Abstract
Dengue is a pantropic public health problem. In children, dengue shock syndrome (DSS) is the most common life-threatening complication. The ability to predict which patients may develop DSS may improve triage and treatment. To this end, we conducted a nested case-control comparison of the early host transcriptional features in 24 DSS patients and 56 sex-, age-, and virus serotype-matched uncomplicated (UC) dengue patients. In the first instance, we defined the “early dengue” profile. The transcriptional signature in acute rather than convalescent samples (≤72 h post-illness onset) was defined by an overabundance of interferon-inducible transcripts (31% of the 551 overabundant transcripts) and canonical gene ontology terms that included the following: response to virus, immune response, innate immune response, and inflammatory response. Pathway and network analyses identified STAT1, STAT2, STAT3, IRF7, IRF9, IRF1, CEBPB, and SP1 as key transcriptional factors mediating the early response. Strikingly, the only difference in the transcriptional signatures of early DSS and UC dengue cases was the greater abundance of several neutrophil-associated transcripts in patients who progressed to DSS, a finding supported by higher plasma concentrations of several canonical proteins associated with neutrophil degranulation (bactericidal/permeability-increasing protein [BPI], elastase 2 [ELA2], and defensin 1 alpha [DEF1A]). Elevated levels of neutrophil-associated transcripts were independent of the neutrophil count and also of the genotype of the infecting virus, as genome-length sequences of dengue virus serotype 1 (DENV-1) (n = 15) and DENV-2 (n = 3) sampled from DSS patients were phylogenetically indistinguishable from those sampled from uncomplicated dengue patients (32 DENV-1 and 9 DENV-2 sequences). Collectively, these data suggest a hitherto unrecognized association between neutrophil activation, pathogenesis, and the development of DSS and point to future strategies for guiding prognosis.
Dengue is the most significant mosquito-borne viral infection of humans. The dengue pandemic has spread to the extent that between 70 and 500 million infections occur each year in over 100 countries, resulting in ∼40 million clinically apparent cases and ∼20,000 deaths (31). There are no licensed vaccines for the prevention of dengue, and treatment of severe cases is limited to supportive care.
Dengue is an acute, self-limiting systemic viral infection that affects mostly children and young adults in settings of dengue endemicity. Dengue manifestations range from nonspecific fever to a more severe syndrome commonly characterized by increased vascular permeability, thrombocytopenia, and a bleeding diathesis. In severe cases, the increased vascular permeability results in circulatory compromise, and the patient may develop potentially life-threatening dengue shock syndrome (DSS). A characteristic feature of DSS is that it manifests clinically between days 4 and 6 of the illness, a time when the viremia is in steep decline and the host immune response is well established. The timing of these events suggests the host proinflammatory response, rather than direct virus-mediated effects, mediates the vascular permeability syndrome leading to DSS. Fortunately, these physiological derangements are transient, and most patients recover fully if supported with parenteral fluid therapy during the period of maximal vascular leakage. Host features implicated in susceptibility to severe disease include genetic variation (8), previous dengue virus (DENV) infection history (7, 13, 33, 39), and individual propensity for capillary leakage, which itself is influenced by age and sex (5). Viral genetic traits might also be important, with some lineages of DENV being virologically (9) and epidemiologically (28) fitter than others and more frequently associated with severe disease.
There are no animal models that fully mimic the dengue vascular leakage syndrome, and therefore, clinical studies are of particular importance. Previous studies of the whole-blood host transcriptional response in Vietnamese, Singaporean, and Thai children and adults have been relatively small and used nonuniform sampling times. Collectively, these studies have suggested that during the febrile phase, transcripts from interferon-stimulated genes are highly prominent (11, 26, 36) but that this signature wanes rapidly with the resolution of infection. However, previous studies have not fully explored the interconnectedness of elements of the transcriptional signature in a way that could yield insights into critical gene regulation pathways and transcriptional factors. Moreover, previous studies have not examined whether the early (≤72 h of illness history) transcriptional signature of dengue is distinct in patients who subsequently develop clinically important complications, e.g., DSS. This is important, as the identification of prognostic markers of severe disease could allow for improved patient triage and management or, in the future, rational treatment with antiviral or immune-modulating therapies.
To this end, the aim of this study was, first, to provide a definitive profile of the early whole-blood transcriptional signature of dengue in a large number of clinically and virologically well-characterized Vietnamese children and young adults with severe and mild dengue. Second, we sought to identify features of the whole-blood transcriptome that were associated with progression to DSS. Intriguingly, our findings identified several neutrophil-associated transcripts as being more abundant in patients who progressed to DSS, suggesting a hitherto unrecognized association among neutrophil activation, pathogenesis, and the development of DSS.
MATERIALS AND METHODS
Study enrollment and investigations.
The study protocol was approved by the Scientific and Ethical Committee of Dong Thap Hospital and the Oxford Tropical Research Ethical Committee. Clinical and hematological assessments were performed daily. Patients underwent screening in the outpatient clinic of Dong Thap Hospital if there was a clinical suspicion of dengue and illness duration of less than 72 h. Plasma samples obtained from screened patients were tested with an NS1 antigen enzyme-linked immunosorbent assay (ELISA) (Bio-Rad, California). Patients that were NS1 ELISA positive were admitted for further observation and participation in the study protocol if written informed consent was obtained from the patients (patients > 14 years) or from the patient's parents/guardians (patients ≤ 14 years). Serial blood samples were collected for isolation of RNA, DNA, and plasma at the time of enrollment (study day 1), on study day 3, and at the time of discharge. Hematological data were recorded daily, and all patients received an ultrasound within 24 h of defervescence. From June 2006 to December 2007, 450 patients were recruited, of whom 35 patients developed DSS, according to WHO criteria (pulse pressure of ≤20 mmHg, with poor peripheral perfusion and rapid, weak pulse). The remaining 415 patients were defined as having uncomplicated (UC) dengue without evidence of cardiovascular compromise. In line with recently revised WHO guidelines (42), we used the term “uncomplicated dengue” to refer to these hospitalized patients, as they did not require any significant clinical interventions and were managed throughout in the general infectious diseases ward. At the acute phase (days −3 and −2 relative to defervescence), 24 samples obtained from DSS patients and 56 samples obtained from UC dengue patients were used for expression microarray, respectively. For controls, 18 and 16 autologous follow-up samples were collected at 2 weeks after discharge from DSS and UC dengue patients, respectively.
Patient diagnosis.
Serological investigations (IgM and IgG capture ELISAs) were performed with paired plasma samples using methods described previously (15). A serological determination of dengue was made if IgM seroconversion occurred between the acute and early convalescent plasma samples. Serology was interpreted as suggestive of secondary infection if DENV-reactive IgG was detected in the capture ELISA in the first week of illness. Plasma concentrations of DENV NS1 were measured by capture ELISA (Bio-Rad, California), using recombinant DENV-1 NS1 protein (Hawaii Biotech, HI) as a standard. DENV serotype-specific quantitative reverse transcription-PCR (qRT-PCR) was performed to determine viral serotype and viremia in the enrollment plasma samples collected from each patient using an established method (36).
DENV genome sequencing.
Viral genomes were sequenced using the Broad Institute's capillary sequencing (Applied Biosystems) directed amplification viral sequencing pipeline (http://www.broadinstitute.org/annotation/viral/Dengue). This sequencing effort was part of the Broad Institute's Genome Resources in Dengue Consortium (GRID) project. Viral RNA was isolated from diagnostic plasma samples (QIAamp viral RNA minikit; Qiagen), and the RNA genome was reverse transcribed to cDNA with SuperScript III reverse transcriptase (Invitrogen), random hexamers (Roche), and a specific oligonucleotide targeting the 3′ end of the target genome sequences. cDNA was then amplified using a high-fidelity DNA polymerase (PfuUltra II; Stratagene) and a pool of specific primers to produce 14 overlapping amplicons of 1.5 to 2kb in size for 2× physical coverage. Amplicons were then sequenced in the forward and reverse directions using primer panels consisting of 96 specific primer pairs, tailed with M13 forward and reverse primer sequences, which produce 500- to 700-bp amplicons from the target viral genome. Amplicons were then sequenced in the forward and reverse directions using the M13 primer. Total coverage delivered after amplification and sequencing was ∼8-fold. The resulting sequence reads were assembled de novo and annotated using the Broad Institute's in-house viral assembly and annotation algorithms. All genome sequences newly determined here have been deposited in GenBank and assigned accession numbers (see Table S1 in the supplemental material).
