Abstract
COX-2 has been implicated in Kaposi's sarcoma-associated herpesvirus (KSHV) latency and pathogenesis (A. George Paul, N. Sharma-Walia, N. Kerur, C. White, and B. Chandran, Cancer Res. 70:3697-3708, 2010; P. P. Naranatt, H. H. Krishnan, S. R. Svojanovsky, C. Bloomer, S. Mathur, and B. Chandran, Cancer Res. 64:72-84, 2004; N. Sharma-Walia, A. G. Paul, V. Bottero, S. Sadagopan, M. V. Veettil, N. Kerur, and B. Chandran, PLoS Pathog. 6:e1000777, 2010; N. Sharma-Walia, H. Raghu, S. Sadagopan, R. Sivakumar, M. V. Veettil, P. P. Naranatt, M. M. Smith, and B. Chandran, J. Virol. 80:6534-6552, 2006). However, the precise regulatory mechanisms involved in COX-2 induction during KSHV infection have never been explored. Here, we identified cis-acting elements involved in the transcriptional regulation of COX-2 upon KSHV de novo infection. Promoter analysis using human COX-2 promoter deletion and mutation reporter constructs revealed that nuclear factor of activated T cells (NFAT) and the cyclic AMP (cAMP) response element (CRE) modulate KSHV-mediated transcriptional regulation of COX-2. Along with multiple KSHV-induced signaling pathways, infection-induced prostaglandin E2 (PGE2) also augmented COX-2 transcription. Infection of endothelial cells markedly induced COX-2 expression via a cyclosporine A-sensitive, calcineurin/NFAT-dependent pathway. KSHV infection increased intracellular cAMP levels and activated protein kinase A (PKA), which phosphorylated the CRE-binding protein (CREB) at serine 133, which probably led to interaction with CRE in the COX-2 promoter, thereby enhancing COX-2 transcription. PKA selective inhibitor H-89 pretreatment strongly inhibited CREB serine 133, indicating the involvement of a cAMP-PKA-CREB-CRE loop in COX-2 transcriptional regulation. In contrast to phosphatidylinositol 3-kinase and protein kinase C, inhibition of FAK and Src effectively reduced KSHV infection-induced COX-2 transcription and protein levels. Collectively, our study indicates that mediation of COX-2 transcription upon KSHV infection is a paradigm of a complex regulatory milieu involving the interplay of multiple signal cascades and transcription factors. Intervention at each step of COX-2/PGE2 induction can be used as a potential therapeutic target to treat KSHV-associated neoplasm and control inflammatory sequels of KSHV infection.
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) is associated with a neoplastic angioproliferative endothelial malignancy and two other AIDS-related lymphoid cell malignancies called primary effusion lymphoma (PEL) and multicentric Castleman disease (64, 78). The KSHV life cycle displays distinct latent and lytic replication events (64, 78). Viral latency contributes to infected-cell survival and proliferation and latency maintenance, whereas the lytic cycle participates in the spread of infection and KS progression (64, 78). KSHV has been shown to utilize a variety of strategies not only to alter host cell metabolism via its signaling proteins but also to hijack cellular signaling pathways, transcription factors, and secreted metabolites to its own advantage, especially to remain latent in the host cell (19, 42, 43, 51, 55, 56).
In our previous studies (19, 55, 56), we reported that COX-2 functions as an important host factor maintaining KSHV latency and pathogenesis. The cyclooxygenases catalyzing the rate-limiting step in the synthesis of prostaglandins (PGs) are also referred to as PG endoperoxide synthases and are known to perform two enzymatic functions. As cyclooxygenases, they convert arachidonic acid to prostaglandin G2 (PGG2), and as peroxidases, they convert PGG2 to PGH2. Two forms of the enzyme, named COX-1 and COX-2, have been shown to be expressed in mammalian tissues. COX-1 is present in most tissues as a housekeeper enzyme, whereas COX-2, the product of an 8.2-kb gene containing 11 exons and 10 introns mapping to 1q25.2-q25.3, is the central enzyme in the PG biosynthetic pathway. The gene for COX-2 is considered an immediate-early gene and is stimulated and transcriptionally active during inflammation or pathophysiological process like carcinogenesis and thus plays an important role in the development of human tumors (34, 63). The COX-2/prostaglandin E2 (PGE2) connection with KSHV pathogenesis makes it an attractive chemotherapeutic target.
Levels of COX-2 are tightly controlled in most tissues, and its gene regulation is exclusively dependent on gene transcription and posttranscriptional events (20). The promoter regions of the human (22), mouse (15), rat (62), and chicken (76) COX-2 genes have been cloned, and their expression is tightly regulated at both the transcriptional and posttranscriptional levels. The COX-2 promoter contains a classical TATA box, an E box, and binding sites for transcription factors such as nuclear factor κB, nuclear factor interleukin-6 (IL-6)/CCAAT enhancer-binding protein, two nuclear factor of activated T cells (NFAT) binding sites (NFAT distal site [dNFAT] and NFAT proximal site [pNFAT]) (25, 26) and cyclic AMP (cAMP) response element (CRE)-binding proteins (25, 26). The dNFAT COX-2 site appears to be a pure NFAT site, as evidenced by the absence of any surrounding predicted AP-1 binding sequences and the lack of competition of an AP-1 consensus oligonucleotide for protein binding to this sequence. In contrast, pNFAT contains a highly homologous AP-1 site adjacent to the NFAT core GGAAA motif. Host cell signaling cascade induction has been shown to mediate the recruitment of specific transcription factors to these elements and thus trigger COX-2 activation in other systems (25, 26). However, the underlying mechanism of COX-2 induction upon KSHV infection of endothelial cells has never been reported to date. Therefore, in the present study, we investigated the role of KSHV-induced transcription factors and signaling pathways leading to COX-2 promoter activation, gene transcription, and PGE2 secretion.
MATERIALS AND METHODS
Cells.
Human microvascular dermal endothelial (HMVEC-d) cells from passages 5 to 7 (CC-2543; Lonza, Walkersville, MD), primary human foreskin fibroblast (HFF) cells (Lonza), and 293 cells were cultured as described before (55). Recombinant green fluorescent protein-KSHV (GFP-KSHV-γKSHV.152)-carrying BCBL-1 cells (72) were cultured, and GFP-KSHV was prepared and assessed for infectivity and mycoplasma and endotoxin contamination as described previously (55). Replication-defective virus (UV-inactivated KSHV) was inactivated with UV light (365 nm) for 20 min at a 10-cm distance (53, 54). KSHV DNA was extracted from live KSHV and UV-inactivated KSHV, and viral copy numbers were quantitated by real-time DNA PCR using primers amplifying the KSHV open reading frame 73 (ORF73) gene as described previously (53-56).
Reagents.
LY294002 [20(4-morphodinyl)-8-phenyl-1(4H)-benzopyran-4-one], heparin, sodium orthovanadate, benzamidine, leupeptin, aprotinin, SB216763 (potent and selective cell-permeating, ATP-competitive inhibitor of glycogen synthase kinase 3 [GSK3], a serine/threonine protein kinase), phorbol 12-myristate 13-acetate, and mouse anti-β-actin monoclonal antibody were from Sigma-Aldrich, St. Louis, MO. U0126 (1,4-diamino-2,3-dicyano-1, 4-bis [2-aminophenylthio] butadiene, MEK-1,2 inhibitor), U0124, rottlerin (protein kinase C-δ [PKC-δ]-specific inhibitor), GFX (GF 109203X, a cell-permeating protein kinase inhibitor inhibiting all isoforms of PKC by acting as a competitive inhibitor of the ATP binding site of PKC), COX-2-specific inhibitor NS-398 [N-(2-cyclohexyloxy-4-nitrophenyl)-methanesulfonamide], actinomycin D (RNA polymerase II inhibitor), ionomycin (100 nM; calcium ionophore), the NFAT antagonist cyclosporine A (CsA), PD980059 (2′-amino-3′-methoxyflavone; MEK-1 inhibitor), SB203580 (10 μM; specific inhibitor of the p38 mitogen-activated protein kinase [MAPK] pathway), JNK inhibitor II (SP600125; selective inhibitor of c-jun N-terminal kinase), and H-89 (selective inhibitor of protein kinase A [PKA]) were from Calbiochem, La Jolla, CA. Goat polyclonal anti-lamin B, anti-cAMP response element (CRE)-binding protein (CREB), anti-rabbit PKC, anti-rabbit extracellular signal-regulated kinase 2 (ERK2), and anti-NFAT antibodies were from Santa Cruz Biotechnology, Inc., Santa Cruz, CA. PGE2, PKA selective inhibitor H-89, and forskolin (adenylate cyclase agonist) were from Cayman Chemical, Ann Arbor, MI. Selective focal adhesion kinase (FAK) inhibitors (FAK inhibitor 14; 1,2,4,5-benzenetetramine tetrahydrochloride) and PF 573228 {3,4-dihydro-6-[[4-[[[3-(methylsulfonyl)phenyl]methyl]amino]-5-(trifluoromethyl)-2- pyrimidinyl]amino]-2(1H)-quinolinone} were obtained from Tocris Bioscience, Ellisville, MO. Anti-mouse COX-1 and COX-2 antibodies were from Cayman Chemicals. Anti-mouse phosphatidylinositol 3-kinase (PI3K) and p-Src antibodies were from BD Biosciences and Calbiochem. Anti-mouse p-ERK 1/2 and anti-rabbit Src, p-PKC, and p-PI3K antibodies were from Cell Signaling Technology, Inc. Anti-rabbit PGE2 antibody was from Abcam, Cambridge, MA.
