Abstract
Positive-strand RNA [(+)RNA] viruses invariably replicate their RNA genomes on modified intracellular membranes. In infected Drosophila cells, Flock House nodavirus (FHV) RNA replication complexes form on outer mitochondrial membranes inside ∼50-nm, virus-induced spherular invaginations similar to RNA replication-linked spherules induced by many (+)RNA viruses at various membranes. To better understand replication complex assembly, we studied the mechanisms of FHV spherule formation. FHV has two genomic RNAs; RNA1 encodes multifunctional RNA replication protein A and RNA interference suppressor protein B2, while RNA2 encodes the capsid proteins. Expressing genomic RNA1 without RNA2 induced mitochondrial spherules indistinguishable from those in FHV infection. RNA1 mutation showed that protein B2 was dispensable and that protein A was the only FHV protein required for spherule formation. However, expressing protein A alone only “zippered” together the surfaces of adjacent mitochondria, without inducing spherules. Thus, protein A is necessary but not sufficient for spherule formation. Coexpressing protein A plus a replication-competent FHV RNA template induced RNA replication in trans and membrane spherules. Moreover, spherules were not formed when replicatable FHV RNA templates were expressed with protein A bearing a single, polymerase-inactivating amino acid change or when wild-type protein A was expressed with a nonreplicatable FHV RNA template. Thus, unlike many (+)RNA viruses, the membrane-bounded compartments in which FHV RNA replication occurs are not induced solely by viral protein(s) but require viral RNA synthesis. In addition to replication complex assembly, the results have implications for nodavirus interaction with cell RNA silencing pathways and other aspects of virus control.
Eukaryotic positive-strand RNA [(+)RNA] virus genome replication universally occurs on rearranged host intracellular membranes (1, 37, 49). Membrane rearrangements used by different viruses include, but are not limited to, membranous webs of vesicles (24, 56), double-membrane vesicles (41), and double-membrane layers (52). Among the most common virus-induced membrane rearrangements are 50- to 80-nm membrane invaginations or spherules which are associated with RNA replication by alphaviruses, bromoviruses, nodaviruses, flaviviruses, tymoviruses, tombusviruses, and other viruses (23, 35, 44, 48, 51, 62).
Such replication-associated membrane rearrangements are often induced by one or a few viral nonstructural proteins. The membranous web formed by hepatitis C virus (HCV) is induced by HCV protein NS4B (19). Double-membrane vesicles formed by the equine arterivirus are induced by the viral nsp2 and nsp3 proteins (55). Endoplasmic reticulum (ER) spherules formed by brome mosaic virus (BMV) are induced by BMV RNA replication protein 1a (51).
To better understand the mechanisms of (+)RNA virus replication complex formation, including membrane rearrangement, we examined Flock House virus (FHV) spherule formation. FHV belongs to the family Nodaviridae and the genus Alphanodavirus, whose members naturally infect insects (9). FHV encapsidates a bipartite, single-stranded, positive-sense RNA genome whose RNAs are capped but not polyadenylated (Fig. 1) (9). RNA1 (3.1 kb) encodes multifunctional viral replicase protein A (112 kDa), the only FHV protein required for RNA replication (8). During replication, RNA1 also produces subgenomic RNA3 (387 nucleotides [nt]), which encodes the RNA interference (RNAi) suppressor protein B2 (12 kDa) (22, 26, 31). Genomic RNA2 (1.4 kb) encodes the capsid precursor protein α (43 kDa) (21).
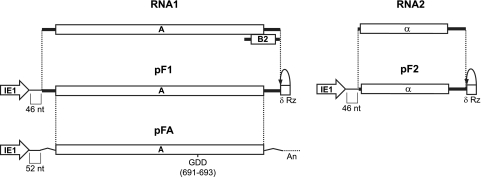
FIG. 1.
Schematic of FHV genome components and the corresponding expression cassettes used in Drosophila expression plasmids. Each FHV component is expressed by a baculovirus IE1 promoter in a plasmid that also contains the baculovirus transactivating hr5 enhancer. Plasmid pF1 expresses RNA1, and pF2 expresses RNA2 with a δRz-mediated authentic 3′ end (δRz). pF1 and pF2 contain 46 nonviral nt between the IE1 transcription start site and the first viral nucleotide. pFA encodes protein A mRNA with nonviral 5′ and 3′ untranslated sequences and a 3′ polyadenylation signal (An). pFA contains 52 nonviral nt between the IE1 transcription start site and the initiating methionine codon.
FHV infection induces the formation of ∼50-nm-diameter membranous vesicles, or spherules, between the mitochondrial outer and inner membranes (35). Three-dimensional electron tomographic imaging shows all such spherules to be invaginations of the outer mitochondrial membrane, with interiors connected to the cytoplasm through ∼10-nm-diameter open necks (28). We previously showed that protein A and FHV RNA synthesis localize to the interiors of these spherules, which thus represent the FHV RNA replication complex (28).
In addition to providing RNA-dependent RNA polymerase and likely capping functions for RNA synthesis (8, 9, 21, 27), protein A has major roles in replication complex assembly. Protein A localizes to mitochondrial outer membranes through an N-terminal mitochondrial targeting and transmembrane sequence (34) and, in a step separable from RNA synthesis, recruits FHV RNA templates to mitochondria (59, 60). Protein A also self-interacts in vivo through multiple domains in ways important for RNA replication (16). Consistent with this, immunogold localization of protein A, biochemical results, and other data show that transmembrane, self-interacting protein A is present at ∼100 molecules per spherule, implying that protein A forms a dense shell-like structure lining the interior spherule membranes (28).
In this study, we examined the requirements for forming the FHV spherule RNA replication compartments. Although protein B2 can interact with protein A (5), we found that B2 and FHV capsid proteins were dispensable for spherule formation. Protein A was required for spherule formation but, unlike the case for many (+)RNA viruses for which one or more nonstructural proteins suffice for replication-associated membrane rearrangements, was insufficient. The results show that spherule formation requires a replication-competent RNA template, occurs downstream of RNA template recruitment, and depends on protein A polymerase activity. Thus, in contrast to many other (+)RNA viruses, the membrane rearrangements associated with FHV RNA replication are tightly linked to viral RNA synthesis. These findings have mechanistic implications for RNA replication, RNA silencing, and virus control.
MATERIALS AND METHODS
Plasmids.
Standard molecular cloning techniques were used (50), and all plasmids and mutations thereof were verified by nucleotide sequencing. All FHV Drosophila expression plasmids were cloned into plasmid pIE1hr/PA, which contains the baculovirus immediate-early 1 (IE1) promoter, the baculovirus transactivating hr5 enhancer, and a polyadenylation signal (11). Plasmids pF1fs, pFA, and pFAD692E were described previously (60). pF1, pF2, and pF3 were made by amplifying RNA1, RNA2, and RNA3, respectively, and a 3′ flanking antigenomic hepatitis delta virus ribozyme (δRz) from a yeast RNA1 or RNA2 expression plasmid (courtesy of D. Miller) and ligating the PstI- and HindIII-digested product into common sites in pIE1hr/PA. p(−15)F1fs was made by amplifying RNA1 with a primer that annealed to nt 16 of RNA1 and a 3′-flanking δRz from pF1fs and ligating the PstI- and HindIII-digested product into common sites in pIE1hr/PA, resulting in a deletion of the first 15 nt of the RNA1 5′ untranslated region (UTR). pF1-3′UTR was made by amplifying δRz using a primer that contained 33 nt of the C-terminal coding region of protein A, including the stop codon sequence and the first 18 nt of δRz and a flanking primer and ligating the BlpI- and HindIII-digested product into common sites in pF1fs. Plasmid pGFP is pEGFP from reference 38. Plasmids used to generate double-stranded RNA (dsRNA) for ago2-specific RNA silencing were created through insertion of part of the open reading frame (ORF; nt 2807 to 3953) into pBS/KS+ (Stratagene). The plasmids used to in vitro transcribe FHV mRNA1 and mRNA2 for generation of FHV clonal stocks have been described previously (7). The nucleotide substitutions in RNA1 to make the protein B2 knockout have been described previously (8, 31).
