Abstract
Cell entry of reovirus requires a series of ordered steps, which include conformational changes in outer capsid protein μ1 and its autocleavage. The μ1N fragment released as a consequence of these events interacts with host cell membranes and mediates their disruption, leading to delivery of the viral core into the cytoplasm. The prototype reovirus strains T1L and T3D exhibit differences in the efficiency of autocleavage, in the propensity to undergo conformational changes required for membrane penetration, and in the capacity for penetrating host cell membranes. To better understand how polymorphic differences in μ1 influence reovirus entry events, we generated recombinant viruses that express chimeric T1L-T3D μ1 proteins and characterized them for the capacity to efficiently complete each step required for membrane penetration. Our studies revealed two important functions for the central δ region of μ1. First, we found that μ1 autocleavage is regulated by the N-terminal portion of δ, which forms an α-helical pedestal structure. Second, we observed that the C-terminal portion of δ, which forms a jelly-roll β barrel structure, regulates membrane penetration by influencing the efficiency of ISVP* formation. Thus, our studies highlight the molecular basis for differences in the membrane penetration efficiency displayed by prototype reovirus strains and suggest that distinct portions of the reovirus δ domain influence different steps during entry.
In comparison to enveloped viruses, the mechanisms by which nonenveloped viruses enter host cells are not well understood. While enveloped viruses use membrane fusion triggered by receptor- or pH-mediated activation of viral membrane protein for entering host cells (63), nonenveloped viruses lack a membrane and therefore must bypass host membranes by mechanisms other than fusion. Nonenveloped viruses from diverse families appear to use a conserved set of cell entry strategies (4, 59). However, a detailed molecular understanding of how their genomic payloads access the interior of host cells has yet to be elucidated. Mammalian reoviruses, which form nonenveloped viruses with two concentric protein shells, are extensively used as model systems to study different aspects of virus-host interactions, including virus entry (12, 17).
Successful entry of reovirus into host cells requires a series of events that culminate in delivery of the inner protein shell, or core, into the host cell cytoplasm (12, 17). Reovirus entry is initiated by binding of the viral attachment protein, σ1, to cell surface carbohydrates (5, 15, 27, 28, 52) and junctional adhesion molecule A (6). Following receptor engagement, reovirus is internalized by β1 integrin-dependent endocytosis (9, 10, 22, 41, 42, 57). Within endosomes, low pH and cathepsin proteases facilitate stepwise disassembly of reovirus to form a metastable intermediate, the infectious subvirion particle (ISVP), which contains a conformationally altered σ1 protein, lacks the σ3 outer capsid protein (which is degraded by proteolysis), and contains a μ1 membrane penetration protein that has been cleaved into three particle-associated fragments (3, 10, 14, 20, 21, 55, 57). Generation of the three μ1 fragments requires autocleavage of μ1 at residue 42, which results in the formation of μ1N and μ1C (34). In addition, μ1C is proteolytically cleaved near residue 580 to generate δ and φ fragments (48). Conformational changes in ISVPs lead to the generation of the next entry intermediate, the ISVP*(11). These rearrangements result in dissociation of μ1N, φ, and σ1 from the ISVP and lead to exposure of hydrophobic domains within the particle-associated δ fragment (11, 33). The μ1N fragments released from the particles are necessary and sufficient to mediate membrane penetration (33, 65). Membrane penetration by μ1N leads to the generation of 4- to 10-nm pores (1, 65), which are thought to facilitate the delivery of the 70-nm cores into the cytoplasm via an as-yet-unknown mechanism.
Prototype reovirus strains, type 1 Lang (T1L) and type 3 Dearing (T3D), display numerous differences in the cell entry pathway described above. For example, T1L and T3D differ in efficiency of autocatalytic μ1 cleavage (49), in the efficiency to form ISVP*s (11) and ultimately in the capacity to permeabilize host cell membranes and evoke hemolysis (11, 39). In each case, T3D is more efficient than T1L and reassortant analysis indicates that the difference in efficiency genetically maps to the M2 gene segment (11, 39, 49), which encodes the membrane penetration protein, μ1 (46). Comparison of the deduced amino acid sequences of our laboratory stocks of T1L and T3D revealed differences at fifteen amino acid residues in the 708-amino-acid μ1 protein, which likely account for the phenotypic differences described above. Although the amino acid sequence of the μ1N fragment was found to be identical between T1L and T3D, nine and six residues were different within the δ and φ domains, respectively (Table 1). The μ1 protein folds into three α-helical domains formed by δ and φ near the base of the μ1 trimer and one jelly-roll β barrel domain formed by δ near its apex (38). How these structural domains contribute to the various functions ascribed to μ1 during cell entry is not known.
TABLE 1.
Location and nature of polymorphic residues in T1L and T3D μ1 proteins
| Polymorphic position | Residue |
|
|---|---|---|
| T1L | T3D | |
| 51 | I | V |
| 96 | N | I |
| 109 | A | T |
| 128 | L | M |
| 305 | V | A |
| 327 | K | R |
| 340 | T | S |
| 342 | G | N |
| 517 | E | D |
| 670 | T | A |
| 682 | N | S |
| 685 | G | S |
| 686 | S | G |
| 688 | P | S |
| 690 | A | N |
In the present study, we used reverse genetics to identify μ1 domains that account for strain-specific differences between T1L and T3D. Using recombinant reoviruses that encode chimeric μ1 proteins derived from portions of T1L and T3D M2 gene segments, we examined the efficiency of μ1-regulated steps during reovirus cell entry. We found that presence of the T3D δ domain increases the efficiency of μ1 autocleavage and ISVP* formation, thereby facilitating membrane penetration. To better define the contribution of the δ domain to these steps, we generated additional viruses that express μ1 proteins with swaps between portions of the T1L and T3D δ domains. Characterization of these viruses indicated a function for the pedestal-forming region in regulating efficiency of autocleavage, whereas a role for the jelly-roll β barrel domain in regulating conformational changes required for membrane penetration. These findings suggest independent regulation of μ1 autocleavage, ISVP* formation, and membrane penetration by distinct portions of the μ1 δ domain during entry of reovirus into host cells.
MATERIALS AND METHODS
Cells.
Murine L929 (L) cells were maintained in Joklik minimal essential medium (MEM; Lonza) supplemented to contain 5% fetal bovine serum (FBS) (Sigma-Aldrich), 2 mM l-glutamine (Invitrogen), 100 U of penicillin (Invitrogen)/ml, 100 μg of streptomycin (Invitrogen)/ml, and 25 ng of amphotericin B (Sigma-Aldrich)/ml. BHK-T7 cells were maintained in Dulbecco modified Eagle medium (Invitrogen) supplemented to contain 5% FBS, 2 mM l-glutamine, 2% MEM amino acid solution (Invitrogen), and 1 mg of Geneticin (Invivogen)/ml in alternate passages.
Plasmids.
pT7-M2T1L and pT7-M2T3D (35, 36) were used to construct plasmids, pT7-M2DL, pT7-M2LD, pT7-M2(LD)L, pT7-M2(LD)D, pT7-M2(DL)L, and pT7-M2(DL)D, which express chimeric μ1 proteins (see Fig. 1 and Fig. 5A). pT7-M2DL and pT7-M2LD were constructed by reciprocally exchanging restriction fragments obtained from digestion of pT7-M2T1L and pT7-M2T3D with NcoI and SpeI. For the construction of pT7-M2(LD)L, pT7-M2(LD)D, pT7-M2(DL)L, and pT7-M2(DL)D, a SnaBI site was introduced in pT7-M2T1L and pT7-M2T3D plasmids by QuikChange (Stratagene) site-directed mutagenesis at nucleotides 963 and 739, respectively, in each case replacing a G for a T. These changes do not lead to a change in the amino acid sequence. pT7-M2(LD)D and pT7-M2(DL)L plasmids were constructed by reciprocally exchanging restriction fragments obtained from digestion of pT7-M2T1L(SnaBI) and pT7-M2T3D(SnaBI) with NcoI and SnaBI. pT7-M2(LD)L and pT7-M2(DL)D plasmids were constructed by reciprocally exchanging restriction fragments obtained from digestion of pT7-M2T1L(SnaBI) and pT7-M2T3D(SnaBI) with SpeI and SnaBI. Chimeric M2 plasmids were confirmed by sequencing.
