Abstract
The interferon (IFN)-induced protein P56 inhibits human papillomavirus (HPV) DNA replication by binding to HPV E1, which has several distinct functions in initiating viral DNA replication. Here, we determined that P56 inhibited HPV type 18 (HPV18) E1's DNA helicase activity, E2 binding, and HPV Ori sequence-specific DNA binding but not nonspecific DNA binding. We observed that deletion of a single amino acid, F399, produced an E1 mutant that could not bind P56. This E1 mutant retained its ability to support Ori DNA replication, but this activity was not inhibited by IFN, demonstrating that P56 is the principal executor of the anti-HPV action of IFN.
The interferon (IFN) system plays a major role in a host's innate immune response to virus infection (4, 19). IFN′s antiviral actions are carried out by the products of the IFN-stimulated genes (ISGs) (3). Among the hundreds of ISG products, some have specific antiviral properties (8, 15, 22, 27). We have been studying the functions of the IFIT1 (ISG56) family members (7, 22), all of whom contain multiple tetratricopeptide repeat (TPR) motifs (18). P56, the most prominent member, binds the “e” subunit of the translation initiation factor eIF-3, causing inhibition of translation initiation (5, 7). The P56 family members have been implicated in IFN′s antiviral actions against hepatitis C virus (HCV), West Nile virus, and lymphocytic choriomeningitis virus (LCMV), presumably by inhibiting viral protein synthesis (24, 25).
Human papillomaviruses (HPVs) are major pathogens (28). They induce hyperproliferative lesions, and the high-risk groups, HPV type 16 (HPV16), HPV18, and HPV31, cause anogenital cancers and cervical carcinoma (9). IFN is effective in treating HPV-induced genital warts; however, the underlying mechanism is not known (14). Turek et al. (23) observed complete elimination of the bovine papillomavirus (BPV) genome upon IFN treatment of BPV-transformed mouse cells, which caused their reversion to a nontransformed phenotype. Similarly, IFN inhibits keratinocyte transformation by HPV16 (12). Long-term treatment with IFN causes growth arrest and apoptosis of HPV-positive squamous carcinoma cells when episomal HPV DNA is eliminated from the surviving cells (2). We recently reported that P56 can inhibit HPV DNA replication by blocking the action of the viral protein E1, which, in conjunction with E2, drives viral DNA replication (22). E1 is a multifunctional protein with ATPase and DNA helicase activities (10). It can bind DNA nonspecifically as well as specifically to the viral Ori sequence, the two activities being mediated by two regions of the protein (17, 20). We reported that the N-terminal region of P56 binds to the C-terminal region of E1 and inhibits E1-mediated HPV DNA replication by inhibiting its DNA helicase activity (22). Here, we studied different biochemical functions of HPV18 E1 and their susceptibilities to inhibition by P56. Moreover, to assess how much P56 contributes to IFN′s inhibitory effects on HPV DNA replication, we generated a functional E1 mutant that does not bind P56.
P56 inhibits select functions of E1.
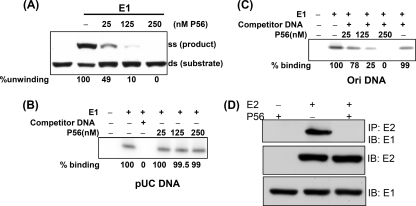
We previously reported that P56 inhibits HPV18 E1's ability to unwind a nonspecific duplex DNA (22). The same was true for unwinding the duplex Ori sequence. The Ori fragment-unwinding assay was carried out as described before (6). Briefly, 32P-duplex Ori, Escherichia coli single-stranded DNA (ssDNA) binding protein, and E1 were incubated in the appropriate buffer for 2 h at 37°C. The substrate and product were electrophoretically separated on a 6% nondenaturing polyacrylamide gel. Increasing amounts of P56 were added at the beginning of the reaction, where indicated. P56 inhibited E1's Ori-dependent helicase activity in