Gene expression microarray.
We used one-color array technology on the Illumina platform (Illumina Inc., San Diego, CA) for gene expression microarray. In brief, whole-blood samples (2.5 ml) were collected directly into PAXgene RNA tubes (Qiagen, Sussex, United Kingdom). RNA extraction was performed using PAXgene RNA kits (Qiagen). Biotinylated amplified cRNA was generated by in vitro transcription (IVT) technology using the Illumina TotalPrep RNA amplification kit (Ambion, Inc., Austin, TX), according to the manufacturer's instructions. After purification, 850 ng of cRNA was hybridized to an Illumina HumanRef-8 v2 BeadChip (containing probes to 23,961 RefSeq gene sequences) at 55°C for 18 h by following the manufacturer's instructions (Illumina, Inc., San Diego, CA). This was followed by washing, blocking, and streptavidin-Cy3 staining steps. Finally, the chip was scanned with an Illumina BeadArray reader confocal scanner.
Normalization of array data and analysis.
Standard normalization procedures in GeneSpring were used (version 10; Agilent, Santa Clara, CA). In brief, array normalization was performed by dividing the mean array intensity value into the 75th percentile value of all arrays. Gene normalization was performed by dividing the mean value of the gene in each array into the 75th percentile value of the gene in all the arrays. Normalized data were then filtered based on expression data, in which only those of transcripts with detection confidence of greater than 0.999 in at least 1 out of 227 samples (total number of samples arrayed) under analysis were used. The detection P value was calculated by BeadStudio software (Illumina). After normalization, 9,780 genes were available for further analyses. Significance analysis of microarray (SAM) was used to detect transcripts that were relatively more or less abundant in one group of samples. Significant genes were those with false-discovery rates (FDRs) of less than 5% and fold differences between 2 groups of at least 2-fold. Normalized intensity data and chip image files have been deposited into GEO (http://www.ncbi.nlm.nih.gov/projects/geo). Network analysis of the differentially expressed genes between acute DSS and acute uncomplicated dengue was performed using Ingenuity Pathway Analysis (IPA) (www.ingenuity.com).
Pathway and GO analyses of DE genes using InnateDB.
Illumina probe identifiers (IDs) were mapped to NCBI Entrez Gene IDs, and these were used to cross-reference and upload differentially expressed (DE) genes to InnateDB (27; www.innatedb.com) along with associated gene expression data. A list of pathways mapping to the uploaded genes was returned, and pathway analysis was undertaken to determine which pathways were significantly overrepresented in the up- and downregulated gene data sets using the hypergeometric test. InnateDB simultaneously tests for overrepresentation of DE genes in more than 3,000 pathways from the KEGG (23), PID (http://pid.nci.nih.gov), INOH (http://www.inoh.org/), NetPath (http://www.netpath.org), and Reactome (21) databases. The Benjamini and Hochberg (BH) FDR correction (3) was applied to correct for multiple testing. Similarly, the InnateDB gene ontology (GO) analysis tool was used to identify gene ontology terms (2) that were significantly associated with DE genes.
InnateDB molecular interaction network analysis.
InnateDB pathway analysis can be a powerful approach to provide insight into which annotated pathways are most significantly associated with DE genes. Network analysis, analyzing the molecular interactions between DE genes and their encoded products, however, provides the opportunity to move beyond investigating signaling pathways as relatively simple linear cascades. Network analysis allows one to potentially identify novel relationships between DE genes and their regulators and to uncover as yet unknown signaling cascades or pathways, functionally relevant subnetworks, and the central molecules, or hubs, of these networks.
InnateDB is one of the most comprehensive databases of all human and mouse experimentally supported molecular interactions but also specifically includes annotation on more than 11,500 manually curated human and mouse innate immunity-relevant interactions, many of which are not present in any other database. InnateDB was used to construct molecular interaction networks consisting of interactions between DE nodes and their interacting partners, with the associated gene expression data overlaid as node attributes. Networks were analyzed using the cyto-Hubba plug-in (25) (http://hub.iis.sinica.edu.tw/cytoHubba/) for Cytoscape 2.6.3 (34) to investigate a variety of properties of a network, including the identification of network hubs and bottlenecks which may represent the key regulatory nodes in the network. The jActiveModules plug-in (19) was also used to identify high-scoring differentially expressed subnetworks.
Transcriptional network analysis.
The networks discussed above consist mainly of protein-protein direct physical or biochemical interactions, with a relatively small number of experimentally verified protein-DNA (transcription factor [TF]-gene) interactions. This is because the production of high-throughput experimental data for protein-DNA interactions has lagged behind the data for protein-protein interactions. Although InnateDB contains ∼13,000 protein-DNA interactions, this number represents less than 3% of all interactions and is a small sample of likely transcription factor-gene interactions. To overcome this limitation, InnateDB now also contains computationally predicted transcription factor interactions using transcription factor binding site analysis data extracted from the cisRED database (www.cisred.org; 31a). InnateDB was used to construct a network of all predicted transcription factor (TF) interactions with DE genes (TF network). Each association between a transcription factor and a gene was inferred as a protein-DNA interaction, and a gene regulatory network was constructed. Given the large number of false positives associated with predicting transcription factor binding sites and therefore transcription factor-gene interactions, identification of the key regulators by ranked degree alone is not sufficient (i.e., a node may have a large number of predicted interactions with DE genes, but this may be expected by chance if that node is annotated to interact with a large number of genes in InnateDB). To overcome this issue, a right-sided Fisher exact test (implemented using the Text::NSP Perl module [http://search.cpan.org/dist/Text-NSP/]) was used to identify nodes which interact with significantly more DE genes than expected by chance.
ELISAs.
The amount of elastase 2 (ELA2) in patient plasma samples was measured using a polymorphonuclear leukocyte (PMN)-elastase ELISA kit (Immundiagnostik, Netherlands). Plasma defensin 1 alpha (DEF1A), bactericidal/permeability-increasing protein (BPI), and myeloperoxidase (MPO) were measured using capture ELISAs (Hycult Biotechnology [Hbt], Netherlands). The limits of detection were 120 pg/ml for ELA2, 50 pg/ml for DEF1A, 250 pg/ml for BPI, and 0.4 ng/ml for MPO. Plasma albumin concentrations were measured by capture ELISA. Briefly, ELISA plates (MaxiSorp; Nunc, Denmark) were coated with anti-albumin pAb (polyclonal rabbit anti-human albumin) (Dako Co.) overnight and then blocked with 2% skim milk (Merck) in 0.05% phosphate-buffered saline-Tween 20 (PBS-T) for 1 h at 37°C. Next, samples and standards were added into the wells, followed by addition of diluted biotinylated anti-albumin antibody solution and incubation for 1 h at 37°C. Diluted streptavidin-horseradish peroxidase (HRP) (P0397; Dako Co., Japan) was added and incubated for 1 h at 37°C before the o-phenylenediamine dihydrochloride (OPD) substrate was added (P9187; Sigma). After the reactions was stopped by addition of diluted sulfuric acid (10%), the optical density (OD) value was measured using a plate reader (Bio-Rad, CA), and the result was analyzed by microplate software (Bio-Rad, CA).
Statistics.
All statistical analysis was performed using Intercooled Stata 9.2 (StataCorp, TX). Significance was assigned at P values of <0.05 for all parameters and were two sided unless otherwise indicated. Categorical variables between groups were compared by Fisher's exact test. The t test was used for continuous variables. Classification analysis by the decision tree forest model (6) was performed to examine whether the set of differentially expressed genes between acute DSS and UC dengue are good predictors of disease outcomes. In brief, expression intensities of the 21 differentially expressed genes were used as “predictors,” and the outcomes are used as “targets.” This model generates an ensemble of 200 trees using randomization of data and predictors. The tree with the most likely predicted value was chosen.
Microarray data accession number.
The GEO accession number for the normalized intensity data and chip image files in GSE25001.
RESULTS
Patient population.
