Abstract
The largest E1A isoform of human adenovirus (Ad) includes a C-4 zinc finger domain within conserved region 3 (CR3) that is largely responsible for activating transcription of the early viral genes. CR3 interacts with multiple cellular factors, but its mechanism of action is modeled primarily on the basis of the mechanism for the prototype E1A protein of human Ad type 5. We expanded this model to include a representative member from each of the six human Ad subgroups. All CR3 domains tested were capable of transactivation. However, there were dramatic differences in their levels of transcriptional activation. Despite these functional variations, the interactions of these representative CR3s with known cellular transcriptional regulators revealed only modest differences. Four common cellular targets of all representative CR3s were identified: the proteasome component human Sug1 (hSug1)/S8, the acetyltransferases p300/CREB binding protein (CBP), the mediator component mediator complex subunit 23 (MED23) protein, and TATA binding protein (TBP). The first three factors appear to be critical for CR3 function. RNA interference against human TBP showed no significant reduction in transactivation by any CR3 tested. These results indicate that the cellular factors previously shown to be important for transactivation by Ad5 CR3 are similarly bound by the E1A proteins of other types. This was confirmed experimentally using a transcriptional squelching assay, which demonstrated that the CR3 regions of each Ad type could compete with Ad5 CR3 for limiting factors. Interestingly, a mutant of Ad5 CR3 (V147L) was capable of squelching wild-type Ad5 CR3, despite its failure to bind TBP, MED23, p300/CBP-associated factor (pCAF), or p300/CBP, suggestive of the possibility that an additional as yet unidentified cellular factor is required for transactivation by E1A CR3.
The adenovirus (Ad) E1A oncoprotein is the first gene expressed upon infection and performs two essential roles in order to initiate the viral replication cycle. E1A uncouples the cell cycle control program of the host cell, driving it into S phase to provide an optimal cellular environment for viral replication. This function can be carried out by the smaller major E1A isoform (243 residues in Ad type 5 [Ad5] E1A) (11, 25). The other function of E1A is to activate transcription of the early viral promoters and is predominantly mediated by the largest E1A isoform (4, 18, 23, 24). The largest E1A isoform, coding for 289 residues in Ad5 E1A, differs from the smaller isoform only by a unique 46-amino-acid C-4 zinc finger domain within conserved region 3 (CR3), which is essential for viral transactivation (4, 18). Single point mutations in CR3 were originally isolated as Ad mutants with a host range limited to HEK 293 cells, which supply wild-type (wt) E1A in trans. These “host range” mutations render E1A unable to transactivate viral promoters, thus preventing virus growth in cells at a low multiplicity of infection (MOI), unless wt E1A is supplied in trans (13, 16).
Transactivation by E1A CR3 has been studied predominantly with Ad5, and this has led to the establishment of a model for CR3 function. The region of Ad5 E1A spanning residues 139 to 204 (which includes CR3) is critical for activating the transcription necessary for virus growth and is sufficient for potent activation of a minimal Gal4-responsive promoter as a Gal4-DNA-binding domain (DBD) fusion (17, 30). This 65-amino-acid region of E1A CR3 can be further subdivided into the three following subdomains: an N-terminal zinc finger, a promoter-targeting region, and a region known as auxiliary region 1 (AR1) (9, 20, 32). The current model is that the N-terminal zinc finger domain of E1A CR3 activates transcription by interacting with cellular TATA binding protein (TBP) and mediator complex subunit 23 (MED23) protein (6, 19, 31). The mediator component MED23 is absolutely critical to E1A CR3 function, as CR3 fails to activate transcription in MED23-null mouse embryonic fibroblasts (MEFs). Furthermore, this requirement is likely shared between the E1A proteins of even very divergent Ads, as mouse adenovirus type 1 is unable to replicate efficiently in MED23-null MEFs (10, 31). In order to nucleate a functional transcription initiation complex at the early viral promoters, the C-terminal promoter-targeting subdomain is required. It confers interaction of E1A CR3 with cellular sequence-specific DNA-binding transcription factors, including members of the ATF family (8, 20, 21). Mutants that delete this promoter-targeting domain function as dominant-negative mutants, unless a second mutation is made in the zinc finger domain of CR3, further underscoring the promoter-targeting activity of this region (20). The precise role of AR1 in E1A CR3-dependent transactivation remains unclear; however, the acidic character of this region is necessary for maximal transactivation by E1A CR3 (32).
Since the initial model was put forth, multiple additional cellular factors have been implicated in transcriptional activation by E1A CR3. The S8 component of the 19S ATPase proteins independent of 20S (APIS), also known as human Sug1 (hSug1), was shown to interact with E1A CR3 and enhance E1A CR3 transactivation (28). A second interaction site for the cellular repressor C-terminal binding protein (CtBP) was also mapped to the CR3 region of Ad5 E1A, and CtBP appears to repress E1A CR3 transactivation (7). The p300/CREB binding protein (CBP) acetyltransferase is critical for Ad5 E1A CR3 transactivation and binds directly to E1A CR3 independently of other interaction motifs in E1A (27). Most recently, the histone acetyltransferase p300/CBP-associated factor (pCAF) was identified to interact with the E1A CR3 region and enhance transactivation (26). Many of these cellular factors interact with other regions of E1A besides CR3, further complicating E1A-mediated activation of transcription.
The vast majority of the work used to build the initial model was done exclusively with Ad5 E1A CR3. Very little is known regarding how E1A CR3s from other Ad types activate transcription. Alignment of the amino acid sequence of E1A CR3 corresponding to Ad5 E1A residues 139 to 204 from representative members of each Ad subgroup demonstrates their high degree of conservation (Fig. 1A). The amino acid sequence identities and similarities to Ad5 E1A CR3 range from 34% to 41% and 41% to 53%, respectively (1). Overall, CR3 is the most conserved domain of E1A, but it is not known if the model for Ad5 E1A CR3 transactivation will apply universally to the entire human Ad family. Our previous work suggests that there may in fact be differences in how specific E1A CR3s activate transcription, as only a subset of the E1A CR3s that we tested interact with and are influenced by pCAF (26).
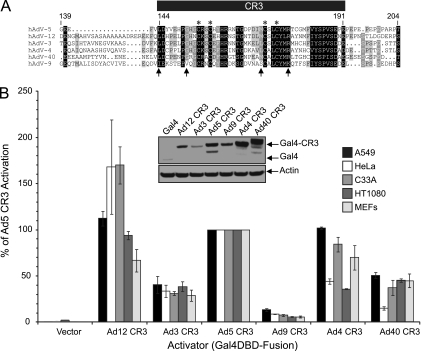
FIG. 1.
Transcriptional activation by E1A CR3s from six human adenovirus subgroups. (A) Sequence alignment of human adenovirus E1A CR3 fragments used from types Ad12 (subgroup A), Ad3 (subgroup B), Ad5 (subgroup C), Ad9 (subgroup D), Ad4 (subgroup E), and Ad40 (subgroup F). Asterisks indicated zinc-coordinating cysteines. Arrows indicate residues in Ad9 E1A CR3 that differ from those in Ad5 E1A CR3 and that are critical for transcriptional activation. (B) Intrinsic transcriptional activation properties of representative E1A CR3s. A549, HeLa, C33A, or HT1080 cells or MEFs were cotransfected with a Gal4-responsive luciferase reporter and vectors expressing the indicated Gal4-DBD fusions. Luciferase activity is expressed as a percentage of the Ad5 CR3 fold activation above that for the empty vector ± SD.
