and protection by the nucleocapsidsa
| Mutant | Mean no. of viral RNA copies/5 μl of isolated RNA |
Relative RNA quantification |
||
|---|---|---|---|---|
| Untreated | Treated | No. of copies in untreated NC/no. in treated NC | No. of copies in treated NC/no. in untreated NC (% of wt) | |
| wt | 8.94 × 107 | 4.92 × 105 | 182 | 5.5 × 10−3 (100) |
| wt-SDSb | 6.87 × 107 | 4.26 × 103 | 16,129 | 6.2 × 10−5 (1) |
| R50 mutant | 2.52 × 107 | 1.32 × 105 | 192 | 5.22 × 10−3 (95) |
| K51 mutant | 3.47 × 107 | 1.66 × 105 | 209 | 4.78 × 10−3 (87) |
| R59 mutant | 3.25 × 107 | 3.45 × 105 | 94 | 1.06 × 10−2 (193) |
| R62 mutant | 7.25 × 107 | 4.33 × 105 | 167 | 5.98 × 10−3 (109) |
| JFH1/ΔE1E2 | 5.38 × 107 | 1.36 × 105 | 396 | 2.52 × 10−3 (46) |
Core protein-associated RNAs were quantified by qRT-PCR, and the ratios in untreated NC/NC treated with RNase A and DNase and in treated NC/untreated NC were calculated. Ratios of values in treated/untreated NC were compared to the wt ratio, and results are expressed as a percentage of the wt, which was set to 100%. This method enabled us to minimize the differences linked to electroporation efficiency and manipulation. Means of the results from duplicate electroporations are shown.
SDS, lysate pretreated with 1% SDS.
Assessment of core protein envelopment by membrane protection assay.
Recently, Ai and coworkers have characterized the HCV core protein interaction with intracellular membranes and core protein envelopment (1). In this study core protein envelopment seems to play a role in the early phase of HCV maturation and morphogenesis (1). To further understand the mechanism by which the mutated basic amino acid residues of the second cluster (R50, K51, R59, and R62) inhibited HCV infectivity, we characterized their effect on core protein envelopment by a membrane protection assay (1). As shown in Fig. 9, comparable levels of core protein from wt and mutants were still resistant to proteolysis by proteinase K in the absence of detergent, which decreased dramatically when postnuclear fractions were treated with detergent and proteinase K. Together, our results suggest that mutating amino acids R50, K51, R59, and R62 to alanines does not interfere with core protein envelopment by intracellular membranes.
FIG. 9.
Assessment of core protein envelopment by membrane protection assay. Electroporated cells were harvested at 72 h postelectroporation, and a membrane protection assay was performed as described in Materials and Methods. Postnuclear fractions were treated with 10 μg/ml proteinase K (PK) in the presence or absence of Triton X-100 (Tx-100) on ice for 1 h (top panel). The density of bands corresponding to the core proteins in each treatment was scanned and quantified. The percentages of core protein levels detected in samples treated with PK alone or with Triton X-100 and PK relative to the untreated sample were determined. A representative graph of experiments from duplicate electroporations with means and standard errors of the means is shown (bottom panel).
DISCUSSION
In the present study we analyzed a series of basic residues at the N terminus of the HCV core protein to determine their potential involvement in HCV infectivity in the context of the HCVcc system (55). Importantly, we identified four basic amino acid residues, R50, K51, R59, and R62, which are essential for the assembly of infectious viral particles. These residues are highly conserved in the HCV core protein (Fig. 1).
As reported previously (9), the N terminus of HCV core protein contains two highly conserved clusters of basic amino acid residues. These two clusters (aa 6 to 23 and 39 to 62) are separated by a neutral linker region. Moreover, the RNA sequence coding the first 62 residues of the core protein has been reported to contain highly conserved structures in the form of two stem-loops: SLV and SLVI (51). These structures are important for RNA translation/replication (35, 54). Among the modifications introduced into the first basic cluster of the core protein, only the R17R18K23 mutant was concerned with the SLV structure. Nucleotide substitutions introduced into SLV decreased replication by about 3-fold and reduced infectious titer by about 5-fold. Indeed, disruption of SLV has been previously reported to reduce viral replication and infectious titer of HCV (54), which can result from disruption of RNA-RNA or RNA-protein interactions.
