Abstract
The high rate of HIV-1 evolution contributes to immune escape, enables the virus to escape drug therapy, and may underlie the difficulty of producing an effective vaccine. Identifying constraints on HIV evolution is therefore of prime importance. To investigate this problem, we examined the relationships between sequence diversity, selection, and protein structure. We found that while there was an increase in sequence diversity over time, this variation had a tendency to be limited to specific structural regions. When individual sites were analyzed, there was, in contrast, substantial and widespread evolutionary constraint over gag and env. This constraint was present even in the highly variable envelope proteins. The evolutionary significance of an individual site is indicated by the change in selection pressure along the time course: increasing entropy indicates that the site is evolving predominantly in a more “clock”-like manner, low entropy values with no increase indicate a high degree of constraint, and high entropy values indicate a lack of constraint. Few sites display high degrees of turnover. Mapping these sites onto the three-dimensional protein structure, we found a significant difference between evolutionary rates for regions buried in the core of the protein and those on the surface. This constraint did not change over the time period analyzed and was not subtype dependent, as similar results were found for subtypes B and C. This link between sequence and structure not only demonstrates the limits of recent HIV-1 evolution but also highlights the origins of evolutionary constraint on viral change.
HIV has a highly diverse viral population both within an infected individual and across populations of hosts (39). This diversity derives from two sources: (i) the rapidly evolving nature of the virus, which continually generates new mutant forms (due to mutation, recombination, and its high rate of replication), and (ii) the active immune response that promotes diversifying selection (13, 49, 53). Significantly, the host immune response is extremely active for the majority of the course of infection (prior to progression to AIDS), and it is the escape mutants, coupled with the emergence of latently infected viruses, that contribute to the infection within an individual (49).
This extreme degree of sequence diversity and the rapid rate of HIV's evolution cause enormous problems for both vaccine design and drug therapy. Indeed, the variability of the HIV population permits repeated immune escape, such that no individual is known to have ever naturally cleared an established infection (22, 32). For HIV, the mutation rate within a single patient has been measured at 5.4 × 10−5 substitutions per site per replication round (18), and it has been argued that this high evolutionary rate could negate any possible vaccine (6, 16, 23), although lack of protective immunity is also a major challenge. This apparently relentless viral evolution, coupled with latency, also leads to escape from drug therapy and the need for continued drug treatment. To date, 93 mutations conferring drug resistance have been identified (8, 41). In some cases, multiple mutations are required to confer resistance to even a single drug (34), and yet, such resistance still emerges.
Despite HIV's unusually high mutability, we might expect that there are significant constraints on how the sequence can change and still code for a replication-competent virus. The analysis of fitness costs of evolving viruses (2, 15, 22, 24, 37) indicates that evolutionary constraints restrict viral change. Furthermore, the HIV genome is extremely compact, with several overlapping open reading frames, such that a mutation that is “silent” in one reading frame may not be in another. These constraints limit the number and location of mutations that can be accepted per replication cycle (21).
Several phylogenetic studies tracing the degree of HIV evolution have been performed with partial gene regions (e.g., C2 and V2 to V5 of env) within subtypes (1, 7, 30, 42). In addition, the relationship between the cytotoxic T lymphocyte (CTL) response, sequence diversity, and protein secondary structure has been studied (55). It may be expected that protein structure constrains evolutionary change in HIV, since this has been demonstrated in other systems (12, 19, 31, 35, 36). For example, it has been shown that the core of a protein is less likely to accept replacements than the surface and that different secondary structure types and hydrogen-bonding classes accept different replacements (19, 35).
Examination of the evolutionary trajectory over time would allow the identification of the evolutionary constraints. Fortunately there is a large amount of sequence data for HIV-1, and the sampling dates are known. Moreover, the structures of all major HIV proteins have been determined. Such a combination of data allows analysis of both the evolution of sequence and the correlation with structure to identify the type and location of evolutionary constraints.
MATERIALS AND METHODS
Data sets.
A total of 1,131 gag and 1,170 env sequences from group M subtypes A to D, F to H, J, and K, excluding known recombinant strains, were collected from the LANL HIV Sequence Database (http://www.hiv.lanl.gov). To examine the evolutionary trajectory of HIV-1, gag and env sequences were sorted into nine groups, based on the year of sampling. Each of these sampling groups was aligned separately using MUSCLE (28). From each alignment, columns containing any gaps were removed. Protein sequences were also downloaded from the HIV Sequence Database site and aligned using the same procedure for DNA.
A subset of these data was chosen to determine the effects of variation of subtypes sampled in the available data. Fifty env sequences were randomly selected (10 subtype A, 20 subtype B, and 20 subtype C) as representative sequences for each time period. Due to the smaller data set size, five time periods were used to obtain statistics. For the comparative analysis of env subtypes B and C, gaps were removed from alignments consisting of 20 random subtype B and C sequences for each time period. Two subtype data sets were separated after this procedure and used for further analysis. These subsets were analyzed in the same way as the main data set.