Between June 2006 and December 2007, 450 patients with dengue and less than 72 h of illness were enrolled into this prospective, hospital-based study. Thirty-five patients subsequently progressed to DSS, as defined by WHO criteria (pulse pressure of ≤20 mmHg, with poor peripheral perfusion and rapid, weak pulse) (42). The remaining 415 patients were defined as having “dengue” by the recently revised WHO criteria (42), as they did not require any clinical interventions and were managed throughout in the general infectious disease ward. Samples collected from 24 DSS patients were available for gene expression microarray analyses. For each DSS patient, 1 to 3 patients with uncomplicated dengue and matched by sex, infecting virus serotype, age (within 2 years), and day of illness were selected as controls for virological investigations and host gene expression profiling (n = 56 in total). The demographic and virological characteristics of DSS and matched uncomplicated dengue patients are summarized in Table 1. The median time of illness was 3 days (range, 2 to 5 days), and the median time to defervescence was 3 days (range, 2 to 3 days) (Table 1). In patients who developed DSS, the median time from enrollment to shock was 2 days (range, 1 to 4 days).
TABLE 1.
Summary characteristics of patients included in the microarray study
| Variable | Value for patient groupb |
P valuec | |
|---|---|---|---|
| DSS | UC dengue | ||
| Median age (range) | 11 (2-30) | 10.5 (2-29) | |
| No. of male patients/total no. of patients (% male) | 16/24 (67) | 34/56 (61) | |
| Median day of illness (range) | 3 (2-5) | 3 (2-5) | |
| Fever day (range)a | −3 (−3 to −2) | −3 (−3 to −2) | |
| No. (%) of patients receiving i.v. fluidsd | 24 (100) | 0 (0) | |
| Median array sample day relative to day of shock (range) | −2 (−3-0) | N/A | |
| No. (%) of patients with: | |||
| Primary infection | 1 (4) | 1 (1.8) | |
| Unknown infection | 1 (4) | 0 | |
| DENV serotype | |||
| 1 | 16 (67) | 41 (73) | |
| 2 | 7 (29) | 15 (27) | |
| 3 | 0 (0) | 0 | |
| 4 | 0 (0) | 0 | |
| Unknown | 1 (4) | 0 | |
| Median level (range) ofe: | |||
| Viremia | 3.47E+7 (3.11E+4-2.15E+9) | 4.18E+07 (1.27E+5-1.46E+9) | 0.7 |
| NS1 (ng/ml) | 1,002 (0.01-3,842) | 117 (0.01-3,415) | 0.048 |
| Neutrophils (103 cells/μl) | 2.75 (0.8-5.5) | 2.48 (0.7-8.4) | 0.7 |
Fever day is the number of days relative to the time point when the patient became afebrile, defined as fever day 0.
A total of 24 DSS patients and 56 UC dengue patients were included in the study. N/A, not applicable.
Mann-Whitney test P values.
i.v., intravenous.
For the DSS and UC dengue groups, the numbers of patients missing from these data are 6 and 14 for viremia, 3 and 17 for NS1, and 1 and 6 for neutrophil levels, respectively.
Virological comparisons between patients with DSS and those with uncomplicated dengue.
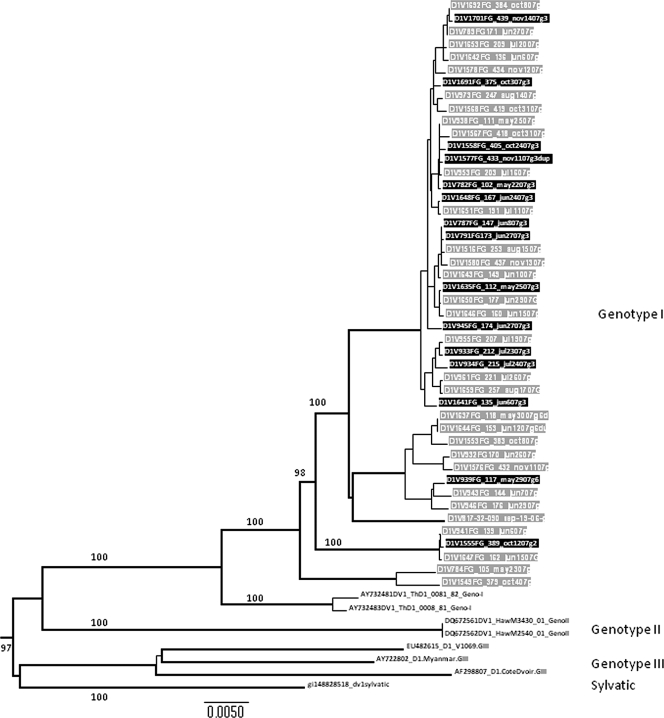
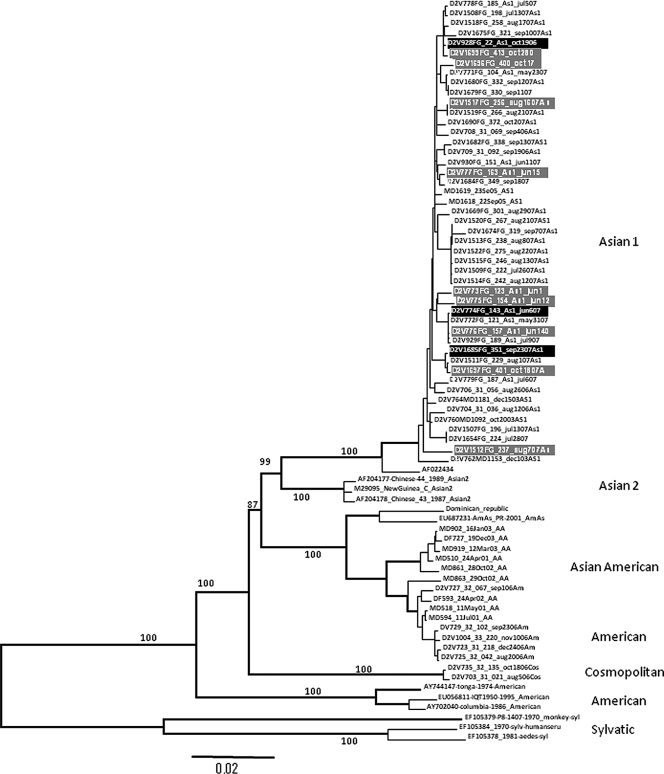
Patients with DSS had significantly higher plasma NS1 concentrations at the time of enrollment than matched uncomplicated dengue controls (Table 1). Plasma viremia levels were not significantly different between the two groups (Table 1). To understand if virus genetic traits were associated with clinical outcome, we attempted to derive the consensus genome sequence of each virus directly from plasma samples collected at study enrollment. From the 24 DSS patients, we derived consensus genome sequences (nucleotides 30 to 10649) of 15 DENV serotype 1 (DENV-1) and 3 DENV-2. These sequences were compared phylogenetically to 32 consensus genome sequences of DENV-1 and 9 consensus genome sequences of DENV-2 sampled from among the 54 patients with uncomplicated dengue (Fig. 1 and 2). All DENV-1 sequences belonged to the genotype I lineage, and there was no evidence of a phylogenetic structure in the neighbor-joining tree that was related to clinical outcome (Fig. 1). All DENV-2 sequences belonged to the Asian 1 lineage, and similarly, consensus genome sequences from DSS patients were not phylogenetically distinct from those from uncomplicated dengue cases (Fig. 2).
FIG. 1.
Phylogenetic tree of DENV-1 consensus genome sequences from patients in this study. The tree (neighbor-joining method) contains consensus genome sequences deduced from plasma samples obtained from 32 DENV-1-infected patients with uncomplicated dengue (gray-highlighted tip labels) and 15 genomes sampled from DENV-1-infected patients with DSS (black-highlighted tip labels). The tree is midpoint rooted and contains sequences from other DENV-1 viruses for reference only (black-highlighted tip labels). Bootstrap values are shown on major branches.
FIG. 2.
Phylogenetic tree of DENV-2 consensus genome sequences from patients in this study. The tree (neighbor-joining method) contains genome sequences deduced from plasma samples obtained from 9 DENV-2-infected patients with uncomplicated dengue (gray-highlighted tip labels) and 3 genomes sampled from DENV-2-infected patients with DSS (black-highlighted tip labels). The tree is midpoint rooted and contains sequences from other DENV-2 viruses for reference only (black-highlighted tip labels). Bootstrap values are shown on major branches.
Differences in host gene transcript abundance between the early febrile phase of dengue and convalescence.
