Abstract
Tetherin and IFITM3 are recently identified interferon-induced cellular proteins that restrict infections by retroviruses and filoviruses and of influenza virus and flaviviruses, respectively. In our efforts to further explore their antiviral activities against other viruses and determine their antiviral mechanisms, we found that the two antiviral proteins potently inhibit the infection of vesicular stomatitis virus (VSV), a prototype member of the Rhabdoviridae family. Taking advantage of this well-studied virus infection system, we show that although both tetherin and IFITM3 are plasma membrane proteins, tetherin inhibits virion particle release from infected cells, while IFITM3 disrupts an early event after endocytosis of virion particles but before primary transcription of incoming viral genomes. Furthermore, we demonstrate that both the N-terminal 21 amino acid residues and C-terminal transmembrane region of IFITM3 are required for its antiviral activity. Collectively, our work sheds light on the mechanisms by which tetherin and IFITM3 restrict infection with rhabdoviruses and possibly other pathogenic viruses.
The interferon (IFN) system is the first line of defense against virus infection in vertebrates. Infection of cells by viruses can be detected by host cellular pattern recognition receptors (PRRs), such as toll-like receptors and RIG-I-like receptors (1, 33, 42). Engagement of the PRRs with virus-associated molecular patterns, such as 5′-triphosphate and/or double-stranded viral RNA, triggers signaling cascades that lead to the synthesis and secretion of type I IFNs, represented by IFN-α and IFN-β (25). Type I IFNs bind to their cognate receptors on the cell surface and activate the receptor-associated Janus kinase 1 (JAK1) and tyrosine kinase 2 (Tyk2) through tyrosine phosphorylation, which in turn stimulates the tyrosine phosphorylation of STAT1 and STAT2. Phosphorylated STAT1 and STAT2, in combination with IRF9, form a trimeric ISGF3 transcription factor that translocates into the nucleus and activates the expression of IFN-stimulated genes (ISGs), whose products limit viral replication, regulate cell proliferation, and/or modulate host innate and adaptive immune responses (34, 36, 39).
Although IFN treatment of cells induces the expression of hundreds of ISGs, only a few ISGs have thus far been demonstrated to instigate an antiviral state in cultured cells, and only four of them, including ISG15 (IFN-stimulated protein of 15 kDa), myxovirus resistance 1 (Mx1), RNase L, and double-stranded RNA-activated protein kinase (PKR), have been shown to play a role in mediating the IFN antiviral response in vivo with gene knockout mouse models (36). While it is most likely that inhibition of the infection of any given virus by IFNs is through induction of multiple ISGs that work cooperatively to disrupt multiple steps of viral replication, identification of individual antiviral ISGs and subsequent elucidation of their modes of action are essential to uncover the antiviral mechanism of IFNs and viral pathogenesis (20, 49, 50).
Recently, several laboratories, including our own, have identified two new families of IFN-induced cellular proteins, tetherin (or BST-2) and three members of the IFN-induced transmembrane family (IFITM) proteins, as novel antiviral ISGs (4, 21, 31). While tetherin inhibits the release of retroviruses and filoviruses by tethering newly budding viral particles on the cell surface, which leads to their subsequent endocytosis and degradation in the lysosome (23, 31, 37), IFITMs disrupt an unidentified early event of influenza virus and flavivirus infection (4, 21). Our studies reported herein for the first time demonstrate that expression of either tetherin or IFITMs in human embryonic kidney 293 (HEK293)-derived cell lines potently inhibits infection by vesicular stomatitis virus (VSV), a well-studied member of the Rhabdoviridae family. As observed for lentiviruses and filoviruses, tetherin does not inhibit VSV RNA replication and protein expression, but rather inhibits the release of virions from infected cells. Conversely, IFITM3, the most potent antiviral IFITM, inhibits the infection of VSV glycoprotein (G)-pseudotyped lentiviruses but does not disrupt the attachment and endocytosis of VSV. However, expression of the ISG significantly reduces viral primary transcription. Hence, it appears that IFITM3 inhibits an early event after endocytosis of virion particles, but at or before primary transcription of incoming viral genomes. Moreover, we provide strong evidence suggesting that both the N-terminal 21 amino acid residues and C-terminal transmembrane region of IFITM3 are required for its antiviral function. Our work thus reveals the distinct antiviral properties of the two newly identified antiviral ISGs and provides a basis for further analyses of the role and mechanisms of tetherin and IFITMs in controlling infection by rhabdoviruses and other pathogenic viruses.
MATERIALS AND METHODS
Cell culture and virus.
The parental FLP-IN T Rex cells (Invitrogen) were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin G, streptomycin, nonessential amino acids, l-glutamine, 10 μg/ml blasticidin, and 100 μg/ml Zeocin. FLP-IN T Rex-derived cell lines expressing the control protein chloramphenicol acetyltransferase (CAT), tetherin, IFITM1, IFITM2, or IFITM3 were established and maintained as described previously (20). Vero cells were grown in DMEM supplemented with 10% FBS, 2 mM l-glutamine, 1.5 g/liter sodium bicarbonate, 50 U/ml of penicillin, and 50 μg/ml of streptomycin. BHK-21 cells were grown in minimal essential medium (MEM; Gibco-BRL) supplemented with 5% FBS, 2 g/liter sodium bicarbonate, 50 U/ml of penicillin, and 50 μg/ml of streptomycin. Wild-type VSV (Indiana serotype) has been described elsewhere (15). Recombinant VSV expressing green fluorescent protein (VSV-GFP) was kindly provided by Matthias J. Schnell (Thomas Jefferson University, Philadelphia, PA) and propagated in BHK-21 cells. VSV titers were determined by plaque assay in Vero cells, essentially as described previously (15).
Immunofluorescence.
FLP-IN T Rex-derived cell lines were left untreated or treated with 1 μg/ml of tetracycline for 24 h followed by fixation with 2% paraformaldehyde and permeabilization of the cell membrane with 0.1% Triton X-100. Cells were then stained with either IFITM3- or tetherin-specific antibody, and the bound antibodies were visualized by using Alexa Fluor 488 goat anti-mouse IgG. Cells were imaged with a Nikon fluorescence microscope and photographed with a charge-coupled-device camera.
Analysis of membrane topologies of tetherin and IFITM3.
Tetherin- and IFITM3-expressing FLP-IN cell lines were seeded into 12-well plates at a density of 5 × 105 cells per well and cultured in the absence or presence of tetracycline (1 μg/ml) for 24 h. Cells were then detached from the wells by treatment with phosphate-buffered saline (PBS) containing 0.5 M EDTA at 37°C for 5 min, followed by suspension in fluorescence-activated cell sorting (FACS) buffer, which was PBS containing 2% bovine serum albumin (BSA). After fixation with PBS containing 1% paraformaldehyde for 30 min at room temperature, cells were extensively washed with PBS and divided into two tubes. While one tube was left untreated, another tube was treated with PBS containing 0.2% Tween 20 at 37°C for 15 min to permeabilize the cells. Both the untreated and permeabilized cells were suspended in FACS buffer containing 1 μg/ml of mouse anti-Flag antibody (Sigma) and incubated on ice for 1 h. After extensive washes with PBS to remove unbound antibodies, the cells were incubated with FACS buffer containing fluorescein 549-labeled rabbit anti-mouse IgG on ice for 30 min. The bound antibodies were analyzed by using a Guava EasyCyte Plus flow cytometer (Millipore). Cells not induced with tetracycline were used as a negative control, along with an isotype-matched primary antibody.
Quantification of viral RNA by real-time RT-PCR.