Plasmids.
Plasmids containing the 5′ flanking region of human COX-1 and COX-2 promoter deletion and mutation constructs linked to luciferase reporter plasmids were obtained from M. Fresno (Madrid, Spain) and have been described previously (25, 26). Plasmids for the human COX-2 promoter with wild-type (WT) CRE (CRE-WT; −59TTCGTCA−53) and mutated CRE (CRE-Mut; TTGAGCT) were obtained from Karsten Schroer (Düsseldorf, Germany). These plasmids were constructed into a promoterless luciferase expression plasmid, pGL3-Basic (Promega). VIVIT, a plasmid containing the NFAT regulatory domain, was obtained from Addgene (2). Plasmids p-CMV-CREB, p-CMV-CREB 133, p-CMV-KCREB, p-CMV-IKβM, and p65 (p65 subunit of NF-κB) were purchased from BD Clontech, Mountain View, CA. p-CMV-CREB constitutively expresses human WT CREB. The p-CMV-CREB 133 vector expresses a mutant variant of human CREB that contains a serine-to-alanine mutation corresponding to amino acid 133 (S133A). This mutation blocks phosphorylation of CREB that blocks transcription. p-CMV-KCREB vector expresses a mutant variant of human CREB that contains mutations in its DNA-binding domain. KCREB acts as a dominant repressor by forming an inactive dimer with CREB, blocking its ability to bind the cAMP-regulated enhancer element CRE.
Cytotoxicity assay.
293, HFF, or HMVEC-d cells were incubated with Dulbecco modified Eagle medium containing different concentrations of various inhibitors for 4 h or 4 days. At different time points, supernatants were collected and assessed for cellular toxicity using a cytotoxicity assay kit (Promega, Madison, WI) as described previously (53, 54, 56).
Transient transfection.
For reporter gene assays, HFF and 293 cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) as described before (53). Low-passage (passages 5 to 7) HMVEC-d cells were transfected using Effectene reagent (Qiagen, Valencia, CA) in accordance with the manufacturer's instructions.
Signaling inhibitor treatment.
For luciferase assays, gene expression, PGE2 secretion, and COX-2/COX-1/β-actin protein measurements, cells were pretreated with either solvent controls or signaling inhibitors. Briefly, target cells grown to confluence in 25-cm2 flasks were serum starved for 10 h and then exposed to signaling inhibitors. MEK inhibitors PD980059, U0126, and U0124 (structural analogue of U0126), the general tyrosine kinase (Tyr-K) inhibitor genistein, FAK inhibitors (FAK inhibitor 14 and PF573228), p38 MAPK inhibitor SB203580, JNK inhibitor II (SP600125), Src kinase inhibitors PP2 and SU6656, phosphatidylinositol 3-kinase (PI-3K) inhibitors LY294002 and wortmannin, and GSK3 inhibitor SB216763 were used for 1 h at 37°C prior to KSHV infection (30 DNA copies/cell). Cells pretreated with the vehicle control (the dimethyl sulfoxide concentration was the same as that used for each specific inhibitor) for 1 h at 37°C prior to KSHV infection were used as a control.
Real-time RT-PCR.
Host (COX-1 and COX-2) and viral (ORF73 and ORF50) gene transcripts were detected by real-time reverse transcription (RT)-PCR using gene-specific real-time primers in accordance with the procedures described previously (56).
Immunofluorescence assay (IFA).
Confluent HMVEC-d cells in eight-well chamber slides (Nalge Nunc International, Naperville, IL) were either left uninfected or infected with 30 DNA copies of KSHV/cell at 37°C for 30 min, 1 h, or 2 h. For NFAT immunostaining, cells were fixed with 4% paraformaldehyde and permeabilized for 10 min with 0.1% Triton X-100. The cells were incubated for 2 h with anti-NFAT antibody (G1-D10; Santa Cruz Biotechnologies), washed extensively with phosphate-buffered saline, and incubated with a secondary antibody (Alexa; Molecular Probes, Eugene, OR) for 1 h at room temperature in the dark. Nuclei were visualized by using 4′,6-diamidino-2-phenylindole (DAPI; Ex358/Em461; Molecular Probes) as a counterstain. Stained cells were viewed with appropriate filters under a fluorescence microscope with the Nikon Metamorph digital imaging system. For immunofluorescence staining to demonstrate translocation of NFAT, HMVEC-d cells were grown and left untreated or pretreated with CsA (1.0 μM) before KSHV infection. ORF59 staining in KSHV-infected HMVEC-d cells was performed using ORF59 antibody generated in Bala Chandran's lab as described before (55).
ELISA for PGE2 detection.
Levels of PGE2 in the supernatants of cell signaling inhibitor-treated HMVEC-d cells either left uninfected or KSHV infected for 2 h were measured with a commercially available enzyme-linked immunoassay (ELISA) as described previously (55).
Luciferase reporter assays.
The effect of KSHV infection on COX-2 full-length promoter (P2-1900), deletion, or mutant constructs (25, 26) and the COX-1 promoter (P2-1009) was measured using a Dual-Luciferase kit according to the manufacturer's protocol (Promega). 293 cells were transfected using methods described before (56). The relative COX-2 promoter activity or number of relative luciferase units (RLU) was normalized to Renilla luciferase protein levels.
Preparation of nuclear extracts.
HMVEC-d or 293 cells were serum starved for 10 and 24 h, respectively, and then infected with 30 DNA copies of KSHV/cell for 10 min, 15 min, 30 min, 1 h, 2 h, or 4 h at 37°C, and nuclear extracts were prepared using a Nuclear Extract kit (Active Motif Corp., Carlsbad, CA) in accordance with the manufacturer's instructions. After measurement of protein concentrations with bicinchoninic acid protein assay reagent (Pierce Biotechnology, Rockford, IL), extracts were aliquoted and stored at −80°C until used. The purity of the nuclear extracts was assessed by immunoblotting using anti-lamin B antibodies, and cytoskeletal contamination was checked for by using an anti-β-actin antibody.
Transcription factor activation assay.
A total of 2 μg of each nuclear extract was used to measure NFAT, CREB, and phospho-CREB activities using ELISA-based TransAM-NFAT and TransAM-CREB kits (Active Motif Corp.) in accordance with the manufacturer's instructions. In this assay, transcription factors bind to the immobilized oligonucleotide containing the consensus sequences specific for the particular transcription factor, which is detected by a sandwich ELISA. A competition assay was done by premixing nuclear extracts for 30 min at 4°C with the WT and mutated (mut) consensus oligonucleotides provided in the kit before addition to the probe immobilized on the plate.
Electrophoretic mobility shift assay (EMSA).
Nuclear protein extracted from uninfected and KSHV-infected cells (HMVEC-d and 293) by the methods described before were stored at −80°C (53). Double-stranded DNA probes (CRE or dNFAT) were synthesized and end labeled with 50 mCi of [γ-32P]ATP using T4 polynucleotide kinase, and EMSA was performed as described previously (53). A competition EMSA was performed by adding a 100-fold molar excess of an unlabeled double-stranded oligonucleotide probe. In the case of a supershift, 1 μl of a 1-μg/μl concentration of the respective specific antibody (NFAT or CREB) was added to the binding reaction mixture before addition of the labeled probe.
Phospho-MAPK antibody arrays.
Lysates from HMVEC-d cells either left uninfected or infected with 30 DNA copies of KSHV/cell for different times or treated with exogenous PGE2 (1 μM) for 2 h were used for analysis of the phosphorylation state of all MAPKs using a human phospho-MAPK array kit, equivalent to immunoprecipitation and Western blot analysis (R&D Systems, Minneapolis, MN). Arrays were incubated overnight at 4°C with 100 μg of lysates. The arrays were washed three times and incubated for 2 h with the provided detection antibody cocktail containing phospho site-specific MAPK biotinylated antibodies. The wash steps were repeated, after which the arrays were exposed to chemiluminescent reagents and film. The data on the developed X-ray film were scanned and quantitated using image analysis software.