We undertook an expansive interrogation of the early host transcriptional signature in dengue by comparing transcriptional profiles of 9,870 genes in samples collected early in the acute phase (fever day −2 or −3) from all 80 acute dengue patients (24 DSS and 56 uncomplicated dengue patients) and 34 convalescent control samples (18 severe dengue and 16 uncomplicated dengue control samples). By SAM with an FDR of <5%, we identified 860 differentially expressed (DE) transcripts (fold change equal or greater than 2; q value ≤ 5%). Of the 860 differentially abundant gene transcripts, 309 were less abundant in acute samples relative to convalescent samples, and 551 were more abundant. InnateDB pathway, ontology, and network analysis of differentially expressed transcripts was performed to identify the molecular pathways, networks, and biological processes that dominate the whole-blood transcriptional profile, and in particular, the major signaling hubs involved in the early immune response.
Pathways and gene ontology terms associated with upregulated genes.
Many of the overabundant DE transcripts are interferon inducible; indeed, of the 551 upregulated genes, 173 (31%) were annotated as type I interferon inducible by the Interferome database (32). Pathways that were significantly overrepresented in upregulated genes after correction for multiple testing (FDR < 5%) included the systemic lupus erythematosus (KEGG database), classical complement (PID BioCarta), and complement and coagulation cascades (KEGG) pathways. The pathway systemic lupus erythematosus is likely significant due to the overlap between this pathway and the complement pathways and because of the large number of histone genes that are upregulated. A number of complement- and coagulation-related genes were found to be upregulated (C1QA, C1QB, C1QC, C2, C3AR1, C5, F9, PLAUR, PROS1, SERPINA1, and SERPING1). Prior to correction for multiple testing (which is conservative, given the large number of pathways tested), a range of pathways was identified as being significantly overrepresented. These pathways included the Toll-like receptor signaling pathway, interleukin-27 (IL-27)-mediated signaling events, IL-12-mediated signaling events, the chemokine signaling pathway, cytokine-cytokine receptor interactions, the lysosome pathway, the RIG-I-like receptor signaling pathway, the gamma interferon (IFN-γ) pathway and the JAK-STAT pathway and regulation pathway. Many of these are likely of biological significance (despite being below the statistical threshold) and are highlighted in Table S2 in the supplemental material. Some of the prominent upregulated transcripts here were associated with recognition of virus ligands and downstream signaling, including Toll-like receptors (TLRs), RIG-I, MDA5, IRF7, AIM2, DAI, and STATs, all of which make contributions to innate antiviral sensing and downstream signaling.
The most significant GO terms associated with upregulated genes (FDR < 5%) included the following: response to virus, immune response, innate immune response, and inflammatory response. A full list of GO terms associated with upregulated genes is described in Table S3 in the supplemental material.
Pathways and GO terms associated with downregulated genes.
Almost all of the significantly downregulated pathways were related to translation (see Table S4 in the supplemental material). These pathways were significant due to the downregulation of genes encoding ribosomal subunit proteins. The hemoglobin's chaperone pathway was also significantly overrepresented due to the downregulation of several genes involved in the heme biosynthesis pathway (ALAD, ALAS2, ERAF, FECH, HBB, and HMBS). GO analysis paints a picture similar to that of the pathway analysis, with a number of terms related to translation all being significant (FDR < 5%) (see Table S5 in the supplemental material).
Network analysis of differential gene expression profiles in acute dengue versus convalescent dengue.
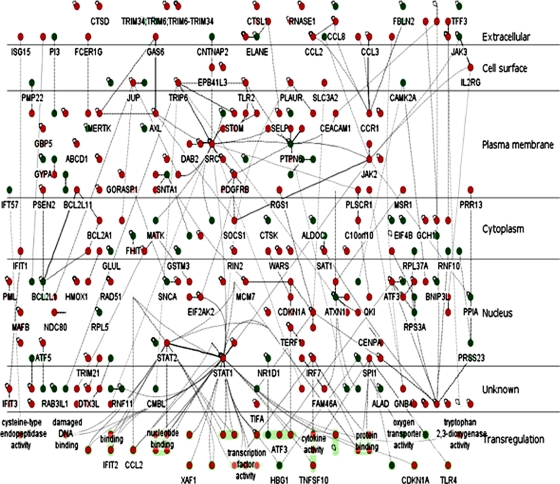
InnateDB (27; www.innatedb.com) was used to generate molecular interaction networks involving DE genes and their encoded products (DE nodes). The first was a network consisting only of the interactions between DE nodes (Fig. 3), while the second expanded upon this network by including all nondifferentially expressed interacting partners of the DE nodes (see the AcuteDengue_All network in Fig. S1 in the supplemental material). The AcuteDengue_DE network has 289 nodes and 429 edges, in comparison to the more extensive AcuteDengue_All network, which has 4,364 nodes and 9,094 edges.
FIG. 3.
AcuteDengue_DE network. Network of known protein-protein and protein-DNA interactions encoded by genes differentially expressed in 80 acute dengue patients in comparison to 34 convalescent samples. Nodes encoded by upregulated genes are shown in red and by downregulated genes are shown in green. This network was generated using InnateDB (www.innateDB.com) and was visualized using the Cerebral 2.0 plug-in for Cytoscape 2.6.2, which was developed as part of the InnateDB project. This network has 289 nodes and 429 edges. The top 5 hubs (i.e., genes/proteins that are highly connected to other DE genes) in this network were identified as the transcription factors STAT1 and STAT2 (2× upregulated), the tyrosine kinase SRC (2× upregulated), PTPN6 (SHP1) (2.5× downregulated), and C1orf103 (2× upregulated).
The AcuteDengue_DE network was analyzed to identify network hubs and bottlenecks which may represent the key regulatory nodes in the network. Using the “Degree” algorithm from the cyto-Hubba plug-in (25), the top 5 hubs (i.e., genes/proteins that are highly connected to other DE genes) in this network were identified as the transcription factors STAT1 and STAT2 (2-fold upregulated), the tyrosine kinase SRC (2-fold upregulated), PTPN6 (SHP1) (2.5-fold downregulated), and C1orf103 (2-fold upregulated). The Hubba software also allows one to predict proteins that act as bottlenecks in the network. Bottlenecks are network nodes that are the key connector proteins in a network and have many “shortest paths” going through them, similar to bridges or tunnels on a highway map (43). Several of the hubs, including STAT1, SRC, and PTPN6 (SHP1), were identified as bottlenecks in the network, further supporting their central role in signaling. TRIP6 and JAK2 were also identified in the top 5 bottlenecks. Analysis of the AcuteDengue_All network, which consists of all interactions involving molecules encoded by DE genes (regardless of whether the interacting molecule is DE), also identified SRC, STAT1, TRIP6, SHP1, C1orf103, and JAK2 in the top 20 hub/bottleneck nodes (see Fig. S2 in the supplemental material). SRC is the highest ranked hub in this network, and SRC and STAT1 are the top 2 bottlenecks.
Two major differentially expressed subnetworks were identified in the AcuteDengue_DE network. The top-ranked network consisted of 23 nodes (including JAK2, JAK3, SRC, TLR2, IL2RG, SOCS1, SHP1, TRIP6, and other JAK/STAT and SRC regulators) (see Fig. S3A in the supplemental material). The second-ranked subnetwork, also consisting of 23 nodes, was enriched for nuclear-localized proteins (13 nodes), includes terms such as transcription corepressor activity (ATF3, DDIT3 [CEBPZ], ID3, and NFIL3), and may represent an important transcriptional regulatory network (see Fig. S3B in the supplemental material).
Combining the transcriptional regulatory network and the physical interaction network.
InnateDB was also used to construct a network of all predicted transcription factor (TF) interactions with DE genes (TF network). This predicted transcriptional regulatory network was then merged with the AcuteDengue_All network. This combined network provides a much more comprehensive picture of the connection between signaling and transcriptional regulation (see Fig. S4 in the supplemental material). Nodes with significantly more interactions with DE genes than expected by chance were highly enriched in transcription factors involved in IFN/NF-κB signaling responses, including STAT1, STAT2, STAT3, IRF7, IRF9, IRF1, CEBPB, and SP1. Notably, this approach enabled the identification of potentially important regulators that were not identified as DE at the time point sampled. IRF1, for example, although not DE itself, is predicted to interact with 100 DE genes. Similarly, CEBPB is predicted to interact with 88 genes and is not itself DE. Gene expression analysis alone would not reveal the multitude of transcription factors that are likely the key regulators driving the host response to DENV infection.