FLP-IN T Rex cells expressing CAT, tetherin, or IFITM3 were cultured in the absence or presence of tetracycline (1 μg/ml) for 24 h, followed by infection with VSV at a multiplicity of infection (MOI) of 5. Cells were harvested prior to being infected or at the indicated time points after infection. Total cellular RNA was extracted with TRIzol reagent by following the directions of the manufacturer (Invitrogen). VSV RNA was measured in a TaqMan reverse transcription-PCR (RT-PCR) assay (40). Briefly, primers and probes for TaqMan RT-PCR detection of VSV RNA were designed and purchased from Integrated DNA Technologies. The sequences of the forward and reverse primers were 5′-GATAGTACCGGAGGATTGACGACTA-3′ and 5′-TCAAACCATCCGAGCCATTC-3′, respectively. The dually labeled probe was 5′-carboxyfluorescein-TGCACCGCCACAAGGCAGAGA-6-carboxytetramethylrhodamine-3′. RT-PCR was performed with a TaqMan One-Step RT-PCR Master Mix reagents kit (Applied Biosystems), as described by the manufacturer, using a 20-μl sample volume with 5 μM (each) primers, 25 μM probe, and 100 ng of total cellular RNA. TaqMan PCR assays were performed using an ABI 7500 Fast real-time PCR system (Applied Biosystems) and the following amplification profile: 1 cycle at 48°C for 30 min, 1 cycle at 95°C for 10 min, and 44 cycles at 95°C for 15 s and 60°C for 1 min. All samples were tested in triplicate.
Western blot assay.
Cell monolayers were washed once with PBS buffer and lysed with 1× Laemmli buffer. A fraction of cell lysate was separated on sodium dodecyl sulfate-12% polyacrylamide gels and electrophoretically transferred onto a polyvinylidene difluoride membrane (Bio-Rad). Membranes were blocked with PBS containing 5% nonfat dry milk and probed with antibodies against Flag tag (Sigma), tetherin (Proteintech Group, Inc.), IFITM3 (Proteintech Group, Inc.), VSV G protein (Rockland, Inc.), or β-actin (Chemicon International). Bound antibodies were revealed by horseradish peroxidase-labeled secondary antibodies and visualized with an enhanced chemiluminescence detection system (Amersham Pharmacia Biotech) according to the protocol of the manufacturer.
FACS analysis of GFP-expressing VSV-infected cells.
FLP-IN T Rex-derived stable cells expressing CAT, tetherin, or IFITM3 were seeded into 12-well plates at a density of 5 × 105 cells per well and cultured in the absence or presence of tetracycline (1 μg/ml) for 24 h, followed by infection with VSV-GFP at an MOI of 5. Four hours after infection, cells were detached from the wells by treatment with PBS containing 0.5 M EDTA for 5 min at 37°C. Cells were resuspended in FACS buffer (PBS plus 2% BSA) and gently pelleted by centrifugation at 2,000 rpm for 5 min. Cells were reuspended in fixative solution (PBS containing 1% paraformaldehyde) and incubated at room temperature for 30 min, followed by two washes with PBS and resuspension in FACS buffer. GFP expression was analyzed by using a Guava EasyCyte Plus flow cytometer (Millipore).
VSV-G-pseudotyped lentivirus infection.
FLP-IN cells expressing CAT, tetherin, or IFITMs were seeded at a density of 5 × 104 cells per well in a 96-well plate. At 24 h postseeding, the cells were cultured in the absence or presence of 1 μg/ml tetracycline for 24 h, followed by infection with a VSV-G protein-pseudotyped lentivirus (SABioscience) that expressed firefly luciferase at an MOI of 2 for 1 h, and cell culturing continued in medium without or with tetracycline for an additional 48 h. Cells were then lysed, and luciferase activity in the cell lysates was quantified by using a Steady-Glo firefly luciferase assay kit (Promega).
Virus binding and entry assays.
FLP-IN T Rex cells induced to express CAT, IFITM3, or tetherin were seeded in 12-well plates at a density of 5 × 105 cells per well and cultured in the absence or presence of tetracycline for 36 h. Cells were infected with VSV at an MOI of 5 for 1 h on ice to allow attachment but impede virus entry. After three washes with PBS, RNA was extracted to measure the amount of cell-bound virus. To assay virus entry into cells via endocytosis, virus inocula were removed after 1 h of binding on ice, cells were washed, prewarmed medium was added, and the cells were incubated for another 10 min at 37°C. Cells were then washed once with PBS followed by treatment with 0.25% trypsin for 10 min and again washed three times with PBS to remove any cell-associated virus which had not entered the cytoplasm. Total cellular RNA was extracted to measure the amount of viral genomes that had entered cells by using a real-time RT-PCR assay.
Northern blot hybridization.
Total cellular RNA was extracted with TRIzol reagent (Invitrogen) by following the manufacturer's directions. Five micrograms of total RNA was fractionated on a 1.5% agarose gel containing 2.2 M formaldehyde and transferred onto nylon membranes. Membranes were hybridized with riboprobes specific for VSV N mRNA under the conditions described previously (14).
Establishment of cell lines expressing mutant IFITMs.
cDNA constructs expressing Flag-tagged IFITM mutant proteins harboring point mutations in the conserved region and chimeric IFITM proteins consisting of structural domains from both IFITM1 and IFITM3 were constructed by overlap extension PCR. The sequences of the primers are available upon request. Briefly, two separate PCRs were performed to amplify two overlapping fragments of the coding region of the targeting molecule by using four primers. The point mutations were introduced by the two middle primers. The final PCR products were cloned into vector pcDNA5/FRT/ΔCAT. The cDNA construct expressing an N-terminally truncated IFITM3 protein was constructed by cloning the PCR products, with amplification with a pair of primers bracketing the desired regions of IFITM3 cDNA, into a pcDNA5/FRT/ΔCAT vector (20). All the resulting DNA clones were sequenced to verify the desired mutation(s). Stable cell lines inducibly expressing the mutant IFITMs were established by cotransfection of FLP-IN T Rex cells with pOG44 and the above-described IFITM mutant protein-expressing plasmids, followed by selection and pooling of hygromycin- and blasticidin-resistant cell colonies, essentially as previously described (20).
RESULTS
Expression of IFITMs and tetherin elicits antiviral activity against VSV in HEK293 cells.
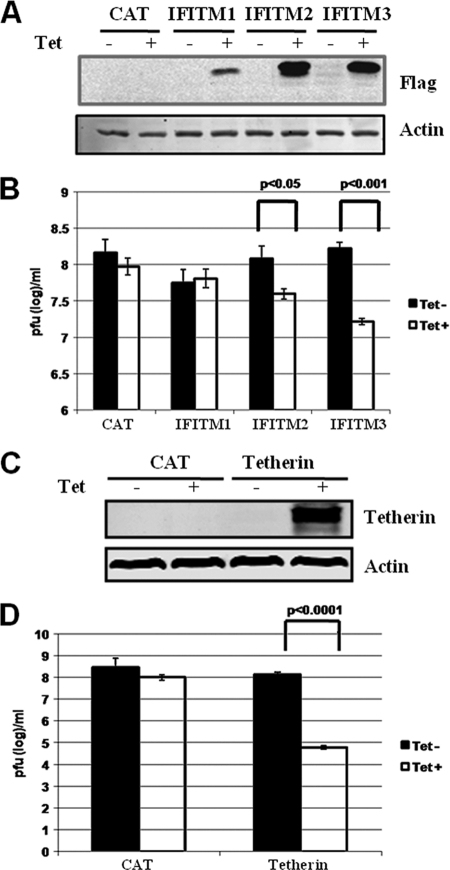
In our recent efforts to systematically identify antiviral ISGs that inhibit hepatitis C virus (HCV) and dengue virus (DENV) infection, we have established 36 FLP-IN T Rex-derived stable cell lines that express individual ISGs in a tetracycline-inducible manner (20, 21). Taking advantage of this unique cell culture system, we tested the antiviral activities of the 36 ISGs against infection with VSV. Aside from PKR and ISG20, which had been reported previously to inhibit VSV infection in cultured cells and in mice in vivo (3, 11, 12, 41), we found that expression of tetherin and two members of the IFITM family of proteins, IFITM2 and IFITM3, reduced VSV yields by over 1,000-, 5-, and 15-fold, respectively (Fig. 1B and D). Inducible expression of each of the three IFITMs and tetherin in FLP-IN T Rex-derived cell lines, upon addition of tetracycline in culture medium, was demonstrated by Western blot analyses (Fig. 1A and C). The observed antiviral profiles of the three IFITMs against VSV were consistent with our previous observations that IFITM3 and to a lesser extent IFITM2, but not IFITM1, elicit an antiviral activity against both West Nile virus (WNV) and DENV infection in these cell lines (21).