We report here on the first comprehensive study of the cellular factors required for E1A CR3 transactivation using representative E1A CR3s from each human Ad subgroup. The representative E1A CR3s in the panel show dramatic differences in their ability to activate transcription as Gal4-DBD fusions which cannot be explained by the existing model of E1A CR3 function. Systematic analysis of the roles of MED23, TBP, hSug1, and p300/CBP, which have been implicated in Ad5 E1A CR3 function, reveals that these interactions are conserved across all human Ad subgroups, and each representative E1A CR3 can compete for a common factor(s) and squelch Ad5 E1A CR3 transactivation. However, the known cellular factors required by E1A CR3 cannot explain the dramatic differences in transactivation observed among these representative E1A CR3s. These results demonstrate that many of the cellular targets utilized by Ad5 E1A CR3 are conserved across the human Ad family, expanding the existing model of E1A CR3 transactivation to encompass all six subgroups. Importantly, these data also indicate that additional factors influencing CR3-dependent transactivation remain to be discovered.
MATERIALS AND METHODS
Cells, cell culture, and transfections.
Human A549, HeLa, C33A, USOS, and HT1080 cells, as well as MEFs and MED23−/− MEFs, were maintained at 37°C and 5% CO2 in Dulbecco modified Eagle medium (Wisent) with 10% fetal bovine serum (Gibco) and 100 U/ml of penicillin-streptomycin (Wisent). A549 cells and MEFs were transfected with the FuGENE HD reagent (Roche), according to the manufacturer's directions, in a ratio of 3 μg to 9 μl per well of a six-well plate. HeLa and HT1080 cells were transfected with the Superfect reagent (Qiagen), according to the manufacturer's directions.
The U2OS stable cell line containing an integrated Gal4-responsive luciferase reporter (U2OS-UAS) was made by cotransfection of U2OS cells with pGL2-(Gal4)6-Luc and pcDNA3.1-Hygro in a 9:1 ratio and selection on 400 μg/ml of hygromycin. Hygromycin-resistant pools were used for all experiments.
Plasmid construction.
The Gal4-responisve luciferase reporter vector pGL2-(Gal4)6-Luc and Gal4-DBD fusions for each hAd E1A CR3 and wt human papillomavirus (HPV) type 16 E7 have been described previously (2, 30). Expression vectors for enhanced green fluorescent protein (EGFP) fusions of hAd E1A CR3s and wt HPV E7 were cloned from their respective Gal4-DBD fusions into pCAN-myc-EGFP with EcoRI and XbaI. The expression vectors for the full-length E1A clones of Ad types 3, 4, 5, 9, 12, and 40 were cloned into pM (Clontech Laboratories Inc.) with EcoRI and SalI. The expression vector for human MED23 (hMED2) (pCS2+-human Sur2) was a gift from A. Berk and described previously (6). Expression vectors for human TBP (pcDNA4HA-hTBP) and hSug1 (pcDNA4HA-hSug1) were described previously (28). The p300 expression vector was described previously (27).
Gal4 fusion activation assay.
At 24 h prior to transfection, 1.5 × 105 A549 cells/well were seeded on six-well plates. Cells were transfected in a 1:1 ratio of reporter vector [pGL2-(Gal4)6-Luc] to activator (either pM or pM-CR3). Cells were harvested 48 h posttransfection in 1× cell culture lysis buffer (Promega) and assayed for luciferase activity using Steady-Glo substrate (Promega). The number of relative light units (RLUs) was normalized to the protein concentration and plotted as the mean fold activation above that achieved with Gal4-DBD alone (pM) ± standard deviation (SD).
Coimmunoprecipitation and Western blot analysis.
Typically, 1.5 × 106 HT1080 cells were seeded into 10-cm plates 24 h prior to transfection. Cells were transfected in a 1:1 ratio of the myc-EGFP fusion and hemagglutinin (HA)-tagged binding partner or myc-EGFP fusion/E1A alone if endogenous binding partners were used. Cells were harvested at 24 h posttransfection by scraping and washed once with 1× phosphate-buffered saline. Cells were lysed in either NP-40 (50 mM Tris, pH 7.8, 150 mM NaCl, 0.1% NP-40) or E1A (50 mM HEPES, pH 6.8, 230 mM NaCl, 0.5% NP-40) lysis buffer supplemented with 1× mammalian protease inhibitor cocktail (Sigma). Typically, 1 mg of cell lysate was mixed with 100 μl of anti-myc hybridoma (clone 9E10) or M73 anti-E1A hybridoma supernatant and 125 μl of a 10% slurry of protein A-Sepharose resin (Sigma) and incubated at 4°C for 1 h with nutating. Immunoprecipitates were washed five times with lysis buffer, resuspended in 1× lithium dodecyl sulfate sample buffer, and boiled for 5 min. Samples were then separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane (GE), and blocked in 5% nonfat milk in Tris-buffered saline-Tween 20. Western blot analysis was carried out with mouse anti-myc hybridoma clone (9E10), rat monoclonal anti-HA (clone 3F10 Roche), monoclonal anti-Rb (clone C36), rabbit antiactin (Sigma), mouse monoclonal anti-TBP (Millipore), rabbit anti-hSug1 (14), or mouse monoclonal anti-p300 (clone Rw128; Millipore), followed by either rabbit anti-mouse horseradish peroxidase (HRP; Jackson Laboratories), goat anti-rat HRP (Pierce), or goat anti-rabbit HRP (Jackson Laboratories).
siRNA knockdown.
Custom small interfering RNA (siRNA) against p300 was used as described previously (27). Silencer select siRNAs against TBP (s13826 siRNA) and hSug1 (s11381 siRNA) were purchased from Ambion. siRNA transfections were performed with siLentFECT reagent (Bio-Rad), according to the manufacturer's directions. Typically, 1.5 × 106 HeLa cells were seeded on 10-cm plates for siRNA transfection. At 24 h after siRNA transfection, cells were reseeded to six-well plates at 2 × 105 cells per well. At 48 h after siRNA transfection, cells were transfected as described above to perform the Gal4 fusion activation assay.
Squelching assay.
At 24 h prior to transfection, 1.5 × 105 A549 cells/well were seeded on six-well plates. Cells were transfected in a 1:1:1 ratio of reporter [pGL2-(Gal4)6-Luc]-activator (either pM or pM-Ad5 CR3)-squelcher (myc-EGFP fusion). Cells were harvested at 48 h posttransfection for use in the luciferase assay, as described above. The number of RLUs was normalized to the protein concentration and plotted as a percentage of the number for the Gal4-Ad5 E1A CR3 wt with EGFP (empty vector) as competitor ± SD. The squelching rescue assay was performed as described above with the addition of pcDNA4HA-hSug1 or empty pcDNA4HA to achieve a 1:1:1:1 ratio.
Quantitative reverse transcription-PCR (qRT-PCR).