Differences in early host gene transcript abundance between DSS and uncomplicated dengue patients.
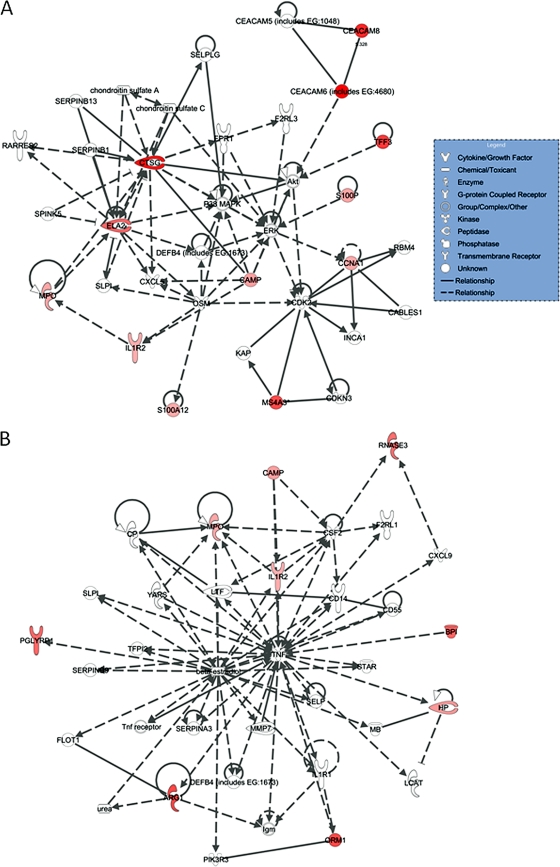
The previous analysis dealt with differences in the transcriptome between acute and convalescent samples. Since the identification of prognostic markers of severe dengue is an important goal, we turned our focus to identifying differences in the early acute transcriptome between severe and uncomplicated dengue cases. To this end, we compared gene transcript abundance on fever day −2 or −3 in the 24 DSS patients and their 56 matched controls. By SAM, 21 transcripts were significantly enriched in DSS patients relative to uncomplicated dengue patients (FDR of <5% and >2-fold difference in abundance). There were no significantly downregulated transcripts in acute DSS patients compared to those in acute uncomplicated dengue patients. Table 2 summarizes the list of 21 differentially abundant transcripts. Prominent among these were transcripts that could be linked to activated neutrophils, with 12 of the 21 differentially expressed genes associated with activated neutrophils in the shape of membrane-bound integrin receptors (CEACAM6 and CEACAM8), a cytokine decoy receptor (interleukin-1 receptor, type II [IL1R2]), secreted proteases (CTSG and ELA2), inflammatory molecules (S100A12), secreted antimicrobial proteins/peptides (DEF1, DEF4, BPI, CAMP, and PGLYRP1), or oxidative enzymes (MPO). We explored possible functional relationships between the 21 differentially expressed genes using InnateDB and separately using the Ingenuity Pathway Analysis (IPA) knowledge base. We describe here the IPA method, as both methods identified networks enriched in neutrophil proteins. Unsupervised network IPA identified two clusters of 35 genes each that included 18 of the 21 differentially expressed genes (Fig. 4). These two network interactions were highly unlikely to have occurred by chance (P = 10−31 and 10−21). The first cluster included 12 differentially expressed genes, and the second cluster included 9 differentially expressed genes. Neutrophil-associated CAMP and MPO, together with the decoy receptor IL1R2, were found in both networks. Cathepsin G (CTSG) and elastase (ELA), which were found on the list of 21 DE genes, were the key elements of the first network, while tumor necrosis factor alpha (TNF-α) and β-estradiol were the key elements of the second network. Strikingly, these data suggest that gene expression profiles from members of two overlapping networks discriminate between patients who progress to DSS and those with an uncomplicated disease course. InnateDB identified the transcription factor CEBPB as a potential important regulator of several of the differentially expressed genes. We performed classification analysis using a decision tree forest model. In the analysis, the intensity signals of all 21 differentially expressed genes were used as “predictors,” and the disease outcomes were used as “targets.” From the total 200 trees built, the best misclassification rate was 15/24 (62.5%) for DSS patients and 10/46 (21.7%) for uncomplicated dengue patients. The result indicates that these differentially expressed genes could not be used as predictors of disease outcomes.
TABLE 2.
Differentially expressed transcripts in acute DSS patients relative to UC dengue patients at fever day −2 or −3
| Transcripta | SAM results |
Results from samples obtained from patients with: |
||||||
|---|---|---|---|---|---|---|---|---|
| Acute DSS |
Acute UC dengue |
|||||||
| Fold change | q value (%) | No. (%) of samples detected | Mean intensity | SD | No. (%) of samples detected | Mean intensity | SD | |
| DEF4A | 6.7 | 0 | 22 (92) | 5.5 | 17.4 | 39 (70) | 0.8 | 0.8 |
| CEACAM6 | 6.5 | 0 | 12 (50) | 8.3 | 24.4 | 13 (23) | 1.3 | 2 |
| CTSG | 6 | 2.6 | 17 (71) | 5.8 | 15.5 | 23 (41) | 1 | 1.1 |
| CEACAM8 | 5.3 | 0 | 22 (92) | 5.5 | 14 | 39 (70) | 1 | 1.3 |
| ARG1 | 4.5 | 0 | 20 (83) | 4.9 | 10.8 | 26 (46) | 1.1 | 0.9 |
| MS4A3 | 4.5 | 0 | 20 (83) | 5 | 13.8 | 37 (66) | 1.1 | 1.2 |
| BPI | 3.8 | 0 | 21 (88) | 4.8 | 12.1 | 37 (66) | 1.2 | 1 |
| ELA2 | 3.8 | 0 | 23 (96) | 3.6 | 7.3 | 48 (86) | 0.9 | 0.8 |
| PGLYRP1 | 3.7 | 0 | 24 (100) | 3.3 | 7.2 | 56 (100) | 0.9 | 0.8 |
| RNASE3 | 3.6 | 0 | 12 (50) | 4 | 8.8 | 30 (54) | 1.1 | 0.7 |
| MS4A3 | 3.2 | 4.4 | 20 (83) | 3.5 | 9.6 | 38 (68) | 1.1 | 0.8 |
| LOC728358 | 2.7 | 0 | 24 (100) | 2.7 | 4.1 | 56 (100) | 1 | 1.2 |
| HP | 2.5 | 0 | 24 (100) | 4.6 | 5.2 | 53 (95) | 1.8 | 1.7 |
| S100P | 2.3 | 2.4 | 24 (100) | 2.9 | 3.4 | 43 (77) | 1.3 | 1.7 |
| CCNA1 | 2.1 | 4.7 | 22 (92) | 25.8 | 38.3 | 46 (82) | 12.2 | 13.4 |
| S100A12 | 2.1 | 0 | 24 (100) | 2.5 | 2.2 | 54 (96) | 1.2 | 1.1 |
| CAMP | 2.4 | 0 | 24 (100) | 2.7 | 3.7 | 56 (100) | 1.1 | 0.9 |
| TFF3 | 4.9 | 0 | 11 (46) | 3.3 | 7.8 | 8 (14) | 0.7 | 0.6 |
| ORM1 | 4.5 | 0 | 9 (38) | 4.3 | 11.1 | 12 (21) | 1 | 2.4 |
| MPO | 2.2 | 4.3 | 24 (100) | 3.1 | 6.3 | 46 (82) | 1.4 | 1 |
| IL1R2 | 2 | 2.1 | 21 (88) | 4.1 | 4.1 | 43 (77) | 2 | 1.8 |
DEF4A, defensin 4 alpha corticostatin; CEACAM6, carcinoembryonic antigen-related cell adhesion molecule 6; CTSG, cathepsin G; CEACAM8, carcinoembryonic antigen-related cell adhesion molecule; BPI, bactericidal/permeability-increasing protein; ELA2, elastase 2; PGLYRP1, peptidoglycan recognition protein 1; RNASE3, RNase A, family 3 (eosinophil cationic protein); DEF1A, defensin 1 alpha; HP, haptoglobin; S100P, S100 calcium binding protein P; CCNA1, cyclin A1; S100A12, S100 calcium binding protein A12; CAMP, cathelicidin antimicrobial peptide; TFF3, trefoil factor 3 (intestinal); ORM1, orosomucoid 1; MPO, myeloperoxidase; IL1R2, interleukin-1 receptor, type II.