a dose-dependent fashion (Fig. 1A), whereas a P56 mutant, M2, that does not bind E1 (22), had no effect, even at the highest concentration (see Fig. 3D). To determine the mechanism of inhibition of the helicase activity, we enquired whether P56 could inhibit nonspecific or Ori-specific DNA binding by E1 (13). The method for measuring the binding of E1 to DNA has been reported previously (21). Briefly, 32P-end-labeled Ori or pUC DNA, 60-bp in length, and His-E1 were incubated in the presence or in the absence of P56 at 37°C for 2 h in binding buffer. Competitor DNA was added at the beginning of the reaction, where indicated. Nickel-nitrilotriacetic acid (Ni-NTA) agarose beads were then added and incubated for 1 h. The complexes that bound to Ni-NTA beads were collected by centrifugation and washed 4 times with binding buffer, incubated 15 min at 65°C in dissociation buffer, extracted with phenol-chloroform, and precipitated with ethanol. The precipitated DNA fragment was then resolved by 12% polyacrylamide gel electrophoresis (PAGE). His-E1 bound to a radiolabeled 60-bp pUC DNA sequence, and this binding was eliminated by an excess unlabeled 60-bp of a different sequence, demonstrating that this DNA binding by E1 was sequence nonspecific (Fig. 1B). More importantly, P56 did not inhibit nonspecific DNA binding by E1, even at a high concentration (Fig. 1B). In contrast, P56 inhibited Ori-specific DNA binding by E1 (Fig. 1C). The HPV18 Ori sequence bound to E1; this binding was specific because it was not competed out by an excess of unlabeled 60-bp DNA that did not contain the Ori sequence. P56 inhibited E1's specific binding to the Ori sequence in a dose-dependent fashion (Fig. 1C). In addition to binding DNA, E1 can also bind directly to the viral protein E2 (1). The effect of P56 on the interaction of E1 and E2 was measured by their ability to coimmunoprecipitate in the presence and in the absence of P56. HT1080 cells were transfected with appropriate plasmids, and cell lysates were immunoprecipitated with anti-Flag antibody to precipitate Flag-E2 and associated proteins. P56 expression strongly inhibited the interaction of E1 and E2 (Fig. 1D). The results presented so far indicate that the binding of P56 to E1 selectively inhibits some, but not all, of its functions. Nonspecific DNA binding was not affected, but both E2 binding and HPV Ori-specific DNA binding were strongly inhibited. These observations provide the biochemical basis of the observed inhibitory effect of P56 on HPV DNA replication.
FIG. 1.
(A) Ori DNA-unwinding activity of E1 was inhibited by P56. E1 helicase assays were performed using Ori containing dsDNA and 25 nM E1. The positions of the substrate and the product and the concentrations of added P56 are indicated. For each lane, the two bands were quantified and the extent of conversion was calculated. The relative unwinding activities are given at the bottom. (B) Binding of E1 to pUC-nonspecific DNA (60-bp) was not inhibited by P56. Increasing amounts of P56 were added; where indicated, a 100-fold excess of 60-bp pcDNA competitor was added. Quantitation of binding is shown at the bottom. (C) Binding of E1 to Ori DNA (60 bp) was inhibited by P56. Increasing amounts of P56 and a 100-fold excess of 60-bp competitor pcDNA were added, where indicated. Binding by E1 alone was set at 100. (D) E1-E2 binding was inhibited by P56. HT1080 cells were transfected with a plasmid expressing His-E1; in addition, Flag-E2, P56, or both were coexpressed. Cell extracts were immunoprecipitated (IP) using Flag antibody and immunoblotted (IB) with His antibody (upper panel). Direct Western blotting with Flag (middle panel) and His (lower panel) antibodies was carried out to show expression of the proteins.
P56 binding requires a specific residue of E1.