FIG. 1.
Antiviral activities of IFITMs and tetherin against VSV in FLP-IN T Rex cells, expressing CAT or the desired ISGs, left untreated or treated with 1 μg/ml of tetracycline for 24 h. (A and C) Cells were harvested, and induction levels of IFITMs and tetherin expression were confirmed by Western blot analyses with antibody against Flag tag or tetherin protein. (B and D) Alternatively, cells were infected with VSV at an MOI of 0.001 and culturing continued in medium without or with tetracycline for an additional 16 h. VSV titers in the supernatants of the infected cells were determined by a plaque assay with Vero cells. Error bars indicate standard deviations of the means from three experiments. Statistical analysis was performed using Student's t test.
Both IFITM and tetherin restrict VSV infection.
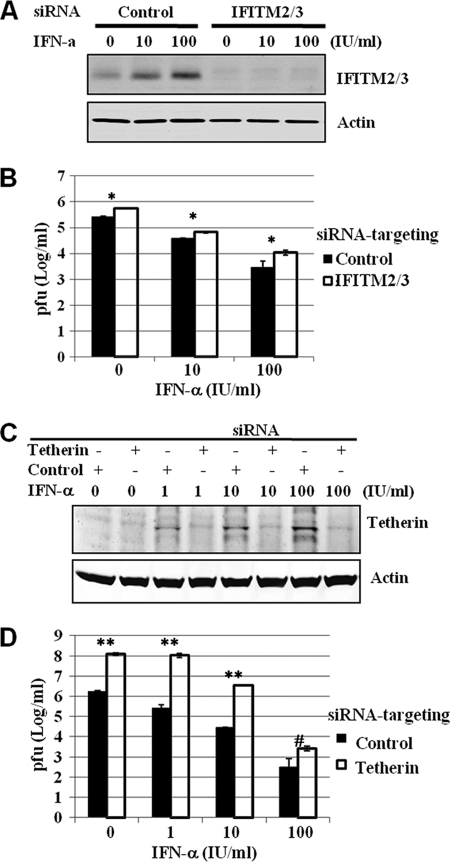
To further investigate the physiological role of the IFITMs and tetherin in controlling VSV infection, we intended to determine the effects of depletion of basal levels and IFN-induced expression of IFITM and tetherin on VSV infection. Compared with HeLa cells transfected with nontargeting small interfering RNA (siRNA; control), transfection of SMARTpool siRNA targeting IFITM2 and -3 (Fig. 2A) or tetherin (Fig. 2C) not only efficiently reduced basal levels but also reduced IFN-α-induced levels of IFITM and tetherin expression, respectively. Interestingly, knockdown of IFITM2/3 expression resulted in an approximately 2- to 3-fold increase of VSV yields in cells mock treated or treated with IFN-α (Fig. 2B). Intriguingly, transfection of siRNA targeting tetherin (Fig. 2D) into HeLa cells drastically increased permissiveness of the cells to VSV infection, as indicated by the approximately 100-fold increase of progeny virus production (Fig. 2D). Moreover, reduction of IFN-induced tetherin expression also significantly attenuated the ability of IFN-α to inhibit VSV infection at lower concentrations (1 to 10 IU/ml). However, in the cells treated with a higher concentration of IFN-α (100 IU/ml), the attenuation effect was decreased, presumably due to compensation by other IFN-activated antiviral pathways under this condition (Fig. 2D). Hence, our results imply that both IFITM2/3 and tetherin not only restrict VSV infection at their basal levels of expression but also are physiological mediators of the IFN-induced antiviral response against VSV.
FIG. 2.
IFITM and tetherin restrict VSV infection. (A) HeLa cells were transfected with SMARTpool siRNA targeting IFITM2 and IFITM3 or nontargeting siRNA (control), following the directions of the manufacturer (Dharmacon). Forty-eight hours posttransfection, cells were left untreated or were treated with the indicated concentrations of IFN-α for 24 h. Cells were harvested for Western blot analysis of IFITM protein in the cell lysates with an antibody recognizing IFITM2 and -3 (Proteintech). β-Actin served as the loading control. (B) Following IFN-α treatment, HeLa cells were infected with VSV at an MOI of 0.01. Virus yields in culture medium samples harvested at 24 h postinfection were determined with a plaque assay and are expressed as means ± standard errors (n = 3). *, P < 0.001. (C) HeLa cells were transfected with SMARTpool siRNA targeting tetherin or nontargeting siRNA (control), following the directions of the manufacturer (Dharmacon). Six hours posttransfection, cells were left untreated or treated with the indicated concentrations of IFN-α for 24 h. Cells were harvested for Western blot analysis of tetherin expression in the cell lysates or, alternatively, infected with VSV at an MOI of 0.01. (D) Virus yields in culture medium samples harvested at 24 h postinfection were determined with a plaque assay and are expressed as means ± standard errors (n = 3). Statistical analysis was performed using Student's t test. **, P < 0.0001; #, P < 0.05.
Characterization of tetherin and IFITM expression and membrane topology in FLP-IN T Rex cells.
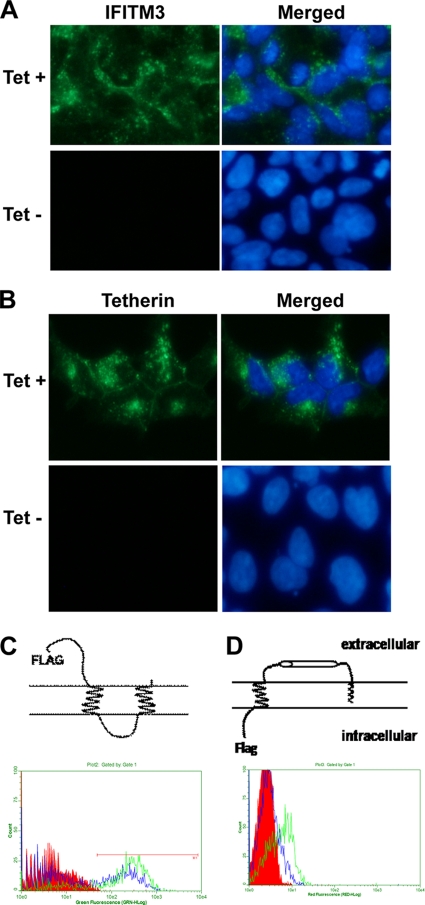
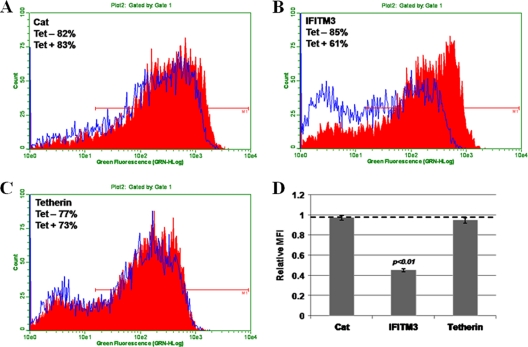
Tetherin and IFITMs are type I IFN-inducible cell membrane proteins (18). Immunofluorescent staining with tetherin- and IFITM3-specific antibodies demonstrated that upon induction by the addition of tetracycline into the culture medium, both IFITM3 and tetherin were detected in the cytoplasm of FLP-IN cells (Fig. 3A and B). The localization of tetherin was enriched in the peripheral zones of cells, most likely on the plasma membrane. Recently, Brass and colleagues showed that IFITM3 resided in endoplasmic reticulum (ER) membranes (4). However, it was also reported previously that IFITMs localize in the plasma membranes of several types of cells (10). Protein sequence analysis with the TMpred program (www.ch.embnet.org) predicted that IFITMs contain two transmembrane regions and a conserved intracellular loop. Such a membrane topology (Fig. 3C, top) suggests that both the N and C termini of IFITM proteins localize extracellularly. In contrast, tetherin is a type II transmembrane protein with a glycosylphosphatidylinositol (GPI) anchor at its C terminus; therefore, its N terminus localizes intracellularly (Fig. 3D, top) (31). Because both tetherin and IFITMs engineered into FLP-IN T Rex cells are N-terminally Flag tagged (20), we performed FACS analysis to detect Flag epitope expression in intact and permeabilized FLP-IN T Rex cells expressing IFITM3 or tetherin. As shown in Fig. 3C and D, while the Flag epitope was detected in both permeabilized IFITM3-expressing and tetherin-expressing cells, it was detected only on intact cells expressing IFITM3 and not those expressing tetherin. These results thus convincingly demonstrate that IFITM3 is expressed on the plasma membrane of FLP-IN cells with the predicted membrane topology.