FIG. 4.
Ingenuity Pathway Analysis (IPA) of differentially expressed genes between 24 DSS and 56 uncomplicated dengue patients. Twenty-one differentially expressed transcripts were analyzed using IPA. The following two significant networks were identified: cancer, cell cycle, and cell-mediated immune response (network 1, score of 31) (A) and antigen presentation, cell-mediated immune response, and humoral immune response (network 2, score of 21) (B). The lines between genes represent known interactions, with solid lines representing direct interactions and dashed lines representing indirect interactions. Differentially expressed genes are highlighted in red, and genes identified by IPA are not highlighted. The high scores associated with these networks indicate they were highly unlikely to be formed by chance.
The differential abundance of neutrophil-associated transcripts in patients who progressed to DSS was not simply a reflection of neutrophil levels in the sample, as the mean absolute counts of neutrophils for each group were not significantly different (median count [95% confidence interval {CI}] in DSS patients was 2.75 × 103 cells/mm3 [2.22 to 3.33] versus that of 2.48 × 103 cells/mm3 [2.27 to 3.10] in uncomplicated dengue cases). These results indicated that neutrophils are phenotypically activated in children who subsequently develop DSS and suggests that certain neutrophil-associated transcripts could have prognostic value by identifying patients at risk of severe disease.
Plasma concentrations of BPI, DEF1A, ELA2, and MPO.
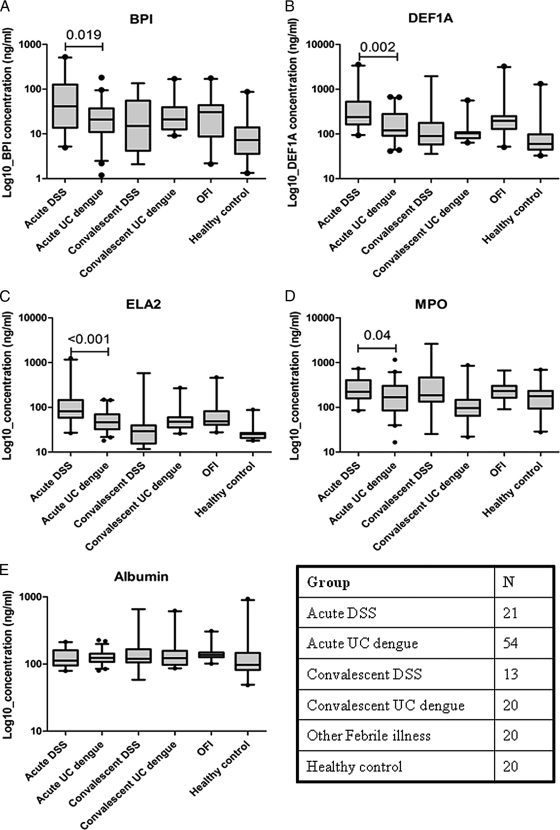
Concentrations of BPI, DEF1A, ELA2, and MPO in plasma samples collected at the same time point as the RNA used for expression array analysis were measured to determine if there was evidence suggestive of neutrophil activation. Consistent with the array findings, plasma concentrations of all four proteins were significantly higher at the time of study enrollment in those children who developed DSS relative to children with uncomplicated dengue, although the absolute difference was small (Fig. 5A to D). Plasma concentrations of BPI, DEF1A, and ELA2 were also significantly higher in samples obtained from children who developed DSS than in convalescent samples and healthy donor samples.
FIG. 5.
Concentrations of secreted neutrophil-associated proteins in plasma samples. Concentrations of BPI (A), DEF1A (B), ELA2 (C), MPO (D), and albumin (E) in acute and convalescent dengue cases and, for reference, in patients with other febrile illnesses and in healthy-donor plasma samples. The box-and-whisker plots represent median and interquartile ranges. OFI, other febrile illnesses.
To understand if elevated concentrations of these neutrophil-associated proteins were independent of early hemoconcentrations in children with DSS (i.e., reduced vascular volume, leading to higher plasma protein concentrations), plasma albumin levels in all samples were measured as a surrogate marker of the plasma protein concentration (Fig. 5E). Levels of BPI, ELA2, and MPO were not significantly correlated with the plasma albumin concentration in the same sample, suggesting their elevated levels in plasma were not merely a reflection of the hemoconcentration at this time point (Pearson's correlation coefficient used; plasma albumin concentration versus levels of BPI (r = −0.04; P = 0.6), ELA2 (r = −0.41; P = 0.6), and MPO (r = −0.008; P = 0.9). In contrast, concentrations of DEF1A were weakly correlated with the plasma albumin concentration (plasma albumin concentration versus level of DEF1A (r = −0.176; P = 0.014), suggesting that at least some component of this measurement might have been influenced by the reduced vascular volume.
DISCUSSION
This case-control study investigated the early whole-blood transcriptional signature in children who subsequently developed DSS, the most common life-threatening complication of dengue in children. This study was rooted in clinical practice by focusing on the dengue syndrome that always requires a clinical intervention, often in the setting of the intensive care unit. Strikingly, we identified, in the first few days of illness, two overlapping gene networks that distinguished patients who developed DSS from those with uncomplicated dengue. Features of these networks were genes associated with neutrophil activation and degranulation, suggesting a hitherto unrecognized association of neutrophils with pathogenesis and expression of the overall disease phenotype.
Previous studies from our groups have described the whole-blood transcriptional signature in dengue patients by microarray analysis (11, 26, 36). The timing of sample collection is clearly a major factor in the transcriptional signature, with samples collected during the febrile phase having characteristic antiviral profiles, e.g., with interferon-stimulated genes being highly prominent (11, 26, 36), while those collected during the afebrile stage having predominantly metabolic profiled (26, 36). Studies of peripheral blood mononuclear cells (PBMC) (i.e., minus the neutrophil population) have also been described (30, 40). In short, however, all previous microarray studies of the dengue host response have used relatively small amounts of samples collected at heterogenous time points and rarely included patients with DSS, the commonest life-threatening complication in children. Against this backdrop, a strength of the current study is the matched case-control design, large sample size, early sampling prior to defervescence and cardiovascular decompensation, and inclusion of detailed genomic scale information on the infecting pathogen.
Previous epidemiological in vitro and in vivo studies have suggested that phenotypic differences can exist between virus lineages of the same serotype (1, 9, 41). We determined consensus virus genome sequences and demonstrated that viruses in DSS patients in this study were not phylogenetically different from those in patients with uncomplicated dengue, implying that host factors were more important determinants of the clinical course. In addition, the viremia (as measured by qRT-PCR) was not significantly higher in children who subsequently developed DSS, although the plasma NS1 concentration was higher, suggesting that antigen burden may be a better correlate of severe clinical outcomes, as has been alluded to previously (24).
Using a large sample size, we defined the major transcriptional features of the acute response to DENV infection during the febrile period. The complement, TLR, and RIG-I signaling pathways, interferon-stimulated genes, and cytokine/chemokines and their receptors were the major features of the transcriptional signature, consistent with previous studies in smaller numbers of febrile dengue patients (11, 26, 40). Utilizing a systems biology approach that investigated transcriptional profiles in the context of their molecular interaction networks, we identified the transcription factors STAT1 and STAT2, the tyrosine kinase SRC, SHP1, TRIP6, and JAK2 as key central molecules in these networks. Furthermore, the top-ranked differentially expressed subnetwork was enriched in molecules involved in cytokine signaling and JAK/STAT pathways (including JAK2, JAK3, SRC, TLR2, IL2RG, SOCS1, SHP1, TRIP6, and other JAK/STAT and SRC regulators). The top hub/bottleneck molecules in these networks form a densely connected network module, with a variety of known interactions between the nodes, which supports this being a core signaling module in the network and a central feature of the host response. Analysis of the transcriptional regulatory network also identified several STAT and IRF transcription factors as the key regulators of the transcriptional response. The importance of the STAT1 pathway in control of DENV replication in mice and mosquitoes has been demonstrated previously (35, 37), and STAT1/2 may also be targets of DENV-mediated interference in the interferon signaling pathway (20, 29).