Human A549 cells were infected with either Ad5 (dl309 or dl312) or wt Ad9 virus at an MOI of 2.0 (or MOIs of 200 and 2,000 for the Ad9 wt). At 16 h postinfection, total RNA was isolated with the Trizol reagent (Invitrogen), according to the manufacturer's directions. For each sample, 1 μg of total RNA was first heated to 70°C for 5 min and subjected to DNase I treatment (Invitrogen), according to the manufacturer's directions. First-strand synthesis was performed using OligodT (Invitrogen) and SuperScriptII enzyme (Invitrogen), according to the manufacturer's directions. Quantitative PCR was performed in triplicate with a 15-μl reaction mixture and 1× iQ-SYBR green SuperMix (Bio-Rad), according to the manufacturer's directions, in a MyiQ real-time PCR instrument (Bio-Rad). The following primers for Ad9 targets were used at a 200 nM final concentration: RTAd913S E1AF (5′-AGCTTTATTTACAGTCCGGTGTCAGA-3′), RTAd913SE1AR (5′-ACACTTGCAGGGGCGTTT-3′), RTAd9E4orf6-7F (5′-CATAATACTGTGACCTTGGAC-3′), and RTAd9E4orf6-7R (5′-TTTCCTGGCGAGCCAAAC-3′). The primers for Ad5 targets and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were described previously (28). Data were analyzed using IQ5 software (Bio-Rad); briefly, E4orf6/7 mRNA levels were normalized to GAPDH levels as an internal control and the respective E1A mRNA level for each sample. E1A and E4orf6/7 mRNA levels in cells infected with mutant dl312 (from which E1A is deleted) were set equal to 1.
RESULTS
E1A CR3-mediated transactivation differs greatly between Ad types.
E1A is a potent activator of transcription, and Ad5 E1A CR3 (residues 139 to 204) is sufficient to activate transcription when it is fused to a heterologous DBD in mammalian and yeast cells (26-30, 34). We tested whether the corresponding E1A CR3 portions of Ad type 3, 4, 9, 12, and 40 E1A proteins, which represent E1A proteins from the other five Ad subgroups, were capable of activating transcription as Gal4-DBD fusions in the A549 human alveolar basal epithelial cell line (Fig. 1B). A549 cells were chosen, as they are commonly used as a diagnostic cell line for clinical Ad infections. All six E1A CR3s activated a Gal4-responsive promoter in A549 cells. Interestingly, there were dramatic differences in their relative activities (Fig. 1B). Given that E1A is the first gene expressed during adenovirus infection and is responsible for activating viral gene expression, the current model would predict that all E1A CR3s should strongly activate transcription. However, there appeared to be three classes of E1A CR3s with respect to activation of transcription: subgroup A (Ad12), C (Ad5), and E (Ad4) E1A CR3s were the most potent activators of transcription and subgroup B (Ad3) and subgroup F (Ad40) E1A CR3s exhibited an intermediate ability, while subgroup D (Ad9) E1A CR3 was a weak activator. To determine if these differences in activity were specific to A549 cells, we repeated these experiments in HeLa and C33A human cervical carcinoma cells, HT1080 human fibrosarcoma cells, and MEFs. Although some differences in activation were seen between cell types, Ad9 CR3 was consistently the weakest activator in all cells tested (Fig. 1B).
Diverse E1A CR3s share common cellular targets.
Since little is known about the mechanism by which the E1A CR3 regions of any Ad type other than Ad5 activate transcription, we first determined if there are common cellular factors targeted by the different E1A CR3s. We designed a competition, or squelching, assay to determine if there is functional overlap of cellular targets required for transactivation. A549 cells were cotransfected with a Gal4-responsive luciferase reporter, an activator, and a competitor. In this assay, the readout is the level of activation by the Gal4-Ad5 E1A CR3 wt above that by Gal4 alone and is expressed as a percentage of that for the Gal4-Ad5 E1A CR3 wt with EGFP (empty vector) as a competitor. If a competitor does not target factors required by the DNA-bound transactivator, the level of transcriptional activation should remain at 100%, regardless of the level of competitor present. However, if the competitor targets a factor(s) required for function by the activator, a dose-dependent reduction of transactivation will be observed as the level of the competitor is increased. As expected, wt Ad5 E1A CR3 fused to EGFP potently squelched the activity of wt Gal4-Ad5 E1A CR3 in a dose-dependent manner (Fig. 2A). This clearly demonstrates that soluble EGFP Ad5 E1A CR3, which is unable to bind the promoter, will sequester limiting factors from promoter-tethered Gal4-Ad5 E1A CR3, reducing its ability to stimulate transcription of the Gal4-responsive luciferase reporter.
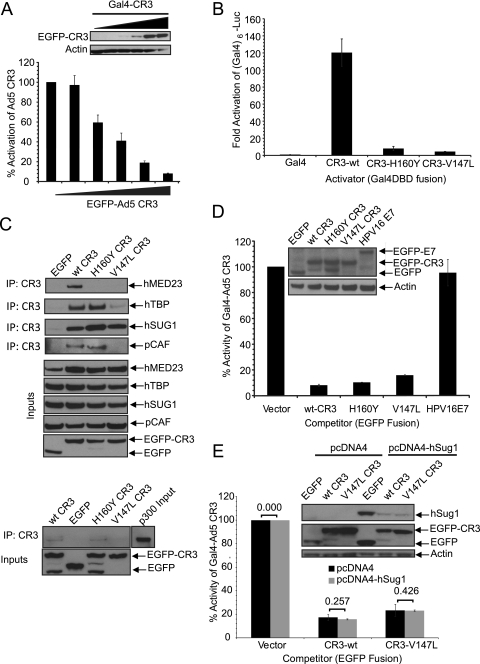
FIG. 2.
Squelching of Ad5 E1A CR3 function. (A) Design and titration of squelching assay. A549 cells were cotransfected with a Gal4-responsive luciferase reporter, vectors expressing Gal4 alone, or Gal4-Ad5 CR3 as activators and either EGFP or increasing amounts (0 ng, 20 ng, 100 ng, 500 ng, and 1,000 ng) of vector expressing the EGFP-Ad5 CR3 fusion as a competitor. Luciferase activity is expressed as a percentage of the fold activation of Gal4-Ad5 CR3 above that for the vector alone with EGFP as the competitor ± SD. (B) Activation of established mutants of E1A CR3 as Gal4 fusions. A549 cells were cotransfected with a Gal4-responsive luciferase reporter and vectors expressing the indicated Gal4 fusions to E1A CR3 or empty vector. Luciferase activity is expressed as a percentage of the fold activation above that for the vector ± SD. (C) Interaction of mutants of Ad5 E1A CR3 with known cellular targets of CR3. E1A CR3 fusions were immunoprecipitated with anti-myc antibody and blotted with anti-HA antibody for the indicated targets. Inputs are probed with anti-myc antibody for CR3s or anti-HA antibody for MED23, TBP, hSug1, and p300. IP, immunoprecipitation. (D) Squelching of E1A CR3-dependent transactivation by Ad5 CR3 mutants. A549 cells were cotransfected as described for panel A but with equal amounts of vectors expressing EGFP fused to the indicated Ad5 CR3 mutant or HPV E7 (1,000 ng). (E) Sequestration of hSug1 is not responsible for transcriptional squelching by CR3 of the V147L mutant. Human A549 cells were cotransfected as described for panel D with either empty vector (pcDNA4) or a hSug1 expression vector (pcDNA4-hSug1) in a 1:1:1:1 ratio. The levels of activation in vector-transfected cells versus those in hSug1-transfected cells were compared by Student's t test, and P values are indicated above the bars.
E1A mutants unable to bind multiple factors still squelch wt E1A activity.