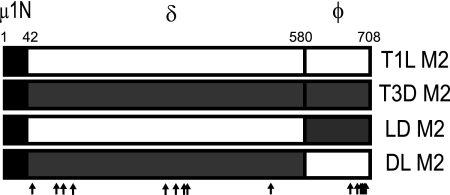
FIG. 1.
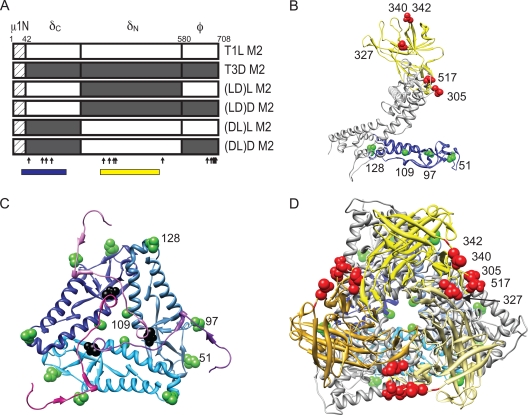
Viruses with chimeric μ1 membrane penetration protein. Schematic diagrams of the μ1-encoding T1L and T3D M2 gene segments are shown, along with cleavage products generated during virus entry. Chimeric M2 gene segments (LD and DL) were constructed by reciprocally exchanging the δ- and φ-encoding portions of T1L and T3D M2 gene segments. The amino acid sequence of μ1N is identical for T1L and T3D and is shown as a hatched box. The locations of polymorphic residues in T1L and T3D μ1 are shown using vertical arrows.
Viruses.
Recombinant strain T1L (rsT1L), rsT1L/T3D M2, rsT1L/DL M2, rsT1L/LD M2, rsT1L/(LD)L M2, rsT1L/(LD)D M2, rsT1L/(DL)L M2, and rsT1L/(DL)D M2, which contain a wild-type or chimeric M2 gene in an otherwise T1L background were generated by using a four- or ten-plasmid-based reverse-genetics strategy (36). To confirm sequences of mutant viruses, viral RNA was extracted from infected cells and subjected to reverse transcription-PCR using three sets of M2-specific primers. PCR products were resolved on Tris-acetate-EDTA agarose gels, purified and subjected to sequence analysis. Purified reovirus virions were generated by using second- or third-passage L-cell lysate stocks of reovirus as described previously (25). Viral particles were extracted from infected cell lysates using Vertrel-XF (Dupont) (43), layered onto 1.2- to 1.4-g/cm3 CsCl gradients, and centrifuged at 187,813 × g for 4 h. Bands corresponding to virions (1.36 g/cm3) (56) were collected and dialyzed in virion-storage buffer (150 mM NaCl, 15 mM MgCl2, 10 mM Tris-HCl [pH 7.4]). The concentration of reovirus virions in purified preparations was determined from an equivalence of one optical density (OD) unit at 260 nm equals 2.1 × 1012 virions (56). Virus titer was determined by plaque assay using L cells (60). ISVPs were generated by incubation of 2 × 1011 virions with 200 μg of N-p-tosyl-l-lysine chloromethyl ketone-treated chymotrypsin/ml in a total volume of 0.1 ml at 37°C for 1 h (47). Proteolysis was terminated by addition of 2 mM phenylmethylsulfonyl fluoride and incubation of reactions on ice. Generation of ISVPs was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie brilliant blue staining.
Growth assay.
L cells (2 × 105) in 24-well plates were adsorbed in triplicate with 2 PFU/cell of reovirus at room temperature for 1 h, washed once with phosphate-buffered saline (PBS), and incubated at 37°C for 0 and 24 h. Cells were frozen and thawed twice prior to determination of the virus titer by plaque assay using L cells (60). Viral yield was calculated according to the following formula: viral yield (log10PFU/ml) = log10(PFU/ml)24 h − log10(PFU/ml)0 h.
Analysis of μ1N-μ1C cleavage efficiency.
Reducing sample buffers containing 125 mM Tris (adjusted to a final pH of 6.8, 8.3, or 9.8), 10% (vol/vol) glycerol, 2% (wt/vol) SDS, 0.01% (wt/vol) bromophenol blue, and 5% β-mercaptoethanol were mixed with an equal volume of dialysis buffer containing 1011 virions. The samples were disrupted by heating at 60°C for 5 min, resolved on SDS-10% PAGE gels and stained using Coomassie brilliant blue. Gels were scanned by using an Odyssey infrared imager (LI-COR), and the intensity of μ1 and μ1C bands was determined by densitometry using Odyssey software. The percent uncleaved μ1 was calculated as described previously (49), according to the following formula: % uncleaved μ1 = [μ1 intensity/(μ1 + μ1C intensity)] × 100.
Analysis of ISVP* generation.
ISVPs (8 × 1010) were incubated with 300 mM CsCl or NaCl at 30°C for 20 min, transferred to ice for 20 min, and incubated with 100 μg of trypsin/ml at 4°C for 30 min. Trypsin digestion was terminated by addition of SDS-PAGE loading buffer and removal of the samples to dry ice. Generation of ISVP*s was confirmed by SDS-PAGE and Coomassie brilliant blue staining.
Measurement of thermal stability.
ISVPs in virion storage buffer were incubated at various temperatures for 15 min and transferred to ice for 1 h. Residual infectivity in triplicate samples was assessed by plaque assay (60).
Hemolysis assay.
Citrated calf red blood cells (RBCs; Colorado Serum Company) were washed extensively with chilled PBS supplemented to contain 2 mM MgCl2 (PBS-Mg) and resuspended at a concentration of 30% (vol/vol) in PBS-Mg. Hemolysis efficiency was analyzed by mixing a 3-μl aliquot of resuspended RBCs with virion-storage buffer containing 0 or 300 mM CsCl-1% Triton X-100 (TX-100) in virion storage buffer or ISVPs in a total volume of 30 μl, followed by incubation at 37°C for 1 h. Samples were placed on ice for 30 min to prevent further hemolysis and centrifuged at 500 × g at 4°C for 3 min. The extent of hemoglobin release was quantified by measuring the A405 of a 1:5 dilution of the supernatant in a microplate reader (Molecular Devices). The percent hemolysis was calculated by using the following formula: % hemolysis = 100 × (Asample − Abuffer)/(ATX-100 − Abuffer).
Assessment of ISVP* formation after exposure to erythrocytes.
Hemolysis reactions were set up as described above. After incubation at 37°C for 40 min, samples were transferred to ice for 20 min and incubated with 100 μg of trypsin/ml at 4°C for 30 min. Trypsin digestion was terminated by the addition of SDS-PAGE loading buffer and removal of the samples to dry ice. Samples were resolved by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked for 1 h in blocking buffer (Tris-buffered saline [TBS] containing 0.05% Tween 20 and 5% milk), followed by incubation with a μ1-specific MAb 4A3 diluted 1:1,000 in blocking buffer at 4°C overnight. Membranes were washed three times for 10 min each time with TBS containing 0.05% Tween 20 and incubated with 1:25,000 dilution of Alexa Fluor-conjugated goat anti-mouse immunoglobulin in blocking buffer. After three washes, membranes were scanned using an Odyssey infrared imager (LI-COR).
RESULTS
Viruses with chimeric μ1 are viable.