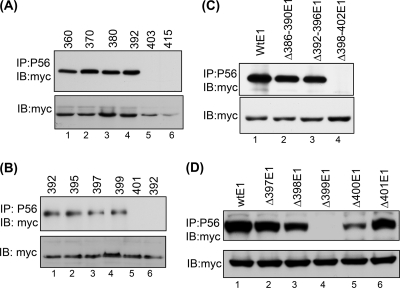
To map the domain of E1 that is required for P56 binding, we carried out a systematic deletion analysis of E1. The N-terminal region of E1 could not bind P56, but its C-terminal region (residues 360 to 657) bound P56 (data not shown). To further narrow down the binding region, we coexpressed P56 and E1 mutants having serial deletions of 10 residues from the N terminus of the fragment containing residues 360 to 657 and performed coimmunoprecipitations (Fig. 2A). Proteins starting at residues 360 (lane 1), 370 (lane 2), 381 (lane 3), and 392 (lane 4) bound P56, but those starting at residue 403 (lane 5) or 415 (lane 6) did not. When additional mutants were tested, the results indicated that residues 399 and 400 were needed for P56 binding (Fig. 2B). In the next experiment, single residues were deleted, demonstrating that residue F399 was absolutely required for this property (data not shown). To confirm the conclusions from the above experiments, we introduced internal deletions to full-length E1. When residues 398 to 402 were deleted from the full-length E1, it did not interact with P56 (Fig. 2C). Deletion of single amino acids from full-length E1 clearly showed that the removal of F399 completely abrogated the capacity of E1 to bind P56 (Fig. 2D, lane 4). Elimination of either of the two neighboring residues, A398 (Fig. 2D, lane 3) or L400 (Fig. 2D, lane 5) also impaired binding. The results presented here led us to conclude that the ΔF399 mutant of HPV18 E1, which is missing only residue 399 of wild-type (Wt) E1, cannot bind P56.
FIG. 2.
Deletion analysis of E1 for P56 interaction. (A) Several nested deletion mutants of a fragment of E1 containing amino acids (aa) 360 to 657 (from aa 360 toward the C terminus) were coexpressed with P56. Cell lysates were immunoprecipitated with P56 antibody and Western blotted with myc antibody. Lane 1, aa 360 to 657; lane 2, aa 370 to 657; lane 3, aa 380 to 657; lane 4, aa 392 to 657; lane 5, aa 403 to 657; and lane 6, aa 415 to 657. (B) Two amino acid deletion mutants of E1 were generated and analyzed for interactions with P56. Lane 1, aa 392 to 657; lane 2, aa 395 to 657; lane 3, aa 397 to 657; lane 4, aa 399 to 657; lane 5, aa 401 to 657, all with P56; and lane 6, aa 392 to 657 without P56. (C) Several internal deletion mutants of full-length myc-E1 were analyzed for interaction with P56. (D) Single-amino-acid internal deletion mutants of full-length myc-E1 were analyzed for interaction with P56.
The ΔF399 E1 mutant is biochemically active.
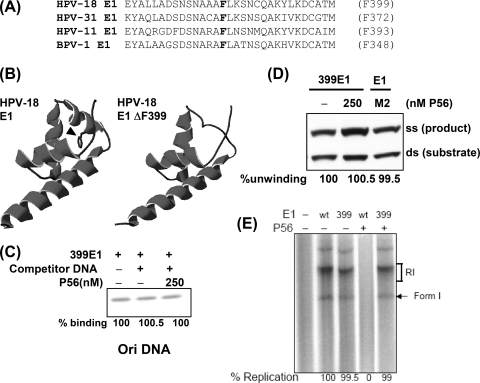
The F399 residue and the surrounding amino acids are well conserved in E1 proteins of several HPV strains and BPV type 1 (BPV1) (Fig. 3A). We used the known structure of BPV E1 (16) and the PHYRE server (11) to model the three-dimensional structure of residues 358 to 426 of HPV18 E1. F399 is located in an α-helix (Fig. 3B, arrow on left), and its deletion disrupts the helical structure (Fig. 3B, right). This indicates that P56 probably recognizes this helix in E1. Indeed, substitution of residue F399 by Ala, which maintains the predicted helix, did not affect P56 interaction (data not shown). Fortunately, deletion of residue F399 did not affect any biochemical property of the protein, indicating that there were no gross changes in the protein structure. The mutant protein could bind Ori DNA (Fig. 3C), unwind double-stranded DNA (dsDNA) (Fig. 3D), and support in vitro Ori DNA replication (Fig. 3E) as competently as Wt E1. More importantly, none of the above properties of the ΔF399 E1 mutant was affected by P56, although for Wt E1, all were inhibited (Fig. 1 and 3). Thus, we identified an HPV18 E1 mutant that is active and resistant to P56.
FIG. 3.
The biochemical activities of the ΔF399 E1 mutant were not inhibited by P56. (A) The amino acid sequences surrounding F399 are well conserved. (B) On the left is the predicted three-dimensional structure of this region of HPV18 E1 (residues 358 to 426). The arrowhead shows F399; on the right is the predicted structure for the F399 deletion mutant. (C) Ori DNA binding by the mutant was not inhibited by P56. (D) The helicase activity of the ΔF399 E1 mutant was not inhibited by P56. In the right lane, the effect of a noninteracting mutant of P56 on Wt E1 helicase activity is shown. (E) The ability to support DNA replication in vitro by the mutant E1 was not inhibited by P56. The quantified results are shown at the bottom.