Cells and plasmid transfection.
Drosophila melanogaster S2 cells were grown at 28°C in express five serum-free medium supplemented with 17 mM l-glutamine (Gibco, Carlsbad, CA). D. melanogaster DL-1 cells were grown at 27°C in Schneider's medium supplemented with 10% fetal bovine serum and 17 mM l-glutamine (Gibco, Carlsbad, CA). DNA plasmids were transfected into cells via a Nucleofector apparatus (Lonza) following the manufacturer-recommended protocols using kit V and the G-030 pulsing parameter. All subsequent assays were performed at 48 h posttransfection.
RNA silencing.
The generation of dsRNA and transfection methods were performed as previously described (30). Cells were transfected with ago2-specific dsRNA in suspension and maintained in monolayer for 1 day at 27°C prior to use.
RNA transfections, FHV generation, and infection.
Full-length FHV RNA1 (wild type [WT] RNA1 or RNA1 with B2 amino acid substitutions) and WT RNA2 were in vitro transcribed from linearized plasmids and capped (AmpliCap-Max T7 High Yield Message Maker kit; Epicentre). ago2-silenced DL-1 cells (2 × 106/ml) were cotransfected with RNA1 and WT RNA2 (200 ng each) that were mixed with 20 μl 1,2-dioleoyl-3-trimethylammonium-propane-dioleyl-phosphatidylethanolamine in serum-free TC-100 growth medium (Invitrogen). After a 4-h incubation at room temperature, the cells were plated and the transfection mixture was replaced with supplemented Schneider's medium. After 2 days of incubation at 27°C, the cells and accompanying growth medium were collected, lysed by freezing-thawing, and clarified by centrifugation (10,000 × g). Fresh ago2-silenced DL-1 cells (2.8 × 107) were incubated with the clarified lysates in suspension for 1 h. The FHV-infected ago2-silenced DL-1 cells were diluted with supplemented Schneider's medium, plated, and incubated at 27°C for 2 days. The cells and accompanying growth medium were collected, treated on ice with 0.5% (vol/vol) Nonidet P-40-0.1% β-mercaptoethanol, and clarified by centrifugation (10,000 × g). FHV was purified by sucrose gradient purification, and infectious FHV titers were determined by plaque assay on ago2-silenced or normal DL-1 cells (53) that were overlaid with 0.6% SeaKem ME agarose in Schneider's medium. Plaques were visualized with 3 mg MTT (Thiazolyl Blue Tetrazolium Bromide; Sigma) per ml of phosphate-buffered saline, pH 6.2. Pass 2 virus was generated in ago2-silenced DL-1 cells at a multiplicity of infection (MOI) of 5. An equal MOI determined on ago2-silenced DL-1 cells was used for subsequent infections. D. melanogaster cells were infected with WT FHV as described previously (28).
RNA extraction and Northern blot analysis.
Total RNA from D. melanogaster cells was isolated and prepared by phenol and chloroform extraction (50) using acidic phenol. Northern blotting was performed as described previously (60), by loading 1 μg RNA per lane. Probes against positive- and negative-strand RNA1, RNA2, and RNA3 have been described previously (43).
Western blotting.
Protein samples were solubilized in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer, separated on Criterion 4 to 15% gradient Tris-HCl acrylamide gels (Bio-Rad) for protein A or 4 to 20% gels for protein B2, transferred to polyvinylidene difluoride membranes, and blotted with antibodies as described previously (35). Prior to probing for actin, blots were stripped with Blot Restore from Millipore (Temecula, CA). Chemiluminescence was detected with a Bio-Rad ChemiDoc XRS.
Antibodies.
A rabbit polyclonal antibody against FHV protein A was described previously (35). A rabbit polyclonal antibody against FHV protein B2 was produced by the expression of a His6/T7-tagged protein in Escherichia coli from pET28 with partial purification over a Co-agarose column. The polyclonal B2 antibody was raised in rabbits by Harlan Bioproducts (Madison, WI). Mouse monoclonal antibody to beta actin was purchased from Abcam (Cambridge, MA). Alkaline phosphatase-conjugated secondary antibodies were purchased from Bio-Rad (Hercules, CA).
EM.
Electron microscopy (EM) was performed as described in reference 35. Briefly, Drosophila cells were fixed with 4% paraformaldehyde and 2% glutaraldehyde in 0.1 M phosphate buffer, postfixed in 1% osmium tetroxide with 1% potassium ferricyanide, stained with 1% uranyl acetate, dehydrated in an ethanol series, and embedded in Spurr's resin. Ultrathin 70-nm sections of the samples were cut, placed on copper grids, and then stained with 8% methanolic uranyl acetate, followed by Reynold's lead citrate.
RESULTS
FHV protein A induces mitochondrial clustering and zippering.
Prior fluorescence microscopy and immunogold EM showed that protein A and FHV RNA synthesis localize to the interior of mitochondrial spherules in FHV-infected cells (28) and that, prior to RNA synthesis, protein A recruits FHV RNA templates to mitochondrial membranes (59). To test whether FHV protein A expression alone is sufficient for spherule formation, we transfected Drosophila S2 cells with previously described protein A expression plasmid pFA (60). pFA expresses WT protein A, but the resulting RNA1-derived transcript is not replicated because the 5′ and 3′ FHV RNA1 UTRs, which include sequences required for RNA replication (8), have been replaced with nonviral sequences (Fig. 1). Total protein analysis by Western blotting with antibodies against protein A showed that pFA directed protein A accumulation to levels similar to those in FHV-infected cells (Fig. 2G). Moreover, micrographs of cells transfected in parallel with a green fluorescent protein (GFP)-expressing plasmid showed that the transfection procedure used to introduce pFA had no noticeable effect on mitochondrial membrane morphology (Fig. 2A). However, in contrast to abundant spherules in cells infected by FHV (Fig. 2B and C) or transfected with the full FHV genome (next section), pFA-transfected cells expressing protein A alone showed no mitochondrial spherule formation, even after examining 2,527 mitochondria in 133 individual cells by EM in three independent experiments (representative results are shown in Fig. 2D and E).
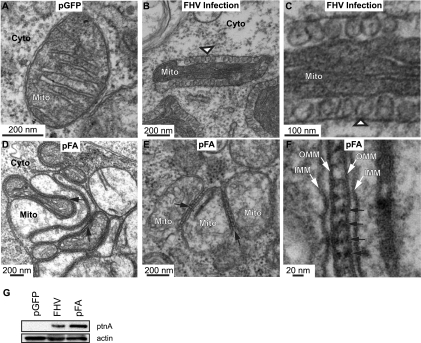
FIG. 2.
Expression of protein A causes mitochondrial zippering. (A) Electron micrograph of a cell expressing GFP showing no apparent alterations in mitochondrial morphology. (B) Electron micrograph of a cell infected with WT FHV at 14 hpi showing formation of mitochondrial spherules (arrowhead) that represent the FHV RNA replication complex. (C) Higher-magnification view of spherules in panel B. (D and E) Electron micrograph of cells expressing pFA, showing mitochondrial aggregation and zippering (arrows) but no spherules. (F) Higher-magnification view of zippered mitochondria, showing structure between mitochondria that may be responsible for zippering effect (arrows). Mitochondria (Mito), cytoplasm (Cyto), inner mitochondrial membrane (IMM), and outer mitochondrial membrane (OMM) are indicated. (G) Total protein was analyzed by Western blotting with antibodies against protein A (ptnA; top) or actin (bottom).