PKA activity assay.
PKA activity measurement in uninfected, KSHV-infected, inhibitor-pretreated, and then KSHV-infected HMVEC-d cells was performed using a PKA activity assay by Assay Designs in accordance with the manufacturer's instructions. This kinase assay is based on a solid-phase ELISA that utilizes a specific synthetic peptide as a substrate for PKA and a polyclonal antibody that recognizes the phosphorylated form of the substrate. Phospho-specific substrate antibody is added to the well and binds specifically to the phosphorylated peptide substrate. The phospho-specific antibody is subsequently bound by a peroxidase-conjugated secondary antibody, the assay is developed with tetramethylbenzidine (TMB) substrate, a color develops in proportion to the PKA phosphotransferase activity, and the intensity of the color is measured in a microplate reader at 450 nm.
Measurement of GSK3β activity by FACE GSK3β ELISA.
Phospho-GSK3β (p-GSK3β) and total GSK3β were measured with a FACE (fast activated cell-based) ELISA kit (Active Motif Corp.) according to the manufacturer's instructions. Briefly, HMVEC-d cells (90% confluence) serum starved for 6 to 8 h were left uninfected or infected with KSHV (30 DNA copies/cell) for various times, fixed in 4% formaldehyde, and assayed for GSK3β.
cAMP assay.
HMVEC-d cells left uninfected, KSHV infected, or exogenous PGE2 treated were lysed and incubated for 30 min at room temperature. Lysed cells were then scraped into microcentrifuge tubes and centrifuged at 1,000 × g for 10 min. The supernatants were collected and subjected to an enzyme immunoassay for cAMP according to Cayman Chemical recommendations.
RESULTS
KSHV de novo infection regulates COX-2 transcription and subsequently PGE2 secretion.
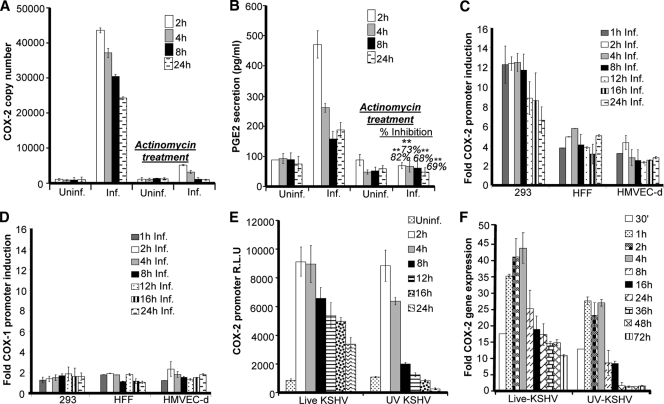
We previously showed by conventional real-time RT-PCR and ELISA that COX-2 mRNA levels and PGE2 secretion are upregulated upon KSHV infection (56). To address whether the levels of COX-2 and PGE2 early upon KSHV de novo infection are regulated at the transcriptional level, we pretreated HMVEC-d cells with the transcriptional inhibitor actinomycin D (1 μg/ml) for 4 h prior to KSHV infection. A noncytotoxic dose of actinomycin D was used for the experiments (see Fig. S1A, a, in the supplemental material). As depicted in Fig. 1A and B, an increase in COX-2 mRNA levels and PGE2 secretion was detected as early as 2 h postinfection (p.i.) and was efficiently maintained until 24 h p.i. Pretreatment of HMVEC-d cells with actinomycin D drastically inhibited the effect of KSHV infection on both COX-2 mRNA levels and PGE2 secretion (Fig. 1A and B). Total RNA polymerase II-dependent transcriptional arrest completely abrogating COX-2 levels clearly indicated that COX-2 gene expression during early times of infection is regulated at the transcriptional level. COX-2 protein levels were also checked by Western blotting in serum-starved, uninfected, and KSHV-infected cells (see Fig. S1C, a and b). KSHV infection of HMVEC-d and 293 cells was confirmed by real-time-PCR for ORF50, ORF73 (see Fig. S1B, a and b), and ORF59 staining by IFA (see Fig. S1B, c). Serum starvation was not cytotoxic (see Fig. S1A, b) and did not induce COX-2 (see Fig. S1C, b). In contrast, COX-2 protein levels were induced upon KSHV infection (see Fig. S1C, a).
FIG. 1.
Effects of KSHV gene expression and PGE2 supplementation on COX-2 promoter activity and gene expression. (A) HMVEC-d cells pretreated with 1 μg/ml actinomycin D for 4 h were infected with KSHV for the indicated times. These cells were washed and lysed, and total RNA was prepared. COX-2 copy numbers were quantitated by real-time RT-PCR as described before (55, 56). (B) PGE2 levels were measured (55, 56) in the supernatants collected from cells treated as described for panel A. Percent inhibition of PGE2 secretion was calculated by taking the PGE2 secretion of cells not pretreated with actinomycin D at the respective points as 100%. Each reaction was done in triplicate, and each point represents the average ± the standard deviation (SD) of three independent experiments. **, statistically significant at P < 0.005. (C and D) Effect of KSHV gene expression on COX-2 and COX-1 promoter activities. The indicated cell types were transfected with control pXP-2, COX-2 promoter (P2-1900), and COX-1 promoter (P1-1009)-luciferase constructs. After 24 h, cells were serum starved, infected (Inf.) for different times, lysed, and assayed for luciferase activity as described previously. Data are expressed as mean numbers of RLU after normalization with the cotransfected Renilla luciferase activity. Each reaction was done in triplicate, and each point represents the average ± SD of three independent experiments. The activation in uninfected (Uninf.) cells at the respective time points was taken as 1-fold for comparison. (E and F) COX-2 promoter activity (E) and gene expression (F) upon UV-inactivated KSHV (30 DNA copies/cell) or live KSHV (30 DNA copies/cell) infection. HMVEC-d cells grown to 80% confluence were serum starved for 10 h and infected with 30 DNA copies of either live or UV-inactivated KSHV per cell for different times. COX-2 promoter activity (as described for panel C) and gene expression (56) were measured in these cells.
COX-2 promoter activity is induced in all of the cell types tested.
We have previously shown that COX-2 gene expression is induced in HFF and HMVEC-d cells upon KSHV infection. Therefore, to further explore the transcriptional regulation of the COX-2 gene, we studied COX-2 promoter activation in transiently transfected 293, HFF, and HMVEC-d cells. Compared to the promoterless vector (pXP-2), the COX-2 full-length promoter construct containing the −1796 to +104 region of the human COX-2 gene (P2-1900) was significantly activated during KSHV infection in all of the cell types tested at the indicated time points compared to the level in the respective uninfected cells (Fig. 1C), which was taken as 1-fold. We did not observe induction of control pXP-2 vector luciferase activity upon KSHV infection (data not shown). Compared with that in 293 cells, COX-2 promoter activity induction was lower in HFF and HMVEC-d cells, which might be due to low transfection efficiency in HFF (50%) and HMVEC-d (20 to 30%) cells than in 293 cells (90%). No induction in the luciferase activity of the COX-1 promoter construct (P1-1009) was observed upon KSHV infection (Fig. 1D) in the cell types tested.
KSHV gene expression is required for sustained levels of COX-2.
To determine whether COX-2 promoter induction requires a KSHV-induced signal cascade and/or viral gene expression, we examined COX-2 promoter activity in HMVEC-d cells transfected with the COX-2 promoter and then infected with 30 DNA copies of KSHV/cell (either live or UV inactivated and replication incompetent) for the indicated times (Fig. 1E). UV-inactivated KSHV binds and enters HMVEC-d and HFF cells as efficiently as live KSHV (56). As reported before (56), UV-inactivated KSHV did not induce any KSHV latent (ORF73) or lytic (ORF50, K8, and K5) gene expression and was replication incompetent (data not shown). Compared to live KSHV, although UV-inactivated KSHV could also induce COX-2 promoter activity (Fig. 1E) and gene expression (Fig. 1F), this induction was low and was not sustainable for longer times (8 to 24 h) p.i. Our results clearly indicate that viral gene expression is indeed necessary for sustained COX-2 levels in infected cells, but KSHV binding and entry events are good enough to upregulate appreciable levels of COX-2 promoter activation and gene expression. Seventeen-fold COX-2 gene expression was observed even at 30 min p.i. Our previous studies have shown (53) that UV-inactivated KSHV induces the p42/p44 ERK1/2 MAPK pathway, which in turn regulates the activation of multiple transcription factors (Jun, c-Fos, ATF-2, STAT-1α, and c-Myc) that could potentially regulate COX-2 transcription.