Protein structures were downloaded from the RCSB Protein Data Bank (9). PDB ID 1hiw (20) and 3gv2 (38) were used for p17 and p24 of gag, respectively, and 1gc1 (27) and 1aik (11) for gp120 and gp41 of env.
To make alignments that match the corresponding structure, the sequence corresponding to the known protein structure was added to the multiple sequence alignment, and the sequences were realigned. All columns with a gap in the sequence were removed from the new sequence alignment. For the time course analysis of sequence variability of individual sites for each data set, alignments of whole sequences were constructed first and then sorted into sampling time groups.
Phylogenetic analysis.
To quantify the degree of viral evolution at each time point, phylogenetic trees were constructed for each data set. The maximum likelihood (ML) method was chosen to infer phylogenetic relationships. Pairwise genetic distances were estimated by using a gamma distribution (general time reversible + I + Γ, where I is the proportion of invariable sites and Γ is the shape parameter of the gamma distribution) model with a transition/transversion ratio of 6 in PAUP* (http://paup.csit.fsu.edu/). To find the optimal ML tree, heuristic search algorithm and nearest-neighbor interchange were selected. ML trees of each data set were visualized with CTree software (4). Subtype grouping and calculation of pairwise distance matrices and mean and standard deviations of genetic pairwise distances between individual taxa of each ML tree were also performed in CTree. Box and whisker plots and comparisons of pairwise distances within time periods were performed with the Wilcoxon test using the R program (available at http://www.r-project.org).
Analysis of selection and diversity.
To compare selection and diversity at individual sites, both nonsynonymous-to-synonymous substitution (dN/dS) ratios and Shannon entropy were used. The ratio of dN to dS substitutions is an indication of selection pressure, with values of <1 indicating purifying selection, values of ≈1 indicating neutral evolution, and values of >1 indicating positive selection. Site-by-site dN and dS values were calculated using the single-likelihood ancestor counting (SLAC) method as implemented in the HyPhy software (26). If the dS value of codon index is 0, the dN/dS ratio was assigned as 0 at that site.
In order to determine sequence variability at individual sites, normalized Shannon entropy scores were calculated. Shannon entropy is a quantitative measure of amino acid variability at individual sites, which takes into account the number of possible amino acids replaced and their frequency. Entropy values for each data set were obtained from ENTROPY-ONE (http://www.hiv.lanl.gov/content/sequence/ENTROPY/entropy.html).
Both dN/dS ratios and entropy were calculated for each site at each time period. The values and their change through time at each site were expressed as the mean and the gradient of the line of least-squares best fit. Mean dN/dS and entropy values for buried and exposed regions were compared and tested for statistical significance with the independent t test. Comparisons of mean dN/dS and entropy values between the three secondary structure groups were performed by the one-way analysis of variance (ANOVA) test. For further analysis between the two possible groups chosen from three structural groups, posthoc multiple comparisons were performed. One-way ANOVA and posthoc comparisons were done with the SPSS program.
Structural analysis of HIV proteins.
Solvent accessibility was calculated using the PSA program, which is an implementation of Connolly's rolling probe algorithm (14). Solvent accessibility values were calculated based upon the biological unit, i.e., in the naturally occurring oligomerization state. Residues with a solvent accessibility value of 20% or above were regarded as surface exposed; otherwise, they were considered buried. Oligomer interface residues were identified using the Probe software (50) after addition of hydrogens with Reduce (51). A residue was counted as being in the interface if at least one atom was closer than the default cutoff of 0.5 Å.
RESULTS
Identifying evolutionary divergence of HIV-1 M group.
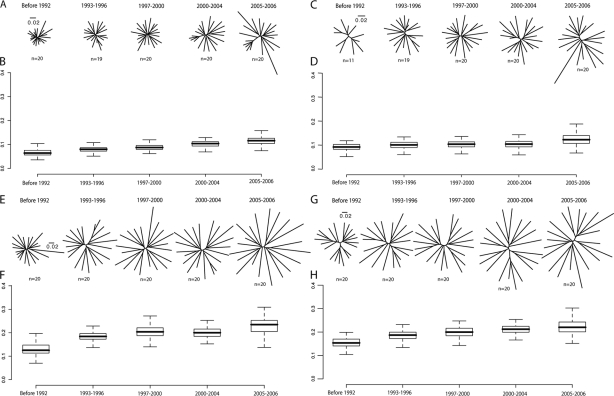
We estimated evolutionary divergence of the HIV-1 B and C subtypes through time by comparing phylogenetic trees of gag and env data sets for different sampling periods. ML trees were reconstructed from the full-length gag and env genes collected in five different periods (from the mid-1980s to 2006) (Fig. 1). There was a small increase in genetic diversity, as indicated by the mean pairwise branch length between all sequences (Fig. 1). Pearson's R values between time of sampling and genetic distances were 0.71 for both subtype B and subtype C.
FIG. 1.