FIG. 3.
Expression and membrane topology of IFITM3 and tetherin in HEK293 cells. FLP-IN T Rex cell lines expressing either IFITM3 or tetherin were left untreated or treated with 1 μg/ml of tetracycline for 24 h. (A and B) Expression levels of IFITM3 (A) and tetherin (B) were visualized by immunofluorescent staining. Cell nuclei were visualized by using 4′,6-diamidino-2-phenylindole staining. (C and D) For FACS analysis, after induction of ISG expression, cells were either left untreated (intact) or permeabilized by incubation with buffer containing 0.1% Triton X-100. Expression of the Flag tag on the surface of intact cells (blue) or in the permeabilized cells (green) was revealed by flow cytometry. Histograms were gated to analyze the population expressing the Flag tag. Diagrams to depict the putative membrane topologies of IFITM3 and tetherin are also presented.
Tetherin and IFITM3 inhibit distinct steps of VSV replication.
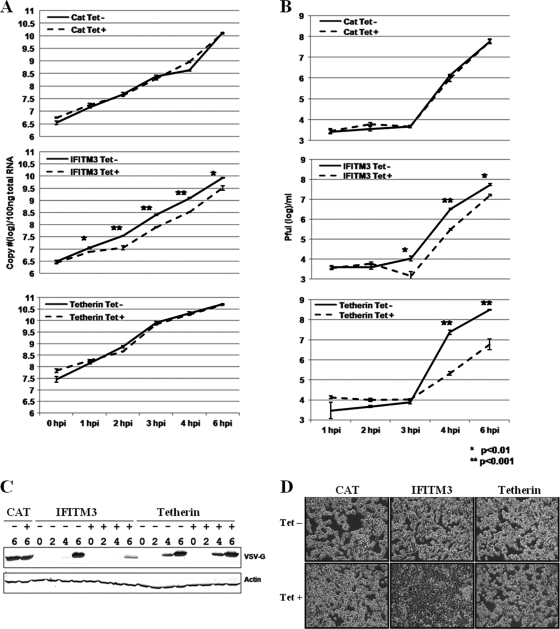
In order to map the viral replication steps targeted by tetherin and IFITM3, a synchronized VSV infection assay was performed. Briefly, the indicated cell lines were cultured in the absence or presence of tetracycline for 24 h followed by infection with VSV at an MOI of 5 on ice for 1 h. Such a multiplicity of infection ensures that at least 99% of cells are infected with a minimum of one infectious viral particle. The virus inocula were then removed, and infected cells were cultured with prewarmed fresh medium at 37°C. At the indicated times after infection, culture medium and cells were harvested to determine the virus yields and the amount of intracellular viral RNA and G protein accumulation. As shown in Fig. 4, expression of the control protein CAT did not affect viral RNA replication, G protein accumulation, or virion production. IFITM3 expression, however, significantly reduced the levels of intracellular viral RNA as early as 1 h postinfection, and similarly, VSV-G protein accumulation was dramatically reduced at 6 h postinfection. As a consequence, virion production was also inhibited by IFITM3. In stark contrast, expression of tetherin did not affect intracellular viral RNA replication or G protein accumulation, but it drastically reduced virion production. Moreover, microscopic inspection indicated that VSV infection caused apparent cytopathic effects (CPE), as demonstrated by cells rounding up and detaching from plates at 6 h postinfection. Interestingly, expression of IFITM3, but not tetherin, efficiently inhibited the VSV-induced CPE (Fig. 4D).
FIG. 4.
Identification of the VSV replication step(s) targeted by IFITM3 and tetherin. FLP-IN T Rex cell lines expressing the control protein CAT or the antiviral proteins IFITM3 or tetherin were left untreated or treated with 1 μg/ml of tetracycline for 24 h prior to infection with VSV at an MOI of 5. At the indicated time points after infection, culture supernatants and cells were harvested. (A) The levels of cell-associated viral RNA were determined in a real-time RT-PCR assay, and the results are expressed as copies per 100 ng of total cellular RNA. (B) Virus yields were measured in a plaque assay. Error bars indicate standard deviations of the means (n = 3). (C) Accumulation of viral G protein was determined by Western blot assay. (D) Cells were fixed with 2% paraformaldehyde and photographed with a Nikon microscope.
To further confirm the distinct antiviral property of tetherin and IFITM3, a synchronized infection assay was performed with a recombinant VSV that expressed GFP. As for all viral proteins, GFP is expressed by the recombinant VSV as an independent transcript unit. Therefore, the level of intracellular GFP can serve as a surrogate marker for viral RNA replication/transcription and protein expression. As shown in Fig. 5, flow cytometry analysis clearly indicated that expression of CAT and tetherin did not affect GFP expression, whereas IFITM3 significantly reduced the percentage of GFP-positive cells as well as the mean fluorescent intensity (MFI) of infected cells.
FIG. 5.
IFITM3, but not tetherin, inhibits the synchronized infection of a recombinant VSV that expresses GFP. FLP-IN T Rex cell lines expressing the control protein CAT (A) or the antiviral proteins IFITM3 (B) or tetherin (C) were left untreated (red) or treated with 1 μg/ml of tetracycline (blue) for 24 h prior to infection with VSV-GFP at an MOI of 5. At 4 h postinfection, cells were fixed and the levels of GFP expression were analyzed by flow cytometry. (D) Ratio of the MFI in cells treated with tetracycline versus that in cells left untreated, plotted as the relative MFI. Error bars indicate standard deviations of the means (n = 3).
Taken together, the results obtained from these experiments suggest that tetherin most likely inhibits VSV release from infected cells, which is in agreement with the mechanism by which ISGs restrict HIV and Ebola virus infection (23, 31). IFITM3, however, most probably inhibits either VSV entry into host cells or viral RNA/protein biosynthesis.
IFITM3 inhibits VSV-G protein-mediated virus entry.
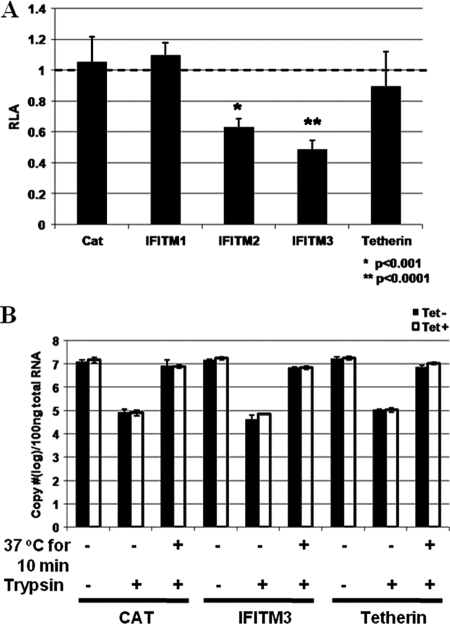
Considering the facts that IFITM3 is a plasma membrane protein and inhibits an unidentified early event of influenza virus and flavivirus infection (4), it is reasonable to envisage that IFITM3 also inhibits an early event of VSV infection. To test this hypothesis, we first determined the effects of the three IFITMs and tetherin on the infection of a VSV-G protein-pseudotyped lentivirus that expressed firefly luciferase upon integration of lentiviral vector DNA into the host cellular chromosome. As shown in Fig. 6A, expression of CAT and tetherin did not affect the pseudovirus infection. However, consistent with the antiviral profiles shown in Fig. 1, expression of IFITM2 and -3, but not IFITM1, significantly reduced the level of the pseudotyped lentivirus-conferred luciferase expression. Because it has been demonstrated independently in several reports that the IFITMs do not inhibit lentiviral infection (4, 31), our results thus most likely indicate that IFITM3 inhibits VSV-G protein-mediated pseudovirus entry into cells but does not disrupt a postentry event of lentiviral replication, such as lentiviral cDNA synthesis, integration, or gene expression.