These results suggested that during early times of KSHV infection, virus binding- and entry-initiated signal transduction cascades might be contributing to COX-2 promoter induction via the activation of various transcription factors. COX-2 expression at the later time points might be due to the cumulative effect of viral gene expression, host cell gene products, autocrine/paracrine signaling by COX-2 metabolite PGE2, and the presence of various growth factors and cytokines in the infected-cell microenvironment.
PGE2 autoregulates COX-2 promoter activation and gene expression.
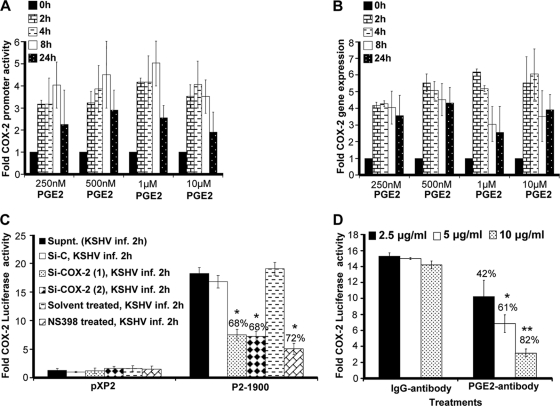
Since KSHV infection-induced PGE2 can initiate multiple signaling cascades (JNK-1, ERK1/2, PKC, PI3K-AKT, HPK1, and Src), second messengers, including cAMP, calcium (Ca2+), and reactive oxygen species, in an autocrine/paracrine manner by binding to its specific G-protein-coupled receptors (EP receptors) (3, 4, 6, 16, 18, 27, 32, 33, 35, 37, 38, 46, 52, 73, 77), we tested the role of exogenous PGE2 in COX-2 promoter and COX-2 gene expression (Fig. 2A and B). Use of biologically relevant concentrations (250 nM, 500 nM, and 1 μM) of exogenous PGE2 could induce COX-2 promoter activation, as well as COX-2 gene expression (Fig. 2A and B). PGE2 concentrations as low as 250 nM could effectively induce COX-2 promoter induction, suggesting that low levels of PGE2 (biologically relevant) in the infected-cell microenvironment are sufficient to stimulate the COX-2-PGE2-COX-2 loop to sustain COX-2. At any given time, PGE2-induced COX-2 promoter activity and COX-2 gene expression levels (Fig. 2A and B) were lower than the levels observed during KSHV de novo infection (Fig. 1E and F). PGE2 supplementation for a similar duration could not change COX-1 promoter activity (data not shown). These results suggest that COX-2 induction upon KSHV infection is the collective outcome of multiple events initiated and maintained by a variety of secreted factors like vascular endothelial growth factor (VEGF), HIF-1α, and IL-1β which are known to induce COX-2 (29, 74). In other words, PGE2 is not the only secreted factor responsible for COX-2 levels in KSHV-infected cells.
FIG. 2.
Effect of exogenous PGE2 supplementation on COX-2 promoter activity and gene expression. (A) 293 cells transfected with a pXP2 (promoterless construct) or a COX-2 promoter (P2-1900) construct were supplemented with various concentrations of PGE2 in their basal growth medium (no serum) for different times as indicated. Promoter activity was measured using luciferase activity in the lysates prepared from unstimulated and PGE2-supplemented cells as described in Materials and Methods. Each reaction was done in duplicate, and each bar represents the average ± SD of three independent experiments. COX-2 promoter activity at 0 h after PGE2 supplementation was taken as 1-fold for comparison. (B) Serum-starved HMVEC-d cells were treated with various doses of exogenous PGE2 for the indicated times, RNA was prepared, and a real-time RT-PCR for COX-2 was performed as described before (55). Each reaction was done in duplicate, and each bar represents the average ± SD of three independent experiments. COX-2 gene expression at 0 h after PGE2 supplementation was taken as 1-fold for comparison. (C and D) Effect of PGE2 in KSHV-infected culture supernatant (Supnt,) on COX-2 promoter activity. Supernatants obtained from HMVEC-d cells treated with COX-2 inhibitor or silenced for COX-2 prior to KSHV infection were used to assess COX-2 promoter activation. 293 cells were transfected with control PXP-2 and COX-2 (P2-1900)-luciferase constructs. After 24 h, cells were serum starved (10 h) and incubated with various supernatants obtained from uninfected or infected cells (as described above). After 2 h of incubation, cells were lysed and assayed for luciferase activity as described previously. The activation in supernatants from uninfected cell was taken as 1-fold. Percent inhibition was calculated by taking as 100% the levels of COX-2 promoter activity in the presence of supernatant obtained from either Si control (Si-C)-treated and KSHV-infected or solvent-treated and KSHV-infected cells. (D) Supernatant (1 ml) from serum-starved (10 h), KSHV-infected HMVEC-d cells (2 h) was incubated at 37°C for 1 h with the indicated concentrations of PGE2 neutralizing antibody or rabbit IgG antibody and used for the COX-2 promoter assay as described for panel C. Percent inhibition was calculated by taking levels of COX-2 promoter activity in the presence of supernatant pretreated with IgG antibody as 100%.
Our previous studies demonstrated that PGE2 production could be diminished upon the treatment of HMVEC-d cells with COX-2 chemical inhibitors prior to KSHV infection or cells silenced for COX-2 and then KSHV infected (55). To evaluate the role of PGE2 present in the KSHV-infected culture supernatant in COX-2 transcriptional regulation, we used supernatants from HMVEC-d cells KSHV infected (solvent control pretreated, COX-2 inhibitor pretreated, and COX-2 silenced) for 2 h (Fig. 2C). Effective reduction (Fig. 2C) was observed in the presence of supernatants obtained from cells pretreated with COX-2 inhibitors (72%) or silenced for COX-2 (68%). Though inhibition of COX-2 primarily affects PGE2 levels, the possibility that it affects the secretion of other factors like VEGF and IL-1β cannot be ruled out. As VEGF and IL-1β have also been shown to modulate COX-2 transcription in other systems, we extended these studies to evaluate the exclusive contribution of PGE2 to COX-2 promoter activation.
To understand the role of PGE2 in regulating KSHV infection-mediated COX-2 promoter transcriptional regulation, we performed neutralization experiments using various concentrations of anti-PGE2 antibody (Fig. 2D). Incubation of KSHV-infected culture supernatant with increasing concentrations (2.5, 5, and 10 μg/ml) of anti-PGE2 neutralizing antibody could effectively reduce COX-2 promoter induction in a dose-dependent fashion (42 to 82%) (Fig. 2D). Therefore, data in Fig. 2 suggest that PGE2 is one of the key components of COX-2 promoter autoregulation and contributes to the continuous loop of COX-2 maintenance in KSHV-infected cells.
KSHV entry-associated signaling molecules regulate COX-2 promoter activity, gene expression, protein levels, and PGE2 secretion.
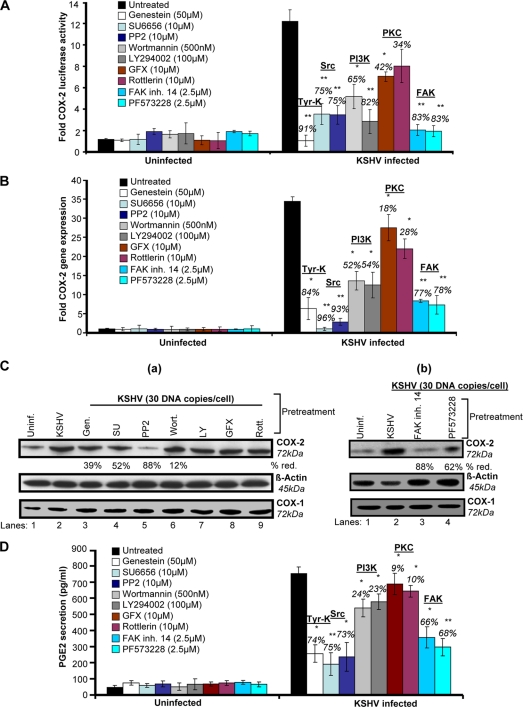
To define the role of signaling molecules induced early during KSHV infection (virus binding and entry) in COX-2 regulation, cells transfected with COX-2 full-length promoter and control Renilla luciferase plasmids were pretreated with noncytotoxic doses of signal cascade inhibitors (see Fig. S2 in the supplemental material) and then infected with KSHV and measured for their luciferase activity (Fig. 3A). Similarly treated cells were used for COX-2 gene expression (Fig. 3B), and their supernatants were analyzed for PGE2 secretion (Fig. 3D). The general Tyr-K inhibitor genistein strongly inhibited COX-2 promoter activity (91%) (Fig. 3A), COX-2 gene expression (84%) (Fig. 3B), and PGE2 secretion (74%) (Fig. 3D). The Src kinase inhibitors PP2 and SU6656 also reduced COX-2 promoter activity (75%) (Fig. 3A), COX-2 gene expression (96 and 93%) (Fig. 3B), and PGE2 secretion (75 and 73%) (Fig. 3D), while the PI3K inhibitors wortmannin and LY294002 inhibited COX-2 promoter activity (65 and 82%) (Fig. 3A), COX-2 gene expression (52 and 54%) (Fig. 3B), and PGE2 secretion (24 and 23%) (Fig. 3D). Pretreatment with the PKC inhibitors GFX and rottlerin could only reduce COX-2 promoter activity by 42 and 34%, respectively (Fig. 3A), COX-2 gene expression by 18 and 28% (Fig. 3B), and PGE2 secretion by only 9 and 10% (Fig. 3D). FAK inhibitors like FAK inhibitor 14 and PF573228 effectively reduced COX-2 promoter activity (83%) (Fig. 3A), COX-2 gene expression (77 and 78%) (Fig. 3B), and PGE2 secretion (66 and 68%) (Fig. 3D). All of these results suggest that the signal molecules, especially FAK and Src, induced upon virus binding and entry are critical for COX-2 promoter activity, gene expression, and PGE2 secretion. Similar to COX-2 gene expression data, we found changes in the levels of COX-2 protein (Fig. 3C, a and b). In contrast to PI3K (wortmannin and LY294002) and PKC (GFX and rottlerin) inhibitors, pretreatment with general Tyr-K (genistein), FAK (FAK inhibitor 14 and PF573228), and Src (SU6656 and PP2) inhibitors effectively inhibited KSHV infection-induced COX-2 protein levels (Fig. 3C).