Two well-characterized mutants of Ad5 E1A CR3 with point mutations H160Y and V147L, which fail to transactivate, were chosen to validate the squelching assay (12). We confirmed that the Gal4 fusions of each of these mutants were unable to activate a Gal4-responsive promoter (Fig. 2B). Ad5 E1A CR3 interacts with multiple cellular transcriptional regulators in order to orchestrate the activation of gene transcription (25). We determined the interaction profiles of these mutants with respect to several of these binding partners. The H160Y mutant bound TBP, hSug1, p300, and pCAF equivalently to wt Ad5 E1A CR3 but did not bind hMED23 (Fig. 2C). The V147L mutant bound hSug1 like wt Ad5 E1A CR3, bound TBP only weakly, and did not bind hMED23, p300, or pCAF (Fig. 2C). Taken together, these mutants represent transactivation-defective mutants that retained selective interactions with factors that might be rate limiting for E1A CR3-dependent activation of transcription. Despite the inability of these mutants to activate transcription or bind key transcriptional components, both effectively squelched activation by the Gal4-Ad5 CR3 wt when they were expressed as soluble EFGP fusions, whereas the human papillomavirus type 16 E7 viral transactivator did not (Fig. 2D). As both the H160Y and V147L mutants remained capable of interacting with hSug1 similarly to wt Ad5 E1A CR3, it remained possible that hSug1 is the limiting factor required by CR3 to activate transcription. We tested whether overexpression of hSug1 would reverse the squelching by the V147L mutant. The overexpression of hSug1 that resulted in no significant change is a result of the squelching activity of the mutant with the V147L point mutation (Fig. 1E). The observation that these mutants lost the ability to interact with multiple cellular factors yet still manage to squelch wt Ad5 E1A CR3 transactivation demonstrates that E1A CR3-dependent activation of transcription is a complex process requiring the concerted action of multiple factors. Importantly, these data provide evidence that additional factors beyond hMED23, hSug1, p300, pCAF, and TBP are critical for Ad5 CR3 transactivation.
E1A CR3s universally squelch Ad5 E1A CR3 function.
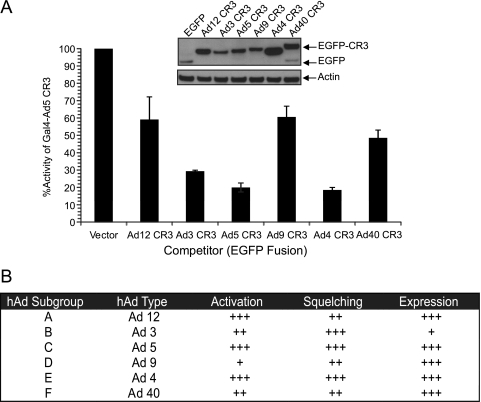
Little is known about the cellular factors required for CR3-dependent transactivation by the E1A proteins of Ad types other than Ad5. We hypothesized that the squelching assay could be applied to determine if the E1A CR3s of other Ad types functioned via interaction with the same set of cellular transcriptional regulators. A549 cells were cotransfected with a Gal4-responsive luciferase reporter and Gal4-Ad5 E1A CR3 wt, as described before, with EGFP fused to the E1A CR3s of chosen representative Ads as competitors. The E1A CR3s of all six Ad serotypes were capable of squelching Ad5 CR3-dependent activation, although to various extents (Fig. 3A). Three classes of squelching ability were observed: Ad5 and Ad4 E1A CR3s were the most potent; Ad3 E1A CR3 appeared to have an intermediate ability; and Ad12, Ad9, and Ad40 E1A CR3s were the least effective squelchers (Fig. 3A). Weaker squelching was not simply due to low levels of expression, as determined by Western blotting (Fig. 3A). Interestingly, the effectiveness of an individual CR3 to squelch was not typically related to its ability to activate transcription as a Gal4-DBD fusion (Fig. 3B).
FIG. 3.
Squelching of Ad5 E1A CR3-dependent activation by the CR3 domains from representative human adenovirus types. (A) Human A549 cells were cotransfected with a Gal4-responsive luciferase reporter, vectors expressing Gal4 alone, or Gal4-Ad5 CR3 as the activator and either EGFP or EGFP fused to each of the indicated E1A CR3s as the competitor. Luciferase activity is expressed as a percentage of the fold activation of Gal4-Ad5 CR3 above that for the vector alone with EGFP as the competitor ± SD. (B) Summary of transcriptional properties of representative E1A CR3s. The transcriptional activation, squelching, and expression levels of each E1A CR3 are summarized relative to those of Ad5 E1A CR3.
On the basis of the findings of the squelching assay, it was clear that the chosen representative E1A CR3s targeted cellular transcriptional regulators also required by Ad5 E1A CR3. We therefore systematically examined their interactions with known coactivators of E1A CR3, including MED23, TBP, hSug1, and p300, as well as the functional requirements for these interactions.
MED23 is required by all E1A CR3s to activate transcription.
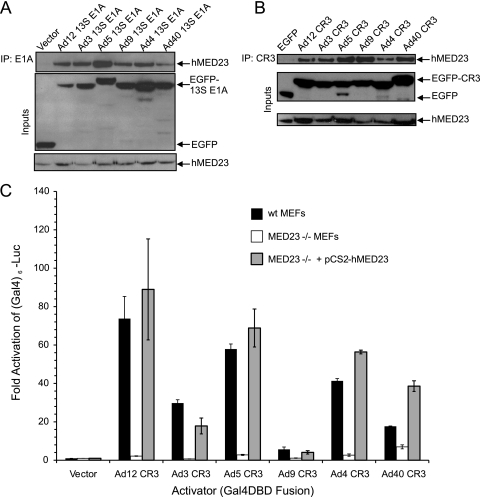
The mediator component MED23 has been implicated to be the most critical cellular coactivator of Ad5 CR3 function (6, 31). This interaction is also required for murine Ad growth, suggesting that this factor may be universally utilized by different adenovirus types (10). We tested the ability of the full-length E1A product from each representative Ad subgroup to interact with hMED23 by coimmunoprecipitation. Each of the different full-length E1A proteins bound hMED23, although none bound it as strongly as the Ad5 E1A (Fig. 4A). Furthermore, the E1A CR3 domains of each representative E1A protein were sufficient for this interaction (Fig. 4B). In order to determine the functional consequences of this hMED23 interaction, the abilities of the representative E1A CR3s to activate transcription in MED23-null (MED23−/−) MEFs were determined. All six E1A CR3s activated transcription in wt MEFs but failed to activate transcription in MED23−/− MEFs (Fig. 4C, black bars and white bars, respectively). The failure to activate transcription was rescued by the expression of exogenous hMED23 in the MED23−/− MEFs (Fig. 4C, gray bars). We conclude that hMED23 is a common target of each of the six representative CR3s and is absolutely required for transactivation.
FIG. 4.
MED23 is targeted by E1A CR3s from multiple Ad types. (A) Coimmunoprecipitation of MED23 and the 13S mRNA-encoded E1A proteins from each human adenovirus subgroup. Human HT1080 cells were cotransfected with a vector expressing HA-tagged human MED23 and vectors expressing either EGFP or an EGFP fused to the indicated E1A proteins. E1A proteins were immunoprecipitated with anti-EGFP antibody and blotted with anti-HA antibody. Inputs were probed with EGFP antibody for E1A proteins or HA antibody for MED23. (B) Coimmunoprecipitation of MED23 with the E1A CR3 domains from each human Ad subgroup. Human HT1080 cells were cotransfected with vectors expressing HA-tagged human MED23 and either myc-EGFP or myc-EGFP fused to the indicated E1A CR3 domain. E1A CR3 domains were immunoprecipitated with anti-myc antibody and blotted with anti-HA antibody. Inputs were probed with anti-myc antibody for CR3s or anti-HA antibody for MED23. (C) E1A CR3 activation in MED23-null MEFs. MED23-null MEFs and wt littermate-derived MEFs were cotransfected with a Gal4-responsive luciferase reporter and vectors expressing Gal4 fused to the indicated E1A CR3 domain (white and black bars, respectively). MED23-null MEFs were also transfected with the Gal4-responsive luciferase reporter, vectors expressing the indicated Gal4-E1A CR3 domain fusions, and an expression vector for human MED23 (gray bars). Luciferase activity is expressed as the fold activation over that for Gal4-DBD alone ± SD.