The ΔF399 E1 mutant is biologically active in cells and is not inhibited by IFN treatment.
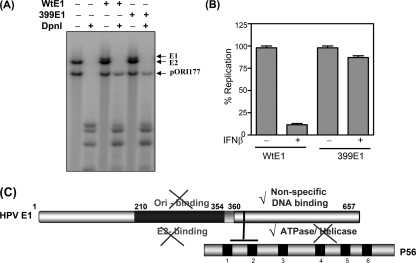
To determine whether the ΔF399 E1 mutant is biologically active in cells, we tested its ability to support HPV18 Ori DNA replication in transfected cells (22). As shown in Fig. 4A, the mutant E1 was competent in supporting Ori DNA replication; quantification of the signals showed that the ΔF399 E1 mutant was as efficient as Wt E1 (data not shown). Although the action of the Wt protein was severely inhibited by IFN treatment of the cells, replication supported by the mutant protein was only marginally affected (Fig. 4B). These results clearly suggest that IFN′s inhibitory effects on HPV DNA replication are primarily, if not entirely, mediated by the interaction of E1 with the IFN-induced protein P56.
FIG. 4.
(A) The ΔF399 E1 mutant could support Ori DNA replication in transfected cells. (A) The plasmid pOri177 (which contains the HPV18 origin) and expression vectors of E2 and wild-type E1 or the ΔF399 E1 mutant were cotransfected into C-33A cells. Replication of Ori DNA was analyzed by Southern blotting of DpnI-digested DNA. Arrows indicate the origin-containing DNA. (B) Ori DNA replication mediated by the ΔF399 E1 mutant was not inhibited by IFN. Cells were transfected as described above and treated with 100 U/ml of beta interferon (IFNβ). Replicated Ori DNA was quantitated and normalized. Data are represented as the means of 3 independent experiments. (C) Schematic of E1 functional and P56-interacting domains. Different functional domains of E1 are displayed. The E1-P56 interaction is mediated by P56 TPR2 and F399 of E1. By binding to E1, P56 inhibits E1 Ori binding, E2 binding, and helicase activity, while the ATPase and nonspecific-DNA binding are not affected. The sketch is not to scale.
Conclusions.
One major conclusion from this study is that only select functions of E1 are inhibited by P56. The linear functional-domain organization of E1 (26) and the binding site of P56 are depicted in Fig. 4C. To obtain a better understanding of the basis of P56's selective inhibitory action on E1, structural information on the E1/P56 complex will be required in the future. The other major conclusion from our study is the identification of P56 as the exclusive effector protein of IFN′s action against HPV DNA replication. Our previous investigation indicated that P56 is a major player; the genetic evidence presented here conclusively establishes P56 as the only relevant antiviral protein in this context.
Acknowledgments
This research was supported by National Institutes of Health grants CA068782 and AI073303.
Footnotes
Published ahead of print on 6 October 2010.