FIG. 3.
Role of signaling molecules induced early during KSHV infection in COX-2 transcriptional regulation/gene expression, protein level, and PGE2 secretion. (A) 293 cells were transfected with COX-2 full-length promoter and control Renilla luciferase plasmids. Transfected cells were serum starved and then treated with nontoxic concentrations of various signaling inhibitors (inh.), as indicated, for 1 h at 37°C and then infected with 30 DNA copies/cell KSHV for 2 h. Cells were lysed, and luciferase activity was measured. Data represent mean numbers of RLU after normalization with the cotransfected Renilla luciferase activity. Each reaction was done in triplicate, and each point represents the average ± SD of five experiments. The activation in uninfected cells was taken as 1-fold for comparison. Percent inhibition was calculated by taking the COX-2 promoter activity in non-inhibitor-treated, KSHV-infected (2 h) cells as 100%. (B) HMVEC-d cells were treated with nontoxic concentrations of various signaling inhibitors for 1 h at 37°C and then infected with KSHV (30 DNA copies/cell) for 2 h. Total RNA was prepared from these cells, and COX-2 gene expression was quantitated as described before. The COX-2 level in uninfected cells was taken as 1-fold for comparison. Percent inhibition was calculated by taking the COX-2 gene expression in non-inhibitor-treated, KSHV-infected (2 h) cells as 100%. (C, a and b). HMVEC-d cells were treated as described for panel B. Total protein was prepared from these cells, and COX-2/COX-1 protein levels were quantitated by Western blotting. Percent reduction (red.) was calculated by taking the COX-2 protein level in non-inhibitor-treated, KSHV-infected (2 h) cells as 100%. β-Actin levels were checked as a loading control. (D) Supernatants collected from the HMVEC-d cells treated as described for panel B were used for PGE2 measurement by ELISA as described before. Data are expressed as mean pg/ml. Each reaction was done in triplicate, and each point represents the average ± SD of five experiments. Percent inhibition was calculated by taking PGE2 secretion from non-inhibitor-treated, KSHV-infected (2 h) cells as 100%. * and **, statistically significant at P < 0.01 and P < 0.005, respectively.
KSHV infection-induced multiple MAPKs regulate COX-2 promoter activity, gene expression, protein levels, and PGE2 secretion.
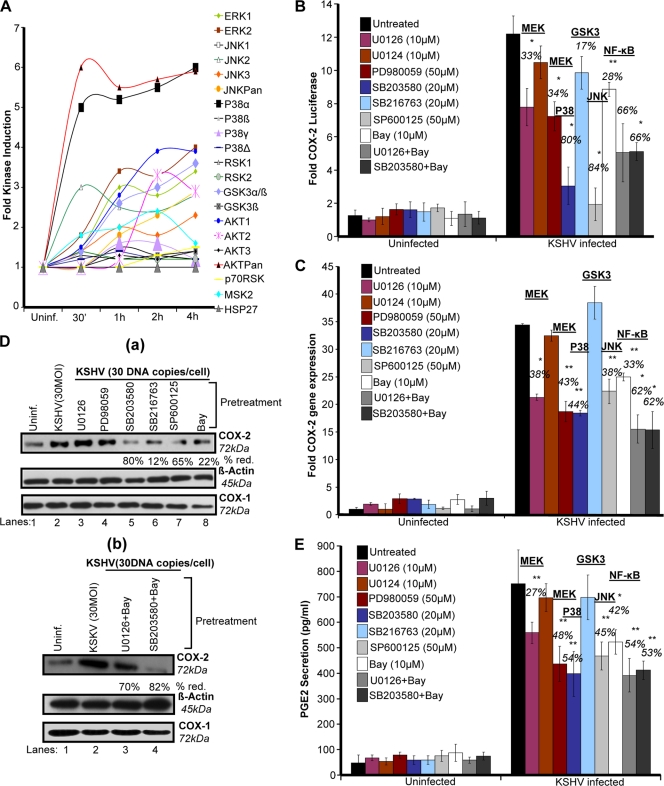
Many receptor Tyr-Ks (RTKs) and MAPKs have been shown to regulate COX-2 promoter activity in other systems (9, 66). In our previous studies, we have shown that virus binding and entry are associated with activation of kinases like FAK, Src, P13K, and MAPKs (42, 51, 53, 54, 70, 71). Therefore, we assessed the majority of MAPKs (Fig. 4A). Phospho-MAPK arrays were used to assess the role of these MAPK phosphorylations at various serine/threonine positions in the context of COX-2 promoter activity in uninfected and KSHV-infected (30 DNA copies/cell) HMVEC-d cell lysates. Of the MAPKs tested (Fig. 4A), KSHV infection induced phosphorylation of ERK1 and -2; JNK2, -3, and -Pan; GSK3α/β and -3α; AKT1 and -2; and MSK2 by ∼3- to 4-fold at 2 h and this induction was time dependent (Fig. 4A). Five- to 6-fold induction of the levels of AKTPan and p38α was observed (Fig. 4A), while very low (1.5- to 2-fold) induction of JNK1; p38β, -γ, and -δ; and RSK1, -2 and -p70 levels was observed (Fig. 4A). These results suggest that MAPKs are induced beginning early in KSHV infection. Similarly, stimulation of HMVEC-d cells by supplementation with 1 μΜ PGE2 also induced phosphorylation of MAPKs (data not shown). We hypothesized that these MAPKs induced early during KSHV infection or PGE2 stimulation might be playing a critical role in the activation of downstream transcription factors and thus controlling COX-2 promoter activity, gene expression, and eventually PGE2 secretion.
FIG. 4.
Role of KSHV infection-induced MAPKs in COX-2 transcriptional regulation/gene expression and PGE2 secretion. (A) Densitometric analysis of MAPK array blots measuring the activation of MAPKs. The values were normalized to identical background levels using the R&D human MAPK antibody array analysis tool. The fold induction of MAPK was calculated by dividing the respective values obtained from KSHV-infected HMVEC-d cell lysates by the values obtained from uninfected-cell lysates. (B) 293 cells were transfected with COX-2 full-length promoter and control Renilla luciferase plasmids. Transfected cells were serum starved and then treated with nontoxic concentrations of various signaling inhibitors as indicated for 1 h at 37°C and then infected with 30 DNA copies/cell KSHV for 2 h. Cells were lysed, and luciferase activity was measured. Data represent mean numbers of RLU after normalization with the cotransfected Renilla luciferase activity. Each reaction was done in triplicate, and each point represents the average ± SD of five experiments. The activation in untreated infected cells was taken as 1-fold for comparison. Percent inhibition was calculated by taking the COX-2 promoter activity in non-inhibitor-treated, KSHV-infected (2 h) cells as 100%. (C) HMVEC-d cells were treated with nontoxic concentrations of various MAPK inhibitors for 1 h at 37°C and then infected with KSHV (30 DNA copies/cell) for 2 h. Total RNA was prepared from these cells, and COX-2 gene expression was quantitated as described before. The COX-2 level in uninfected cells was taken as 1-fold for comparison. Percent inhibition was calculated by taking the COX-2 promoter activity in non-inhibitor-treated, KSHV-infected (2 h) cells as 100%. (D, a and b). HMVEC-d cells were treated as described for panel C. Total protein was prepared from these cells, and COX-2/COX-1 protein levels were quantitated by Western blotting. Percent inhibition was calculated by taking the COX-2 protein level in non-inhibitor-treated, KSHV-infected (2 h) cells as 100%. β-Actin levels were checked as a loading control. (E) Supernatants collected from HMVEC-d cells treated as described for panel C were used for PGE2 measurement by ELISA as described before. Data are expressed as mean pg/ml. Each reaction was done in triplicate, and each point represents the average ± SD of five experiments. Percent inhibition was calculated by taking PGE2 secretion from non-inhibitor-treated, KSHV-infected (2 h) cells as 100%. * and **, statistically significant at P < 0.01 and P < 0.005, respectively.