Genetic distances and sequence variability of HIV-1 gag and env subtype B and C sequences through time. Maximum likelihood phylogenetic trees of HIV-1 group M gag subtype B (A), gag subtype C (C), env subtype B (E), and env subtype C (G) sequences sampled in denoted time periods. The length between each node represents the relative genetic diversity (or distance) within each sequence. The scale bar denotes 0.02 nucleotide substitutions per site. Pairwise distances (mean number of nucleotide substitutions per site) of each strain were calculated from ML trees of corresponding time points for gag subtype B (B), gag subtype C (D), env subtype B (F), and env subtype C (H). The horizontal bar in the box depicts the median of pairwise distance values. The top of the box depicts the 75% percentile, and the bottom shows the 25% percentile. The whiskers stand for either maximum and minimum or 1.5 times the interquartile (range of the box).
We found that the increase in diversity over time for gag was 0.003 and 0.002 (substitutions per site per year for groups B and C, respectively) and for env was 0.005 and 0.004 (substitutions per site per year for groups B and C, respectively). These values are similar to those estimated by Korber et al. (25). Analysis was also performed for the combined group M data, and results, although exhibiting smaller increases in overall diversity through time for env (0.003 substitutions per site per year for gag and 0.0018 for env), were qualitatively similar and so are not shown.
Sequence variation during HIV evolution.
In order to analyze selection pressure on a site-specific basis, the SLAC method was used to calculate dN/dS ratios. This method uses maximum likelihood for the reconstruction of ancestral sequences and calculates dN and dS substitution based on the phylogenetic relationship. Both gag and env were used, as these are the two most variable proteins (Fig. 2). As expected, the majority of sites for both subtypes B and C in both regions were prone to purifying selection (dN/dS ratio < 1), while in gag 13.8% and 15% of sites, for subtypes B and C, respectively, were prone to positive selection, and of those, 54% and 50% were the same sites, respectively. In env, 30.7% and 30% of sites, for subtypes B and C, respectively, were prone to positive selection, and of those, 63% and 65% were the same sites, respectively.
FIG. 2.
Correlation of dN/dS values of gag and env subtypes B and C. (A) Each point represents the dN/dS value of gag subtype B (x axis) and of gag subtype C (y axis) for individual amino acid sites. (B) Scatter plot comparing dN/dS values of env subtypes B and C.
Shannon entropy values were also calculated as a measure of diversity at each site, as this represents the degree of variation at each position in the sequence alignment. Shannon entropy and dN/dS ratios are correlated with sequence entropy in these data sets (see Fig. S1 in the supplemental material) (Pearson's R is 0.78 for gag and 0.66 for env).
Both the mean and gradient of the dN/dS ratios over the time period are shown (Fig. 3). Sequence evolution at individual sites can be divided into four groups: (i) purifying selection resulting in dN/dS and entropy starting and remaining low; (ii) positive selection leading to an increase in dN/dS and entropy; (iii) constant turnover of amino acids, resulting in sequence diversity at a site that may start and remain high; and (iv) sequence diversity decreasing. These four groups have the following characteristics. Sites under purifying selection will have low mean dN/dS values (zero gradient over time), sites under positive selection will have intermediate mean dN/dS values (positive gradient), and sites exhibiting turnover will have high mean dN/dS values (zero gradient); where sequence diversity decreases at a site, the mean value will be intermediate and the gradient negative.
FIG. 3.
Sequence variation and extent of sequence variability within individual sites of Gag and Env subtypes B and C. Histogram of gradients of dN/dS change for Gag p17 and p24 for subtype B (A) and subtype C (C). Env includes gp120, gp41, and variable loops for subtype B (E) and subtype C (G). Scatter plots of gradient and mean dN/dS values for subtype B of Gag p17 and p24 (B), subtype C of Gag p17 and p24 (D), subtype B of Env gp120, gp41, and variable loops (F), and subtype C of Env gp120, gp41, and variable loops (H). For panels A to H, the sites buried in the core of the protein (solvent accessibility values of <20) are shown in blue, and those exposed to solvent (solvent accessibility values of ≥20) in red. Variable loops (V1/V2 and V3) of gp120 are shown in green.
The histograms of gradient indicate that for these proteins the majority of sites show little or no change in the dN/dS value over time (Fig. 3B, D, F, and H). All histograms show some skew in the direction of increasing positive selection, due to an increase at a minority of sites. This lack of increase over time at most sites could be explained by either constraint or turnover, i.e., either a high degree of site-specific purifying selection or an absence of selection leading to a high degree of change at specific sites.
Plots of the gradient versus the mean sequence entropy allow us to distinguish constraint from turnover of residues. Most of the sites with low gradient values (∼−0.5 to +0.5 slope) also have low mean dN/dS values, with few displaying the combination of low-gradient and high-mean dN/dS values (Fig. 3A, C, E, and G). This indicates that a high degree of turnover of multiple amino acids at a single site is rare. In contrast, most sites are evolutionarily constrained. We find similar results when sequence entropy is analyzed (see Fig. S2 in the supplemental material).
Structural constraints acting on viral evolution of HIV.