FIG. 6.
IFITM3, but not tetherin, inhibits VSV-G protein-mediated virus entry. (A) FLP-IN T Rex cells expressing the control protein CAT or the indicated ISGs were left untreated or treated with 1 μg/ml of tetracycline for 24 h. Cells were then infected with VSV-G-pseudotyped lentiviral particles at an MOI of 1. Forty-eight hours postinfection, intracellular levels of firefly luciferase expressed by the recombinant lentiviral vector were determined. Results represent the means ± standard deviations (n = 6) of the ratios of light units obtained from wells cultured in the presence of tetracycline versus those obtained from wells that were cultured in the absence of tetracycline. (B) FLP-IN T Rex cells induced to express CAT, IFITM3, or tetherin were cultured in the absence or presence of tetracycline for 36 h. Cells were infected with VSV at an MOI of 5 for 1 h on ice to allow attachment. After three washes with PBS, RNA was extracted to measure the amount of cell-bound virus. To quantify virus entry, virus inocula were removed after 1 h of binding on ice, and cells were either directly subjected to trypsin treatment or incubated for another 10 min at 37°C, followed by trypsin treatment to remove any cell-associated virus that had not entered the cytoplasm. Intracellular viral RNA was measured in a real-time RT-PCR assay, and the results are expressed as copies per 100 ng of total cellular RNA. Error bars indicate standard deviations of the means (n = 3).
To further validate this observation and determine the precise VSV entry step that is inhibited by the antiviral ISG, a classical virus binding and entry assay was performed (16). Briefly, FLP-IN T Rex cells expressing CAT, tetherin, or IFITM3 were cultured in the absence or presence of tetracycline for 24 h. For the attachment assay, these cells were infected with VSV at an MOI of 5 for 1 h on ice, followed by extensive washing steps with cold PBS. Cellular-associated viral RNA was measured to quantify virus attachment onto the cells. To determine virus entry, cells were infected on ice as described above, and after extensive washing with cold PBS to remove unattached virions, prewarmed medium was added and cells were incubated at 37°C for another 10 min. The time interval chosen to monitor VSV entry was based on previous studies that showed that the endocytosis of VSV virion particles is extremely efficient and takes less than 5 min (9, 22). In fact, similar results were obtained by harvesting cells between 10 and 30 min of incubation (data not shown). After incubation at 37°C, cells were treated with trypsin and washed three times with complete DMEM to remove virions that had not entered cells. As shown in Fig. 6B, this procedure efficiently removed approximately 99% of bound virions (based on comparison with the number of viral RNA copies obtained from attachment to cells immediately treated with trypsin without the temperature shift). Total cellular RNA was extracted, and viral RNA was quantified in a real-time RT-PCR assay. The results showed that expression of neither control proteins (CAT and tetherin) nor IFITM3 affected VSV attachment and entry into cells (Fig. 6B).
Expression of IFITM3 reduces primary transcription of incoming VSV genomes.
While the pseudovirus infection assay results suggested that IFITM3 inhibits VSV-G protein-mediated virus entry (Fig. 6A), it appears that the ISG does not inhibit the attachment and endocytosis of VSV (Fig. 6B). It is therefore possible that IFITM3 disrupts the intracellular trafficking of the endosome and/or fusion of virion and endosomal membranes. However, due to technical limitations, we were unable to follow endosome trafficking and membrane fusion in VSV-infected cells. Instead, in order to further confirm the notion that IFITM3 inhibits an early event of VSV infection, but not viral RNA replication or protein translation, we sought to determine the effect of IFITM3 on the primary transcription of incoming viral genomes. As a negative-stranded RNA virus, upon release of viral nucleocapsids into the cytoplasm through membrane fusion the first biosynthetic step in VSV replication cycle is primary transcription, which is mediated by virion-associated RNA-dependent RNA polymerase. Because all the essential proteins for the primary transcription of viral mRNA are packaged within the nucleocapsid, primary transcription does not require synthesis of viral or additional host cellular proteins (7, 35).
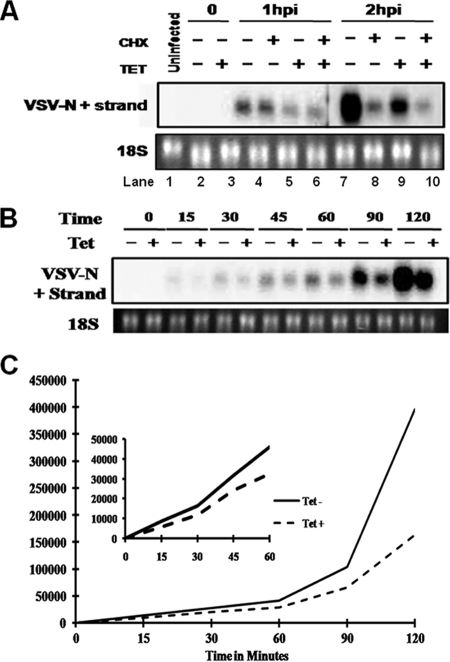
FLP-IN/IFITM3 cells were left untreated or treated with tetracycline for 24 h, followed by infection with VSV at an MOI of 1, on ice, for 1 h. Cells were then washed twice with PBS to remove unattached viruses and refed with prewarmed complete medium without or with cycloheximide, to inhibit protein translation. Cells were harvested immediately after infection (0 h) or at 1 or 2 h postinfection. The levels of viral nucleocapsid protein (N) mRNA were measured by Northern blot hybridization. As shown in Fig. 7A, treatment of cells with cycloheximide did not affect the accumulation of VSV N mRNA at 1 h after infection (compare lanes 3 and 4) but significantly reduced viral mRNA accumulation at 2 h after infection (compare lanes 7 and 8), suggesting that within the first hour postinfection, viral mRNA is exclusively derived from primary transcription, which does not require new protein synthesis. Interestingly, expression of IFITM3 significantly reduced the amount of VSV N mRNA from primary transcription (compare lanes 3 and 5 or lanes 4 and 6). Moreover, as shown in an additional time course study (Fig. 7B and C), VSV N mRNA was readily detectable at 30 min postinfection, and expression of IFITM3 reduced the levels of viral mRNA at all the time points tested. Taken together, our results imply that IFITM3 either directly inhibits VSV primary transcription or, more likely, an early event after endocytosis but before primary transcription.
FIG. 7.
IFITM3 reduces primary transcription of viral mRNA. (A) FLP-IN/IFITM3 cells were cultured in the absence or presence of tetracycline for 24 h. Cells were infected with VSV at an MOI of 1 for 1 h on ice, followed by culturing at 37°C in the absence or presence of cycloheximide for 1 or 2 h. The levels of VSV N protein mRNA were determined by Northern blot hybridization. 18S rRNA served as a loading control. hpi, hours postinfection. (B) FLP-IN/IFITM3 cells were cultured in the absence or presence of tetracycline for 24 h, followed by infection with VSV at an MOI of 1 for 1 h on ice. The infected cells were either immediately harvested or harvested at the indicated time points after being cultured at 37°C. (C) The levels of VSV N mRNA were determined by Northern blot hybridization and quantified using the Quantity One phosphorimaging system.
Identification of the structural domains essential for the antiviral function of IFITM3.