While no spherules formed, transfection with protein A-expressing plasmid pFA caused mitochondria to cluster tightly with outer membranes “zippered” together in 70% of cell sections showing mitochondria (Fig. 2D to F, arrows). While a degree of mitochondrial clustering and localized appression of mitochondrial membranes has been observed previously for FHV infection (35), such extended tight zippering of the outer mitochondrial membranes has not. The space between the zippered outer mitochondrial membranes was ∼20 nm, and the intervening space contained some regular structure with a periodicity of ∼20 nm (Fig. 2E and F). This membrane zippering may result from the fact that protein A is an integral outer mitochondrial membrane protein that self-interacts (16). As considered further in the Discussion, similar closely appressed membranes are induced by certain membrane-associated RNA replication proteins of other (+)RNA viruses and by many self-interacting host membrane proteins. FHV infection causes progressive degradation in organization of the mitochondrial matrix, with early time points showing a nearly normal, condensed matrix with well-defined cristae (Fig. 2B and C) and later time points showing disintegration of the matrix (35). In general, crista morphology is variable and highly dependent on the pathophysiological state of the cell (25), and throughout this work we observed some variations in crista morphology even within single cells. Nevertheless, on average, the mitochondrial matrix was less electron dense with fewer cristae in cells transfected with the protein A expression plasmid (Fig. 2D to F) than in GFP-transfected or even FHV-infected cells (Fig. 2A to C). This lower matrix electron density may be associated with the extensive mitochondrial clustering induced by expressing protein A alone, which reduces the mitochondrial surface area available for exchange with the cytoplasm. In addition, in infected cells, the dilation of the intermembrane space associated with spherule formation appears to condense the matrix to a higher density than in uninfected cells (compare Fig. 2B to A).
FHV genomic RNA1 induces mitochondrial membrane spherules.
Since protein A was not sufficient to induce mitochondrial spherules, we generated plasmids to express FHV genomic RNA1 (pF1) and RNA2 (pF2), the full FHV genome, from the baculovirus IE1 promoter (Fig. 1). The resulting transcripts each had WT viral 3′ ends produced by self-cleavage of a hepatitis delta virus antigenomic ribozyme (δRz) and 46 nonviral nt at the 5′ end. In keeping with prior results that even longer 5′ extensions did not inhibit RNA1 and RNA2 replication (29), cotransfecting pF1 plus pF2 into Drosophila S2 cells resulted in full FHV RNA replication, as indicated by the presence of positive- and negative-strand RNA1 and RNA3 (Fig. 3A and B) and RNA2 (Fig. 3C and D). As expected, pF1 alone directed RNA1 replication (Fig. 3, lane 4) whereas pF2 did not induce RNA2 replication in the absence of pF1 (Fig. 3, lane 5). Since RNA2 suppresses RNA3 replication (17), pF1 induced higher levels of RNA3 replication in the absence of pF2 (Fig. 3A, lanes 3 and 4). Lesser differences in the RNA3/RNA1 ratio between cells transfected with pF1 plus pF2 and cells infected with FHV virions may relate to differing levels of RNA1 and RNA2 transcripts expressed from pF1 and pF2.
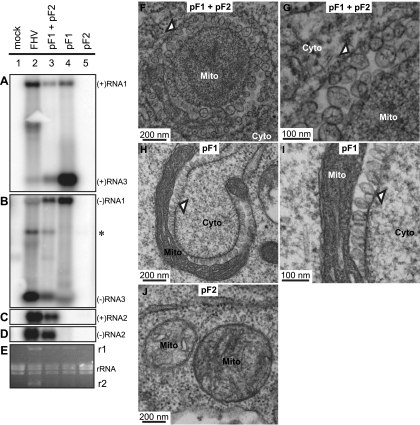
FIG. 3.
Expression of replication-competent FHV RNA1 is sufficient to induce mitochondrial spherules. (A to E) Total RNA and total protein were isolated from Drosophila cells transfected with pGFP (mock) or pF1 and pF2 together or alone or infected with FHV. Total RNA was analyzed by Northern blotting with 32P-labeled cRNA probes for positive-strand RNA1 and RNA3 (A), negative-strand RNA1 and RNA3 (B), positive-strand RNA2 (C), and negative-strand RNA2 (D). (E) Ethidium bromide (EtBr)-stained rRNA is shown as a loading control. FHV RNA1 (r1) and RNA2 (r2) replicate to rRNA levels and can be observed by EtBr staining. The asterisk in panel B represents cross-reactivity of the negative-strand RNA1 probe with RNA2. The band that occurs between RNA1 and RNA3 in lane 2 of panel B may represent a defective interfering RNA (63) or an RNA2-RNA3 heterodimer (4). A similar band is also visible in Fig. 4B. (F) Electron micrograph of a cell expressing pF1 and pF2, showing the formation of mitochondrial spherules (arrowhead) similar to those observed for FHV infection (Fig. 2B and C). (G) Higher-magnification view of spherules in panel F. (H) Electron micrograph of a cell expressing pF1 alone, showing spherule formation (arrowhead). (I) Higher-magnification view of spherules in panel H. (J) Electron micrograph of a cell expressing pF2. Mitochondria (Mito) and cytoplasm (Cyto) are indicated. It should be noted that, as previously established by EM tomography, mitochondria bearing FHV spherules are frequently cup shaped, so that two-dimensional images of these mitochondria can appear quite different, depending on whether the plane of sectioning is perpendicular (Fig. 3F) or parallel (Fig. 3H) to the axis around which the mitochondrion is cupped. See Fig. 1 in reference 28 for further explanation.
We performed transmission EM (TEM) of Drosophila cells transfected with pF1 and pF2 to investigate if they were capable of inducing spherule membrane rearrangements. Transfecting pF1 plus pF2 produced mitochondrial spherules (Fig. 3F and G) equivalent in size and appearance to those observed for FHV infection (Fig. 2B and C). Additionally, the WT mitochondrial spherule phenotype was reproduced in cells transfected with pF1 alone (Fig. 3H and I) but not in cells transfected with pF2 (Fig. 3J). Thus, FHV RNA1 is necessary and sufficient to produce mitochondrial spherules in Drosophila cells, while RNA2 and its protein α capsid precursor product are dispensable.
FHV protein B2 is not required for mitochondrial spherule formation.
In addition to protein A, FHV RNA1 encodes the RNAi suppressor protein B2 (31), which is translated from subgenomic RNA3 (22). Protein B2 is crucial for FHV RNA replication and accumulation in cells with robust RNAi responses, such as normal Drosophila cells (31), but not in cells where RNAi is suppressed or absent (31, 42). Recent work showed that protein B2 interacts with protein A in vivo (5), suggesting that B2 could be involved in spherule formation.
To test if protein B2 is required for spherule formation, we first generated infectious FHV virions bearing a viral genome that did not express protein B2 (FHVΔB2) due to two previously documented mutations (Fig. 4A) (8, 31) that change the B2 initiating methionine to a threonine (M1T) and serine 58 to a stop codon (S58stop). These nucleotide substitutions could be made without affecting the protein A amino acid sequence because B2 is in a +1 reading frame with respect to the protein A ORF. FHV RNA1 with these B2 mutations and WT RNA2 were in vitro transcribed and cotransfected into Drosophila cells whose RNAi machinery was inhibited by depleting the RNAi effector Argonaute2 (ago2) by RNA silencing, which allows FHV RNA replication in the absence of protein B2 (31). FHVΔB2 virions were collected from the transfected cells, and titers were determined on ago2-silenced Drosophila cells. Drosophila DL-1 cells were used for these experiments because their extensive lysis provides a more accurate titer by plaque assay than S2 cells (13).