REFERENCES
- 1.Berg, M., and A. Stenlund. 1997. Functional interactions between papillomavirus E1 and E2 proteins. J. Virol. 71:3853-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, Y. E., L. Pena, G. C. Sen, J. K. Park, and L. A. Laimins. 2002. Long-term effect of interferon on keratinocytes that maintain human papillomavirus type 31. J. Virol. 76:8864-8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fensterl, V., and G. C. Sen. 2009. Interferons and viral infections. Biofactors 35:14-20. [DOI] [PubMed] [Google Scholar]
- 5.Fensterl, V., C. L. White, M. Yamashita, and G. C. Sen. 2008. Novel characteristics of the function and induction of murine p56 family proteins. J. Virol. 82:11045-11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goetz, G. S., F. B. Dean, J. Hurwitz, and S. W. Matson. 1988. The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J. Biol. Chem. 263:383-392. [PubMed] [Google Scholar]
- 7.Guo, J., D. J. Hui, W. C. Merrick, and G. C. Sen. 2000. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 19:6891-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haller, O., M. Frese, and G. Kochs. 1998. Mx proteins: mediators of innate resistance to RNA viruses. Rev. Sci. Tech. 17:220-230. [DOI] [PubMed] [Google Scholar]
- 9.Howley, P. M., and D. R. Lowy. 2001. Papillomaviruses and their replication, p. 2197-2229. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 10.Hughes, F. J., and M. A. Romanos. 1993. E1 protein of human papillomavirus is a DNA helicase/ATPase. Nucleic Acids Res. 21:5817-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelley, L. A., and M. J. Sternberg. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363-371. [DOI] [PubMed] [Google Scholar]
- 12.Khan, M. A., W. H. Tolleson, J. D. Gangemi, and L. Pirisi. 1993. Inhibition of growth, transformation, and expression of human papillomavirus type 16 E7 in human keratinocytes by alpha interferons. J. Virol. 67:3396-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, X., S. Schuck, and A. Stenlund. 2010. Structure-based mutational analysis of the bovine papillomavirus E1 helicase domain identifies residues involved in the nonspecific DNA binding activity required for double trimer formation. J. Virol. 84:4264-4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowy, D. R., and P. M. Howley. 2001. Papillomaviruses, p. 2231-2264. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 15.Sadler, A. J., and B. R. Williams. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanders, C. M., O. V. Kovalevskiy, D. Sizov, A. A. Lebedev, M. N. Isupov, and A. A. Antson. 2007. Papillomavirus E1 helicase assembly maintains an asymmetric state in the absence of DNA and nucleotide cofactors. Nucleic Acids Res. 35:6451-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuck, S., and A. Stenlund. 2005. Assembly of a double hexameric helicase. Mol. Cell 20:377-389. [DOI] [PubMed] [Google Scholar]
- 18.Sen, G. C., and S. N. Sarkar. 2007. The interferon-stimulated genes: targets of direct signaling by interferons, double-stranded RNA, and viruses. Curr. Top. Microbiol. Immunol. 316:233-250. [DOI] [PubMed] [Google Scholar]
- 19.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 20.Stenlund, A. 2003. E1 initiator DNA binding specificity is unmasked by selective inhibition of non-specific DNA binding. EMBO J. 22:954-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun, Y., H. Han, and D. J. McCance. 1998. Active domains of human papillomavirus type 11 E1 protein for origin replication. J. Gen. Virol. 79:1651-1658. [DOI] [PubMed] [Google Scholar]
- 22.Terenzi, F., P. Saikia, and G. C. Sen. 2008. Interferon-inducible protein, P56, inhibits HPV DNA replication by binding to the viral protein E1. EMBO J. 27:3311-3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turek, L. P., J. C. Byrne, D. R. Lowy, I. Dvoretzky, R. M. Friedman, and P. M. Howley. 1982. Interferon induces morphologic reversion with elimination of extrachromosomal viral genomes in bovine papillomavirus-transformed mouse cells. Proc. Natl. Acad. Sci. U. S. A. 79:7914-7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wacher, C., M. Muller, M. J. Hofer, D. R. Getts, R. Zabaras, S. S. Ousman, F. Terenzi, G. C. Sen, N. J. King, and I. L. Campbell. 2007. Coordinated regulation and widespread cellular expression of interferon-stimulated genes (ISG) ISG-49, ISG-54, and ISG-56 in the central nervous system after infection with distinct viruses. J. Virol. 81:860-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, C., J. Pflugheber, R. Sumpter, Jr., D. L. Sodora, D. Hui, G. C. Sen, and M. Gale, Jr. 2003. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J. Virol. 77:3898-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White, P. W., A. Pelletier, K. Brault, S. Titolo, E. Welchner, L. Thauvette, M. Fazekas, M. G. Cordingley, and J. Archambault. 2001. Characterization of recombinant HPV6 and 11 E1 helicases: effect of ATP on the interaction of E1 with E2 and mapping of a minimal helicase domain. J. Biol. Chem. 276:22426-22438. [DOI] [PubMed] [Google Scholar]
- 27.Zhao, C., T. Y. Hsiang, R. L. Kuo, and R. M. Krug. 2010. ISG15 conjugation system targets the viral NS1 protein in influenza A virus-infected cells. Proc. Natl. Acad. Sci. U. S. A. 107:2253-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.zur Hausen, H. 1991. Viruses in human cancers. Science 254:1167-1173. [DOI] [PubMed] [Google Scholar]