Entry of reovirus into host cells consists of a series of events which include μ1 autocleavage, ISVP formation, ISVP-to-ISVP* conversion, and membrane penetration. Prototype reovirus strains T1L and T3D display differences in efficiency at various steps in this pathway and these differences segregate with the μ1-encoding M2 gene segment (11, 39, 49). To define μ1 domains that contribute to the strain-specific differences in efficiency of cell entry between T1L and T3D, we constructed M2 chimeric gene segments by reciprocally exchanging δ and φ domains of T1L and T3D M2 gene segments (Fig. 1). Using plasmid-based reverse genetics, we generated recombinant reoviruses that express either T1L, T3D, or chimeric μ1 proteins in an otherwise T1L background (36). These viruses were designated rsT1L, rsT1L/T3D M2, rsT1L/LD M2 (containing δ from T1L, φ from T3D), and rsT1L/DL M2 (containing δ from T3D and φ from T1L). To evaluate whether the differences in μ1 sequences affect viral growth over a single cycle of reovirus replication, we infected L cells with each of the viruses at a multiplicity of infection (MOI) of 2 PFU/cell and quantified virus titers at 0 and 24 h after infection. At 24 h after infection, each virus strain showed an equivalent (∼100-fold) increase in virus titer (data not shown), indicating that the μ1 chimeric viruses replicate as efficiently as the wild-type virus.
The δ domain is a determinant of μ1N-μ1C cleavage efficiency.
The μ1 protein undergoes autocleavage during virus assembly or cell entry to form μ1N and μ1C (40, 49, 58). This cleavage event affects the release of μ1N and serves an essential role in controlling membrane penetration and virion infectivity (1, 33, 50). When reovirus particles are resolved on denaturing acrylamide gels under standard conditions, the μ1 protein is predominantly found in its autocleaved form, μ1C. However, increasing the pH of the denaturing sample buffer used for resolving viruses for electrophoresis leads to a greater recovery of nonautocleaved μ1 (49). Using this strategy, it has been determined that T3D μ1 is more susceptible to autocleavage in comparison to T1L μ1 (49). Moreover, these strain-specific differences in cleavage efficiency map to the M2 gene segment (49). Although experimental data for this is as yet lacking, we think it possible that differences in autocleavage efficiency observed for T1L and T3D μ1 proteins in vitro also correlate with the extent of autocleavage during cell entry and that similar regions of μ1 may regulate autocleavage under both conditions.
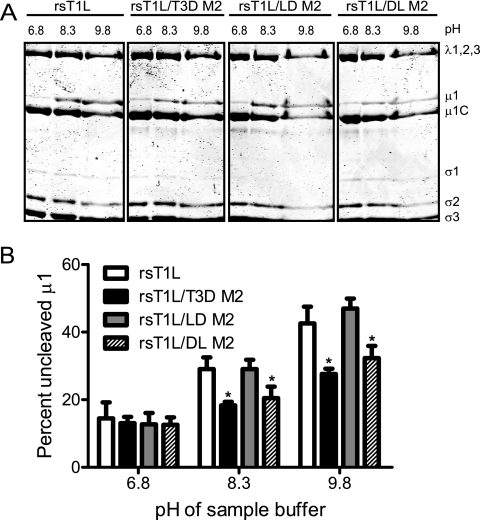
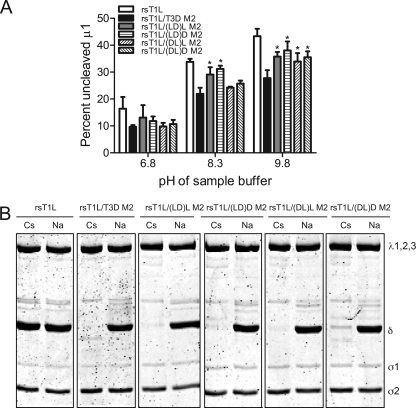
To define regions in μ1 that influence the efficiency of μ1 autocleavage, we compared the capacity of μ1 proteins from rsT1L/LD M2 and rsT1L/DL M2 to undergo autocleavage to rsT1L and rsT1L/T3D M2 (Fig. 2A). For these experiments, virus particles from each virus strain were electrophoresed using sample buffers at pH 6.8, 8.3, or 9.8, and the percentage of μ1 that remained uncleaved was determined by densitometric analysis (Fig. 2B). For samples resolved under standard conditions of pH 6.8, the percentage of uncleaved μ1 was found to be approximately equal for all virus strains tested. As expected, we found that as the pH was increased, a progressively greater proportion of μ1 protein from rsT1L was found in the uncleaved form in comparison to μ1 from rsT1L/T3D M2. When chimeric μ1 viruses were analyzed, we found that while μ1 from rsT1L/LD M2 showed autocleavage efficiency similar to rsT1L, μ1 from rsT1L/DL M2 displayed autocleavage characteristic similar to rsT1L/T3D M2. These data indicate that differences between T1L and T3D in efficiency of μ1N-μ1C cleavage are influenced by the δ, but not the φ domain.
FIG. 2.
The δ domain affects μ1N-μ1C cleavage efficiency. (A) CsCl-purified preparations of the indicated virus strains were mixed with reducing sample buffers at pH 6.8, 8.3, or 9.8. After incubation at 60°C for 5 min, the samples were resolved on SDS-10% PAGE gels and stained with Coomassie brilliant blue. (B) The intensities of the μ1 and μ1C bands were determined by densitometric analysis using a LI-COR Odyssey scanner. The fraction of uncleaved μ1 was determined by dividing the intensity of the μ1 band by the total intensity of the μ1 and μ1C bands. The results are expressed as the mean percent uncleaved μ1 quantified from three independent experiments. Error bars indicate the standard deviations (SD). *, P < 0.05, as determined by using the Student t test in comparison to rsT1L at the respective sample buffer pH.
The δ domain affects efficiency of ISVP* formation.
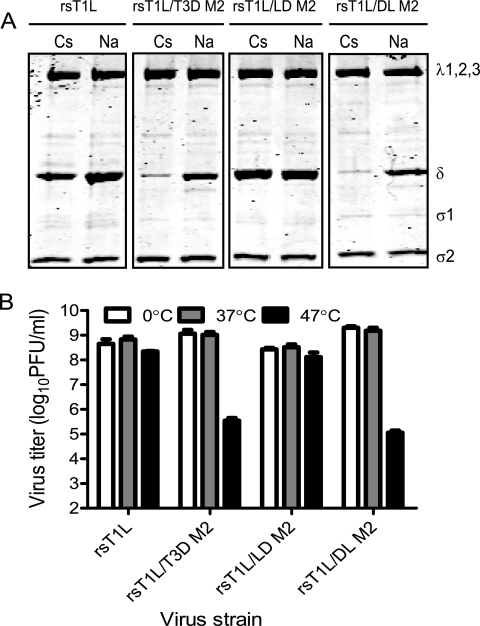
Membrane penetration by ISVPs requires conversion to ISVP*s, which are characterized by the release of μ1N and φ fragments and conformational changes in the particle-associated δ fragment (11, 13, 33). Although the physiologic trigger for ISVP-to-ISVP* conversion is not known, in vitro studies using ISVPs exposed to high concentrations of Cs or K ions are thought to mimic conditions required for ISVP* formation. Under these conditions, T3D forms ISVP*s more readily than T1L with the difference in efficiency mapping to the M2 gene segment (11). To identify the μ1 domain that affects ISVP* formation, we incubated in vitro-generated ISVPs of each virus strain with 300 mM CsCl or with 300 mM NaCl as a control (11) (Fig. 3A). Conformational changes in particle-associated δ were examined by assessing its protease sensitivity to trypsin digestion. Consistent with previous observations (11), we observed minimal protease-mediated loss of T1L δ after treatment of rsT1L with CsCl (Fig. 3A). In contrast, the T3D-derived δ fragment of rsT1L/T3D M2 was lost upon protease treatment in the presence of CsCl. Analysis of chimeric μ1 viruses indicated that δ from rsT1L/LD M2 displayed protease resistance analogous to rsT1L, whereas δ from rsT1L/DL M2 showed protease sensitivity similar to rsT1L/T3D M2 (Fig. 3A). These findings indicate that the capacity to undergo ISVP-to-ISVP* conversion is also controlled by the δ domain and that the presence of T1L residues within this domain renders this process inefficient.