In order to investigate the source of constraint, we colored the structures of gag p17 and env gp120 according to the dN/dS ratio (Fig. 4). “Cooler” colors (blue and purple) indicate low dN/dS ratios, and “hotter” colors (red, yellow, and white) indicate high values. For both proteins and for both subtypes, the ranges of the dN/dS ratios for each site have remained largely unchanged over time. This indicates that sites are initially conserved or variable and that they remain so over the period of time for which we have data. Moreover, this observation is independent of molecule or subtype. Similar results were found when entropy was analyzed (see Fig. S3 in the supplemental material).
FIG. 4.
Sequence variability of Gag p17 and Env gp120 for subtypes B and C. Ribbon representation of gag p17 (A and B) and env gp120 (C and D) (Protein Data Bank accession code 1HIW and 1GC1, respectively), colored as follows: dN/dS ≤ 0.1, blue; dN/dS > 0.1 and ≤ 0.2, blue/purple; dN/dS > 0.2 and ≤ 0.25, purple; dN/dS > 0.25 and ≤ 0.4, pink; dN/dS > 0.4 and ≤ 0.5, red; dN/dS > 0.5 and ≤ 0.75, orange; dN/dS > 0.75 and ≤ 1.0, bright orange; dN/dS > 0.1 and ≤ 1.5, yellow; dN/dS > 1.5 and ≤ 2.5, pale yellow; dN/dS > 2.5, white, for subtype B (A and C) and subtype C (B and D). The figure was generated using the program PyMOL (available at http://www.pymol.org/). Although single chains are shown for protein structures, the correct oligomeric state was used for all analyses.
Lower dN/dS values tend to be found within the protein core, qualitatively suggesting stronger purifying selection in the core of the protein. Figure 3 is also annotated according to solvent accessibility. Solvent accessibility was calculated using the appropriate biological oligomeric state, resulting in the majority of residues in protein interaction interfaces being counted as being inaccessible to solvent. We found that many of the sites with an increase or decrease in entropy over time (slope ≥ 0.5 and < 0.5) were found in solvent-exposed areas of the protein. For gp120, the variable loops (V1 to V4) were likely to be completely solvent exposed. However, they were missing from the crystal structure and were therefore treated separately. Those sites with both a high gradient (>0.35) and a high mean dN/dS ratio (>1.0) were found frequently in variable loops (11 out of 16) or solvent-exposed areas (5 out of 11) (Table 1). These observations suggest that structural features, such as solvent accessibility, are correlated with sequence variability for individual amino acids.
TABLE 1.
List of gp120 amino acids with high mean entropy (>1.0) and high gradient (>0.35)
| Amino acid | Structural context |
|---|---|
| 128 Gly | Exposed |
| 194 Gly | Exposed |
| 295 Asn | Exposed |
| 344 Gln | Exposed |
| 448 Asn | Exposed |
| 136 Asn | V1 |
| 139 Asn | V1 |
| 141 Asn | V1 |
| 143 Ser | V1 |
| 150 Glu | V1 |
| 152 Gly | V1 |
| 170 Glu | V2 |
| 185 Asp | V2 |
| 186 Asn | V2 |
| 302 Trp | V3 |
| 393 Ser | V4 |
To determine whether these trends hold for other HIV proteins, we calculated dN/dS values for all gag and env proteins. Amino acid residues were divided into those in the surface of the proteins (solvent exposed) and those in the core (buried from solvent). Comparisons of mean dN/dS values between the two regions, implemented by an independent t test, indicated that mean dN/dS values were higher for exposed regions of the protein structures (Table 2). Among domains for the gene products of gag and env, dN/dS values for buried and exposed regions were significantly different (P < 0.05) on six proteins. In the case of the p24 C-terminal domain, the difference was not significant, probably due to the small number of residues in the “buried” class. Also, the dN/dS ratios of exposed regions among four domains were higher than those of buried regions.
TABLE 2.
Statistical significance of difference between mean dN/dS ratios of buried and exposed regions of the protein domains
| Protein | Gene product | Mean dN/dS ratio (n)a |
P value | |
|---|---|---|---|---|
| Buried (SA < 20%) | Exposed (SA ≥ 20%) | |||
| Gag | p17 | 0.5237 (45) | 0.8280 (70) | 0.049 |
| p24 | 0.1617 (37) | 0.3051 (183) | 0.009 | |
| Env | gp120 (with V1/V2 and V3) | 0.4611 (118) | 1.125 (281) | <0.001 |
| gp41 | 0.0859 (12) | 0.6765 (58) | 0.103 | |
SA, solvent accessibility.
Similarly, those residues found in the protein-protein interaction interfaces also showed signs of being constrained (Fig. 5). In contrast, when we examined the effects of other structural characteristics, such as secondary structure, we found that there were no significant differences (Table 3).
FIG. 5.
Sequence variation and extent of sequence variability of oligomer interface sites of Gag and Env subtypes B and C. Scatter plots of gradient and mean dN/dS for Gag with p17 and p24 for subtype B (A) and subtype C (B) and Env with gp120 and gp41 for subtype B (C) and subtype C. Interface sites are highlighted in red.
TABLE 3.