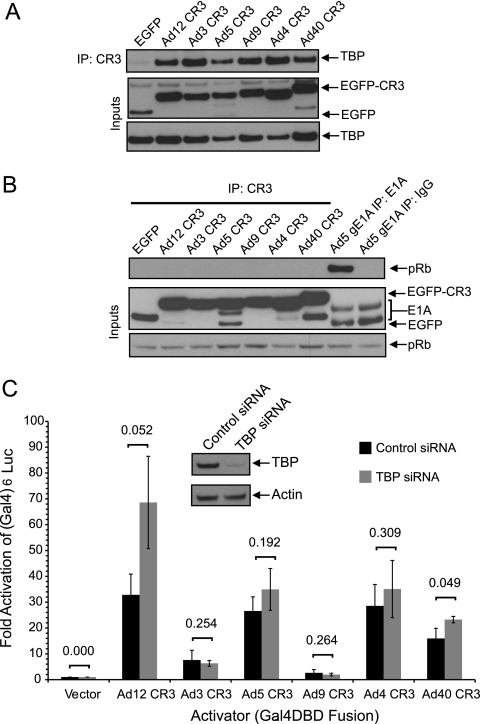
TBP binds all E1A CR3s but is not required for transcriptional activation.
The first cellular protein shown to interact with CR3 of Ad5 E1A was TBP (19). We tested if the interaction with TBP was conserved among the different E1A CR3s and also determined the requirement for TBP in E1A CR3-dependent activation of transcription. Ad5 E1A CR3 was sufficient to coimmunoprecipitate TBP, and the five other E1A CR3s interacted with TBP at least as strongly as Ad5 E1A CR3 (Fig. 5A). As a negative control for these interaction studies, we assessed the abilities of the E1A CR3 fusions to coimmunoprecipitate pRb, which binds E1A primarily via CR2. As expected, none of the E1A CR3 fusion proteins bound pRb (Fig. 5B). To determine the functional role of TBP in E1A CR3 transactivation, we depleted TBP in HeLa cells by RNA interference (RNAi) and then examined the ability of each E1A CR3 to activate transcription as a Gal4 fusion in control siRNA-treated cells and TBP-specific siRNA-treated cells. siRNA knockdown of TBP did not reduce transactivation by any of the six CR3s tested (Fig. 5C). On the basis of this observation, it can be concluded that although TBP was a conserved target of the six CR3s, it does not appear to be nearly as critical as hMED23 for transactivation.
FIG. 5.
TBP is a conserved cellular target of E1A CR3 from multiple Ad types. (A) Coimmunoprecipitation of TBP with representative E1A CR3s. HT1080 cells were cotransfected with a vector expressing HA-tagged TBP and vectors expressing the indicated E1A CR3s fused to EGFP. E1A CR3s were immunoprecipitated with 9E10 antibody and blotted for HA. (B) Negative coimmunoprecipitation of pRb with representative E1A CR3s. HT1080 cells were cotransfected with a vector expressing the indicated E1A CR3s fused to EGFP or genomic E1A. E1A CR3s were immunoprecipitated with anti-myc antibody, and E1A proteins were immunoprecipitated with M73 and blotted for endogenous pRb. (C) siRNA knockdown of TBP and the effect on transcriptional activation by E1A CR3. HeLa cells were transfected with 5 nM siRNA (negative control or TBP specific) and at 2 days after siRNA transfection were retransfected with a Gal4-reponsive luciferase reporter and an expression vector for the indicated Gal4-CR3 fusions. At 48 h posttransfection (120 h after siRNA transfection), cells were harvested and assayed for luciferase activity. Luciferase activity is expressed as the fold above that for Gal4-DBD alone ± SD. The levels of activation in control versus TBP-knockdown cells were compared by Student's t test, and P values are indicated. (Inset) Levels of TBP in knockdown and control cells.
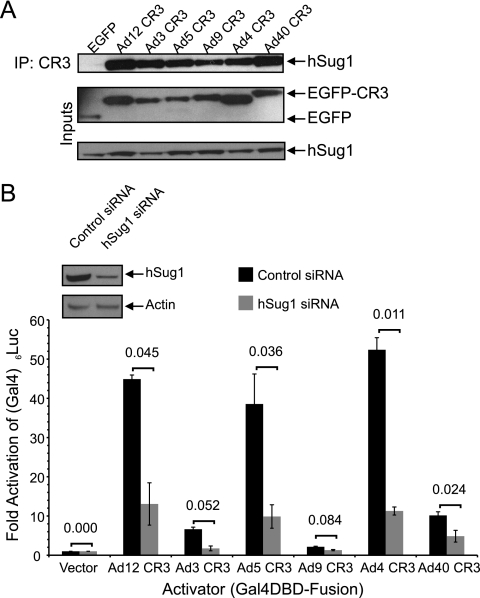
hSug1 binds all E1A CR3s and contributes to transcriptional activation.
The proteasome is a crucial cellular coactivator of Ad5 E1A, and CR3 binds the proteasome via the hSug1 or S8 ATPase component (28). All six representative E1A CR3s interacted with hSug1, as determined by coimmunoprecipitation (Fig. 6A). RNAi-directed knockdown of hSug1 resulted in a reduction of transactivation by all six different E1A CR3s. In particular, the E1A CR3s capable of potent activation of transcription (the Ad12, Ad5, Ad4, and Ad40 CR3s) showed a significant loss of activity in hSug1 siRNA-treated cells relative to that for the control (Fig. 6B).
FIG. 6.
Human Sug1 is a conserved target of E1A CR3 from multiple Ad types. (A) Coimmunoprecipitation of hSug1 with representative Ad E1A CR3s. HT1080 cells were cotransfected with pcDNA4HA-hSug1 and vectors expressing the indicated E1A CR3s fused to myc-EGFP. E1A CR3s were coimmunoprecipitated with anti-myc antibody and blotted for HA. (B) siRNA knockdown of hSug1 and the effect on transcriptional activation by E1A CR3. HeLa cells were transfected with 5 nM siRNA (negative control or Sug1 specific) and at 2 days posttransfection were retransfected with a Gal4-reponsive luciferase reporter and an expression vector for the indicated Gal4-CR3 fusions. At 48 h posttransfection (120 h after siRNA transfection), cells were harvested and assayed for luciferase activity. Luciferase activity is expressed as the fold above that for Gal4-DBD alone ± SD. The levels of fold activation of control versus hSug1 siRNA-treated cells were compared by Student's t test, and P values are indicated above the bars. (Inset) Levels of hSug1 in knockdown and control cells.