We identified 21 genes as differentially expressed (more abundant) in patients who developed DSS compared to those in matched control patients. Remarkably, almost all of these genes belonged to one of two overlapping networks in which some of the interconnecting elements have immune response functions. That TNF-α should be a central hub in the second network is striking, given that TNF-α has been repeatedly implicated in the pathogenesis of severe capillary leakage (4, 14). Although TNF-α is a major hub in the network analysis, we did not measure TNF-α plasma levels because TNF-α transcripts were not significantly different between DSS and uncomplicated dengue patients at the time of enrollment. The suggestion from our work is instead that there may only be small differences in the acute response that leads to a “tipping point” and the physiological derangement of normal vascular endothelial cell functions. Furthermore, these data shed light on previously unrecognized aspects of the acute response to DENV infection and provide an impetus for further investigations of the role of neutrophils in pathogenesis. Clearly, further studies are needed to understand why these two networks, and some of their differentially expressed members, are associated with progression to DSS.
Neutrophil activation and degranulation were most prominent themes in the DSS-associated differentially expressed gene list. A gene signature consistent with neutrophil activation has also been recently described in whole-blood samples obtained from Cambodian children with DSS (10). We verified that plasma protein concentrations of CTSG, BPI, ELA2, and MPO were also higher in early DSS patients than in control patients, albeit the absolute difference was small and unlikely to be useful for prognosis. Of the differentially expressed genes associated with neutrophil degranulation, ELA2, CTSG, and the defensins (DEF1A and DEF4A) are of particular interest. ELA2 and CTSG are serine proteases that can cleave vascular endothelial cadherin and thereby compromise the integrity of the vascular endothelium (18). These proteases might conceivably play similar roles in perturbing the endothelium in capillary beds during DENV infection. Accordingly, ELA2 has also been detected previously at higher concentrations in sera of patients with DSS than in those of patients without shock (22). DEF1A and DEF4A are neutrophil-associated defensins with antiviral activity (12, 16, 17). These innate antimicrobial peptides may also functionally participate in the innate antiviral response to DENV infection.
The dengue capillary leakage syndrome begins in children with secondary infections within the first 1 to 2 days of illness and can be measured by ultrasound as early as the third day of illness (38). Commencement of capillary leakage in infants with primary dengue likely occurs with similar kinetics and can lead to DSS between illness days 4 and 6 (36). The triggering of capillary permeability early in the disease evolution in both of these clinical settings might be mediated by robust innate immune responses rather than acquired responses, particularly since preexisting immunity does not exist in infants with primary infection. We propose that activated, degranulating peripheral blood neutrophils could contribute to an early triggering of capillary permeability. In this model, neutrophils (which express Fc receptors) are activated by immune complexes and/or by high early virus antigen burdens in blood and tissues, where they secrete cytokines and chemotactic molecules. High viral antigen burdens in vivo could be arrived at by antibody-dependent enhancement in secondary infection or in primary infection of infants born to dengue immune mothers. Neutrophil activation and degranulation alone are highly unlikely to be sufficient to drive capillary leakage to the extent seen in patients with DSS. Instead, neutrophil adherence to endothelial cells and the secretion of soluble mediators of vascular permeability may represent a small step in the inflammatory cascade that synergizes with other adaptive host responses to mediate capillary permeability in severe secondary infection. In infants with primary infection and severe dengue, an innate response that includes neutrophil activation, together with an inherently permeable vascular endothelium, might be sufficient to trigger clinically significant vascular leakage. Further studies of neutrophils and their responses to DENV infection are warranted.
Supplementary Material
Acknowledgments
This work was supported by the Wellcome Trust and A*STAR Singapore. InnateDB is funded by Genome Canada and Genome British Columbia through the Pathogenomics of Innate Immunity (PI2) project and by the Foundation for the National Institutes of Health and the Canadian Institutes of Health Research under the Grand Challenges in Global Health Research Initiative (Grand Challenges ID 419). Continued development of InnateDB is also supported by Teagasc (project RMIS 6018).
We have no financial or commercial conflicts of interest.
Footnotes
Published ahead of print on 13 October 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Anderson, J. R., and R. Rico-Hesse. 2006. Aedes aegypti vectorial capacity is determined by the infecting genotype of dengue virus. Am. J. Trop. Med. Hyg. 75:886-892. [PMC free article] [PubMed] [Google Scholar]
- 2.Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, and G. Sherlock. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25:25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple Testing. J. R. Stat. Soc. Series B Stat. Methodol. 57:289-300. [Google Scholar]
- 4.Bethell, D. B., K. Flobbe, X. T. Cao, N. P. Day, T. P. Pham, W. A. Buurman, M. J. Cardosa, N. J. White, and D. Kwiatkowski. 1998. Pathophysiologic and prognostic role of cytokines in dengue hemorrhagic fever. J. Infect. Dis. 177:778-782. [DOI] [PubMed] [Google Scholar]
- 5.Bethell, D. B., J. Gamble, P. L. Pham, M. D. Nguyen, T. H. Tran, T. H. Ha, T. N. Tran, T. H. Dong, I. B. Gartside, N. J. White, and N. P. Day. 2001. Noninvasive measurement of microvascular leakage in patients with dengue hemorrhagic fever. Clin. Infect. Dis. 32:243-253. [DOI] [PubMed] [Google Scholar]
- 6.Breiman, L. 2001. Decision tree forests. Mach. Learn. 45:5-32. [Google Scholar]
- 7.Burke, D. S., A. Nisalak, D. E. Johnson, and R. M. Scott. 1988. A prospective study of dengue infections in Bangkok. Am. J. Trop. Med. Hyg. 38:172-180. [DOI] [PubMed] [Google Scholar]
- 8.Coffey, L. L., E. Mertens, A. C. Brehin, M. D. Fernandez-Garcia, A. Amara, P. Despres, and A. Sakuntabhai. 2009. Human genetic determinants of dengue virus susceptibility. Microbes Infect. 11:143-156. [DOI] [PubMed] [Google Scholar]
- 9.Cologna, R., P. M. Armstrong, and R. Rico-Hesse. 2005. Selection for virulent dengue viruses occurs in humans and mosquitoes. J. Virol. 79:853-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devignot, S., C. Sapet, V. Duong, A. Bergon, P. Rihet, S. Ong, P. T. Lorn, N. Chroeung, S. Ngeav, H. J. Tolou, P. Buchy, and P. Couissinier-Paris. 2010. Genome-wide expression profiling deciphers host responses altered during dengue shock syndrome and reveals the role of innate immunity in severe dengue. PLoS One 5:e11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fink, J., F. Gu, L. Ling, T. Tolfvenstam, F. Olfat, K. C. Chin, P. Aw, J. George, V. A. Kuznetsov, M. Schreiber, S. G. Vasudevan, and M. L. Hibberd. 2007. Host gene expression profiling of dengue virus infection in cell lines and patients. PLoS Negl. Trop. Dis. 1:e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furci, L., F. Sironi, M. Tolazzi, L. Vassena, and P. Lusso. 2007. Alpha-defensins block the early steps of HIV-1 infection: interference with the binding of gp120 to CD4. Blood 109:2928-2935. [DOI] [PubMed] [Google Scholar]
- 13.Graham, R. R., M. Juffrie, R. Tan, C. G. Hayes, I. Laksono, C. Ma'roef Erlin, Sutaryo, K. R. Porter, and S. B. Halstead. 1999. A prospective seroepidemiologic study on dengue in children four to nine years of age in Yogyakarta, Indonesia I. studies in 1995-1996. Am. J. Trop. Med. Hyg. 61:412-419. [DOI] [PubMed] [Google Scholar]