In the first cluster of basic residues, mutants K6R9K10 and K12R13 did not disrupt SLV structure. The K6R9K10 mutant decreased viral replication by about 12-fold and the infectivity titer by about 7-fold, whereas the K12R13 mutant had replication and infectivity levels comparable to those of the wt. The data obtained from these three mutants suggest that the reduced infectivity could be due to the decreased levels of translation/replication. Moreover, a larger modification of basic residues as generated in the RK/8A mutant leads to a strict translation inhibition and to an unviable mutant. While most IRES sequences require only the 5′ NTR, the HCV IRES seems to be dependent on a sequence downstream of the initiating AUG codon (45, 56). Furthermore, a cellular RNA-binding protein has been identified that modulates HCV translation by interacting with the core protein-coding sequence. Indeed, modification of the NSAP1-binding site, which is composed of an adenosine-rich region, has been shown to reduce the HCV IRES activity in an in vitro translation assay (24). Replacing the basic residues with alanines led to modifications of the first two nucleotides of each codon in the RK/8A mutant, which changed several adenosines to cytosine and guanine. This probably prevents NSAP1 interaction with RNA and consequently reduces its translation. In cellulo, the lower viral replication and production result consequently from a lower level of translation. Altogether, our data suggest that no specific residues or motifs affecting virus infectivity are present in the first basic cluster of core protein.
Some mutations of basic amino acid residues of the second cluster extend their nucleotide modifications to the SLVI RNA structure. The R39R40R43 and R47R50K51 mutants, which disrupt the upper and middle parts of SLVI, led to a decrease in replication by about 6- to 8-fold compared to the wt. R55R59 and R61R62 mutants had replication levels comparable to the level of the wt although R55 disrupted the lower part of SLVI. This is in agreement with previous reports (35). However, despite stable core protein expression, these mutants produced neither intracellular nor extracellular infectious particles. Among the single mutants, only the R39 and R47 mutants decreased replication levels by about 6- to 8-fold, and these levels are comparable to those of the R39R40R43 and R47R50K51 mutants, respectively. Interestingly, in the predicted structure of SLVI, these modifications affect pairing bases at the same positions but in opposite strands (Fig. 1). Altogether, our data indicate that only modifications in this part of SLVI affect RNA replication. The remaining mutants had replication levels comparable to the level of the wt.
Analyses of single mutants generated with mutations in the second cluster revealed that 6 out of 10 mutants (R39, R40, R43, R47, R55, and R61) were infectious with some attenuation. In contrast, the infectivity of the R50, K51, R59, and R62 mutants was abolished. No infectious viral particles were detected intracellularly or extracellularly. Our data suggest that the absence of infectious viral particles is not due to the instability of the core protein since this protein was detected at 72 h postelectroporation. In a study by Murray and coworkers, alanine scanning mutagenesis of HCV core protein revealed numerous residues essential for the production of infectious virus (40). Mutants of J6/JFH1 with quadruple alanine substitutions in the core protein (C57/60A and C61/64A) could not be rescued by compensatory mutations located in the viral genome (40). These mutants contained the basic residues R59 and R62, which were characterized in our study as crucial for HCV infectivity. In addition, as reported previously, amino acid insertions between aa 23 to 107 of the core protein were lethal (2). Furthermore, these mutations did not disrupt the subcellular colocalization of the core protein with the LDs and NS5A protein, which have been reported to be essential for virus production (7, 32, 36). Moreover, viral infectivity of a deleted core mutant, Δcore (62-160) could not be rescued by trans-complementation with any of the four mutant core proteins. In contrast, the wt core protein could rescue these mutants, indicating that the effect is due only to amino acid changes and not to a change of nucleotides, which would be involved in RNA encapsidation.
Compared to the wt, analysis by sucrose gradient ultracentrifugation showed that all mutants were able to assemble into intermediate multimeric complexes, which might correspond to nucleocapsid-like structures. Previously reported data suggest that a deletion of 10 basic residues in cluster 2 reduces the ability of the capsid to assemble efficiently (25). Furthermore, when the number of mutated basic residues was increased, a progressive decrease in the ability of core protein assembly was observed (25). In our study, individual core mutations did not interfere with core assembly and multimerization. Moreover, all mutant nucleocapsids were able to protect the viral genome from the RNase treatment to a similar extent as the wt nucleocapsid, suggesting that core-RNA interactions were not altered. Therefore, this suggests that these mutations inhibit viral assembly at a stage downstream of the oligomerization of the core protein and the encapsidation of RNA. A recent report (1) reveals that a cascade of core protein-membrane interactions is important in HCV morphogenesis. A deletion of aa 42 to 68 of the core protein, which contains the four residues identified in our study, decreased its envelopment by cellular membranes (1). In our study, when core protein envelopment was assessed by a membrane protection assay, mutated core proteins were protected by membranes to a similar extent as the wt core protein, suggesting that these residues do not alter core protein envelopment by intracellular membranes.