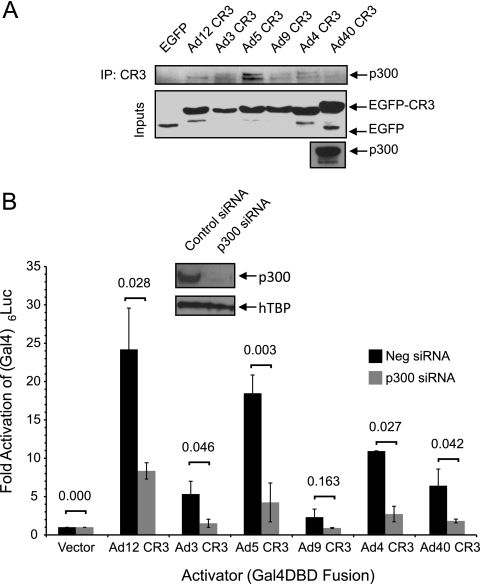
p300/CBP is required by all E1A CR3s to activate transcription.
We have previously shown that the p300/CBP acetyltransferases also function as critical coactivators of Ad5 E1A CR3 function (27). All six representative E1A CR3s interacted with p300, as determined by coimmunoprecipitation, although Ad5 E1A CR3 showed the strongest interaction (Fig. 7A). Depletion of p300 by siRNA resulted in a greater than 50% reduction in CR3 transactivation for all six E1A CR3s (Fig. 7B), and this was statistically significant for all but the Ad9 E1A CR3, the weakest activator.
FIG. 7.
p300 is a conserved target of E1A CR3s from multiple Ad types. (A) Interaction of p300 with the E1A CR3 domains of different Ad types. Human HT1080 cells were cotransfected with an expression vector for HA-tagged p300 and expression vectors for the indicated E1A CR3 myc-EGFP fusions. EGFP fusions were immunoprecipitated with a cocktail of anti-myc and anti-GFP antibodies and blotted with HA. (B) Effect of siRNA depletion of p300 on E1A CR3-dependent activation. HeLa cells were transfected with 20 nM custom siRNA directed against p300 or control siRNA and at 3 days posttransfection were retransfected with a Gal4-reponsive luciferase reporter and an expression vector for the indicated Gal4-CR3 fusions. At 48 h posttransfection (120 h after siRNA transfection), cells were harvested and assayed for luciferase activity. Luciferase activity is expressed as the fold above that for Gal4-DBD alone ± SD. The levels of fold activation of control versus p300 siRNA-treated cells were compared by Student's t test, and P values are indicated above the bars. (Inset) Levels of p300 in knockdown and control cells.
Taken together, these data indicate that each of the six different Ad E1A CR3s shares these four cellular transcriptional regulators as common targets. Importantly, the relative differences between the abilities of each of these E1A CR3s to activate transcription or squelch Ad5 E1A CR3 activity cannot be explained simply by the differences in their association with these factors.
Transcriptional activation by full-length E1A proteins.
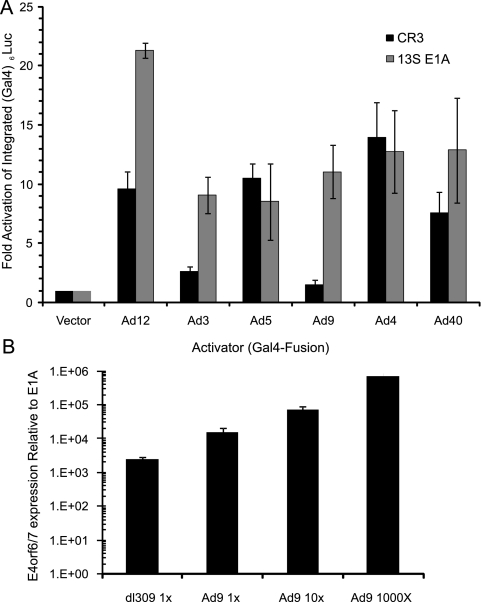
We reasoned that the surprisingly large differences in the intrinsic abilities of the different CR3s to activate transcription could be functionally compensated for by activities present in other portions of these proteins. Indeed, it is well established that the N terminus/CR1 of Ad5 E1A functions as a strong activator of transcription when it is tethered to a heterologous DNA-binding domain (5, 30). We directly compared the abilities of the different CR3s with the corresponding full-length E1A proteins to activate transcription of a Gal4-responsive luciferase reporter stably integrated into the genome of U2OS human osteosarcoma cells (Fig. 8A). While there were again marked differences in the activities of the different E1A CR3s, the full-length E1A proteins were all equivalent or superior activators with respect to Ad5 E1A. This was particularly pronounced for Ad9, suggesting that other regions of this E1A protein may compensate for the weak intrinsic transcriptional activation function of Ad9 E1A CR3. We looked further at the activity of the Ad9 E4 promoter in the context of viral infection by qRT-PCR, as E4 is a well-documented target of E1A transactivation. During infection, the relative expression level of E4orf6/7 mRNA was greatly elevated in the presence of wt Ad5 E1A compared to that during infection with an Ad5 from which E1A was deleted (∼1,500-fold increase). This is expected because E4 expression is highly responsive to full-length E1A (22). The relative expression level of the E4orf6/7 mRNA in Ad9-infected cells exceeds that in wt Ad5-infected cells, similar to what was seen with the integrated reporter assay (Fig. 8B). The expression level of E4orf6/7 in Ad9-infected cells increased in a dose-dependent manner with increased viral inoculum (Fig. 8B). Assuming that the Ad9 E4orf6/7 mRNA is similarly regulated by E1A, it is clear that the Ad9 early genes are potently activated upon infection. This supports our observation for an integrated reporter gene (Fig. 8A) that full-length Ad9 E1A retains strong transactivation function, despite the weak activity intrinsic to CR3 alone.
FIG. 8.
Transactivation by full-length E1A. (A) Transactivation by full-length E1A proteins in the context of chromatin. U2OS-UAS cells, which contain an integrated Gal4-responsive luciferase reporter, were transfected with expression vectors for either the indicated E1A CR3 domains or the indicated full-length 13S E1A proteins. Luciferase activity is expressed as the fold activation above that for Gal4 alone ± SD. (B) Transactivation of the Ad5 and Ad9 E4 promoters during infection. At 16 h postinfection with the indicated viruses, the expression level of E4orf6/7 mRNA was determined by qRT-PCR. The expression level of E4orf6/7 relative to the levels for GAPDH and E1A is indicated, and the expression level for cells infected with dl312 was set equal to 1.
DISCUSSION
The CR3 portion of Ad5 E1A is a potent transcriptional activation module and serves as a paradigm of viral transactivation (3, 11, 25). CR3 is the most highly conserved of the four conserved regions within E1A (Fig. 1A) (1). Given this similarity between E1A proteins and their essential role in activating virus early gene expression, one would predict that all E1A CR3s would function as potent activators of transcription. However, this is not the case; there are dramatic differences in the potencies of representative E1A CR3s to activate transcription (Fig. 1B). In the experiments described here, it was critical to utilize E1A CR3s fused to Gal4-DBD. Direct tethering of the E1A CR3 activation domain to the transcriptional reporter via fusion to Gal4-DBD allows a direct comparison of the transactivation function by bypassing any differences in affinity between the various E1A CR3s and the sequence-specific DNA-binding transcription factors that normally recruit it to the transcriptional template (20, 21). Indeed, our initial experiments revealed that none of the largest E1A products from any of the Ad subgroups (except Ad5) could stimulate transcription of an Ad5 E4 promoter-driven reporter, presumably due to an inability to be targeted to that reporter (unpublished results).