KSHV infection-induced MAPK pathways regulate COX-2 gene transcription, protein levels, and PGE2 secretion.
To define the role of MAPKs induced early during infection in COX-2 regulation, cells transfected with the COX-2 full-length promoter and control Renilla luciferase plasmids were pretreated with signal cascade inhibitors and then infected with KSHV and their luciferase activity was determined (Fig. 4B). Similarly treated cells were used for COX-2 gene expression (Fig. 4C), and their supernatants were assayed for PGE2 (Fig. 4E). MEK inhibitors U0126 and PD980059 inhibited COX-2 promoter activity (33 and 34%) (Fig. 4B), COX-2 gene expression (38 and 43%) (Fig. 4C), and PGE2 secretion (27 and 48%) (Fig. 4E). p38 inhibitor SB203580 repressed COX-2 promoter activity (80%) (Fig. 4B), COX-2 gene expression (44%) (Fig. 4C), and PGE2 secretion (54%) (Fig. 4E), while JNK inhibitor SP-600125 diminished COX-2 promoter activity (84%) (Fig. 4B), COX-2 gene expression (38%) (Fig. 4C), and PGE2 secretion (45%) (Fig. 4E). NF-κB inhibitor Bay11-7082 decreased COX-2 promoter activity (28%) (Fig. 4B), COX-2 gene expression (33%) (Fig. 4C), and PGE2 secretion (42%) (Fig. 4E). The ERK and NF-κB or p38 and NF-κB combination of inhibitors inhibited COX-2 promoter activity (66%) (Fig. 4B), COX-2 gene expression (62%) (Fig. 4C), and PGE2 secretion (55%) (Fig. 4E).
Of the MAPKs tested, JNK and p38 appeared to control COX-2 promoter activity. The ERK or NF-κB inhibitor alone did not noticeably inhibit COX-2 promoter activity, gene expression, protein levels, or PGE2 secretion (Fig. 4B to E) compared to the NF-κB or p38 and NF-κB combination of inhibitors. Since we observed a 3- to 4-fold induction of the phosphorylation of GSK3 isoforms (α, 51 kDa; β, 47 kDa) (Fig. 4A), we used a GSK3 inhibitor. GSK3 inhibitor SB216763 pretreatment could not inhibit COX-2 promoter activity (Fig. 4B) like the other MAPK inhibitors; rather, we observed slightly increased COX-2 gene expression (Fig. 4C). GSK3 inhibitor SB216763 pretreatment downregulated COX-2 protein levels by 12% (Fig. 4D, a).
Since early binding and entry events of KSHV infection effectively modulated COX-2 transcription and PGE2 secretion, we hypothesized that these signal cascades are critical for the activation of various transcription factors involved in COX-2 promoter regulation. Hence, we next studied the mechanism of KSHV infection-induced COX-2 transcription utilizing various COX-2 promoter mutations and deletion constructs.
cis-acting element dNFAT regulates KSHV-induced human COX-2 promoter activity.
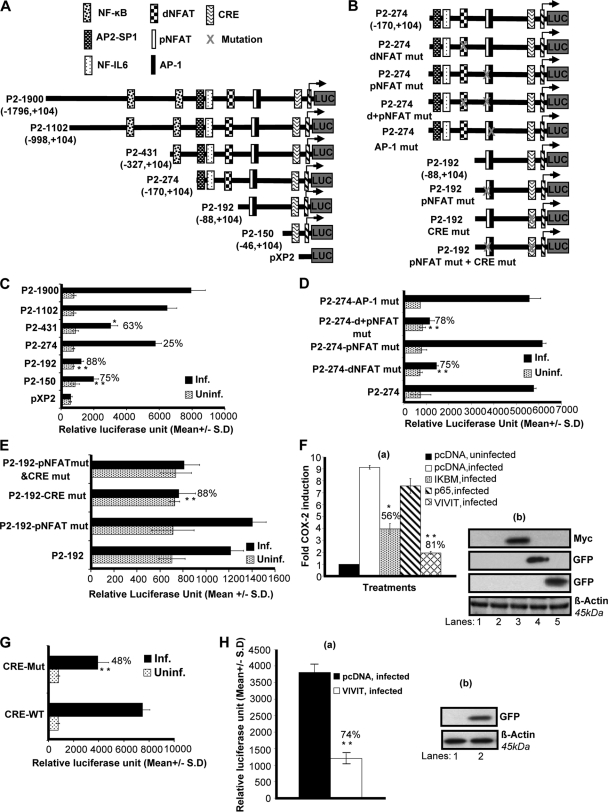
It is well known that COX-2 promoter activity is regulated in other systems by several transcription factors, including NF-κB, nuclear factor IL-6, AP-1, CRE, and NFAT (25, 26). To understand the roles of various transcription factors and their involvement in COX-2 upregulation upon KSHV infection, we studied the role of KSHV de novo infection in cells transiently transfected with deletion (Fig. 5A) and mutation (Fig. 5B) constructs of the human COX-2 promoter. Details of different COX-2 promoter luciferase constructs have been described previously (25, 26), and their maps are given in Fig. 5A and B. The COX-2 promoter has two NFAT cis-acting elements, named COX-2-dNFAT and -pNFAT, which are highly conserved among different animal species (25, 26).
FIG. 5.
Role of cis-acting factors in transcriptional regulation of the COX-2 promoter by KSHV. (A and B) Schematics of COX-2 promoter deletion (A) and mutation (B) constructs. cis-acting consensus sequences are denoted by boxes. The extents of 5′ deletions are shown, with the numbers indicating their lengths relative to the transcription start site. Transcription factor binding sites are represented on the promoter, while the promoter regions relative to the transcription initiation start site are shown in parentheses. (C) 293 cells were transfected with control pXP2 or various COX-2 promoter deletion constructs (P2-1900-luc, P2-1102-luc, P2-431-luc, P2-274-luc, P2-192-luc, and P2-150-luc). At 24 h after transfection, cells were serum starved and then left uninfected or infected with 30 DNA copies of KSHV/cell for 2 h. These cells were harvested, lysed, and assayed for RLU as described before. (D) 293 cells were transfected with P2-274 or P2-274 promoter mutant constructs (P2-274 dNFAT mut, P2-274 pNFAT mut, P2-274 d+pNFAT mut, and P2-274-AP1 mut). After 24 h, cells were serum starved for 10 h and infected for 2 h and a promoter assay was performed as described for panel C. (E) 293 cells were transfected with P2-192 promoter mutant constructs (P2-192 pNFAT mut, P2-192 CRE mut, and P2-192 pNFAT mut+CRE mut). After 24 h, cells were serum starved for 10 h and infected for 2 h and a promoter assay was performed as described for panel C. Data in panels C, D, and E represent the mean number of RLU after normalization with the cotransfected Renilla luciferase activity. Each reaction was done in triplicate, and each point represents the average ± SD of three experiments. (F, a) 293 cells were cotransfected with a COX-2 promoter full-length construct (P2-1900) along with 1 μg of pcDNA, IκβM, p65, or VIVIT expression plasmid and left uninfected or KSHV infected for 2 h. Promoter induction in uninfected cells was taken as 1-fold. Percent inhibition of COX-2 induction was calculated with reference to fold induction in pcDNA-transfected and KSHV-infected (2 h) cells as 100%. * and **, statistically significant at P < 0.01 and P < 0.005, respectively. (F, b) Expression of IκβM, p65, or VIVIT at protein level. Lysates prepared from IκβM-, p65-, or VIVIT-transfected cells were prepared and Western blotted using Myc or GFP antibodies. (G) 293 cells were transfected with CRE-WT and CRE-Mut promoter constructs. After 24 h, cells were serum starved for 10 h and left uninfected or infected for 2 h and a promoter assay was performed as described for panel C. Percent inhibition of COX-2 induction was calculated with reference to induction in CRE-WT plasmid-transfected and then infected cells, which was taken as 100%. (H, a) 293 cells were cotransfected with a COX-2 promoter construct with a CRE-Mut construct along with 1 μg of a pcDNA or VIVIT expression plasmid. After 24 h, cells were serum starved for 10 h and then infected for 2 h and a promoter assay was performed as described for panel C. Percent inhibition of COX-2 promoter induction was calculated with reference to fold induction in pcDNA-transfected and KSHV-infected (2 h) cells, which was taken as 100%. * and **, statistically significant at P < 0.01 and P < 0.005, respectively. (H, b) Expression of VIVIT at protein level. Lysates prepared from pcDNA- or VIVIT-transfected cells were prepared and Western blotted using GFP antibody.