In conclusion, a comprehensive mutagenesis study of basic amino acid residues located at the N terminus of HCV core protein allowed the identification of four amino acids, R50, K51, R59 and R62, that are essential for HCV morphogenesis. None of these amino acids affects core protein colocalization with NS5A and LDs, RNA encapsidation, core protein oligomerization, or envelopment by intracellular membranes. Understanding the detailed mechanism by which these mutations can modulate infectious virus production will help broaden our knowledge of viral particle assembly and further the identification of new potential therapeutic targets.
Acknowledgments
We thank Laurence Cocquerel and Philip Meuleman for critically reading the manuscript and Sophana Ung for the preparation of the figures. Some data were generated with the Imaging Core Facility of the Calmette campus (MICPaL). We thank Steve S. L. Chen (Graduate Institute of Life Sciences, Taipei, Taiwan) for his advice in the membrane protection assay.
This work was supported in part by a grant from the Agence Nationale de Recherche sur le SIDA et les hépatites virales (ANRS) to Y.R. P.-Y.D. was supported by a predoctoral fellowship from MRT. K.A. was supported by a Syrian predoctoral fellowship. J.D. is an international scholar of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 13 October 2010.
REFERENCES
- 1.Ai, L. S., Y. W. Lee, and S. S. Chen. 2009. Characterization of hepatitis C virus core protein multimerization and membrane envelopment: revelation of a cascade of core-membrane interactions. J. Virol. 83:9923-9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arumugaswami, V., R. Remenyi, V. Kanagavel, E. Y. Sue, T. Ngoc Ho, C. Liu, V. Fontanes, A. Dasgupta, and R. Sun. 2008. High-resolution functional profiling of hepatitis C virus genome. PLoS Pathog. 4:e1000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barba, G., F. Harper, T. Harada, M. Kohara, S. Goulinet, Y. Matsuura, G. Eder, Z. Schaff, M. J. Chapman, T. Miyamura, and C. Brechot. 1997. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl. Acad. Sci. U. S. A. 94:1200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumert, T. F., S. Ito, D. T. Wong, and T. J. Liang. 1998. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 72:3827-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumert, T. F., S. A. Rogers, K. Hasegawa, and T. J. Liang. 1996. Two core promotor mutations identified in a hepatitis B virus strain associated with fulminant hepatitis result in enhanced viral replication. J. Clin. Invest. 98:2268-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulant, S., R. Montserret, R. G. Hope, M. Ratinier, P. Targett-Adams, J. P. Lavergne, F. Penin, and J. McLauchlan. 2006. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 281:22236-22247. [DOI] [PubMed] [Google Scholar]
- 7.Boulant, S., P. Targett-Adams, and J. McLauchlan. 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 88:2204-22013. [DOI] [PubMed] [Google Scholar]
- 8.Boulant, S., C. Vanbelle, C. Ebel, F. Penin, and J. P. Lavergne. 2005. Hepatitis C virus core protein is a dimeric alpha-helical protein exhibiting membrane protein features. J. Virol. 79:11353-11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukh, J., R. H. Purcell, and R. H. Miller. 1994. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc. Natl. Acad. Sci. U. S. A. 91:8239-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castelain, S., V. Descamps, V. Thibault, C. Francois, D. Bonte, V. Morel, J. Izopet, D. Capron, P. Zawadzki, and G. Duverlie. 2004. TaqMan amplification system with an internal positive control for HCV RNA quantitation. J. Clin. Virol. 31:227-234. [DOI] [PubMed] [Google Scholar]
- 11.Delgrange, D., A. Pillez, S. Castelain, L. Cocquerel, Y. Rouille, J. Dubuisson, T. Wakita, G. Duverlie, and C. Wychowski. 2007. Robust production of infectious viral particles in Huh-7 cells by introducing mutations in hepatitis C virus structural proteins. J. Gen. Virol. 88:2495-2503. [DOI] [PubMed] [Google Scholar]
- 12.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan, Z., Q. R. Yang, J. S. Twu, and A. H. Sherker. 1999. Specific in vitro association between the hepatitis C viral genome and core protein. J. Med. Virol. 59:131-134. [PubMed] [Google Scholar]