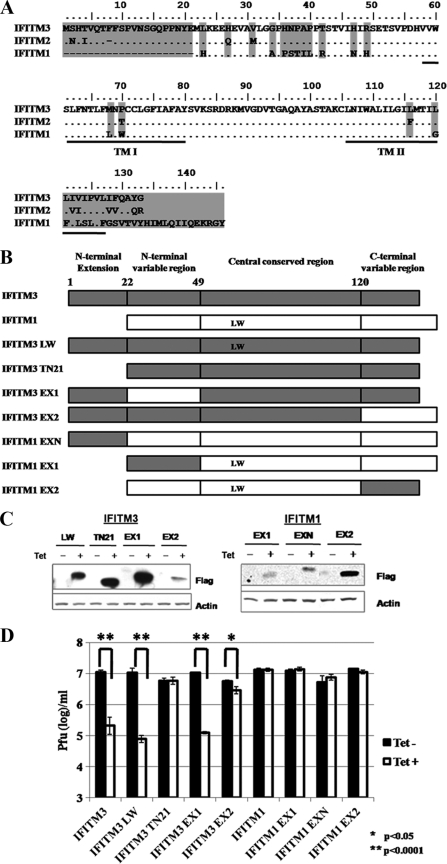
Polypeptide sequence alignment showed that IFITM2 and IFITM3 share 91% amino acid identity and differ at only 12 amino acid residues that are scattered along the 123-amino-acid polypeptides (Fig. 8A). In contrast, IFITM1 shares only 70% amino acid identity with IFITM3, with the major differences lying at the N and C termini. The IFITM1 sequence lacks the first 21 amino acids of IFITM3 and differs markedly over the carboxy-terminal 27 amino acids, being both different in sequence and containing 13 additional amino acids in this region (27) (Fig. 7A). While Brass and colleagues reported that all three IFITM proteins, upon overexpression in A549 cells, inhibited the infection of influenza virus, WNV, and dengue virus (4), our results reported herein and those published previously (21) demonstrate that expression of only IFITM3 and IFITM2, but not IFITM1, in FLP-IN T Rex cells inhibits VSV, WNV, and dengue virus infection. Taking advantage of the differential antiviral activities and distinct structural features of IFITM1 and IFITM3, a panel of mutant IFITM proteins, including proteins with point mutations in transmembrane region 1, an N-terminal truncation of IFITM3, and chimeric IFITM1 and IFITM3 proteins (Fig. 8B), were engineered. FLP-IN T Rex cell lines expressing each of the mutant IFITMs were established. The expression of the desired mutant proteins was verified in a Western blot assay (Fig. 8C). The abilities of the mutant IFITMs, in comparison with the wild-type IFITM3 and IFITM1, to reduce VSV yields were determined in a plaque assay.
FIG. 8.
Structural function analysis of IFITM3. (A) Amino acid sequence alignment of the three IFITM family proteins studied. Differences are indicated by amino acid codes. TM I and TM II, transmembrane domains. (B) Schematic representation of the structures of IFITMs and their mutants. At the top are the overall structural organizations of IFITM3 and IFITM1. The lower panels highlight the nature of the mutations in seven mutant IFITMs (see text for details). (C) Inducible expression of mutant IFITMs in stable FLP-IN T Rex cell lines. Cells were cultured in the absence or presence of tetracycline for 24 h, and the levels of Flag-tagged IFITM1, IFITM3, or their mutants in cell lysates were determined by Western blot analysis with a monoclonal antibody against the Flag tag. β-Actin served as a loading control and was detected by using a monoclonal antibody against human β-actin. (D) Stable cell lines that inducibly express CAT or wild-type or mutant IFITMs were left untreated or treated with 1 μg/ml tetracycline for 24 h prior to VSV infection at an MOI of 0.001. Sixteen hours later, culture medium samples were harvested. VSV titers in the media of the infected cells were determined by a plaque assay with Vero cells. Error bars indicate standard deviations of the means from three experiments. Statistical analysis was performed using Student's t test.
As shown in Fig. 8D, the results revealed the following. First, substitution of the two amino acid residues in transmembrane region 1 of IFITM3 with the amino acid residues at the same positions from IFITM1 (IFITM3M68L/P70W) did not affect the antiviral activity of IFITM3. Second, deletion of the N-terminal 21 amino acid residues from IFITM3 (IFITM3TN21) completely abolished its antiviral activity. Third, replacement of N-terminal variable region of IFITM3 with the corresponding region of IFITM1 (IFITM3EX1) did not impair the antiviral activity of IFITM3, whereas replacement of the C-terminal variable region of IFITM3 with the corresponding region of IFITM1 (IFITM3EX2) significantly compromised the antiviral activity of IFITM3. These results thus suggest that both the N-terminal 21-amino-acid extension and C-terminal (but not N-terminal) variable region of IFITM3 are important for its antiviral activity. In agreement with this conclusion, neither addition of the IFITM3 N-terminal 21-amino-acid extension to IFITM1 (IFITM1EXN) nor replacement of the C-terminal variable region of IFITM1 with the corresponding region of IFITM3 (IFITM1EX2) was able to confer antiviral activity. Interestingly, because the only difference between the central conserved regions of IFITM1 and IFITM3 is the two amino acid residues (L and W [Fig. 8A and B]), which have been shown not to be responsible for the differential antiviral activity of the two proteins, the chimeric protein IFITM3EX1 was structurally equivalent to that produced by simultaneously adding the N-terminal extension of IFITM3 to the N terminus of IFITM1 and replacing the C-terminal variable region of IFITM1 with the corresponding C-terminal region of IFITM3. As shown in Fig. 8D, such a chimeric protein was fully functional in inhibition of VSV infection.
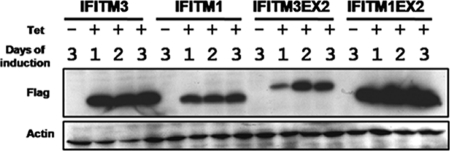
C-terminal variable regions of IFITMs determine steady-state protein levels.
While we consistently observed that IFITM3 and IFITM2, but not IFITM1, were able to inhibit virus infection in FLP-IN T Rex cells, we also noticed that upon induction, both IFITM2 and IFITM3 were expressed at a higher level than IFITM1 (Fig. 1). It is, therefore, possible that the inability of IFITM1 to inhibit virus infection is due to its relatively lower level of expression in this cell line. Interestingly, in our mutagenesis studies presented in the previous section, we noticed that exchange of the C-terminal variable regions between IFITM1 and IFITM3 significantly decreased the steady-state level of IFITM3 (Fig. 8C, compare the mutant IFITM3EX2 with the other IFITM3 mutants) but increased the steady-state level of IFITM1 (Fig. 8C, compare the mutant IFITM3EX2 with the other IFITM3 mutants). To further validate this observation, FLP-IN T Rex cells expressing wild-type IFITM3, IFITM1, and the two C-terminal region-exchanged mutants were left untreated or treated with tetracycline. Cells were harvested at 1, 2, and 3 days postinduction. Levels of IFITM protein expression were determined by Western blot assay. As shown in Fig. 9, such a side-by-side comparison convincingly demonstrated that the C-terminal regions of IFITMs determine their steady-state protein levels. Because the ISG cDNA is integrated into the host chromosome via the single FLP recombination target (FRT) site in FLP-IN T Rex cells and transcribed under the same tetracycline-inducible promoter (20), it is unlikely that the observed differences in the levels of IFITM protein expression were due to differences in transcription. Instead, the most probable explanation is that the C-terminal transmembrane regions of IFITMs determine the stability of the proteins. Nevertheless, although expressed at a much higher level than IFITM1, IFITM1EX2 does not gain any detectable antiviral activity (Fig. 8C). These results thus indicate that failure to inhibit virus infection by IFITM1 is more likely due to its lack of essential functional domains, and not its relatively lower level of expression.
FIG. 9.
The C-terminal regions of IFITMs are responsible for their intracellular steady-state levels. The indicated stable cell lines were cultured in the absence of tetracycline for 3 days or in the presence of tetracycline for 1, 2, or 3 days. The levels of Flag-tagged IFITM1, IFITM3, or their mutants in cell lysates were determined by Western blot analysis with a monoclonal antibody against the Flag tag. β-Actin served as a loading control and was detected by using a monoclonal antibody against human β-actin.