FIG. 3.
The δ domain influences the capacity to undergo ISVP-to-ISVP* conversion. (A) ISVPs of the indicated viruses were treated with CsCl or NaCl at 30°C for 20 min, chilled on ice for 20 min, and treated with trypsin at 4°C for 30 min. Samples were resolved by SDS-PAGE and stained using Coomassie brilliant blue. The positions of reovirus capsid proteins are shown. (B) ISVPs of the indicated viruses were incubated at 37, or 47°C for 15 min. Residual infectivity was assessed by plaque assay. The results are shown as virus titers for triplicate samples. Error bars indicate the SD.
Previous studies indicate that analogous to CsCl treatment, heat treatment of ISVPs also promotes a conformational change that increases the protease sensitivity of the δ fragment (45). Heat-induced conformational changes also result in decreased viral infectivity (16, 44). As an independent measure of the capacity to generate ISVP*s, we assessed the thermal sensitivity of our viruses. For these studies, ISVPs of each virus strain were incubated at 0, 37, and 47°C for 15 min, and the residual infectivity of the viruses was determined by plaque assay (Fig. 3B). While the infectivity of rsT1L and rsT1L/LD M2 was unaffected after high-temperature incubation, the infectivity of rsT1L/T3D M2 and rsT1L/DL M2 was diminished by ∼4 orders of magnitude after incubation at 47°C. These findings further support the idea that viruses containing the δ domain from T1L are less capable of undergoing conformational changes that resemble ISVP-to-ISVP* conversion.
Strain-specific differences in membrane penetration are affected by δ.
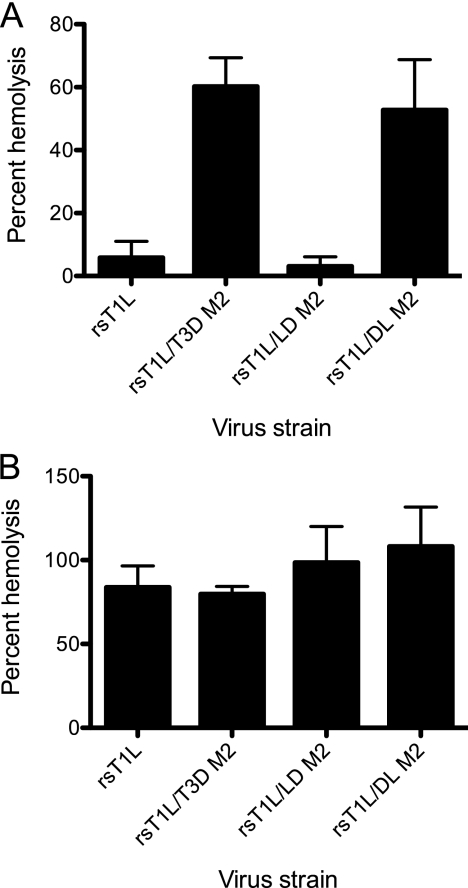
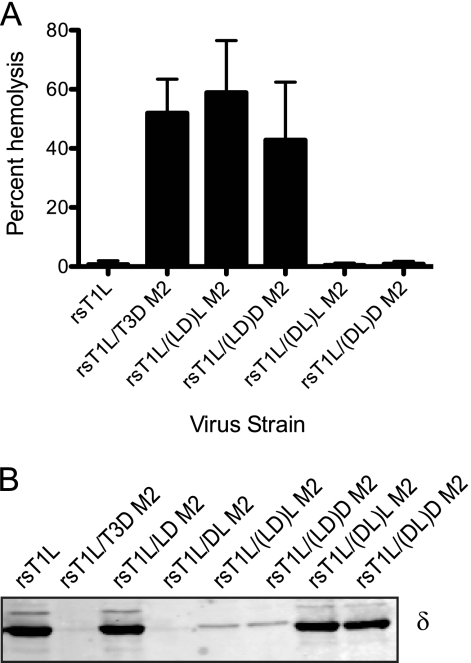
The capacity of reovirus ISVPs to disrupt erythrocyte membrane integrity and produce hemolysis correlates with endosomal membrane penetration (13, 16, 18, 44). Reovirus strains T1L and T3D differ in their membrane-penetration capacities, with T3D being more efficient than T1L, and this efficiency difference has been mapped to the M2 gene segment (11). To identify μ1 domains that affect membrane penetration capacity of reovirus, we tested ISVPs generated from wild-type and chimeric viruses for the capacity to lyse erythrocytes (Fig. 4A). As expected, while rsT1L failed to produce hemolysis, rsT1L/T3D M2 efficiently lysed erythrocytes. Analysis of chimeric μ1 viruses under these conditions showed that rsT1L/LD M2 is incapable of erythrocyte lysis, whereas rsT1L/DL M2 is capable of eliciting erythrocyte lysis. These findings suggest that residues within the δ domain of T3D M2 are required for efficient membrane penetration by reovirus.
FIG. 4.
The δ domain modulates membrane penetration. A 3% (vol/vol) solution of bovine erythrocytes was incubated with 4.8 × 1010 ISVPs of the indicated viruses in virion-storage buffer containing no CsCl (A) or 300 mM CsCl (B) at 37°C for 1 h. Hemolysis was quantified by determining absorbance of the supernatant at 405 nm. Hemolysis after treatment of an equal number of cells with virion-storage buffer or virion-storage buffer containing 1% TX-100 was considered to be 0 or 100%, respectively. The results are expressed as the mean percent hemolysis for triplicate samples. Error bars indicate the SD.
Two possible mechanisms may explain why rsT1L and rsT1L/LD M2 fail to elicit hemolysis. First, the decreased efficiency may be a consequence of a diminished capacity to generate ISVP* as described in Fig. 3 above. Second, reduced hemolytic capacity of rsT1L and rsT1L/LD M2 may be related to a decrease in efficiency of pore formation. To distinguish between these possibilities, we performed hemolysis reactions in the presence of CsCl. Addition of CsCl to the reaction allows hemolysis by T1L (11), likely by promoting conformational changes that are required for the release and exposure of the μ1N fragment (33, 65). Consistent with previous studies, rsT1L produced efficient hemolysis in the presence of 300 mM CsCl to levels that were equivalent to those produced by rsT1L/T3D M2 (Fig. 4B). Similarly, both rsT1L/LD M2 and rsT1L/DL M2 also were capable of erythrocyte lysis in the presence of CsCl. These findings indicate that the diminished capacity of rsT1L/LD M2 to undergo ISVP-to-ISVP* conversion likely contributes to the diminished capacity of this virus to elicit hemolysis.
Viruses expressing chimeric δ domains are viable.
The findings presented thus far indicate a function for δ domain in μ1 autocleavage and in ISVP-to-ISVP* conversion. The nine polymorphic differences in the δ domain form two clusters, one each toward the N- and C-terminal ends of δ, henceforth designated δN and δC, respectively (Fig. 5A). Four polymorphic residues (residues 51, 96, 109, and 128) that lie within δN are all found within domain I, as described in the crystal structure of μ1 (encompassing amino acids 30 to 186), which forms an α-helical pedestal at the base of the μ1 trimer (38) (Fig. 5B). The remaining five residues (residues 305, 327, 340, 342, and 517) that are found in δC lie within or proximal to domain IV, as described in the crystal structure of μ1 (encompassing amino acids 306 to 514), which forms a jelly-roll β barrel structure at the apex of the trimer (38) (Fig. 5B). To define which portion of δ determines strain-specific differences in μ1N-μ1C cleavage and ISVP* formation, we constructed chimeric gene segments by reciprocally exchanging cDNA fragments corresponding to the δN and δC portions of T1L and T3D μ1 (Fig. 5A). These constructs were used to generate four additional recombinant viruses in a T1L background, rsT1L/(LD)L M2 (containing δN from T1L, δC from T3D, and φ from T1L), rsT1L/(LD)D M2 (containing δN from T1L, δC from T3D, and φ from T3D), rsT1L/(DL)L M2 (containing δN from T3D, δC from T1L, and φ from T1L), and rsT1L/(DL)D M2 (containing δN from T3D, δC from T1L, and φ from T3D) (Fig. 5A). To determine whether the chimeric δ viruses were capable of efficient replication, we infected L cells with rsT1L and chimeric viruses at an MOI of 2 PFU/cell, and we quantified the virus titers at 0 and 24 h after infection. At 24 h after infection, each virus strain showed an equivalent (∼100-fold) increase in virus titer, indicating that the chimeric δ viruses are not compromised for growth and replicate as efficiently as the wild-type virus (data not shown).