Statistical significance of difference between mean entropy values of different secondary structure subtypes
| Protein | Gene product | Mean entropy value (n) |
P value (one-way ANOVA) | P value (posthoc) | ||
|---|---|---|---|---|---|---|
| Helix | Strand | Coil (others) | ||||
| Gag | p17 | 0.4566 (71) | 0.5659 (9) | 0.4590 (35) | 0.796 | 0.782 |
| p24-N | 0.1199 (84) | 0.3014 (16) | 0.1838 (35) | 0.04 | 0.038 | |
| p24-C | 0.1558 (44) | 0.2407 (3) | 0.1508 (26) | 0.845 | 0.843 | |
| Env | gp120 | 0.5874 (38) | 0.3878 (153) | 0.4740 (106) | 0.128 | 0.134 |
DISCUSSION
We found that for HIV-1 subtypes B and C there was a small increase in sequence diversity from the mid-1980s to the mid-2000s. When examined on a site-by-site basis, we identified more sites with increasing diversity than with decreasing diversity, whereas the majority of the sites showed a negligible increase in sequence diversity (Fig. 3). Clearly there has been an increase in HIV sequence diversity at some point in the past and especially subsequent to founder effects (5, 40). Such an increase in diversity is key to attempts to date the transmission of HIV to humans, with the current estimate of the origin of human HIV-1 infection being near the beginning of the 20th century (52). Even though we have analyzed sequences collected over a 20-year period, this time scale is relatively short compared to the time since the most recent common ancestor of group M, estimated to have originated between 1884 and 1924 (52). In part this explains why the dating of the transmission to humans based on data from the recent epidemic is challenging (17, 25, 43, 44) and underlines the value of sequences collected early in the pandemic (52, 56).
Interestingly a high proportion of the same sites, more than would be expected by chance (13), are prone to positive selection in both subtypes in gag and env, indicating similar selection pressures on both HIV-1 lineages, although this pressure may differ in strength (13). Clade-specific differences in selection have previously been identified (46), and it has been suggested that these may be due to functional changes within clades. As with this previous work, we found that these sites are in the minority. The overwhelming majority of sites in gag and env are under evolutionary constraint, i.e., subject to purifying selection. While expected, this result highlights the difference between genetic change and functional change, with most HIV variation being selected against. We find that even the envelope proteins display substantial evolutionary constraint, despite their high rates of evolution and structural and evolutionary plasticity. Moreover, analysis of the sequence entropy and dN/dS in the context of protein structure demonstrates that limited selection correlates with the regions expected to be under evolutionary constraint from protein structure. That is, residues with increased positive selection are likely to be located in the relatively solvent-exposed parts of the structure, whereas conserved residues are found mainly in the protein core. This is in line with previous work showing increased amino acid diversity at sites that are on the protein surface and are positively selected (53).
In the case of gp120 structures used in our study, variable loops, such as V1/V2, V3, and V4, are omitted due to the technical limitations in elucidating extended loop structure through X-ray crystallography. These variable loops protrude from the core of gp120, and so, the omitted loops are likely to be exposed to solvent. These loops appear to have a degree of constraint similar to that of other solvent-exposed residues. However, insertions and deletions are common in the hypervariable regions. It is likely that the extreme flexibility allows accommodation of a much larger degree of variability than is indicated by the analysis of substitutions. The unusual structural and evolutionary characteristics of the variable loops may indicate that over a long enough time period almost any substitution may be observed here.
Since we observed that there is a correlation between evolutionary constraint and protein structure, we conclude that protein structure is likely to be the primary origin of this constraint. In addition to structural constraints associated with solvent accessibility, we examined the possibility that secondary structure may also alter replacement patterns (Table 3). Although there are differences between helix and strand regions, none were statistically significant. The immune response might select for mutations that confer immune resistance but is unlikely to directly give rise to an increase in sequence diversity.
Any region of the viral genome may have many roles that lead to evolutionary constraint. Individual component proteins within the virus must perform regulatory, functional, and structural roles in the course of its life cycle. Viral proteins must fold to a stable three-dimensional structure. This structure must be protease resistant, must not aggregate, and in some cases must be inserted into the membrane. The required binding sites and active sites must form efficiently in three dimensions to have the correct specificity and affinity. Any required conformational changes must occur. These constraints will restrict the possibilities for change in viral sequence space. Moreover, constraints are not uniform over the genome. Amino acid residues that are directly involved in interaction sites are under much stronger selection pressure than those that are not (29); indeed, we observe that protein-protein interaction interface regions are among the most constrained.
Structural constraints have long been discussed in the context of other molecules (10, 31). These differing constraints on evolution have been characterized and quantified in the form of environment-specific substitution tables (35, 36). Constraints from structure include the necessity of forming correct secondary structure, disulfide bonds and specific hydrogen bonds. The combination of these effects can be more than additive, with the most conserved residues on average being those that are both in the protein core and forming hydrogen bonds (35). Evolutionary constraints due to structure and function can be not only identified but distinguished from each other (12). Even in the case of analogous proteins, sites that exhibit “conservatism of conservatism” have been identified (33). These sites may represent key residues required to specify the fold. Small numbers of these sites are found in some protein folds and may explain a subset of the structure-related conservation seen here.