- 14.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fromentin, R., N. Majeau, M. E. Laliberte Gagne, A. Boivin, J. B. Duvignaud, and D. Leclerc. 2007. A method for in vitro assembly of hepatitis C virus core protein and for screening of inhibitors. Anal. Biochem. 366:37-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gastaminza, P., S. B. Kapadia, and F. V. Chisari. 2006. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 80:11074-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goueslain, L., K. Alsaleh, P. Horellou, P. Roingeard, V. Descamps, G. Duverlie, Y. Ciczora, C. Wychowski, J. Dubuisson, and Y. Rouille. 2010. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J. Virol. 84:773-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffin, S. D., L. P. Beales, D. S. Clarke, O. Worsfold, S. D. Evans, J. Jaeger, M. P. Harris, and D. J. Rowlands. 2003. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett. 535:34-38. [DOI] [PubMed] [Google Scholar]
- 20.Hijikata, M., N. Kato, Y. Ootsuyama, M. Nakagawa, and K. Shimotohno. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. U. S. A. 88:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houghton, M. 1996. Hepatitis C viruses, p. 1035-1058. In D. M. Knipe, B. Roizman, P. M. Howley, T. P. Monath, S. E. Straus, R. M. Chanock, J. L. Melnick, and B. N. Fields (ed.), Fields virology, 3rd ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 22.Hussy, P., H. Langen, J. Mous, and H. Jacobsen. 1996. Hepatitis C virus core protein: carboxy-terminal boundaries of two processed species suggest cleavage by a signal peptide peptidase. Virology 224:93-104. [DOI] [PubMed] [Google Scholar]
- 23.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-18017. [DOI] [PubMed] [Google Scholar]
- 24.Kim, J. H., K. Y. Paek, S. H. Ha, S. Cho, K. Choi, C. S. Kim, S. H. Ryu, and S. K. Jang. 2004. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol. Cell. Biol. 24:7878-7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein, K. C., S. R. Dellos, and J. R. Lingappa. 2005. Identification of residues in the hepatitis C virus core protein that are critical for capsid assembly in a cell-free system. J. Virol. 79:6814-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein, K. C., S. J. Polyak, and J. R. Lingappa. 2004. Unique features of hepatitis C virus capsid formation revealed by de novo cell-free assembly. J. Virol. 78:9257-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopp, M., C. L. Murray, C. T. Jones, and C. M. Rice. 2010. Genetic analysis of the carboxy-terminal region of the hepatitis C virus core protein. J. Virol. 84:1666-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koschel, M., D. Oed, T. Gerelsaikhan, R. Thomssen, and V. Bruss. 2000. Hepatitis B virus core gene mutations which block nucleocapsid envelopment. J. Virol. 74:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 30.Major, M. E., B. Rehermann, and S. M. Feinstone. 2001. Hepatitis C viruses, p. 1127-1161. In B. N. Fields, D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 31.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 32.Masaki, T., R. Suzuki, K. Murakami, H. Aizaki, K. Ishii, A. Murayama, T. Date, Y. Matsuura, T. Miyamura, T. Wakita, and T. Suzuki. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964-7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto, M., S. B. Hwang, K. S. Jeng, N. Zhu, and M. M. Lai. 1996. Homotypic interaction and multimerization of hepatitis C virus core protein. Virology 218:43-51. [DOI] [PubMed] [Google Scholar]
- 34.McLauchlan, J., M. K. Lemberg, G. Hope, and B. Martoglio. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMullan, L. K., A. Grakoui, M. J. Evans, K. Mihalik, M. Puig, A. D. Branch, S. M. Feinstone, and C. M. Rice. 2007. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc. Natl. Acad. Sci. U. S. A. 104:2879-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089-1097. [DOI] [PubMed] [Google Scholar]
- 37.Moradpour, D., C. Englert, T. Wakita, and J. R. Wands. 1996. Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology 222:51-63. [DOI] [PubMed] [Google Scholar]