The unexpected and dramatic differences between the abilities of the different E1A CR3 domains to activate transcription suggest that the Ad life cycle can initiate and progress efficiently even if the E1A protein is a relatively weak activator. Indeed, 3 of the 5 other E1A CR3s that we tested were less than 50% as active as the prototype Ad5 E1A CR3, with Ad9 E1A CR3 being by far the weakest (Fig. 1B). In agreement with this, previous work using a panel of E1A CR3 mutants found that growth of Ad5 was not significantly reduced unless E1A-dependent transactivation was reduced by 5- to 20-fold, which translates to the suggestion that a minimum cutoff of approximately 20% of Ad5 E1A CR3 function is critical to virus growth (16).
To understand the molecular basis for the differences in transactivation between Ad types, we initially utilized a transcriptional squelching assay. Despite the inability of the Ad5 V147L E1A CR3 mutant to interact with multiple cellular proteins targeted by E1A CR3 (MED23, TBP, pCAF, and p300), it behaved nearly like wt Ad5 E1A CR3 in the squelching assay. This is highly indicative that this mutant retains binding to additional limiting factors necessary for E1A CR3-dependent transactivation that remain to be identified. When expression was tested in the squelching assay, expression of each of the five other representative E1A CR3s as fusions to EGFP reduced the level of activation by Gal4-Ad5 E1A CR3 (Fig. 3A). These results confirmed that each of the distinct Ad E1A CR3 domains was capable of competing with Gal4-Ad5 E1A CR3 for at least one critical factor. Interestingly, there did not appear to be any correlation between the potency of a given E1A CR3 to transactivate (Fig. 1B) and the ability to squelch Ad5 E1A CR3 (Fig. 3B). These results suggest that the mechanism of E1A CR3 transactivation is a complex hierarchy of binding kinetics and that multiple cellular factors required by E1A CR3 are limiting in vivo. Simply put, the extent to which any given E1A CR3 squelches Gal4-Ad5 E1A CR3 is based on its ability to sequester one or more of these targets. Furthermore, the ability of Ad12 E1A CR3 to activate as strongly as Ad5 E1A CR3 yet squelch Gal4-Ad5 E1A CR3 poorly indicates that important mechanistic differences in transactivation exist between at least these two E1A proteins. One explanation for these observations could be that Ad12 E1A CR3 targets additional factors not utilized by Ad5 E1A CR3 that contribute to its strong transactivation function (15).
We directly tested the ability of each of the six E1A CR3 domains from the different Ad types to bind MED23 (Fig. 4B), TBP (Fig. 5A), hSug1 (Fig. 6A), and p300 (Fig. 7A) and the role that these interacting proteins had on their ability to transactivate (Fig. 4C, 5B, 6B, and 7B, respectively). These data demonstrated, for the first time in most cases, that each of the different E1A CR3s had the ability to interact with these cellular factors, although to various extents with respect to that of the Ad5 E1A CR3 prototype. These data also demonstrate that MED23, hSug1, and p300 play vital roles in transactivation by most, if not all, of the different E1A CR3s, as previously described for the Ad5 E1A CR3 prototype (27, 28, 31). In contrast, the interaction with TBP is not necessary for transcriptional activation by the different CR3s, at least in the context of the Gal4-CR3 fusions.
The binding data demonstrated that Ad9 E1A CR3 interacted to some degree with all of the known cellular targets of E1A CR3 tested (Fig. 4B to 7B). Indeed, it bound MED23 (Fig. 4A) and pCAF far better than all other E1A CR3s except Ad5 (26). On the basis of the binding data, it is not surprising that Ad9 E1A CR3 squelched Ad5 E1A CR3-dependent activation (Fig. 3). However, it is surprising that it was the weakest activator of transcription of the six E1A CR3s tested. The existing model of E1A CR3 function cannot explain this phenomenon. According to what is currently known about the E1A CR3 function, Ad9 E1A CR3 should be able to potently activate transcription as a Gal4 fusion because it can strongly interact with all of the known cellular coactivators of E1A CR3 so far identified. This line of evidence may indicate that the weak activity of the Ad9 E1A CR3 results from the involvement of an as yet unidentified cellular cofactor required by E1A CR3 to activate transcription. Alternatively, if E1A CR3 serves as a scaffold to assemble all the factors required for transcriptional activation, Ad9 E1A CR3 on its own may not properly organize them spatially or temporally.
Interestingly, the Ad9 E1A CR3 sequence has the least identity with the Ad5 E1A CR3 sequence (47%) compared to its identity with the Ad type 12, 3, 4, and 40 E1A CR3 sequences (60%, 60%, 56%, and 54%, respectively). The key residues essential for transcriptional activation by Ad5 CR3 have been systematically identified (33). On the basis of that analysis, inspection of the CR3 sequence of Ad9 E1A reveals that multiple residues expected to be critical for activation are different. Specifically, individual conservative changes in L144, G151, M170, and R177 significantly impair Ad5 E1A CR3 activation, and all these residues differ in the Ad9 E1A CR3 sequence (Fig. 1A). Despite the pronounced defect in Ad9 E1A CR3-dependent activation, the full-length Ad9 E1A protein is a very potent activator (Fig. 8A) and the Ad9 E4 promoter is highly active upon infection (Fig. 8B), suggesting that other regions of the protein can effectively complement the deficiency in CR3 to activate viral early gene expression.
From the work presented here, it is clear that there are multiple conserved interactions among the representative E1A CR3s with cellular cofactors that are involved in activating transcription. Beyond these conserved interactions, our current work and previous work (26) also provide growing evidence that there are selective cellular targets required by the E1A CR3 domains of some Ad types and not others. These may be particularly important for infection of specific tissue types. Perhaps it is the subtle differences in accessory factors rather than the conserved coactivators that ultimately regulate the potency of a given E1A CR3 to activate transcription. Further studies of these potent transcriptional activation domains may lead to the identification of additional cellular transcriptional regulators and provide novel insight into their mechanism of action.
Acknowledgments
We are grateful to Arnie Berk for the gift of MEFs.
J.N.G.A. was supported in part by an OGSST award. A.F.Y. and P.P. were supported in part by awards from the Strategic Training Program in Cancer Research and Technology Transfer. This work was supported by a grant from the Canadian Institutes of Health Research awarded to J.S.M.
Footnotes
Published ahead of print on 29 September 2010.