KSHV infection induced the activity of the P2-1900 and P2-1102 promoter luciferase constructs similarly (Fig. 5C). Promoter analysis results indicated that promoter elements between −88 and −170 are essential for KSHV-induced COX-2 promoter activity, as an 88% reduction in promoter activity was observed upon the deletion of this region (Fig. 5C). These results suggest that dNFAT/NF-IL-6/AP-2-SP1 is likely to be responsible for promoter activation following KSHV infection. The possibility of NF-κB involvement cannot be ruled out either, as we observed a 63% reduction in promoter activity of the construct with one κB site removed (P2-431), whereas the loss of both of the sites binding NF-κB (P2-274) relieved this inhibition (25%), suggesting the involvement of NF-κB in the regulation of COX-2 promoter activity upon KSHV infection (Fig. 5C). The P2-150 construct containing CRE and the TATA box was appreciably activated by KSHV infection compared to P2-192, suggesting a potential role for CRE in COX-2 promoter activity (Fig. 5C). These results also suggested that pNFAT and AP-1 in P2-192 might be negatively regulating COX-2 promoter activity (Fig. 5C).
To investigate the role of both NFAT sites in detail, we used P2-274-AP-1 mut, P2-274 dNFAT mut, P2-274 pNFAT mut, and P2-274 d+pNFAT mut constructs to transfect 293 cells. The transfected cells were serum starved and then infected with KSHV for 2 h. Mutation of dNFAT resulted in a 75% reduction of COX-2 promoter activity with respect to P2-274, suggesting the involvement of dNFAT in the regulation of COX-2 (Fig. 5D). Mutation of pNFAT did not result in the downregulation of COX-2, ruling out the possibility of pNFAT involvement in COX-2 transcriptional regulation and verifying the results depicted in Fig. 5C. Mutation of both the proximal and distal NFAT sites resulted in a 78% reduction (Fig. 5D). Since the proximal NFAT site is not purely NFAT but is always bound to AP-1, we used P2-274-AP-1 mut to study COX-2 promoter activity. AP-1 mut did not affect COX-2 promoter activation further, verifying the results shown in Fig. 5C. Collectively, the data shown in Fig. 5D strongly suggest the involvement of dNFAT in KSHV-induced COX-2 promoter activity. P2-274-AP-1 mut did not show any reduction in luciferase activity, ruling out the possible involvement of AP-1 in COX-2 transcriptional regulation (Fig. 5D).
CRE recognition sequences in the human COX-2 promoter are important for KSHV infection-induced transcriptional activity.
To investigate the role of CRE, we used P2-192 pNFAT mut, P2-192 CRE mut, and P2-192 pNFAT mut + CRE mut constructs to transfect 293 cells. The transfected cells were serum starved and then infected with KSHV for 2 h. Mutated pNFAT's failure to inhibit COX-2 promoter activity further ruled out the involvement of pNFAT in KSHV-induced COX-2 (Fig. 5E). Mutation of CRE resulted in an 88% reduction in COX-2 promoter activity compared to that obtained with the P2-192 construct (Fig. 5E). The promoter luciferase activity of the P2-192 pNFAT mut + CRE mut construct was similar to that of P2-192 CRE mut, suggesting a role for CRE but not pNFAT in COX-2 promoter induction (Fig. 5E). Collectively, COX-2 promoter deletion and mutation analyses showed that dNFAT and CRE recognition sequences present in the human COX-2 promoter are important for transcriptional activation upon KSHV infection.
Calcineurin (Cn)/NFAT pathway regulates KSHV-induced COX-2 promoter activity.
To further investigate the involvement of NF-κB and NFAT in COX-2 promoter induction, we measured COX-2 full-length promoter activity in the presence of p65, IκBM, and VIVIT expression plasmids. As NFAT activity is controlled by Cn phosphatase, we cotransfected 293 cells with COX-2 promoter construct P2-1900 and a plasmid expressing the peptide VIVIT, which competes with NFAT for Cn binding and selectively inhibits NFAT activation without disrupting other Cn-dependent pathways (Fig. 5F). COX-2 luciferase reporter activity was dramatically lowered (81%) in the presence of a plasmid expressing the peptide VIVIT, suggesting a role for Cn binding in KSHV-induced NFAT activity (Fig. 5F). We also assessed the effect of NF-κB by using the plasmid for IκBM (dominant negative mutant IκB), which blocks NF-κB activation (Fig. 5F). Cotransfection with IκBM inhibited COX-2 promoter activation by 56%, whereas the p65 plasmid did not change COX-2 promoter activity appreciably (Fig. 5F, a). Expression of IκBM, p65, and VIVIT plasmids was checked by Western blotting (Fig. 5F, b) using antibodies to the respective tags (Myc and GFP). p65 and VIVIT expression in the transfected cells was also checked by observing GFP expression under a microscope. These results supported the data in Fig. 5C suggesting that, along with NF-κB, there are other factors regulating KSHV infection-induced COX-2 transcription, of which Cn/NFAT appeared to be especially critical.
CRE and NFAT play obligatory roles in KSHV infection-induced COX-2 promoter activity.
To further verify the involvement of CRE and NFAT in COX-2 promoter induction, we measured COX-2 promoter activity using COX-2 CRE-WT and COX-2 CRE-Mut constructs in uninfected and KSHV-infected (2 h) cells. KSHV infection induced COX-2 CRE-WT activation compared to the CRE-Mut construct (Fig. 5G). A 48% reduction was observed in COX-2 CRE-Mut promoter activity upon KSHV infection compared to COX-2-CRE-WT (Fig. 5G).
To evaluate the combined effect of CRE mutation and NFAT blocking on COX-2 promoter activity, a cotransfection experiment was performed using the pCDNA or VIVIT expression plasmid along with the CRE-Mut construct. Cotransfection with VIVIT further reduced COX-2 CRE-Mut activity by 74%, suggesting critical roles for CRE and NFAT in KSHV infection-induced COX-2 transcriptional regulation (Fig. 5H, a). Expression of VIVIT was confirmed by Western blotting (Fig. 5H, b).
KSHV infection induces NFAT.
NFAT belongs to the family of transcription factors which comprises four classical members: NFAT1, NFAT2, NFAT3, and NFAT4. All of the members of the NFAT family are Ca2+ responsive and are regulated by the Ca2+/Cn signaling pathway (7, 13, 23, 28, 36, 45, 47). NFAT activation is controlled via nuclear import and export involving dephosphorylation by Cn phosphatase, optimal DNA binding, and regulation of the transactivating activity. In resting cells, NFATs are phosphorylated at a cluster of serine residues located in the regulatory domain which effectively mask the nuclear localization signal and thereby retain NFAT in an inactive conformation in the cytoplasm (44, 47). Upon stimulation with agonists which increase intracellular Ca2+, NFATs are dephosphorylated by the phosphatase Cn and translocate to the nucleus. In the nucleus, NFATs interact with fos and Jun family members to cooperatively bind to and transactivate NFAT sites on the promoter regions of target genes (36, 58, 61). When the cells return to their unstimulated state, NFAT becomes rephosphorylated and is exported out of the nucleus (47).