DISCUSSION
Our work identified that IFITM2/3 and tetherin, two families of structurally distinct IFN-inducible cell membrane proteins, inhibited VSV infection upon overexpression in HEK293 cells. Based upon the data presented above, we conclude that IFITM proteins inhibit a postendocytosis event of virus entry, whereas tetherin impairs a late step in the VSV replication cycle, most likely viral particle (virion) release. Our findings not only expand the antiviral spectrum of the two families of ISGs but also demonstrate that the two ISGs restrict VSV infection and are physiological mediators of the IFN-induced antiviral response against the virus. Hence, our work provides an explanation for previous observations indicating that IFN inhibits VSV entry and release from infected cells (7, 44, 45).
The IFITM proteins were identified more than 2 decades ago based on their inducible expression by type I and, to a lesser extent, type II IFNs in neuroblastoma cells (46), and they were originally termed the 1-8 gene family (27). The three human IFITM genes lie adjacent on chromosome 11 and share very similar gene structures, consisting of an interferon-stimulated response element (ISRE) in the immediate 5′-flanking sequence and two exons (27). This family of proteins has been implicated to play a role in cell adhesion, oncogenesis, germ cell homing, and maturation, as well as regulation of endocytosis (10, 26, 43, 46, 47). In addition to being induced by IFNs, IFITM3 expression has been reportedly upregulated in certain types of cancer cells (2, 48). Furthermore, it was also reported that IFITM3 was responsible for IFN-induced cell growth inhibition (5, 47). However, in agreement with Brass et al. (4), even prolonged expression (for more than 3 weeks) of the IFITMs in FLP-IN T Rex cells did not disturb proliferation of the cells (data not shown).
In our previous studies we discovered that while expression of IFITM3 in FLP-IN T Rex cells efficiently inhibited the infection of both West Nile virus and DENV virus-like particles (VLP), the antiviral protein was not able to inhibit replication of the viral replicon when the replicon RNA was directly delivered into cells via transfection. We thus concluded that IFITM3 inhibited a replication step of the flavivirus life cycle prior to translation and replication of incoming viral RNA genomes, i.e., virus entry and/or uncoating (21). This notion is consistent with a recent report that elegantly showed that overexpression of IFITM proteins in several types of cells inhibited the infection of influenza virus envelope protein-pseudotyped retroviral particles (4). In this report, we used a VSV infection system to further dissect the antiviral mechanism of IFITM3 and provided evidence suggesting that IFITM3 inhibits a VSV entry step after endocytosis, but before primary transcription, with the most likely targets being endosome trafficking and/or fusion of virion and endosomal membranes. In fact, this proposed function of IFITM3 is consistent with the observed subcellular localization of the protein. As demonstrated herein and reported recently by Brass and colleagues, in addition to its plasma membrane localization, IFITM3 also associates with the ER membrane and multivesicular compartments that contain endosome-like organelles (4, 5, 47). To further determine the relationship between the intracellular localization and antiviral function of IFITM3, we added a classical ER retrieval signal KDEL sequence at the C terminus of IFITM3, and we also introduced two point mutations (T118Y and P125S) in the C-terminal transmembrane region in an attempt to withhold the protein within the ER compartment and prevent its endosomal and plasma membrane localization (28, 38). However, FACS analysis suggested that both of the mutant proteins were still expressed on the plasma membrane at a similar level as wild-type IFITM3-expressing cells. Not surprisingly, both the mutant proteins elicited an antiviral response against VSV (data not shown). In the future, it will be interesting to further explore the relationship between the subcellular localization and antiviral activity of IFITM3 by using both genetic and cell imaging technologies. In addition, a recent report showed that IFITM1 localizes in caveolae of the plasma membrane and could interact with CAV-1. Deletion mutagenesis clearly revealed that the hydrophobic transmembrane domains are responsible for the interaction between IFITM1 and CAV-1 (46). It will be of great interest to test the hypothesis that failure of IFITM1 to inhibit both VSV and flavivirus infection in HEK293 cells is due its distinct intracellular localization.
Tetherin is an interferon-induced, transmembrane, and GPI-anchored protein that restricts the release of numerous enveloped viruses, including all retroviruses tested as well as members of the Arenavirus (Lassa virus) and Filovirus (Ebola and Marburg viruses) families (23, 31, 37). In this report, we further extended its antiviral activity to VSV, a member of the Rhabdoviridae family. However, we found that tetherin was not able to inhibit the release of DENV, a member of the Flaviviridae family (data not shown). Although we could not rule out the possibility that, like many other viruses (24, 29), DENV encodes proteins that antagonize the function of tetherin, these observations are in agreement with the notion that tetherin is localized in cholesterol-enriched microdomains of the plasma membrane and thus inhibits the release of virions that bud from such membrane microdomains (13, 17, 32). In contrast, the flaviviruses presumably bud from the ER membrane and, thus, could potentially elude tetherin's antiviral activity (6, 8, 19, 30). Moreover, consistent with that observed in HIV infection, elimination of the GPI anchor of tetherin by addition of a V5 tag sequence at its C terminus severely impaired its antiviral activity against VSV (data not shown), suggesting that its GPI anchor is necessary to inhibit the release of VSV and, thus, occurs via a similar mechanism as that for tethering HIV and other viruses (31).
In conclusion, our work presented herein advances our understanding of the two newly identified antiviral proteins. Further studies to decipher the structural and cell biological bases of the antiviral activities of IFITM and tetherin may ultimately lead to the development of a broad-spectrum antiviral therapeutic approach against enveloped virus infection by enhancing their antiviral functions or disarming the antagonists of the viruses against the IFN-induced antiviral proteins.
Acknowledgments
We thank Haitao Guo for suggestions during this study and critical reading of the manuscript.
This work was supported by a grant from the National Institutes of Health (AI061441) and by the Hepatitis B Foundation through an appropriation of the Commonwealth of Pennsylvania. Ju-Tao Guo is the Bruce Witte Scholar of Hepatitis B Foundation.
Footnotes
Published ahead of print on 13 October 2010.