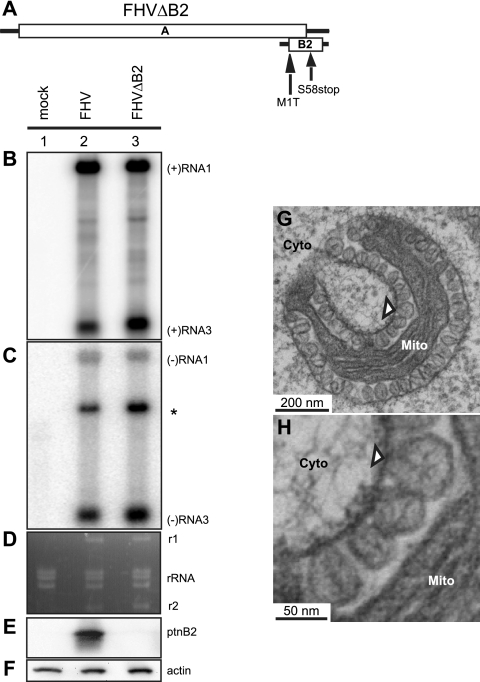
FIG. 4.
FHV protein B2 is not required for spherule formation. (A) Schematic of RNA1 showing mutations in the B2 ORF. Total RNA and total protein were isolated from Argonaute2 (ago2)-silenced Drosophila cells left uninfected (mock) or infected with WT FHV or FHV with mutations in the B2 ORF to block B2 expression (FHVΔB2). Total RNA was analyzed by Northern blotting with 32P-labeled cRNA probes for positive-strand RNA1 and RNA3 (B) or negative-strand RNA1 and RNA3 (C). The asterisk represents cross-reactivity of the negative-strand RNA1 probe with RNA2. (D) EtBr-stained rRNA is shown as a loading control and also shows that RNA1 (r1) and RNA2 (r2) are replicated to rRNA levels. Total protein was analyzed by Western blotting with antibodies against protein B2 (E) or actin (F) showing that cells infected with FHVΔB2 do not express detectable levels of protein B2. (G) Electron micrograph of ago2-silenced cells infected with FHVΔB2, showing spherule formation (arrowhead) in the absence of protein B2 expression. (H) Higher-magnification view of spherules in panel G. Mitochondria (Mito) and cytoplasm (Cyto) are indicated.
Next, Drosophila cells were transfected with dsRNA to ago2, and 24 h later, the ago2-silenced cells were infected with WT FHV or FHVΔB2 virions at an MOI of 10. At 14 h postinfection (hpi), infected cells were harvested and total RNA was isolated and analyzed by Northern blotting with probes that recognize either positive- or negative-strand RNA1 and RNA3 (Fig. 4B and C). Both WT FHV and FHVΔB2 replicated to similar levels in the ago2-silenced Drosophila cells. We also analyzed total protein by Western blotting with an antibody against protein B2, which confirmed that B2 was not expressed in the FHVΔB2-infected cells (Fig. 4E). Lastly, we examined the FHVΔB2-infected cells by TEM and observed mitochondrial spherules that were similar in size and appearance to WT FHV-infected cells (Fig. 4G and H). Similar to FHV-infected cells, the majority of the FHVΔB2-infected cells contained spherule-bearing mitochondria. Thus, FHV protein B2 is not required for mitochondrial spherule formation.
Coexpressing a replicatable RNA template with WT protein A induces spherule formation.
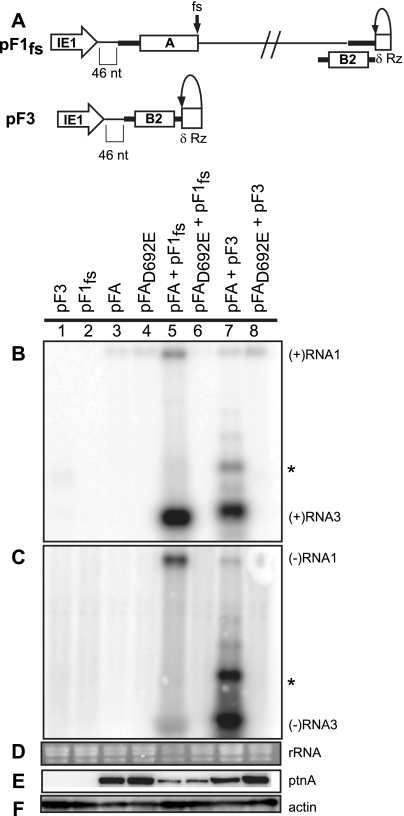
The above results show that protein A is the only FHV protein required to induce mitochondrial spherules (Fig. 3 and 4) but is not sufficient to induce spherules (Fig. 2). Since no FHV proteins beyond protein A are required, we next addressed whether an RNA template might be involved in spherule formation. To this end, we used a previously described trans replication system (32) that allows independent manipulation and functional testing of protein A and RNA templates. As one replication template, we used full-length RNA1 derivative RNA1fs (pF1fs), in which a frameshift blocks protein A expression (Fig. 5A) (32). Since RNA3 is replicated by protein A in the absence of full-length RNA1 (17), we also constructed plasmid pF3, expressing the complete RNA3 sequence (Fig. 5A). Both of these RNA templates had 5′ nonviral sequences equivalent to pF1.
FIG. 5.
trans replication of FHV RNA templates. (A) Schematic of plasmid-directed RNA1fs and RNA3 expression in Drosophila cells. pF1fs is the same as pF1, except that a 4-nt sequence causes a frameshift (fs) that prevents full-length protein A production. pF3 expresses RNA3 from the IE1 promoter with a δRz-mediated authentic 3′ end (δRz). The 5′ ends of pF1fs and pF3 have 46 nonviral nt between the transcription start site and the first viral nucleotide, as was shown for pF1 and pF2. Drosophila cells were transfected with pF3, pF1fs, pFA, pFAD692E, pFA plus pF1fs, pFAD692E plus pF1fs, pFA plus pF3, or pFAD692E plus pF3. (B to D) Total RNA was analyzed by Northern blotting with 32P-labeled cRNA probes for positive-strand RNA1 and RNA3 (B) or negative-strand RNA1 and RNA3 (C). Note that in panel B generally low but somewhat varying levels of the expected pFA- or pFAD692E-generated, nonreplicatable protein A mRNA transcripts are visible in lanes 3 to 8 at a position slightly above the replicated positive-strand RNA1 band in lane 5. As noted in an earlier study (59), the similarly sized, weak band near RNA1 in panel C, lane 7, appears to represent detection of an incompletely denatured hybrid between negative-strand RNA3 and the positive-strand protein A mRNA. This band does not represent negative-strand RNA1 since, despite its high position in the gel, the band is only detected with a strand-specific probe against the common sequences of negative-strand RNA3 and RNA1, but not with a strand-specific probe against the unique 3′-proximal portion of negative-strand RNA1, outside of the sequences shared with RNA3 (59). (D) EtBr-stained rRNA is shown as a loading control. Total protein was analyzed by Western blotting with antibodies against protein A (E) or actin (F). The asterisk represents a slower-migrating form of RNA3 that has been previously observed.