Although protein structure somewhat restricts the successful residue replacement at a site, even some buried sites display evolutionary change. A key mechanism that allows this is compensatory change, or coevolution (54). For example, such properties appear to restrict recombination in HIV's envelope (3, 45). If one residue in a buried part of the structure undergoes sequence change, sequence change may also occur on a neighboring residue in order to avoid disruption of the protein structure. It has been suggested that these correlated sequence evolutions could be a way of maintaining optimal structural and functional integrity (47). Alternatively, small structural rearrangements, or a combination of coevolution and structural change (48), may allow change at these buried sites.
In contrast to evolutionary constraint, we find little evidence for repeated replacements at a single site, i.e., evolutionarily dynamic turnover of amino acids. Even in Env, we find very few sites where there is a high degree of variability and replacements are saturated. It is possible that at a given site sequence diversity would “toggle” between a small number of amino acids. Such sites would also have low sequence entropy that does not increase over time; thus, there may be hidden evolutionary dynamics. However, if there are such a small number of residues seen at a given site, this is probably also due to an evolutionary constraint, albeit one that can be satisfied by more than one residue.
In conclusion, evolutionary constraints from protein structure, and from other sources, place limits on the space of sequences that HIV can explore. Despite the high rate of mutation, we find that comparatively few sequences are sampled. This observation offers promise when trying to account for escape mutations; although HIV can sample a large number of sequences, the available evolutionary trajectory is by no means infinite. Indeed, the overwhelming majority of sites display only limited numbers of replacements, even in the most variable proteins. Our understanding of constraints on viral diversity and evolution offers the possibility of understanding the evolutionary trajectory of HIV. Such knowledge could lead to a reliable prediction of HIV evolution, of relevance to escape from drug therapy and potential vaccine strategies.
Supplementary Material
Acknowledgments
We thank John Archer for help with data processing.
J.W. is funded by an Overseas Research Studentship award from the University of Manchester. We also thank the Apple Research & Technology Support scheme for support.
Footnotes
Published ahead of print on 29 September 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abebe, A., V. V. Lukashov, G. Pollakis, A. Kliphuis, A. L. Fontanet, J. Goudsmit, and T. F. de Wit. 2001. Timing of the HIV-1 subtype C epidemic in Ethiopia based on early virus strains and subsequent virus diversification. AIDS 15:1555-1561. [DOI] [PubMed] [Google Scholar]
- 2.Allen, T. M., M. Altfeld, X. G. Yu, K. M. O'Sullivan, M. Lichterfeld, S. Le Gall, M. John, B. R. Mothe, P. K. Lee, E. T. Kalife, D. E. Cohen, K. A. Freedberg, D. A. Strick, M. N. Johnston, A. Sette, E. S. Rosenberg, S. A. Mallal, P. J. Goulder, C. Brander, and B. D. Walker. 2004. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J. Virol. 78:7069-7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer, J., J. W. Pinney, J. Fan, E. Simon-Loriere, E. J. Arts, M. Negroni, and D. L. Robertson. 2008. Identifying the important HIV-1 recombination breakpoints. PLoS Comput. Biol. 4:e1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archer, J., and D. L. Robertson. 2007. CTree: comparison of clusters between phylogenetic trees made easy. Bioinformatics 23:2952-2953. [DOI] [PubMed] [Google Scholar]
- 5.Archer, J., and D. L. Robertson. 2007. Understanding the diversification of HIV-1 groups M and O. AIDS 21:1693-1700. [DOI] [PubMed] [Google Scholar]
- 6.Barouch, D. H. 2008. Challenges in the development of an HIV-1 vaccine. Nature 455:613-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bello, G., M. L. Guimaraes, and M. G. Morgado. 2006. Evolutionary history of HIV-1 subtype B and F infections in Brazil. AIDS 20:763-768. [DOI] [PubMed] [Google Scholar]
- 8.Bennett, D. E., R. J. Camacho, D. Otelea, D. R. Kuritzkes, H. Fleury, M. Kiuchi, W. Heneine, R. Kantor, M. R. Jordan, J. M. Schapiro, A. M. Vandamme, P. Sandstrom, C. A. Boucher, D. van de Vijver, S. Y. Rhee, T. F. Liu, D. Pillay, and R. W. Shafer. 2009. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 4:e4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blundell, T. L., and S. P. Wood. 1975. Is the evolution of insulin Darwinian or due to selectively neutral mutation? Nature 257:197-203. [DOI] [PubMed] [Google Scholar]