REFERENCES
- 1.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124:783-801. [DOI] [PubMed] [Google Scholar]
- 2.Andreu, P., S. Colnot, C. Godard, P. Laurent-Puig, D. Lamarque, A. Kahn, C. Perret, and B. Romagnolo. 2006. Identification of the IFITM family as a new molecular marker in human colorectal tumors. Cancer Res. 66:1949-1955. [DOI] [PubMed] [Google Scholar]
- 3.Balachandran, S., P. C. Roberts, L. E. Brown, H. Truong, A. K. Pattnaik, D. R. Archer, and G. N. Barber. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13:129-141. [DOI] [PubMed] [Google Scholar]
- 4.Brass, A. L., I. C. Huang, Y. Benita, S. P. John, M. N. Krishnan, E. M. Feeley, B. J. Ryan, J. L. Weyer, L. van der Weyden, E. Fikrig, D. J. Adams, R. J. Xavier, M. Farzan, and S. J. Elledge. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brem, R., K. Oraszlan-Szovik, S. Foser, B. Bohrmann, and U. Certa. 2003. Inhibition of proliferation by 1-8U in interferon-alpha-responsive and non-responsive cell lines. Cell. Mol. Life Sci. 60:1235-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brinton, M. A. 2002. The molecular biology of West Nile virus: a new invader of the western hemisphere. Annu. Rev. Microbiol. 56:371-402. [DOI] [PubMed] [Google Scholar]
- 7.Carey, B. L., M. Ahmed, S. Puckett, and D. S. Lyles. 2008. Early steps of the virus replication cycle are inhibited in prostate cancer cells resistant to oncolytic vesicular stomatitis virus. J. Virol. 82:12104-12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clyde, K., J. L. Kyle, and E. Harris. 2006. Recent advances in deciphering viral and host determinants of dengue virus replication and pathogenesis. J. Virol. 80:11418-11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cureton, D. K., R. H. Massol, S. Saffarian, T. L. Kirchhausen, and S. P. Whelan. 2009. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 5:e1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deblandre, G. A., O. P. Marinx, S. S. Evans, S. Majjaj, O. Leo, D. Caput, G. A. Huez, and M. G. Wathelet. 1995. Expression cloning of an interferon-inducible 17-kDa membrane protein implicated in the control of cell growth. J. Biol. Chem. 270:23860-23866. [DOI] [PubMed] [Google Scholar]
- 11.Durbin, R. K., S. E. Mertz, A. E. Koromilas, and J. E. Durbin. 2002. PKR protection against intranasal vesicular stomatitis virus infection is mouse strain dependent. Viral Immunol. 15:41-51. [DOI] [PubMed] [Google Scholar]
- 12.Espert, L., G. Degols, C. Gongora, D. Blondel, B. R. Williams, R. H. Silverman, and N. Mechti. 2003. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J. Biol. Chem. 278:16151-16158. [DOI] [PubMed] [Google Scholar]
- 13.Fitzpatrick, K., M. Skasko, T. J. Deerinck, J. Crum, M. H. Ellisman, and J. Guatelli. 2010. Direct restriction of virus release and incorporation of the interferon-induced protein BST-2 into HIV-1 particles. PLoS Pathog. 6:e1000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo, J. T., J. Hayashi, and C. Seeger. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 79:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Habjan, M., N. Penski, V. Wagner, M. Spiegel, A. K. Overby, G. Kochs, J. T. Huiskonen, and F. Weber. 2009. Efficient production of Rift Valley fever virus-like particles: the antiviral protein MxA can inhibit primary transcription of bunyaviruses. Virology 385:400-408. [DOI] [PubMed] [Google Scholar]
- 17.Hammonds, J., J. J. Wang, H. Yi, and P. Spearman. 2010. Immunoelectron microscopic evidence for Tetherin/BST2 as the physical bridge between HIV-1 virions and the plasma membrane. PLoS Pathog. 6:e1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayashi, J., R. Stoyanova, and C. Seeger. 2005. The transcriptome of HCV replicon expressing cell lines in the presence of alpha interferon. Virology 335:264-275. [DOI] [PubMed] [Google Scholar]
- 19.Huang, H., F. Sun, D. M. Owen, W. Li, Y. Chen, M. Gale, Jr., and J. Ye. 2007. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 104:5848-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, D., H. Guo, C. Xu, J. Chang, B. Gu, L. Wang, T. M. Block, and J. T. Guo. 2008. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J. Virol. 82:1665-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, D., J. M. Weidner, M. Qing, X. B. Pan, H. Guo, C. Xu, X. Zhang, A. Birk, J. Chang, P. Y. Shi, T. M. Block, and J. T. Guo. 2010. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J. Virol. 84:8332-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johannsdottir, H. K., R. Mancini, J. Kartenbeck, L. Amato, and A. Helenius. 2009. Host cell factors and functions involved in vesicular stomatitis virus entry. J. Virol. 83:440-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jouvenet, N., S. J. Neil, M. Zhadina, T. Zang, Z. Kratovac, Y. Lee, M. McNatt, T. Hatziioannou, and P. D. Bieniasz. 2009. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J. Virol. 83:1837-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaletsky, R. L., J. R. Francica, C. Agrawal-Gamse, and P. Bates. 2009. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 106:2886-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawai, T., and S. Akira. 2007. Antiviral signaling through pattern recognition receptors. J. Biochem. 141:137-145. [DOI] [PubMed] [Google Scholar]
- 26.Lange, U. C., D. J. Adams, C. Lee, S. Barton, R. Schneider, A. Bradley, and M. A. Surani. 2008. Normal germ line establishment in mice carrying a deletion of the Ifitm/Fragilis gene family cluster. Mol. Cell. Biol. 28:4688-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewin, A. R., L. E. Reid, M. McMahon, G. R. Stark, and I. M. Kerr. 1991. Molecular analysis of a human interferon-inducible gene family. Eur. J. Biochem. 199:417-423. [DOI] [PubMed] [Google Scholar]
- 28.Lewis, M. J., and H. R. Pelham. 1992. Ligand-induced redistribution of a human KDEL receptor from the Golgi complex to the endoplasmic reticulum. Cell 68:353-364. [DOI] [PubMed] [Google Scholar]
- 29.Miyagi, E., A. J. Andrew, S. Kao, and K. Strebel. 2009. Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface down-modulation and intracellular depletion. Proc. Natl. Acad. Sci. U. S. A. 106:2868-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089-1097. [DOI] [PubMed] [Google Scholar]
- 31.Neil, S. J., T. Zang, and P. D. Bieniasz. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425-430. [DOI] [PubMed] [Google Scholar]
- 32.Perez-Caballero, D., T. Zang, A. Ebrahimi, M. W. McNatt, D. A. Gregory, M. C. Johnson, and P. D. Bieniasz. 2009. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139:499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pichlmair, A., and C. Reis e Sousa. 2007. Innate recognition of viruses. Immunity 27:370-383. [DOI] [PubMed] [Google Scholar]
- 34.Platanias, L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5:375-386. [DOI] [PubMed] [Google Scholar]
- 35.Rose, J. K., and M. A. Whitt. 2001. Rhabdoviridae: the viruses and their replication, p. 1221-1277. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 36.Sadler, A. J., and B. R. Williams. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakuma, T., T. Noda, S. Urata, Y. Kawaoka, and J. Yasuda. 2009. Inhibition of Lassa and Marburg virus production by tetherin. J. Virol. 83:2382-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato, K., M. Sato, and A. Nakano. 2003. Rer1p, a retrieval receptor for ER membrane proteins, recognizes transmembrane domains in multiple modes. Mol. Biol. Cell 14:3605-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 40.Simon, I. D., J. Publicover, and J. K. Rose. 2007. Replication and propagation of attenuated vesicular stomatitis virus vectors in vivo: vector spread correlates with induction of immune responses and persistence of genomic RNA. J. Virol. 81:2078-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stojdl, D. F., N. Abraham, S. Knowles, R. Marius, A. Brasey, B. D. Lichty, E. G. Brown, N. Sonenberg, and J. C. Bell. 2000. The murine double-stranded RNA-dependent protein kinase PKR is required for resistance to vesicular stomatitis virus. J. Virol. 74:9580-9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeuchi, O., and S. Akira. 2008. MDA5/RIG-I and virus recognition. Curr. Opin. Immunol. 20:17-22. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka, S. S., Y. L. Yamaguchi, B. Tsoi, H. Lickert, and P. P. Tam. 2005. IFITM/Mil/fragilis family proteins IFITM1 and IFITM3 play distinct roles in mouse primordial germ cell homing and repulsion. Dev. Cell 9:745-756. [DOI] [PubMed] [Google Scholar]
- 44.Trottier, M. D., Jr., B. M. Palian, and C. S. Reiss. 2005. VSV replication in neurons is inhibited by type I IFN at multiple stages of infection. Virology 333:215-225. [DOI] [PubMed] [Google Scholar]
- 45.Whitaker-Dowling, P. A., D. K. Wilcox, C. C. Widnell, and J. S. Youngner. 1983. Interferon-mediated inhibition of virus penetration. Proc. Natl. Acad. Sci. U. S. A. 80:1083-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu, Y., G. Yang, and G. Hu. 2009. Binding of IFITM1 enhances the inhibiting effect of caveolin-1 on ERK activation. Acta Biochim. Biophys. Sin. (Shanghai) 41:488-494. [DOI] [PubMed] [Google Scholar]
- 47.Yang, G., Y. Xu, X. Chen, and G. Hu. 2007. IFITM1 plays an essential role in the antiproliferative action of interferon-gamma. Oncogene 26:594-603. [DOI] [PubMed] [Google Scholar]
- 48.Yang, Y., J. H. Lee, K. Y. Kim, H. K. Song, J. K. Kim, S. R. Yoon, D. Cho, K. S. Song, Y. H. Lee, and I. Choi. 2005. The interferon-inducible 9-27 gene modulates the susceptibility to natural killer cells and the invasiveness of gastric cancer cells. Cancer Lett. 221:191-200. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, Y., C. W. Burke, K. D. Ryman, and W. B. Klimstra. 2007. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 81:11246-11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou, A., J. M. Paranjape, S. D. Der, B. R. Williams, and R. H. Silverman. 1999. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology 258:435-440. [DOI] [PubMed] [Google Scholar]