As expected, Drosophila S2 cells transfected with RNA template-expressing plasmid pF1fs or pF3 alone did not show any protein A expression or RNA replication (Fig. 5B to F, lanes 1 and 2). Cotransfecting pFA with pF1fs or pF3 in each case induced RNA replication (Fig. 5B and C, lanes 5 and 7). Primer extension data indicate that the slightly slower migration of RNA3 derived from pF3 was due to the retention, under the conditions and duration of this assay, of the 5′ nonviral sequences (P. Van Wynsberghe, personal communication). The much slower-migrating form (*) visible upon high-level RNA3 replication (Fig. 5B and C, lane 7) has been observed previously and may represent dsRNA3 or head-to-tail RNA3 dimers (4, 32). TEM examination showed that cells transfected with only pF1fs or pF3 did not contain spherules (Fig. 6A and B). However, in cells transfected with either of these RNA template plasmids and pFA, mitochondrial spherules formed (Fig. 6C to F, arrowheads) that were equivalent in appearance to those in WT FHV infection (Fig. 2B and C). Thus, since pFA alone did not form spherules (Fig. 2D to F), an RNA template is crucial for FHV spherule formation. Interestingly, the spherules were similar in size and appearance regardless of whether the RNA template provided was RNA1fs (3.1 kb) or RNA3 (0.3 kb), showing that even a 10-fold change in the template RNA size did not influence spherule dimensions. Also, use of the trans replication system indicates that protein A does not need to be translated from a viral RNA replication product, and the FHV RNA replication components do not need to be provided in cis to form spherules.
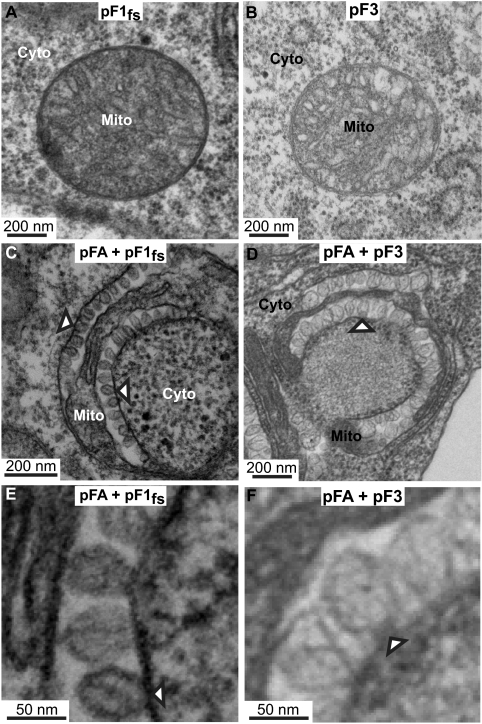
FIG. 6.
Coexpressing FHV RNA templates with WT protein A induces mitochondrial spherules. (A) Electron micrograph of a cell expressing pF1fs alone, showing no mitochondrial spherules. (B) Electron micrograph of a cell expressing pF3 alone, showing no mitochondrial spherules. (C) Electron micrograph of a cell expressing pFA plus pF1fs in which trans replication of RNA1fs is occurring (Fig. 5, lane 5), showing mitochondrial spherule formation (arrowheads). (D) Electron micrograph of a cell expressing pFA plus pF3 in which trans replication of RNA3 is occurring (Fig. 5, lane 7), showing mitochondrial spherule formation (arrowheads). (E and F) Higher-magnification views of spherules from panels C and D, respectively. Mitochondria (Mito) and cytoplasm (Cyto) are indicated.
Protein A's polymerase activity is required for spherule formation.
To test if RNA synthesis is necessary for spherule formation, we used a protein A derivative with a polymerase-inactivating D-to-E (amino acid 692) mutation at the first aspartate residue in the conserved GDD motif of the polymerase domain (pFAD692E). This previously described mutation eliminates protein A's ability to replicate viral RNA yet retains the other known functions of protein A, including the recruitment of the viral RNA into a membrane-associated state at mitochondria (60). As expected, when pFAD692E was cotransfected into cells with a replication-competent RNA template (pF1fs or pF3), RNA1 and RNA3 replication was inhibited (Fig. 5B and C, compare lane 5 to lane 6 and lane 7 to lane 8). Western blot analysis also showed similar protein A levels between the trans-replicating and nonreplicating cases (Fig. 5E, compare lane 5 to lane 6 and lane 7 to lane 8). When we used TEM to examine 1,960 mitochondria in 111 individual cells cotransfected with pFAD692E plus the replicatable RNA template pF1fs in three independent experiments, no mitochondrial spherules were observed, although mitochondria were clustered and zippered (Fig. 7A) as when WT protein A was expressed alone. Cells cotransfected with pFAD692E plus the replicatable RNA template pF3 also did not show any spherule formation (Fig. 7B). This finding implies that protein A-RNA template interactions are not enough to drive spherule formation. The possibility exists that since there is no replication there are not enough RNA templates available to form an amount of spherules that can be observed by TEM. However, a previously published real-time RT-PCR analysis (60) showed that, under equivalent conditions, pFAD692E directs the selective recruitment to mitochondrial membranes of ∼8,000 FHV RNA1fs templates per Drosophila cell (Van Wynsberghe, personal communication). These ∼8,000 FHV RNA1fs templates per Drosophila cell, which have been visualized to be highly localized to mitochondrial membranes (59), are comparable in number to our previous calculation of ∼20,000 spherules and ∼16,000 negative-strand RNA1 molecules per FHV-infected cell (28). If even 10% of the RNA1 template so recruited formed spherules, this would have been readily observed by our TEM analysis. Thus, spherule formation is dependent on FHV RNA polymerase activity and/or FHV RNA polymerization.
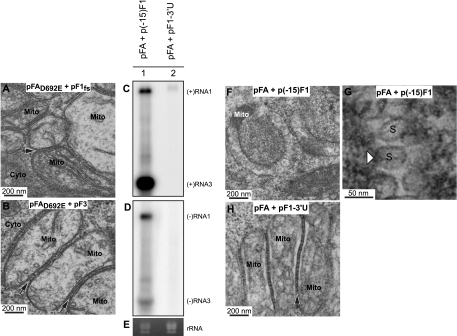
FIG. 7.
Spherule formation requires protein A polymerase activity and a replicatable RNA template. (A) Electron micrograph of a cell expressing pFAD692E plus pF1fs in which no replication is occurring (Fig. 5, lane 6), showing no mitochondrial spherule formation but mitochondrial zippering (arrow), as occurs when expressing pFA alone (Fig. 2D to F). (B) Electron micrograph of a cell expressing pFAD692E plus pF3 in which no replication is occurring (Fig. 5, lane 8), showing no mitochondrial spherule formation but mitochondrial zippering (arrows), as occurs in panel A and when expressing pFA alone (Fig. 2D to F). Drosophila cells were transfected with pFA plus an RNA1fs template with a truncation of the first 15 nt [p(−15)F1] or a deletion of the 3′ UTR [pF1-3′U]. Total RNA was analyzed by Northern blotting with 32P-labeled cRNA probes for positive-strand RNA1 and RNA3 (C) or negative-strand RNA1 and RNA3 (D). (E) EtBr-stained rRNA. (F) Electron micrograph of cells transfected with pFA plus p(−15)F1. (G) Higher-magnification view of spherules from panel F. (H) Electron micrograph of cells transfected with pFA plus pF1-3′U, showing mitochondrial zippering but no spherules. Mitochondria (Mito), cytoplasm (Cyto), and spherules (S) are indicated.