- 11.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 12.Chelliah, V., L. Chen, T. L. Blundell, and S. C. Lovell. 2004. Distinguishing structural and functional restraints in evolution in order to identify interaction sites. J. Mol. Biol. 342:1487-1504. [DOI] [PubMed] [Google Scholar]
- 13.Choisy, M., C. H. Woelk, J. F. Guegan, and D. L. Robertson. 2004. Comparative study of adaptive molecular evolution in different human immunodeficiency virus groups and subtypes. J. Virol. 78:1962-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connolly, M. L. 1983. Solvent-accessible surfaces of proteins and nucleic acids. Science 221:709-713. [DOI] [PubMed] [Google Scholar]
- 15.Crawford, H., J. G. Prado, A. Leslie, S. Hue, I. Honeyborne, S. Reddy, M. van der Stok, Z. Mncube, C. Brander, C. Rousseau, J. I. Mullins, R. Kaslow, P. Goepfert, S. Allen, E. Hunter, J. Mulenga, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2007. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J. Virol. 81:8346-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desrosiers, R. C. 2004. Prospects for an AIDS vaccine. Nat. Med. 10:221-223. [DOI] [PubMed] [Google Scholar]
- 17.Drummond, A., O. G. Pybus, and A. Rambaut. 2003. Inference of viral evolutionary rates from molecular sequences. Adv. Parasitol. 54:331-358. [DOI] [PubMed] [Google Scholar]
- 18.Gao, F., Y. Chen, D. N. Levy, J. A. Conway, T. B. Kepler, and H. Hui. 2004. Unselected mutations in the human immunodeficiency virus type 1 genome are mostly nonsynonymous and often deleterious. J. Virol. 78:2426-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong, S., and T. L. Blundell. 2008. Discarding functional residues from the substitution table improves predictions of active sites within three-dimensional structures. PLoS Comput. Biol. 4:e1000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. U. S. A. 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes, E. C. 2003. Error thresholds and the constraints to RNA virus evolution. Trends Microbiol. 11:543-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iversen, A. K., G. Stewart-Jones, G. H. Learn, N. Christie, C. Sylvester-Hviid, A. E. Armitage, R. Kaul, T. Beattie, J. K. Lee, Y. Li, P. Chotiyarnwong, T. Dong, X. Xu, M. A. Luscher, K. MacDonald, H. Ullum, B. Klarlund-Pedersen, P. Skinhoj, L. Fugger, S. Buus, J. I. Mullins, E. Y. Jones, P. A. van der Merwe, and A. J. McMichael. 2006. Conflicting selective forces affect T cell receptor contacts in an immunodominant human immunodeficiency virus epitope. Nat. Immunol. 7:179-189. [DOI] [PubMed] [Google Scholar]
- 23.Johnston, M. I., and A. S. Fauci. 2007. An HIV vaccine—evolving concepts. N. Engl. J. Med. 356:2073-2081. [DOI] [PubMed] [Google Scholar]
- 24.Kent, S. J., C. S. Fernandez, C. J. Dale, and M. P. Davenport. 2005. Reversion of immune escape HIV variants upon transmission: insights into effective viral immunity. Trends Microbiol. 13:243-246. [DOI] [PubMed] [Google Scholar]
- 25.Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789-1796. [DOI] [PubMed] [Google Scholar]
- 26.Kosakovsky Pond, S. L., and S. V. Muse. 2005. HyPhy: hypothesis testing using phylogenies, p. 1-57. In R. Nielsen (ed.), Statistical methods in molecular evolution, vol. XII. Springer, New York, NY. [Google Scholar]
- 27.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 29.Lichtarge, O., H. R. Bourne, and F. E. Cohen. 1996. An evolutionary trace method defines binding surfaces common to protein families. J. Mol. Biol. 257:342-358. [DOI] [PubMed] [Google Scholar]
- 30.Liu, S., H. Xing, X. He, R. Xin, Y. Zhang, J. Zhu, and Y. Shao. 2008. Dynamic analysis of genetic diversity of gag and env regions of HIV-1 CRF07_BC recombinant in intravenous drug users in Xinjiang Uvghur Autonomous Region, China. Arch. Virol. 153:1233-1240. [DOI] [PubMed] [Google Scholar]
- 31.Lüthy, R., J. U. Bowie, and D. Eisenberg. 1992. Assessment of protein models with three-dimensional profiles. Nature 356:83-85. [DOI] [PubMed] [Google Scholar]
- 32.McMichael, A. J. 2006. HIV vaccines. Annu. Rev. Immunol. 24:227-255. [DOI] [PubMed] [Google Scholar]
- 33.Mirny, L. A., and E. I. Shakhnovich. 1999. Universally conserved positions in protein folds: reading evolutionary signals about stability, folding kinetics and function. J. Mol. Biol. 291:177-196. [DOI] [PubMed] [Google Scholar]
- 34.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 35.Overington, J., D. Donnelly, M. S. Johnson, A. Sali, and T. L. Blundell. 1992. Environment-specific amino acid substitution tables: tertiary templates and prediction of protein folds. Protein Sci. 1:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Overington, J., M. S. Johnson, A. Sali, and T. L. Blundell. 1990. Tertiary structural constraints on protein evolutionary diversity: templates, key residues and structure prediction. Proc. Biol. Sci. 241:132-145. [DOI] [PubMed] [Google Scholar]