- 14.Green, S., S. Pichyangkul, D. W. Vaughn, S. Kalayanarooj, S. Nimmannitya, A. Nisalak, I. Kurane, A. L. Rothman, and F. A. Ennis. 1999. Early CD69 expression on peripheral blood lymphocytes from children with dengue hemorrhagic fever. J. Infect. Dis. 180:1429-1435. [DOI] [PubMed] [Google Scholar]
- 15.Hang, V. T., N. M. Nguyet, D. T. Trung, V. Tricou, S. Yoksan, N. M. Dung, T. Van Ngoc, T. T. Hien, J. Farrar, B. Wills, and C. P. Simmons. 2009. Diagnostic accuracy of NS1 ELISA and lateral flow rapid tests for dengue sensitivity, specificity and relationship to viraemia and antibody responses. PLoS Negl. Trop. Dis. 3:e360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartshorn, K. L., M. R. White, T. Tecle, U. Holmskov, and E. C. Crouch. 2006. Innate defense against influenza A virus: activity of human neutrophil defensins and interactions of defensins with surfactant protein D. J. Immunol. 176:6962-6972. [DOI] [PubMed] [Google Scholar]
- 17.Hazrati, E., B. Galen, W. Lu, W. Wang, Y. Ouyang, M. J. Keller, R. I. Lehrer, and B. C. Herold. 2006. Human alpha- and beta-defensins block multiple steps in herpes simplex virus infection. J. Immunol. 177:8658-8666. [DOI] [PubMed] [Google Scholar]
- 18.Hermant, B., S. Bibert, E. Concord, B. Dublet, M. Weidenhaupt, T. Vernet, and D. Gulino-Debrac. 2003. Identification of proteases involved in the proteolysis of vascular endothelium cadherin during neutrophil transmigration. J. Biol. Chem. 278:14002-14012. [DOI] [PubMed] [Google Scholar]
- 19.Ideker, T., O. Ozier, B. Schwikowski, and A. F. Siegel. 2002. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics 18(Suppl. 1):S233-S240. [DOI] [PubMed] [Google Scholar]
- 20.Jones, M., A. Davidson, L. Hibbert, P. Gruenwald, J. Schlaak, S. Ball, G. R. Foster, and M. Jacobs. 2005. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J. Virol. 79:5414-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joshi-Tope, G., M. Gillespie, I. Vastrik, P. D'Eustachio, E. Schmidt, B. de Bono, B. Jassal, G. R. Gopinath, G. R. Wu, L. Matthews, S. Lewis, E. Birney, and L. Stein. 2005. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 33:D428-D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juffrie, M., G. M. van Der Meer, C. E. Hack, K. Haasnoot Sutaryo, A. J. Veerman, and L. G. Thijs. 2000. Inflammatory mediators in dengue virus infection in children: interleukin-8 and its relationship to neutrophil degranulation. Infect. Immun. 68:702-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa, M., M. Araki, S. Goto, M. Hattori, M. Hirakawa, M. Itoh, T. Katayama, S. Kawashima, S. Okuda, T. Tokimatsu, and Y. Yamanishi. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36:D480-D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Libraty, D. H., P. R. Young, D. Pickering, T. P. Endy, S. Kalayanarooj, S. Green, D. W. Vaughn, A. Nisalak, F. A. Ennis, and A. L. Rothman. 2002. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J. Infect. Dis. 186:1165-1168. [DOI] [PubMed] [Google Scholar]
- 25.Lin, C. Y., C. H. Chin, H. H. Wu, S. H. Chen, C. W. Ho, and M. T. Ko. 2008. Hubba: hub objects analyzer—a framework of interactome hubs identification for network biology. Nucleic Acids Res. 36:W438-W443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long, H. T., M. L. Hibberd, T. T. Hien, N. M. Dung, T. V. Ngoc, J. Farrar, B. Wills, and C. P. Simmons. 2009. Patterns of gene transcript abundance in the blood of children with severe or uncomplicated dengue highlight differences in disease evolution and host response to dengue virus infection. J. Infect. Dis. 199:537-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynn, D. J., G. L. Winsor, C. Chan, N. Richard, M. R. Laird, A. Barsky, J. L. Gardy, F. M. Roche, T. H. Chan, N. Shah, R. Lo, M. Naseer, J. Que, M. Yau, M. Acab, D. Tulpan, M. D. Whiteside, A. Chikatamarla, B. Mah, T. Munzner, K. Hokamp, R. E. Hancock, and F. S. Brinkman. 2008. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 4:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Messer, W. B., D. J. Gubler, E. Harris, K. Sivananthan, and A. M. de Silva. 2003. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg. Infect. Dis. 9:800-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munoz-Jordan, J. L., M. Laurent-Rolle, J. Ashour, L. Martinez-Sobrido, M. Ashok, W. I. Lipkin, and A. Garcia-Sastre. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nascimento, E. J., U. Braga-Neto, C. E. Calzavara-Silva, A. L. Gomes, F. G. Abath, C. A. Brito, M. T. Cordeiro, A. M. Silva, C. Magalhaes, R. Andrade, L. H. Gil, and E. T. Marques, Jr. 2009. Gene expression profiling during early acute febrile stage of dengue infection can predict the disease outcome. PLoS One 4:e7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pediatric Dengue Vaccine Initiative. 2009. Global burden of dengue. Pediatric Dengue Vaccine Initiative, Seoul, Korea. http://www.pdvi.org/about_dengue/GBD.asp.
- 31a.Robertson, G., M. Bilenky, K. Lin, A. He, W. Yuen, M. Dagpinar, R. Varhol, K. Teague, O. L. Griffith, X. Zhang, Y. Pan, M. Hassel, M. C. Sleumer, W. Pan, E. D. Pleasance, M. Chuang, H. Hao, Y. Y. Li, N. Robertson, C. Fjell, B. Li, S. B. Montgomery, T. Astakhova, J. Zhou, J. Sander, A. S. Siddiqui, and S. J. Jones. 2006. cisRED: a database system for genome-scale computational discovery of regulatory elements. Nucleic Acids Res. 34:D68-D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samarajiwa, S. A., S. Forster, K. Auchettl, and P. J. Hertzog. 2009. INTERFEROME: the database of interferon regulated genes. Nucleic Acids Res. 37:D852-D857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sangkawibha, N., S. Rojanasuphot, S. Ahandrik, S. Viriyapongse, S. Jatanasen, V. Salitul, B. Phanthumachinda, and S. B. Halstead. 1984. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am. J. Epidemiol. 120:653-669. [DOI] [PubMed] [Google Scholar]
- 34.Shannon, P., A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski, and T. Ideker. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shresta, S., K. L. Sharar, D. M. Prigozhin, H. M. Snider, P. R. Beatty, and E. Harris. 2005. Critical roles for both STAT1-dependent and STAT1-independent pathways in the control of primary dengue virus infection in mice. J. Immunol. 175:3946-3954. [DOI] [PubMed] [Google Scholar]
- 36.Simmons, C. P., S. Popper, C. Dolocek, T. N. Chau, M. Griffiths, N. T. Dung, T. H. Long, D. M. Hoang, N. V. Chau, T. T. Thao le, T. T. Hien, D. A. Relman, and J. Farrar. 2007. Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever. J. Infect. Dis. 195:1097-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Souza-Neto, J. A., S. Sim, and G. Dimopoulos. 2009. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc. Natl. Acad. Sci. U. S. A. 106:17841-17846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srikiatkhachorn, A., A. Krautrachue, W. Ratanaprakarn, L. Wongtapradit, N. Nithipanya, S. Kalayanarooj, A. Nisalak, S. J. Thomas, R. V. Gibbons, M. P. Mammen, Jr., D. H. Libraty, F. A. Ennis, A. L. Rothman, and S. Green. 2007. Natural history of plasma leakage in dengue hemorrhagic fever: a serial ultrasonographic study. Pediatr. Infect. Dis. J. 26:283-290. [DOI] [PubMed] [Google Scholar]
- 39.Thein, S., M. M. Aung, T. N. Shwe, M. Aye, A. Zaw, K. Aye, K. M. Aye, and J. Aaskov. 1997. Risk factors in dengue shock syndrome. Am. J. Trop. Med. Hyg. 56:566-572. [DOI] [PubMed] [Google Scholar]
- 40.Ubol, S., P. Masrinoul, J. Chaijaruwanich, S. Kalayanarooj, T. Charoensirisuthikul, and J. Kasisith. 2008. Differences in global gene expression in peripheral blood mononuclear cells indicate a significant role of the innate responses in progression of dengue fever but not dengue hemorrhagic fever. J. Infect. Dis. 197:1459-1467. [DOI] [PubMed] [Google Scholar]
- 41.Watts, D. M., K. R. Porter, P. Putvatana, B. Vasquez, C. Calampa, C. G. Hayes, and S. B. Halstead. 1999. Failure of secondary infection with American genotype dengue 2 to cause dengue haemorrhagic fever. Lancet 354:1431-1434. [DOI] [PubMed] [Google Scholar]
- 42.World Health Organization. 2009. Dengue: guidelines for treatment, prevention and control. WHO Press, Geneva, Switzerland. [PubMed]
- 43.Yu, H., P. M. Kim, E. Sprecher, V. Trifonov, and M. Gerstein. 2007. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput. Biol. 3:e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.