Deleting 3′-terminal sequences from FHV RNA1, including just the last 5 nt, blocks negative-strand synthesis and RNA replication (7, 8). Accordingly, to verify that the lack of spherule formation was not due to an unexpected effect of the D692E polymerase mutation, we expressed WT protein A and blocked RNA replication by deleting the entire 71-nt 3′ UTR of RNA1 from pF1fs, creating pF1-3′U. Cells transfected with pFA plus pF1-3′U showed accumulation of the modified positive-strand RNA1 primary transcript but, as expected, did not show any detectable negative-strand RNA1 synthesis or positive-strand RNA1 amplification (Fig. 7C to E). Nevertheless, nt 40 to 190 of RNA1 were retained in this 3′-truncated RNA1 and are sufficient for efficient, protein A-mediated, selective recruitment of RNA1 derivatives into a membrane-associated state (59). However, TEM examination of cells transfected with pFA plus pF1-3′U showed no detectable spherules, even after examination of sections of 1,382 mitochondria in 107 cells over three independent experiments (Fig. 7H). Thus, in combination, the Fig. 7 findings on inactivating mutations in FHV RNA polymerase or in FHV RNA templates show that mitochondrial spherule formation requires protein A with polymerase activity and a replication-competent viral RNA template.
In parallel with pF1-3′U, we also constructed and tested p(−15)F1 by deleting the 5′-terminal 15 nt of RNA1 from pF1fs. Consistent with prior findings that the replication template activity of RNA1 is more tolerant of 5′ than 3′ mutations (7, 8), cells transfected with pFA plus p(−15)F1 showed normal levels of RNA1 replication (Fig. 7C to E) and spherule formation (Fig. F and G). These results further extend the close, consistent linkage between FHV RNA synthesis and spherule formation.
DISCUSSION
This study reveals that while FHV and some other (+)RNA viruses such as BMV replicate their RNA genomes in membrane compartments of similar architecture, they generate these compartments by very different RNA- and polymerase-dependent or -independent pathways (Fig. 8). Below, we discuss the relationships of these findings to other features of nodavirus RNA replication, to other viruses, and to viral interaction with host innate immune defenses.
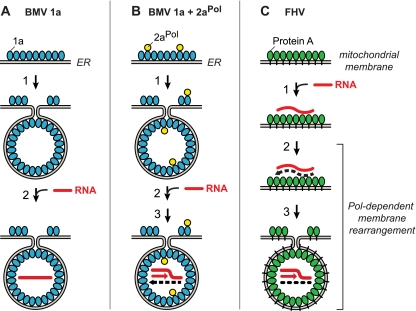
FIG. 8.
Polymerase independence and dependence of BMV and FHV spherule membrane rearrangements and replication complex assembly. (A) BMV spherule generation by multifunctional BMV RNA replication factor 1a (blue). In the absence of BMV RNA-dependent RNA polymerase 2aPol, 1a localizes to ER membranes, self-interacts, and induces the formation of ∼70-nm invaginations or spherules (39, 40, 51). In a separable subsequent reaction dependent on the activity of the C-proximal 1a NTPase/helicase domain, 1a transfers genomic RNA (red line) to the spherule interior (61). (B) In the presence of 1a and 2aPol (yellow), 1a also recruits 2aPol to ER membranes and directs spherule formation and genomic RNA recruitment as in panel A. 2aPol then synthesizes negative-strand RNA (dashed black line) that is retained in the spherule and repeatedly used as a template to synthesize new positive strands. (C) Protein A (green), the sole FHV-encoded RNA replication factor, localizes to mitochondrial outer membranes (34) and self-interacts (16) but does not induce spherule formation unless replication-competent FHV RNA templates are present and protein A's RNA polymerase domain is active (Fig. 2 to 7). Active-site mutations abolishing protein A polymerase activity or deletion of 3′ RNA replication signals still allow protein A to recognize specific 5′-proximal elements in viral RNA templates and recruit them to mitochondrial membranes (59, 60) but block RNA synthesis and invagination of FHV spherules (Fig. 7). See Discussion for additional details, including the role of FHV RNA interference suppressor B2.
Relationship to RNA replication compartment formation by other viruses.
FHV and BMV, e.g., replicate their RNAs in 50- to 70-nm, virus-induced membrane invaginations or spherules on mitochondrial and ER membranes, respectively (28, 51). Each FHV or BMV spherule contains one to a few hundred copies of a membrane-associated, self-interacting viral replication factor, FHV protein A or BMV 1a, which is required for spherule formation and appears to form a shell lining the spherule interior (16, 28, 39, 40, 51). Each spherule also contains one or two genomic RNA replication intermediates composed of positive- and negative-strand RNAs and primarily synthesizes (+)RNAs that are exported to the cytoplasm for translation and encapsidation (28, 51). Based in part on these results, similar models have been proposed for other viral RNA replication complexes (46, 61, 62).
Despite such similarities, we find that FHV and BMV membrane spherules are generated by highly distinct mechanisms. BMV 1a is the sole viral component required to induce BMV replication spherules, with no requirement for viral RNA templates or the BMV RNA polymerase, 2apol (51) (Fig. 8A and B). Similarly, for many other (+)RNA viruses, one or two nonpolymerase RNA replication proteins suffice to induce the membrane rearrangements associated with RNA replication (18, 19, 36, 47, 55).
These results and the above parallels with BMV 1a protein in spherule architecture suggested that FHV protein A was likely sufficient to form spherules. However, in this study, we found that protein A, although the only viral protein required, was insufficient for spherule formation (Fig. 2 and 4). Forming FHV spherules also required functional viral RNA templates and protein A's RNA polymerase activity (Fig. 8C). Multiple results indicated that RNA synthesis was required. In the presence of WT protein A, spherule formation was supported by genomic RNA1 or subgenomic RNA3, which are both replication templates (Fig. 3 and 6), but was blocked by the deletion of 3′ replication signals (Fig. 7). Similarly, in the presence of WT FHV RNA replication templates, spherule formation was abolished by an active-site mutation that blocks protein A RNA polymerase activity but not protein A's binding to mitochondrial membranes or recruitment of viral RNA (60) (Fig. 5 and 7).
Viral RNA and polymerase activity might contribute to FHV spherule formation by inducing a necessary conformational change or posttranslational modification of protein A or because it produces negative-strand or dsRNA, which might recruit a necessary host factor or provide an RNA structure equivalent to an encapsidation initiation site for triggering assembly of the capsid-like protein A shell. The latter possibility could have similarities to initiation of retrovirus RNA packaging by an RNA signal dependent on viral RNA dimerization (15). Further experiments are in progress to define the nature of the requirement for RNA synthesis.
Relationship to RNA replication template recruitment.
These differences in BMV and FHV membrane spherule formation parallel and appear potentially related to differences in RNA template recruitment. FHV genomic RNA templates are recruited to mitochondrial membranes by protein A in a step that is independent of protein A polymerase activity (59, 60). Thus, since our results above show that protein A polymerase activity and RNA templates are essential for FHV spherule formation (Fig. 2 and 5 to 7), such RNA recruitment precedes FHV spherule formation (Fig. 8C, step 1).
In contrast, BMV spherule formation not only is independent of viral RNA (51) but is required for normal RNA template recruitment (33, 61). BMV protein 1a recruits viral RNA templates into a membrane-associated, nuclease-resistant state that multiple results imply to be the interior of the 1a-induced spherules (1, 51). Accordingly, 1a and host cell mutations that inhibit or stimulate spherule formation modulate RNA recruitment in parallel (14, 33). Among other functions, 1a has a nucleoside triphosphatase (NTPase)/helicase domain (2, 3, 61). Mutations throughout the conserved helicase motifs show that the activity of this NTPase/helicase domain is not required for spherule formation but is essential for transferring RNA templates into a nuclease-resistant state (Fig. 8A, steps 1 and 2), suggesting that this NTPase/helicase activity may be involved in translocating viral RNAs into preformed spherules (61).