FIG. 5.
Viruses with chimeric δ fragments. (A) Schematic diagram of the μ1-encoding T1L and T3D M2 gene segments are shown, along with cleavage products generated during virus entry. Chimeric M2 gene segments (LD)L, (LD)D, (DL)L, and (DL)D were constructed by reciprocally exchanging the δN- and δC-encoding portions of T1L and T3D M2 gene segments. The locations of polymorphic residues in T1L and T3D μ1 are indicated by vertical arrows. Regions spanning domain I and domain IV are indicated by blue and yellow boxes, respectively. (B) A side view of a μ1C monomer rendered using UCSF chimera from the crystal structure of μ1 (PDB:1JMU) is shown with domain I in blue and domain IV in yellow (38, 53). The positions of the polymorphic residues within domains I and IV are shown in green and red, respectively. Residue 97 is labeled since residue 96 was not resolved in the crystal structure and is not in the PDB file. (C) A close-up top view of the region near the autocleavage site is shown, with the domain I comprising δ in shades of blue and the μ1N fragment in shades of magenta. Polymorphic residues on one of the three chains are labeled as described for panel B. The autocleavage site is shown in black. (D) A top view of a μ1C trimer is shown, with domain I in shades of blue and domain IV in shades of yellow. Polymorphic residues on one of the three chains are labeled as described for panel B.
Distinct portions of the δ domain regulate autocleavage and ISVP* formation.
To determine the contribution of the δN and δC domains to regulation of μ1 autocleavage, we incubated purified preparations of each virus with sample buffers of increasing pHs and compared the fraction of uncleaved μ1 present in each preparation using densitometric analysis (Fig. 6A). Analogous to parental viruses rsT1L and rsT1L/T3D M2, we found that the fraction of uncleaved μ1 for each virus in sample buffer of pH 6.8 was equivalent. As the pH of the sample buffer was increased to 8.3, a substantially greater fraction of T1L μ1 was found in the uncleaved form in comparison to T3D μ1. Analysis of δ chimeric viruses indicated that at pH 8.3, rsT1L/(LD)L M2 and rsT1L/(LD)D M2 also showed a statistically higher percentage of uncleaved μ1 in comparison to rsT1L/T3D M2. In contrast, a minimal increase in uncleaved μ1 was observed for rsT1L/(DL)L M2 and rsT1L/(DL)D M2. Similar results were obtained when ISVPs of each of these viruses were incubated in sample buffer at pH 9.8, with rsT1L/(LD)L M2 and rsT1L/(LD)D M2 displaying a higher fraction of uncleaved μ1 than rsT1L/T3D M2. In addition to these viruses, the fraction of uncleaved μ1 in rsT1L/(DL)L and rsT1L/(DL)D M2 also was found to be greater than that present in rsT1L/T3D M2 at pH 9.8. These findings suggest that the presence of the δN portion of T3D increases the efficiency of μ1 autocleavage and hints at a possible contribution from the δC domain in controlling autocleavage.
FIG. 6.
Different portions of δ modulate autocleavage efficiency and ISVP* formation. (A) CsCl-purified preparations of the indicated virus strains were mixed with reducing sample buffers at pH 6.8, 8.3, or 9.8. After incubation at 60°C for 5 min, the samples were resolved on SDS-10% PAGE gels and stained using Coomassie brilliant blue. The intensities of the μ1 and μ1C bands were determined by densitometric analysis using the LI-COR Odyssey scanner. The fraction of uncleaved μ1 was determined by dividing the intensity of the μ1 band by the total intensity of the μ1 and μ1C bands. The results are expressed as mean percent uncleaved μ1 for four independent experiments. Error bars indicate the SD. *, P < 0.05, as determined by using the Student t test in comparison to rsT1L/T3D M2 at the respective sample buffer pH. (B) ISVPs of the indicated viruses were treated with CsCl or NaCl at 30°C for 20 min, chilled on ice for 20 min, and treated with trypsin at 4°C for 30 min. Samples were resolved by SDS-PAGE and stained using Coomassie brilliant blue. The positions of reovirus capsid proteins are shown.
To define which portion of δ affects the capacity for ISVP-to-ISVP* transition, ISVPs from each virus strain were incubated with either CsCl or NaCl and assessed for protease susceptibility of the particle-associated δ fragment, which is characteristic of ISVP* formation (Fig. 6B). As expected, the δ domain from the parental viruses rsT1L and rsT1L/T3D M2, respectively, displayed protease resistance and protease susceptibility in the presence of CsCl. When chimeric δ viruses were similarly examined, we found that the δ fragment of all viruses was converted to a protease-sensitive form in the presence of CsCl, suggesting that these viruses were capable of forming ISVP*s under conditions in which rsT1L was not. These results suggest that the presence of either the δN or the δC from T3D is sufficient to confer to T1L μ1 the capacity for efficient ISVP-to-ISVP* conversion.
Sequences within δC regulate the efficiency of membrane penetration.
To determine how δN and δC influence membrane penetration by reovirus, we compared the hemolytic capacities of the chimeric δ viruses to those of the parental viruses (Fig. 7A). We found that rsT1L/(LD)L M2 and rsT1L(LD)D M2 were capable of hemolysis analogous to rsT1L/T3D M2. In contrast, rsT1L/(DL)L M2 and rsT1L/(DL)D M2 were unable to lyse bovine erythrocytes similar to rsT1L. Because rsT1L/(DL)L M2 and rsT1L/(DL)D M2 μ1 proteins are efficiently autocleaved (Fig. 6A) and because these viruses are capable of ISVP* formation when treated with Cs (Fig. 6B), their reduced capacity for hemolysis suggest a function for the δ domain subsequent to ISVP* formation. An alternate, equally likely possibility is that despite their capacity to undergo conformational changes that resemble ISVP* formation after incubation with CsCl in vitro, these viruses fail to undergo functional conformational changes that result in ISVP* formation and μ1N release during interaction with membranes, as would occur during hemolysis. To test this idea, trypsin sensitivity of δ in hemolysis reactions was assessed by using immunoblots. Consistent with previous results (11), the δ fragments from rsT1L and rsT1L/T3D, respectively, displayed trypsin resistance and trypsin susceptibility. Similar examination of our chimeric viruses indicated that δ fragments of nonhemolytic viruses rsT1L/LD M2, rsT1L/(DL)L M2, and rsT1L/(DL)D M2 remained trypsin resistant, whereas those from hemolysis-competent viruses rsT1L/DL M2, rsT1L/(LD)L M2, and rsT1L/(LD)D M2, were protease sensitive (Fig. 7B). Based on these findings we conclude that one or more residues within T3D-derived δC promote hemolysis by increasing the efficiency of ISVP* formation.
FIG. 7.