REFERENCES
- 1.Avvakumov, N., A. E. Kajon, R. C. Hoeben, and J. S. Mymryk. 2004. Comprehensive sequence analysis of the E1A proteins of human and simian adenoviruses. Virology 329:477-492. [DOI] [PubMed] [Google Scholar]
- 2.Avvakumov, N., J. Torchia, and J. S. Mymryk. 2003. Interaction of the HPV E7 proteins with the pCAF acetyltransferase. Oncogene 22:3833-3841. [DOI] [PubMed] [Google Scholar]
- 3.Berk, A. J. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24:7673-7685. [DOI] [PubMed] [Google Scholar]
- 4.Berk, A. J., F. Lee, T. Harrison, J. Williams, and P. A. Sharp. 1979. A pre-early adenovirus 5 gene product regulates synthesis of early messenger RNAs. Cell 17:935-944. [DOI] [PubMed] [Google Scholar]
- 5.Bondesson, M., M. Mannervik, G. Akusjarvi, and C. Svensson. 1994. An adenovirus E1A transcriptional repressor domain functions as an activator when tethered to a promoter. Nucleic Acids Res. 22:3053-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer, T. G., M. E. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. [DOI] [PubMed] [Google Scholar]
- 7.Bruton, R. K., P. Pelka, K. L. Mapp, G. J. Fonseca, J. Torchia, A. S. Turnell, J. S. Mymryk, and R. J. Grand. 2008. Identification of a second CtBP binding site in adenovirus type 5 E1A conserved region 3. J. Virol. 82:8476-8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatton, B., J. L. Bocco, M. Gaire, C. Hauss, B. Reimund, J. Goetz, and C. Kedinger. 1993. Transcriptional activation by the adenovirus larger E1a product is mediated by members of the cellular transcription factor ATF family which can directly associate with E1a. Mol. Cell. Biol. 13:561-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Culp, J. S., L. C. Webster, D. J. Friedman, C. L. Smith, W.-J. Huang, F. Y.-H. Wu, M. Rosenberg, and R. P. Ricciardi. 1988. The 289-amino acid E1A protein of adenovirus binds zinc in a region that is important for trans-activation. Proc. Natl. Acad. Sci. U. S. A. 85:6450-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang, L., J. L. Stevens, A. J. Berk, and K. R. Spindler. 2004. Requirement of Sur2 for efficient replication of mouse adenovirus type 1. J. Virol. 78:12888-12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frisch, S. M., and J. S. Mymryk. 2002. Adenovirus-5 e1a: paradox and paradigm. Nat. Rev. Mol. Cell Biol. 3:441-452. [DOI] [PubMed] [Google Scholar]
- 12.Geisberg, J. V., W. S. Lee, A. J. Berk, and R. P. Ricciardi. 1994. The zinc finger region of the adenovirus E1A transactivating domain complexes with the TATA box binding protein. Proc. Natl. Acad. Sci. U. S. A. 91:2488-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glenn, G. M., and R. P. Ricciardi. 1985. Adenovirus 5 early region 1A host range mutants hr3, hr4, and hr5 contain point mutations which generate single amino acid substitutions. J. Virol. 56:66-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grand, R. J., A. S. Turnell, G. G. Mason, W. Wang, A. E. Milner, J. S. Mymryk, S. M. Rookes, A. J. Rivett, and P. H. Gallimore. 1999. Adenovirus early region 1A protein binds to mammalian SUG1—a regulatory component of the proteasome. Oncogene 18:449-458. [DOI] [PubMed] [Google Scholar]
- 15.Guan, H., J. F. Williams, and R. P. Ricciardi. 2009. Induction of neuronal and tumor-related genes by adenovirus type 12 E1A. J. Virol. 83:651-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison, T., F. Graham, and J. Williams. 1977. Host range mutants of adenovirus type 5 defective for growth in HeLa cells. Virology 77:319-329. [DOI] [PubMed] [Google Scholar]
- 17.Jelsma, T. N., J. A. Howe, C. M. Evelegh, N. F. Cunniff, M. H. Skiadopoulos, M. R. Floroff, J. E. Denman, and S. T. Bayley. 1988. Use of deletion and point mutants spanning the coding region of the adenovirus 5 E1A gene to define a domain that is essential for transcriptional activation. Virology 163:494-502. [DOI] [PubMed] [Google Scholar]
- 18.Jones, N., and T. Shenk. 1979. An adenovirus type 5 early gene function regulates expression of other early viral genes. Proc. Natl. Acad. Sci. U. S. A. 76:3665-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, W. S., C. C. Kao, G. O. Bryant, X. Liu, and A. J. Berk. 1991. Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell 67:365-376. [DOI] [PubMed] [Google Scholar]
- 20.Liu, F., and M. R. Green. 1994. Promoter targeting by adenovirus E1a through interaction with different cellular DNA-binding domains. Nature 368:520-525. [DOI] [PubMed] [Google Scholar]
- 21.Martin, K. J., J. W. Lillie, and M. R. Green. 1990. Evidence for interaction of different eukaryotic transcriptional activators with distinct cellular targets. Nature 346:147-152. [DOI] [PubMed] [Google Scholar]
- 22.Montell, C., E. F. Fisher, M. H. Caruthers, and A. J. Berk. 1982. Resolving the functions of overlapping viral genes by site-specific mutagenesis at a mRNA splice site. Nature 295:380-384. [DOI] [PubMed] [Google Scholar]
- 23.Moran, E., T. Grodzicker, R. J. Roberts, M. B. Mathews, and B. Zerler. 1986. Lytic and transforming functions of individual products of the adenovirus E1A gene. J. Virol. 57:765-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran, E., B. Zerler, T. M. Harrison, and M. B. Mathew. 1986. Identification of separate domains in the adenovirus E1A gene for immortalization activity and the activation of virus early genes. Mol. Cell. Biol. 6:3470-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelka, P., J. N. Ablack, G. J. Fonseca, A. F. Yousef, and J. S. Mymryk. 2008. Intrinsic structural disorder in adenovirus E1A: a viral molecular hub linking multiple diverse processes. J. Virol. 82:7252-7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelka, P., J. N. Ablack, M. Shuen, A. F. Yousef, M. Rasti, R. J. Grand, A. S. Turnell, and J. S. Mymryk. 2009. Identification of a second independent binding site for the pCAF acetyltransferase in adenovirus E1A. Virology 391:90-98. [DOI] [PubMed] [Google Scholar]
- 27.Pelka, P., J. N. Ablack, J. Torchia, A. S. Turnell, R. J. Grand, and J. S. Mymryk. 2009. Transcriptional control by adenovirus E1A conserved region 3 via p300/CBP. Nucleic Acids Res. 37:1095-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasti, M., R. J. Grand, A. F. Yousef, M. Shuen, J. S. Mymryk, P. H. Gallimore, and A. S. Turnell. 2006. Roles for APIS and the 20S proteasome in adenovirus E1A dependent transcription. EMBO J. 25:2710-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shuen, M., N. Avvakumov, J. Torchia, and J. S. Mymryk. 2003. The E1A proteins of all six human adenovirus subgroups target the p300/CBP acetyltransferases and the SAGA transcriptional regulatory complex. Virology 316:75-83. [DOI] [PubMed] [Google Scholar]
- 30.Shuen, M., N. Avvakumov, P. G. Walfish, C. J. Brandl, and J. S. Mymryk. 2002. The adenovirus E1A protein targets the SAGA but not the ADA transcriptional regulatory complex through multiple independent domains. J. Biol. Chem. 277:30844-30851. [DOI] [PubMed] [Google Scholar]
- 31.Stevens, J. L., G. T. Cantin, G. Wang, A. Shevchenko, A. Shevchenko, and A. J. Berk. 2002. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296:755-758. [DOI] [PubMed] [Google Scholar]
- 32.Strom, A. C., P. Ohlsson, and G. Akusjarvi. 1998. AR1 is an integral part of the adenovirus type 2 E1A CR3 transactivation domain. J. Virol. 72:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Webster, L. C., and R. P. Ricciardi. 1991. trans-Dominant mutants of E1A provide genetic evidence that the zinc finger of the trans-activating domain binds a transcription factor. Mol. Cell. Biol. 11:4287-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yousef, A. F., C. J. Brandl, and J. S. Mymryk. 2009. Requirements for E1A dependent transcription in the yeast Saccharomyces cerevisiae. BMC Mol. Biol. 10:32. [DOI] [PMC free article] [PubMed] [Google Scholar]