Unlike BMV 1a, FHV protein A has no identifiable NTPase or helicase motifs or activities (27). Lacking the ability to translocate RNA into a preformed spherule, FHV may be obliged to trigger the formation of its replication compartments around its RNA templates or replication intermediates. Coupling such membrane rearrangements with RNA synthesis further ensures that a replication-competent viral template is enclosed within the replication compartment (Fig. 8C).
Interestingly, BMV and certain other (+)RNA viruses require viral RNA templates and, in some cases, RNA synthesis at later stages of replication complex formation, after membrane rearrangement. For example, membrane extracts from cells expressing BMV RNA replication factors 1a and 2apol contain normal levels of viral proteins and spherules but cannot copy added viral RNAs in vitro unless functional BMV RNA templates were coexpressed in vivo (45). Thus, in vivo, BMV RNA activates the complex for later copying of other RNAs in vitro, such as by recruiting a required host factor or altering the state of viral replication factors. Similarly, forming active poliovirus RNA replication complexes requires viral RNA templates and RNA synthesis (18). Again, unlike FHV, this block occurs after membrane rearrangement since, in the absence of viral RNA synthesis, poliovirus nonstructural proteins induce membranous structures indistinguishable from those associated with natural infection (10, 18).
Protein A-induced clustering of mitochondria.
While expressing FHV protein A alone in Drosophila cells induced no detectable spherules, it caused mitochondria to cluster with outer membranes that appeared zippered together (Fig. 2E and F). This zippering of mitochondrial membranes was similar to the ability of many self-interacting ER membrane proteins, when overexpressed, to induce closely appressed ER membrane layers called karmellae. Since karmella formation depends on self-interaction of the inducing proteins (54), mitochondrial clustering appears consistent with the ability of transmembrane protein A to self-interact through multiple domains (16).
Similar close appression of membranes is observed in cells infected with some other (+)RNA viruses or cells expressing certain membrane-linked viral RNA replication proteins. Turnip yellow mosaic virus infection, e.g., induces spherule RNA replication compartments on the outer membranes of chloroplasts, and such chloroplasts also cluster in a fashion similar to protein A-induced mitochondrial clustering (44). As another example, expressing picornavirus membrane-associated RNA replication protein 2C or 2BC in the absence of other viral proteins rearranges ER membranes into stacked layers of closely appressed or zippered membranes (12, 57).
The results presented here show that viral RNA synthesis or its products must modulate protein A's membrane or protein interactions to favor invagination of single mitochondrial membranes into spherules over protein A-induced zippering between distinct mitochondria. Similarly, membrane-bound, self-interacting BMV protein 1a can be modulated to either invaginate ER membranes into spherules or zipper them together into stacks of double membrane layers, depending on the level of 1a interaction with BMV 2a polymerase (52). Thus, the potential for such varied ultrastructural outcomes may be inherent in the balance of protein-protein, -membrane, and -RNA interactions required for RNA replication by many (+)RNA viruses.
Relationship to host RNAi defenses.
One potential function of membrane-bound replication compartments is to sequester viral RNA replication intermediates from dsRNA-triggered innate immune responses such as RNAi (1). However, our results show that formation of FHV spherules requires FHV RNA synthesis. Thus, there must be a stage in the FHV replication cycle, prior to spherule formation, when dsRNA replication intermediates are potentially accessible to the siRNA-producing Dicer nuclease and other RNAi components (Fig. 8). These results provide an explanation for recent findings that virus-specific siRNAs produced in FHV infection are derived from both strands, implying that dsRNA replication intermediates are the predominant source of these siRNAs (5, 20, 58). Once spherules are formed, they may contribute to protecting viral replication intermediates from RNAi recognition. Another major contributor to such protection is FHV's RNAi suppressor protein B2. B2 interacts with protein A in vivo, which may facilitate B2 interaction with and protection of newly synthesized FHV RNAs (5). Nevertheless, despite B2's interaction with protein A and its role in protecting viral RNA from RNAi, spherule formation was unaffected by the presence or absence of B2 (Fig. 4).
Overall, our results show that FHV RNA polymerase activity plays important roles in generating membrane-bound viral RNA replication compartments, as well as in their later function. RNA-dependent RNA polymerases are common targets of antiviral drugs for several (+)RNA viruses (6). While inhibiting polymerase activity can attenuate the level of viral replication and virus produced after RNA replication has started, our results suggest that for some (+)RNA viruses, polymerase inhibitors may also prevent fundamental steps in the assembly of critical RNA replication compartments, providing a further, previously unrecognized, mode of action for this important class of antivirals.
Acknowledgments
We thank Brandi Gancarz, Arturo Diaz, and other members of our laboratory for helpful discussions.
This work was supported by National Institutes of Health grant GM35072 (to P.A.). E.W.S. was supported by National Institutes of Health grant AI25557 (to P.D.F.). P.A. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 13 October 2010.
REFERENCES
- 1.Ahlquist, P. 2006. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 4:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahola, T., and P. Ahlquist. 1999. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J. Virol. 73:10061-10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahola, T., J. A. den Boon, and P. Ahlquist. 2000. Helicase and capping enzyme active site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping. J. Virol. 74:8803-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albariño, C. G., B. D. Price, L. D. Eckerle, and L. A. Ball. 2001. Characterization and template properties of RNA dimers generated during Flock House virus RNA replication. Virology 289:269-282. [DOI] [PubMed] [Google Scholar]
- 5.Aliyari, R., Q. Wu, H. W. Li, X. H. Wang, F. Li, L. D. Green, C. S. Han, W. X. Li, and S. W. Ding. 2008. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe 4:387-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Appleby, T., I. Shih, and W. Zhong. 2009. Viral RNA polymerase inhibitors, p. 527-548. In C. E. Cameron, M. Gotte, and K. D. Raney (ed.), Viral genome replication. Springer Science+Business Media, New York, NY.
- 7.Ball, L. A. 1994. Replication of the genomic RNA of a positive-strand RNA animal virus from negative-sense transcripts. Proc. Natl. Acad. Sci. U. S. A. 91:12443-12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ball, L. A. 1995. Requirements for the self-directed replication of Flock House virus RNA 1. J. Virol. 69:720-727. (Erratum, 69:2722.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ball, L. A., and K. L. Johnson. 1998. Nodaviruses of insects, p. 225-267. In L. K. Miller and L. A. Ball (ed.), The insect viruses. Plenum Publishing Corp., New York, NY.