- 38.Moradpour, D., M. J. Evans, R. Gosert, Z. Yuan, H. E. Blum, S. P. Goff, B. D. Lindenbach, and C. M. Rice. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morota, K., R. Fujinami, H. Kinukawa, T. Machida, K. Ohno, H. Saegusa, and K. Takeda. 2009. A new sensitive and automated chemiluminescent microparticle immunoassay for quantitative determination of hepatitis C virus core antigen. J. Virol. Methods 157:8-14. [DOI] [PubMed] [Google Scholar]
- 40.Murray, C. L., C. T. Jones, J. Tassello, and C. M. Rice. 2007. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J. Virol. 81:10220-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogino, T., H. Fukuda, S. Imajoh-Ohmi, M. Kohara, and A. Nomoto. 2004. Membrane binding properties and terminal residues of the mature hepatitis C virus capsid protein in insect cells. J. Virol. 78:11766-11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pavlovic, D., D. C. Neville, O. Argaud, B. Blumberg, R. A. Dwek, W. B. Fischer, and N. Zitzmann. 2003. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. U. S. A. 100:6104-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poynard, T., M. F. Yuen, V. Ratziu, and C. L. Lai. 2003. Viral hepatitis C. Lancet 362:2095-2100. [DOI] [PubMed] [Google Scholar]
- 44.Ray, R. B., and R. Ray. 2001. Hepatitis C virus core protein: intriguing properties and functional relevance. FEMS Microbiol. Lett. 202:149-156. [DOI] [PubMed] [Google Scholar]
- 45.Reynolds, J. E., A. Kaminski, H. J. Kettinen, K. Grace, B. E. Clarke, A. R. Carroll, D. J. Rowlands, and R. J. Jackson. 1995. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 14:6010-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rocha-Perugini, V., M. Lavie, D. Delgrange, J. Canton, A. Pillez, J. Potel, C. Lecoeur, E. Rubinstein, J. Dubuisson, C. Wychowski, and L. Cocquerel. 2009. The association of CD81 with tetraspanin-enriched microdomains is not essential for Hepatitis C virus entry. BMC Microbiol. 9:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rouille, Y., F. Helle, D. Delgrange, P. Roingeard, C. Voisset, E. Blanchard, S. Belouzard, J. McKeating, A. H. Patel, G. Maertens, T. Wakita, C. Wychowski, and J. Dubuisson. 2006. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J. Virol. 80:2832-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russell, R. S., J. C. Meunier, S. Takikawa, K. Faulk, R. E. Engle, J. Bukh, R. H. Purcell, and S. U. Emerson. 2008. Advantages of a single-cycle production assay to study cell culture-adaptive mutations of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 105:4370-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santolini, E., G. Migliaccio, and N. La Monica. 1994. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol. 68:3631-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimoike, T., S. Mimori, H. Tani, Y. Matsuura, and T. Miyamura. 1999. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J. Virol. 73:9718-9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, D. B., and P. Simmonds. 1997. Characteristics of nucleotide substitution in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J. Mol. Evol. 45:238-246. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka, Y., T. Shimoike, K. Ishii, R. Suzuki, T. Suzuki, H. Ushijima, Y. Matsuura, and T. Miyamura. 2000. Selective binding of hepatitis C virus core protein to synthetic oligonucleotides corresponding to the 5′ untranslated region of the viral genome. Virology 270:229-236. [DOI] [PubMed] [Google Scholar]
- 53.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vassilaki, N., P. Friebe, P. Meuleman, S. Kallis, A. Kaul, G. Paranhos-Baccala, G. Leroux-Roels, P. Mavromara, and R. Bartenschlager. 2008. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J. Virol. 82:11503-11515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, T. H., R. C. Rijnbrand, and S. M. Lemon. 2000. Core protein-coding sequence, but not core protein, modulates the efficiency of cap-independent translation directed by the internal ribosome entry site of hepatitis C virus. J. Virol. 74:11347-11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu, K. L., S. I. Jang, and J. C. You. 2009. Identification of in vivo interaction between Hepatitis C Virus core protein and 5′ and 3′ UTR RNA. Virus Res. 145:285-292. [DOI] [PubMed] [Google Scholar]
- 58.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]