- 37.Peyerl, F. W., H. S. Bazick, M. H. Newberg, D. H. Barouch, J. Sodroski, and N. L. Letvin. 2004. Fitness costs limit viral escape from cytotoxic T lymphocytes at a structurally constrained epitope. J. Virol. 78:13901-13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pornillos, O., B. K. Ganser-Pornillos, B. N. Kelly, Y. Hua, F. G. Whitby, C. D. Stout, W. I. Sundquist, C. P. Hill, and M. Yeager. 2009. X-ray structures of the hexameric building block of the HIV capsid. Cell 137:1282-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rambaut, A., D. Posada, K. A. Crandall, and E. C. Holmes. 2004. The causes and consequences of HIV evolution. Nat. Rev. Genet. 5:52-61. [DOI] [PubMed] [Google Scholar]
- 40.Rambaut, A., D. L. Robertson, O. G. Pybus, M. Peeters, and E. C. Holmes. 2001. Human immunodeficiency virus. Phylogeny and the origin of HIV-1. Nature 410:1047-1048. [DOI] [PubMed] [Google Scholar]
- 41.Rhee, S. Y., M. J. Gonzales, R. Kantor, B. J. Betts, J. Ravela, and R. W. Shafer. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 31:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robbins, K. E., P. Lemey, O. G. Pybus, H. W. Jaffe, A. S. Youngpairoj, T. M. Brown, M. Salemi, A. M. Vandamme, and M. L. Kalish. 2003. U.S. human immunodeficiency virus type 1 epidemic: date of origin, population history, and characterization of early strains. J. Virol. 77:6359-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salemi, M., K. Strimmer, W. W. Hall, M. Duffy, E. Delaporte, S. Mboup, M. Peeters, and A. M. Vandamme. 2001. Dating the common ancestor of SIVcpz and HIV-1 group M and the origin of HIV-1 subtypes using a new method to uncover clock-like molecular evolution. FASEB J. 15:276-278. [DOI] [PubMed] [Google Scholar]
- 44.Sharp, P. M., E. Bailes, F. Gao, B. E. Beer, V. M. Hirsch, and B. H. Hahn. 2000. Origins and evolution of AIDS viruses: estimating the time-scale. Biochem. Soc. Trans. 28:275-282. [DOI] [PubMed] [Google Scholar]
- 45.Simon-Loriere, E., R. Galetto, M. Hamoudi, J. Archer, P. Lefeuvre, D. P. Martin, D. L. Robertson, and M. Negroni. 2009. Molecular mechanisms of recombination restriction in the envelope gene of the human immunodeficiency virus. PLoS Pathog. 5:e1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Travers, S. A., M. J. O'Connell, G. P. McCormack, and J. O. McInerney. 2005. Evidence for heterogeneous selective pressures in the evolution of the env gene in different human immunodeficiency virus type 1 subtypes. J. Virol. 79:1836-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Travers, S. A., D. C. Tully, G. P. McCormack, and M. A. Fares. 2007. A study of the coevolutionary patterns operating within the env gene of the HIV-1 group M subtypes. Mol. Biol. Evol. 24:2787-2801. [DOI] [PubMed] [Google Scholar]
- 48.Williams, S. G., and S. C. Lovell. 2009. The effect of sequence evolution on protein structural divergence. Mol. Biol. Evol. 26:1055-1065. [DOI] [PubMed] [Google Scholar]
- 49.Wolinsky, S. M., B. T. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Cao, D. D. Ho, and J. T. Safrit. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]
- 50.Word, J. M., S. C. Lovell, T. H. LaBean, H. C. Taylor, M. E. Zalis, B. K. Presley, J. S. Richardson, and D. C. Richardson. 1999. Visualizing and quantifying molecular goodness of fit: small-probe contact dots with explicit hydrogen atoms. J. Mol. Biol. 285:1711-1733. [DOI] [PubMed] [Google Scholar]
- 51.Word, J. M., S. C. Lovell, J. S. Richardson, and D. C. Richardson. 1999. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 285:1735-1747. [DOI] [PubMed] [Google Scholar]
- 52.Worobey, M., M. Gemmel, D. E. Teuwen, T. Haselkorn, K. Kunstman, M. Bunce, J. J. Muyembe, J. M. Kabongo, R. M. Kalengayi, E. Van Marck, M. T. Gilbert, and S. M. Wolinsky. 2008. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455:661-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, W., J. P. Bielawski, and Z. Yang. 2003. Widespread adaptive evolution in the human immunodeficiency virus type 1 genome. J. Mol. Evol. 57:212-221. [DOI] [PubMed] [Google Scholar]
- 54.Yeang, C. H., J. F. Darot, H. F. Noller, and D. Haussler. 2007. Detecting the coevolution of biosequences—an example of RNA interaction prediction. Mol. Biol. Evol. 24:2119-2131. [DOI] [PubMed] [Google Scholar]
- 55.Yusim, K., C. Kesmir, B. Gaschen, M. M. Addo, M. Altfeld, S. Brunak, A. Chigaev, V. Detours, and B. T. Korber. 2002. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J. Virol. 76:8757-8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, T., B. T. Korber, A. J. Nahmias, E. Hooper, P. M. Sharp, and D. D. Ho. 1998. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature 391:594-597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.