- 10.Belov, G. A., Q. Feng, K. Nikovics, C. L. Jackson, and E. Ehrenfeld. 2008. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog. 4:e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cartier, J. L., P. A. Hershberger, and P. D. Friesen. 1994. Suppression of apoptosis in insect cells stably transfected with baculovirus p35: dominant interference by N-terminal sequences p35(1-76). J. Virol. 68:7728-7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. [DOI] [PubMed] [Google Scholar]
- 13.Dasgupta, R., B. Selling, and R. Rueckert. 1994. Flock House virus: a simple model for studying persistent infection in cultured Drosophila cells. Arch. Virol. Suppl. 9:121-132. [DOI] [PubMed] [Google Scholar]
- 14.Diaz, A., X. Wang, and P. Ahlquist. 2010. Membrane-shaping host reticulon proteins play crucial roles in viral RNA replication compartment formation and function. Proc. Natl. Acad. Sci. U. S. A. 107:16291-16296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Souza, V., and M. F. Summers. 2004. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature 431:586-590. [DOI] [PubMed] [Google Scholar]
- 16.Dye, B. T., D. J. Miller, and P. Ahlquist. 2005. In vivo self-interaction of nodavirus RNA replicase protein a revealed by fluorescence resonance energy transfer. J. Virol. 79:8909-8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckerle, L. D., C. G. Albariño, and L. A. Ball. 2003. Flock House virus subgenomic RNA3 is replicated and its replication correlates with transactivation of RNA2. Virology 317:95-108. [DOI] [PubMed] [Google Scholar]
- 18.Egger, D., N. Teterina, E. Ehrenfeld, and K. Bienz. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 74:6570-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flynt, A., N. Liu, R. Martin, and E. C. Lai. 2009. Dicing of viral replication intermediates during silencing of latent Drosophila viruses. Proc. Natl. Acad. Sci. U. S. A. 106:5270-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friesen, P., and R. R. Rueckert. 1981. Synthesis of black beetle virus proteins in cultured Drosophila cells—differential expression of RNAs 1 and 2. J. Virol. 37:876-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friesen, P. D., and R. R. Rueckert. 1982. Black beetle virus: messenger for protein B is a subgenomic viral RNA. J. Virol. 42:986-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griparic, L., and A. M. van der Bliek. 2001. The many shapes of mitochondrial membranes. Traffic 2:235-244. [DOI] [PubMed] [Google Scholar]
- 26.Guarino, L. A., A. Ghosh, B. Dasmahapatra, R. Dasgupta, and P. Kaesberg. 1984. Sequence of the black beetle virus subgenomic RNA and its location in the viral genome. Virology 139:199-203. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, K. N., K. L. Johnson, R. Dasgupta, T. Gratsch, and L. A. Ball. 2001. Comparisons among the larger genome segments of six nodaviruses and their encoded RNA replicases. J. Gen. Virol. 82:1855-1866. [DOI] [PubMed] [Google Scholar]
- 28.Kopek, B. G., G. Perkins, D. J. Miller, M. H. Ellisman, and P. Ahlquist. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krishna, N. K., D. Marshall, and A. Schneemann. 2003. Analysis of RNA packaging in wild-type and mosaic protein capsids of Flock House virus using recombinant baculovirus vectors. Virology 305:10-24. [DOI] [PubMed] [Google Scholar]
- 30.Lannan, E., R. Vandergaast, and P. D. Friesen. 2007. Baculovirus caspase inhibitors P49 and P35 block virus-induced apoptosis downstream of effector caspase DrICE activation in Drosophila melanogaster cells. J. Virol. 81:9319-9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 32.Lindenbach, B. D., J. Y. Sgro, and P. Ahlquist. 2002. Long-distance base pairing in Flock House virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J. Virol. 76:3905-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, L., W. M. Westler, J. A. den Boon, X. Wang, A. Diaz, H. A. Steinberg, and P. Ahlquist. 2009. An amphipathic alpha-helix controls multiple roles of brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathog. 5:e1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, D. J., and P. Ahlquist. 2002. Flock House virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J. Virol. 76:9856-9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, D. J., M. D. Schwartz, and P. Ahlquist. 2001. Flock House virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 75:11664-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, S., S. Kastner, J. Krijnse-Locker, S. Buhler, and R. Bartenschlager. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 282:8873-8882. [DOI] [PubMed] [Google Scholar]
- 37.Miller, S., and J. Krijnse-Locker. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olson, V. A., J. A. Wetter, and P. D. Friesen. 2002. Baculovirus transregulator IE1 requires a dimeric nuclear localization element for nuclear import and promoter activation. J. Virol. 76:9505-9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Reilly, E. K., J. D. Paul, and C. C. Kao. 1997. Analysis of the interaction of viral RNA replication proteins by using the yeast two-hybrid assay. J. Virol. 71:7526-7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Reilly, E. K., Z. Wang, R. French, and C. C. Kao. 1998. Interactions between the structural domains of the RNA replication proteins of plant-infecting RNA viruses. J. Virol. 72:7160-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1-a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price, B. D., M. Roeder, and P. Ahlquist. 2000. DNA-directed expression of functional Flock House virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J. Virol. 74:11724-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price, B. D., R. Rueckert, and P. Ahlquist. 1996. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 93:9465-9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prod'homme, D., S. Le Panse, G. Drugeon, and I. Jupin. 2001. Detection and subcellular localization of the turnip yellow mosaic virus 66K replication protein in infected cells. Virology 281:88-101. [DOI] [PubMed] [Google Scholar]
- 45.Quadt, R., M. Ishikawa, M. Janda, and P. Ahlquist. 1995. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc. Natl. Acad. Sci. U. S. A. 92:4892-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinkert, D., R. Bartenschlager, and V. Lohmann. 2005. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 79:13594-13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roosendaal, J., E. G. Westaway, A. Khromykh, and J. M. Mackenzie. 2006. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J. Virol. 80:4623-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubino, L., F. Weber-Lotfi, A. Dietrich, C. Stussi-Garaud, and M. Russo. 2001. The open reading frame 1-encoded (′36K') protein of Carnation Italian ringspot virus localizes to mitochondria. J. Gen. Virol. 82:29-34. [DOI] [PubMed] [Google Scholar]
- 49.Salonen, A., T. Ahola, and L. Kaariainen. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 51.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 52.Schwartz, M., J. Chen, W. M. Lee, M. Janda, and P. Ahlquist. 2004. Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc. Natl. Acad. Sci. U. S. A. 101:11263-11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Selling, B. H., and R. R. Rueckert. 1984. Plaque assay for black beetle virus. J. Virol. 51:251-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Snapp, E. L., R. S. Hegde, M. Francolini, F. Lombardo, S. Colombo, E. Pedrazzini, N. Borgese, and J. Lippincott-Schwartz. 2003. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 163:257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snijder, E. J., H. van Tol, N. Roos, and K. W. Pedersen. 2001. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 82:985-994. [DOI] [PubMed] [Google Scholar]
- 56.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teterina, N. L., K. Bienz, D. Egger, A. E. Gorbalenya, and E. Ehrenfeld. 1997. Induction of intracellular membrane rearrangements by HAV proteins 2C and 2BC. Virology 237:66-77. [DOI] [PubMed] [Google Scholar]
- 58.van Rij, R. P., and E. Berezikov. 2009. Small RNAs and the control of transposons and viruses in Drosophila. Trends Microbiol. 17:163-171. [DOI] [PubMed] [Google Scholar]
- 59.Van Wynsberghe, P. M., and P. Ahlquist. 2009. 5′ cis elements direct nodavirus RNA1 recruitment to mitochondrial sites of replication complex formation. J. Virol. 83:2976-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Wynsberghe, P. M., H. R. Chen, and P. Ahlquist. 2007. Nodavirus RNA replication protein a induces membrane association of genomic RNA. J. Virol. 81:4633-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, X., W. M. Lee, T. Watanabe, M. Schwartz, M. Janda, and P. Ahlquist. 2005. Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J. Virol. 79:13747-13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Welsch, S., S. Miller, I. Romero-Brey, A. Merz, C. K. Bleck, P. Walther, S. D. Fuller, C. Antony, J. Krijnse-Locker, and R. Bartenschlager. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhong, W., and R. R. Rueckert. 1993. Flock House virus: down-regulation of subgenomic RNA3 synthesis does not involve coat protein and is targeted to synthesis of its positive strand. J. Virol. 67:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]