The jelly-roll β barrel domain affects efficiency of membrane penetration. (A) A 3% (vol/vol) solution of bovine erythrocytes was incubated with 4.8 × 1010 ISVPs of the indicated viruses in virion-storage buffer at 37°C for 1 h. Hemolysis was quantified by determining absorbance of the supernatant at 405 nm. Hemolysis after treatment of an equal number of cells with virion-storage buffer or virion-storage buffer containing 1% TX-100 was considered to be 0 or 100%, respectively. The results are expressed as mean percent hemolysis for six samples. Error bars indicate the SD. (B) A 3% (vol/vol) solution of bovine erythrocytes was incubated with 4.8 × 1010 ISVPs of the indicated viruses in virion-storage buffer at 37°C for 40 min, chilled on ice for 20 min, and treated with trypsin at 4°C for 30 min. Samples were resolved by SDS-PAGE and transferred to nitrocellulose membranes. The samples were probed with a μ1-specific MAb and visualized by using a LI-COR Odyssey scanner.
DISCUSSION
Membrane penetration by reovirus is an exquisitely regulated, multistep process controlled by the μ1 outer capsid protein (12, 17). Successful membrane penetration requires priming of the membrane-penetration machinery through μ1 autocleavage (1, 33, 49, 50). In addition, conformational changes in the μ1 protein are required to expose domains capable of membrane interaction and pore formation (11, 13, 16). Although the primary sequence of the 708-amino-acid μ1 proteins of prototype reovirus strains T1L and T3D differ by only fifteen amino acids, these viruses exhibit differences in the efficiency of autocleavage (49), the propensity to undergo conformational changes required for membrane penetration (11), and the capacity of these strains to permeabilize host cell membranes (11, 39). To better understand how each of these cell entry steps is regulated by μ1, we generated and characterized recombinant viruses that express chimeric T1L-T3D μ1 proteins. We found that the efficiency of μ1 autocleavage is regulated by the α-helical pedestal domain formed by the N-terminal region of δ (δN). In addition, we found that the C-terminal portion of δ, which forms the jelly-roll β barrel (δC), regulates the membrane penetration efficiency of reovirus by controlling the efficiency of ISVP* formation. Collectively, our studies highlight how amino acid polymorphisms within reovirus μ1 affect differences in efficiency of cell entry displayed by prototype reovirus strains and indicate that distinct portions of the reovirus δ domain influence different steps during entry.
Autocleavage of μ1 resulting in the formation of μ1N and μ1C is required for membrane penetration (1, 33, 49, 50). Recent biochemical evidence along with structural information about μ1 from high-resolution cryo-electron microscopic (cryoEM) structures indicate that μ1 exists in its unautocleaved form in assembled virions and suggest that autocleavage occurs at an early stage during entry (49, 66). Conformational changes in μ1δ occurring as a consequence of ISVP-to-ISVP* transition have been proposed as a possible trigger for autocleavage (49). However, it is also possible that autocleavage occurs prior to this step. In support of this idea, a recent 3.3-Å reconstruction of related aquareovirus indicates that VP5, the μ1 homolog, exists in its autocleaved form in the ISVP (67). The autocleavage site between Asn42 and Pro43 lies within a pocket on the upper surface of domain I (Fig. 5C), and it is hypothesized that residues on the same or adjacent μ1 molecules that are in close proximity to the cleavage site may facilitate autocleavage (38). The data presented here offer experimental evidence to support this idea. We found that viruses containing the pedestal forming domain from T3D display higher μ1 autocleavage efficiency in comparison to those that contain the similar region from T1L. Based on structural information, residues Thr104, Glu105, and His106 on the helix formed by the same μ1 subunit and residue Lys113 on the helix formed by the neighboring μ1 subunit have been implicated in facilitating this process (38). The pedestal region sequences of T1L and T3D μ1 differ at residues 51, 96, 109, and 128, suggesting that one or more of these residues influences the efficiency of autocleavage. Whether these residues directly participate in the nucleophilic reaction required for μ1N-μ1C cleavage or whether they affect the efficiency of autocleavage by altering the local structure of μ1 in that region remains unknown. Comparison of the subatomic-resolution cryoEM structures of ISVPs expressing T1L, T3D, or chimeric μ1 may be helpful in better understanding how the δN domain affects autocleavage.
Our results indicate that virus strains expressing a T1L-derived pedestal [rsT1L/(LD)L M2 and rsT1L/(LD)D M2] retain the capacity for hemolysis despite having a lower autocleavage efficiency than those containing a T3D-derived pedestal (rsT1L/T3D M2 or rsT1L/DL M2). These findings suggest that differences in autocleavage efficiency observed in parental strains T1L and T3D (49) do not contribute to the differences in membrane penetration efficiency observed by hemolysis or chromium release assays (11, 39). Additional studies with recombinant viruses that encode μ1 mutants with significantly lower autocleavage efficiency are needed to quantify cleavage efficiency required for the release of a sufficient number of μ1N molecules to mediate membrane penetration.
The μ1N fragment liberated from μ1 by autocleavage remains buried in the pedestal domain formed by δ and is not functional for membrane penetration in ISVPs (38). The exposure and release of μ1N requires conversion of ISVPs to ISVP*s, which leads to massive conformation changes in the particle-associated δ fragment, as evidenced by exposure of new δ epitopes and enhanced protease sensitivity (11). Although the precise nature of the conformational change is not understood, amino acid substitutions in the jelly-roll β barrel domain of δ that enhance thermal stability of reovirus particles (44) or render them resistant to 33% ethanol (32, 62) prevent conformational changes in δ, implicating the δ jelly-roll β barrel domain in regulating ISVP* formation. The findings presented here provide additional evidence for the function of the jelly-roll β-barrel domain in regulating the efficiency of membrane penetration. There are five polymorphic differences between T1L and T3D in this region of the μ1 protein (Table 1). Two of these residues lie in positions where they can affect interaction between two μ1 monomers within a trimer (residues 305 and 327) (Fig. 5D). The remaining three residues (residues 340, 342, and 517) are found exposed to the solvent on the outer surface of the μ1 trimer (Fig. 5D), where they may influence interactions between adjacent μ1 trimers. Interestingly, mutant residues in ethanol- and heat-resistant reoviruses also are found within the same regions described above that can potentially affect interaction between μ1 monomers or between μ1 trimers (38, 44, 62). Thus, we think that residues in the inter- and intratrimer interface both regulate the efficiency of ISVP* formation.
In addition to domain IV, structural studies also implicate domain I in regulating ISVP-to-ISVP* conversion (38, 66). Residues 72 to 96, which are disordered in the crystal structure of μ1 (38), form a loop that projects sideways and interacts with two other different subunits on an adjacent μ1 trimer (66). The 72-96 loop contacts one subunit between residues 126 and 129 and one subunit between residues 57 and 65 (66). In addition to participating in μ1-μ1 interactions, domain I also is thought to interact with the core particle via a loop formed by amino acids 51 to 62 (66). Three of the four amino acid residues that differ between T1L and T3D (residues 51, 96, and 128) lie within these predicted regions of interaction. Thus, it is possible that differences in the efficiency of these viruses to form ISVP* is related to amino acid differences in domain I. Consistent with this idea, characterization of rsT1L/(DL)D M2 and rsT1L/(DL)L M2 indicated that the presence of the T3D-derived pedestal region was sufficient to promote ISVP* in the presence of CsCl. However, this region did not promote ISVP* formation and hemolysis when ISVPs of rsT1L/(DL)D M2 and rsT1L/(DL)L M2 were exposed to erythrocytes, suggesting that polymorphisms within the pedestal domain do not account for differences in the efficiency of T1L and T3D μ1 to regulate functional conformational changes that are required for ISVP-to-ISVP* conversion and pore formation upon interaction with the membranes.
The cell entry mechanism of reovirus parallels other related and unrelated nonenveloped viruses in many aspects. For example, the capsid protein needs to be primed for entry by cleavage for closely related rotavirus and also more distantly related viruses, such as adenovirus, Flock House virus, and poliovirus. In some cases, these cleavage events are protease mediated (e.g., cleavage of adenovirus proprotein VI and rotavirus VP4) (23, 29), whereas in others capsid protein cleavage is autocatalytic, similar to reovirus (e.g., cleavage of Flock house virus α, poliovirus, and rhinovirus VP0) (7, 26, 31, 37, 54). Although these cleavage events are critical regulators of viral infectivity, where and when this cleavage occurs during virus infection remains unclear. In addition to priming cleavage, conformational changes that expose membrane-interacting hydrophobic domains also are required for cell entry for these viruses. In some cases, interaction with a host cell receptor is the trigger for conformational change (e.g., Flock house virus and poliovirus) (2, 24, 61); in others, the low pH environment of the cellular endosomes modulates conformational changes (e.g., adenovirus) (30, 64). Since the known receptors and endosomal pH appear not to be required to induce conformational changes in reovirus, the cellular trigger that promotes conformational changes needed for cell entry remain unknown. Analogous to the release of reovirus μ1N (1, 33, 65), adenovirus protein VI (64), Flock house virus γ peptide (8, 51), and poliovirus VP4 (19) also need to be released to mediate the disruption of host cell membranes. Much remains to be elucidated about the mechanisms that govern the function of these released peptides. Through mutational analysis of reovirus μ1 coupled with biochemical and structural studies, we hope to enhance our understanding of how nonenveloped viruses enter host cells.
Acknowledgments
We thank members of our laboratory, Karl Boehme and Tuli Mukhopadhyay, for helpful suggestions and review of the manuscript. We thank Terry Dermody for sharing plasmids used for generating reovirus mutants and Ulla Buchholz for BHK-T7 cells.
This research was supported by American Heart Association Midwest Affiliate Award 09SDG2140019 (P.D.).
Footnotes
Published ahead of print on 13 October 2010.
REFERENCES
- 1.Agosto, M. A., T. Ivanovic, and M. L. Nibert. 2006. Mammalian reovirus, a nonfusogenic nonenveloped virus, forms size-selective pores in a model membrane. Proc. Natl. Acad. Sci. U. S. A. 103:16496-16501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arita, M., S. Koike, J. Aoki, H. Horie, and A. Nomoto. 1998. Interaction of poliovirus with its purified receptor and conformational alteration in the virion. J. Virol. 72:3578-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer, G. S., and T. S. Dermody. 1997. Mutations in reovirus outer-capsid protein σ3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J. Virol. 71:4921-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee, M., and J. E. Johnson. 2008. Activation, exposure and penetration of virally encoded, membrane-active polypeptides during non-enveloped virus entry. Curr. Protein Peptide Sci. 9:16-27. [DOI] [PubMed] [Google Scholar]
- 5.Barton, E. S., J. L. Connolly, J. C. Forrest, J. D. Chappell, and T. S. Dermody. 2001. Utilization of sialic acid as a coreceptor enhances reovirus attachment by multistep adhesion strengthening. J. Biol. Chem. 276:2200-2211. [DOI] [PubMed] [Google Scholar]
- 6.Barton, E. S., J. C. Forrest, J. L. Connolly, J. D. Chappell, Y. Liu, F. Schnell, A. Nusrat, C. A. Parkos, and T. S. Dermody. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441-451. [DOI] [PubMed] [Google Scholar]
- 7.Basavappa, R., R. Syed, O. Flore, J. P. Icenogle, D. J. Filman, and J. M. Hogle. 1994. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: structure of the empty capsid assembly intermediate at 2.9 Å resolution. Protein Sci. 3:1651-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bong, D. T., C. Steinem, A. Janshoff, J. E. Johnson, and M. Reza Ghadiri. 1999. A highly membrane-active peptide in Flock House virus: implications for the mechanism of nodavirus infection. Chem. Biol. 6:473-481. [DOI] [PubMed] [Google Scholar]
- 9.Borsa, J., B. D. Morash, M. D. Sargent, T. P. Copps, P. A. Lievaart, and J. G. Szekely. 1979. Two modes of entry of reovirus particles into L cells. J. Gen. Virol. 45:161-170. [DOI] [PubMed] [Google Scholar]
- 10.Borsa, J., M. D. Sargent, P. A. Lievaart, and T. P. Copps. 1981. Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology 111:191-200. [DOI] [PubMed] [Google Scholar]
- 11.Chandran, K., D. L. Farsetta, and M. L. Nibert. 2002. Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein μ1 mediates membrane disruption. J. Virol. 76:9920-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandran, K., and M. L. Nibert. 2003. Animal cell invasion by a large nonenveloped virus: reovirus delivers the goods. Trends Microbiol. 11:374-382. [DOI] [PubMed] [Google Scholar]
- 13.Chandran, K., J. S. Parker, M. Ehrlich, T. Kirchhausen, and M. L. Nibert. 2003. The delta region of outer-capsid protein μ1 undergoes conformational change and release from reovirus particles during cell entry. J. Virol. 77:13361-13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, C. T., and H. J. Zweerink. 1971. Fate of parental reovirus in infected cell. Virology 46:544-555. [DOI] [PubMed] [Google Scholar]
- 15.Chappell, J. D., J. L. Duong, B. W. Wright, and T. S. Dermody. 2000. Identification of carbohydrate-binding domains in the attachment proteins of type 1 and type 3 reoviruses. J. Virol. 74:8472-8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danthi, P., C. M. Coffey, J. S. Parker, T. W. Abel, and T. S. Dermody. 2008. Independent regulation of reovirus membrane penetration and apoptosis by the mu1 phi domain. PLoS Pathog. 4:e1000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danthi, P., K. M. Guglielmi, E. Kirchner, B. Mainou, T. Stehle, and T. S. Dermody. 2010. From touchdown to transcription: the reovirus cell entry pathway. Curr. Top. Microbiol. Immunol. 2100:91-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danthi, P., T. Kobayashi, G. H. Holm, M. W. Hansberger, T. W. Abel, and T. S. Dermody. 2008. Reovirus apoptosis and virulence are regulated by host cell membrane penetration efficiency. J. Virol. 82:161-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danthi, P., M. Tosteson, Q. H. Li, and M. Chow. 2003. Genome delivery and ion channel properties are altered in VP4 mutants of poliovirus. J. Virol. 77:5266-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dermody, T. S., M. L. Nibert, J. D. Wetzel, X. Tong, and B. N. Fields. 1993. Cells and viruses with mutations affecting viral entry are selected during persistent infections of L cells with mammalian reoviruses. J. Virol. 67:2055-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebert, D. H., J. Deussing, C. Peters, and T. S. Dermody. 2002. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 277:24609-24617. [DOI] [PubMed] [Google Scholar]
- 22.Ehrlich, M., W. Boll, A. Van Oijen, R. Hariharan, K. Chandran, M. L. Nibert, and T. Kirchhausen. 2004. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell 118:591-605. [DOI] [PubMed] [Google Scholar]
- 23.Estes, M. K., D. Y. Graham, and B. B. Mason. 1981. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J. Virol. 39:879-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fricks, C. E., and J. M. Hogle. 1990. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol. 64:1934-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furlong, D. B., M. L. Nibert, and B. N. Fields. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallagher, T. M., and R. R. Rueckert. 1988. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J. Virol. 62:3399-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gentsch, J. R., and A. F. Pacitti. 1987. Differential interaction of reovirus type 3 with sialylated receptor components on animal cells. Virology 161:245-248. [DOI] [PubMed] [Google Scholar]
- 28.Gentsch, J. R., and A. F. Pacitti. 1985. Effect of neuraminidase treatment of cells and effect of soluble glycoproteins on type 3 reovirus attachment to murine L cells. J. Virol. 56:356-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greber, U. F., P. Webster, J. Weber, and A. Helenius. 1996. The role of the adenovirus protease on virus entry into cells. EMBO J. 15:1766-1777. [PMC free article] [PubMed] [Google Scholar]
- 30.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477-486. [DOI] [PubMed] [Google Scholar]
- 31.Hindiyeh, M., Q. H. Li, R. Basavappa, J. M. Hogle, and M. Chow. 1999. Poliovirus mutants at histidine 195 of VP2 do not cleave VP0 into VP2 and VP4. J. Virol. 73:9072-9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hooper, J. W., and B. N. Fields. 1996. Role of the μ1 protein in reovirus stability and capacity to cause chromium release from host cells. J. Virol. 70:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivanovic, T., M. A. Agosto, L. Zhang, K. Chandran, S. C. Harrison, and M. L. Nibert. 2008. Peptides released from reovirus outer capsid form membrane pores that recruit virus particles. EMBO J. 27:1289-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jayasuriya, A. K., M. L. Nibert, and B. N. Fields. 1988. Complete nucleotide sequence of the M2 gene segment of reovirus type 3 Dearing and analysis of its protein product μ1. Virology 163:591-602. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi, T., A. A. Antar, K. W. Boehme, P. Danthi, E. A. Eby, K. M. Guglielmi, G. H. Holm, E. M. Johnson, M. S. Maginnis, S. Naik, W. B. Skelton, J. D. Wetzel, G. J. Wilson, J. D. Chappell, and T. S. Dermody. 2007. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 1:147-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi, T., L. S. Ooms, M. Ikizler, J. D. Chappell, and T. S. Dermody. 2010. An improved reverse genetics system for mammalian orthoreoviruses. Virology 398:194-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, W.-M., S. S. Monroe, and R. R. Rueckert. 1993. Role of maturation cleavage in infectivity of picornaviruses: activation of an infectosome. J. Virol. 67:2110-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liemann, S., K. Chandran, T. S. Baker, M. L. Nibert, and S. C. Harrison. 2002. Structure of the reovirus membrane-penetration protein, μ1, in a complex with its protector protein, σ3. Cell 108:283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lucia-Jandris, P., J. W. Hooper, and B. N. Fields. 1993. Reovirus M2 gene is associated with chromium release from mouse L cells. J. Virol. 67:5339-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mabrouk, T., and G. Lemay. 1994. The sequence similarity of reovirus sigma 3 protein to picornaviral proteases is unrelated to its role in mu 1 viral protein cleavage. Virology 202:615-620. [DOI] [PubMed] [Google Scholar]
- 41.Maginnis, M. S., J. C. Forrest, S. A. Kopecky-Bromberg, S. K. Dickeson, S. A. Santoro, M. M. Zutter, G. R. Nemerow, J. M. Bergelson, and T. S. Dermody. 2006. β1 integrin mediates internalization of mammalian reovirus. J. Virol. 80:2760-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maginnis, M. S., B. A. Mainou, A. Derdowski, E. M. Johnson, R. Zent, and T. S. Dermody. 2008. NPXY motifs in the β1 integrin cytoplasmic tail are required for functional reovirus entry. J. Virol. 82:3181-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendez, I. I., L. L. Hermann, P. R. Hazelton, and K. M. Coombs. 2000. A comparative analysis of Freon substitutes in the purification of reovirus and calicivirus. J. Virol. Methods 90:59-67. [DOI] [PubMed] [Google Scholar]
- 44.Middleton, J. K., M. A. Agosto, T. F. Severson, J. Yin, and M. L. Nibert. 2007. Thermostabilizing mutations in reovirus outer-capsid protein μ1 selected by heat inactivation of infectious subvirion particles. Virology 361:412-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Middleton, J. K., T. F. Severson, K. Chandran, A. L. Gillian, J. Yin, and M. L. Nibert. 2002. Thermostability of reovirus disassembly intermediates (ISVPs) correlates with genetic, biochemical, and thermodynamic properties of major surface protein mu1. J. Virol. 76:1051-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mustoe, T. A., R. F. Ramig, A. H. Sharpe, and B. N. Fields. 1978. Genetics of reovirus: identification of the dsRNA segments encoding the polypeptides of the μ and σ size classes. Virology 89:594-604. [DOI] [PubMed] [Google Scholar]
- 47.Nibert, M. L., J. D. Chappell, and T. S. Dermody. 1995. Infectious subvirion particles of reovirus type 3 Dearing exhibit a loss in infectivity and contain a cleaved σ1 protein. J. Virol. 69:5057-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nibert, M. L., and B. N. Fields. 1992. A carboxy-terminal fragment of protein μ1/μ1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J. Virol. 66:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nibert, M. L., A. L. Odegard, M. A. Agosto, K. Chandran, and L. A. Schiff. 2005. Putative autocleavage of reovirus μ1 protein in concert with outer-capsid disassembly and activation for membrane permeabilization. J. Mol. Biol. 345:461-474. [DOI] [PubMed] [Google Scholar]
- 50.Odegard, A. L., K. Chandran, X. Zhang, J. S. Parker, T. S. Baker, and M. L. Nibert. 2004. Putative autocleavage of outer capsid protein μ1, allowing release of myristoylated peptide μ1N during particle uncoating, is critical for cell entry by reovirus. J. Virol. 78:8732-8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Odegard, A. L., M. H. Kwan, H. E. Walukiewicz, M. Banerjee, A. Schneemann, and J. E. Johnson. 2009. Low endocytic pH and capsid protein autocleavage are critical components of Flock House virus cell entry. J. Virol. 83:8628-8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paul, R. W., A. H. Choi, and P. W. K. Lee. 1989. The α-anomeric form of sialic acid is the minimal receptor determinant recognized by reovirus. Virology 172:382-385. [DOI] [PubMed] [Google Scholar]
- 53.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 54.Schneemann, A., W. Zhong, T. M. Gallagher, and R. R. Rueckert. 1992. Maturation cleavage required for infectivity of a nodavirus. J. Virol. 66:6728-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silverstein, S. C., C. Astell, D. H. Levin, M. Schonberg, and G. Acs. 1972. The mechanism of reovirus uncoating and gene activation in vivo. Virology 47:797-806. [DOI] [PubMed] [Google Scholar]
- 56.Smith, R. E., H. J. Zweerink, and W. K. Joklik. 1969. Polypeptide components of virions, top component and cores of reovirus type 3. Virology 39:791-810. [DOI] [PubMed] [Google Scholar]
- 57.Sturzenbecker, L. J., M. L. Nibert, D. B. Furlong, and B. N. Fields. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61:2351-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tillotson, L., and A. J. Shatkin. 1992. Reovirus polypeptide σ3 and N-terminal myristoylation of polypeptide μ1 are required for site-specific cleavage to μ1C in transfected cells. J. Virol. 66:2180-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsai, B. 2007. Penetration of nonenveloped viruses into the cytoplasm. Annu. Rev. Cell Dev. Biol. 23:23-43. [DOI] [PubMed] [Google Scholar]
- 60.Virgin, H. W., IV, R. Bassel-Duby, B. N. Fields, and K. L. Tyler. 1988. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing). J. Virol. 62:4594-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walukiewicz, H. E., J. E. Johnson, and A. Schneemann. 2006. Morphological changes in the T=3 capsid of Flock House virus during cell entry. J. Virol. 80:615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wessner, D. R., and B. N. Fields. 1993. Isolation and genetic characterization of ethanol-resistant reovirus mutants. J. Virol. 67:2442-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White, J. M., S. E. Delos, M. Brecher, and K. Schornberg. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiethoff, C. M., H. Wodrich, L. Gerace, and G. R. Nemerow. 2005. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 79:1992-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, L., M. A. Agosto, T. Ivanovic, D. S. King, M. L. Nibert, and S. C. Harrison. 2009. Requirements for the formation of membrane pores by the reovirus myristoylated micro1N peptide. J. Virol. 83:7004-7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, X., Y. Ji, L. Zhang, S. C. Harrison, D. C. Marinescu, M. L. Nibert, and T. S. Baker. 2005. Features of reovirus outer capsid protein μ1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Å resolution. Structure 13:1545-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, X., L. Jin, Q. Fang, W. H. Hui, and Z. H. Zhou. 2010. 3.3 Å cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell 141:472-482. [DOI] [PMC free article] [PubMed] [Google Scholar]