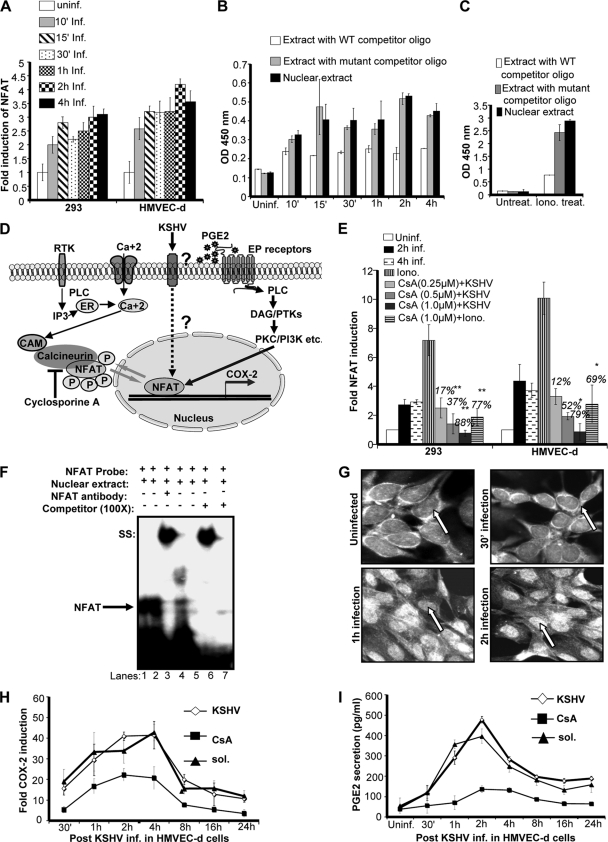
As COX-2 promoter analysis showed NFAT involvement in COX-2 promoter regulation, we examined whether KSHV de novo infection induces NFAT transcription factor activation in 293 and HMVEC-d cells. Using a transcription factor ELISA, nuclear extracts from uninfected and infected 293 or HMVEC-d cells were assessed for the ability of host transcription factors to bind to their respective WT DNA sequences. Infection of 293 or HMVEC-d cells with KSHV increased NFAT activation in a time-dependent manner (Fig. 6A), although NFAT activation was lower in 293 cells than in HMVEC-d cells (Fig. 6A). Results in Fig. 6B show a representative example of HMVEC-d cells infected with KSHV for different times. NFAT activity was barely detected in the serum-starved uninfected cells (Fig. 6B). To determine the specificity of the above transcription factor detection, uninfected and infected nuclear extracts were preincubated for 30 min with WT or mutated consensus sequences for the respective factors before application to the plate-bound WT DNA sequences. In this experiment, transcription factors prebound to the WT oligonucleotides will be unable to bind to the plate-bound DNA and hence will be removed in the washing steps. The ability of transcription factors to bind their respective target sequences was significantly inhibited by preincubation with WT oligonucleotides, while no effect was seen with mutated oligonucleotides (Fig. 6B), demonstrating the specificity of the assay. Nuclear extracts prepared from the ionomycin (NFAT agonist)-stimulated cells showed dramatic (28-fold) induction in NFAT activity and was used as a positive control (Fig. 6C). Altogether, these data suggested a link between NFAT activity and COX-2 induction upon KSHV infection.
FIG. 6.
Role of KSHV de novo infection-induced activation of Cn/NFAT-dependent COX-2 gene expression and PGE2 secretion. (A) KSHV de novo infection of HMVEC-d and 293 cells induces NFAT activation. Relative activation was calculated by taking the activation in uninfected cells as 1-fold at all of the indicated times. (B) To monitor the specificity of NFAT activation, a competition assay using soluble oligonucleotides was performed. Briefly, nuclear extracts prepared from HMVEC-d cells left uninfected or infected with KSHV for different times were incubated with either the WT consensus oligonucleotides (competitor for NFAT binding which will prevent NFAT binding to the probe immobilized on the plate) or mutant oligonucleotides for 30 min prior to their addition to the plate with immobilized oligonucleotides, and an ELISA was performed with an antibody specific to the NFAT transcription factor in accordance with the manufacturer's instructions. Optical density (OD) values are presented. (C) Nuclear lysates prepared from ionomycin-treated cells were used as a positive control, and the specificity of the assay was verified as shown in Fig. 5B. Data represent the average ± SD of three experiments. (D) Schematic of the possible pathways of NFAT induction by KSHV infection and PGE2 secreted in the infected-cell microenvironment. (E) Histogram depicting NFAT fold induction in nuclear extracts prepared from HMVEC-d cells left untreated or pretreated with various concentrations of CsA and then infected with KSHV for 2 h. Percent inhibition of NFAT activation upon CsA pretreatment was calculated with respect to the DNA-binding activities in KSHV-infected HMVEC-d cells without CsA pretreatment, which were taken as 100%. Ionomycin-treated nuclear lysates were used as a positive control. Data represent the average ± SD of three experiments. (F) EMSA for nuclear extracts prepared from HMVEC-d cells using the dNFAT probe. This EMSA experiment shows the binding of nuclear extracts prepared from virus-infected and uninfected HMVEC-d cells left untreated or pretreated with CsA to the dNFAT probe. The specificity of the DNA-protein interaction was assessed by competition (using a 100-fold molar excess of unlabeled double-stranded oligonucleotide NFAT probe as a competitor) and a supershift EMSA using anti-NFAT antibody. Each EMSA is representative of at least three independent experiments. (G) NFAT's subcellular localization upon KSHV infection. HMVEC-d cells left uninfected or KSHV infected at the different times indicated were labeled with anti-NFAT antibody and then examined under a microscope. Each image shown corresponds to one of three independent experiments. (H) Effect of CsA on COX-2 gene expression in HMVEC-d cells. cDNA prepared from serum-starved (10 h) uninfected, KSHV-infected (2 h), solvent-pretreated, and then KSHV-infected (2 h) or CsA-treated and then KSHV-infected (2 h) HMVEC-d cells was used for COX-2 gene expression analysis. COX-2 gene expression in uninfected cells was taken as 1-fold. Data represent the average ± SD of three experiments. (I) Effect of CsA on PGE2 secretion in HMVEC-d cells. Supernatants collected from serum-starved (10 h), uninfected, KSHV-infected (2 h), solvent-pretreated, and then KSHV-infected (2 h) or CsA-treated and then KSHV-infected (2 h) cells were used for PGE2 quantitation by ELISA, and levels in pg/ml are presented. Data represent the average ± SD of three experiments. * and **, statistically significant at P < 0.01 and P < 0.005, respectively.
Multiple scenarios can be hypothesized for KSHV- or PGE2-induced NFAT activation (Fig. 6D). In other systems, RTKs have been shown to induce IP3, which can lead to Ca2+ release from the endoplasmic reticulum, and this Ca2+ can induce Cn/NFAT signaling, which can dephosphorylate NFAT and augment nuclear translocation of NFAT.
The remarkable (81%) reduction in COX-2 promoter activity upon cotransfection with NFAT peptide inhibitor plasmid VIVIT (Fig. 5F) suggested the importance of Cn signaling in COX-2 transcription regulation. Cn signaling has been implicated in a broad spectrum of infections and pathological conditions, and there is evidence that NFAT activation is mediated by a Ca2+/Cn-dependent pathway. To evaluate the contribution of Cn in KSHV infection-induced COX-2 expression, CsA, a specific inhibitor of Cn/NFAT, was employed. First, we checked the role of CsA in KSHV infection-induced NFAT activity (Fig. 6E). Pretreatment of HMVEC-d or 293 cells with various concentrations of CsA (0.25 μM, 0.5 μM, and 1 μM) was able to effectively inhibit NFAT induction in a dose-dependent manner, as indicated in Fig. 6E. All of the doses of CsA used were noncytotoxic, while the 1 μM concentration was most effective in NFAT inhibition in both of the cell types tested (Fig. 6E). CsA pretreatment also drastically inhibited ionomycin-induced NFAT activity in both of the cell types tested (293 and HMVEC-d), indicating the specificity of NFAT inhibition (Fig. 6E).
KSHV-induced COX-2 promoter activity is preferentially regulated via dNFAT binding to the human COX-2 promoter.
To further confirm a functional activation of NFAT by KSHV, dNFAT DNA binding was assessed by EMSAs with an oligonucleotide containing a consensus dNFAT binding site as a probe (Fig. 6F). Nuclear extracts prepared from HMVEC-d cells left uninfected (Fig. 6F, lane 5) or KSHV infected for 1 h (Fig. 6F, lane 1) or 2 h (Fig. 6F, lane 2) were incubated with consensus oligonucleotides for the dNFAT binding site used as labeled probes. Compared with uninfected cells (Fig. 6F, lane 5), KSHV infection markedly induced the formation of a complex with the consensus dNFAT probe (Fig. 6F, lanes 1 and 2). Compared to nuclear extracts prepared from infected cells (Fig. 6F, lanes 1 and 2), CsA-treated and then KSHV-infected cell nuclear lysates (Fig. 6F, lane 4) showed a marked reduction in complex formation with the labeled dNFAT probe. A competition assay with an excess of unlabeled probe (COX-2 dNFAT) completely abolished complex formation with the labeled probe, suggesting that the complex is specific to COX-2 dNFAT (Fig. 6F, lane 7). The specificity of in vitro DNA binding was also confirmed by supershift EMSA using nuclear extracts prepared from HMVEC-d cells infected for 1 h (Fig. 6F, lane 3) or 2 h (Fig. 6F, lane 6).
Since NFAT activity is intimately correlated with its nuclear localization, we investigated the subcellular localization of this transcription factor after KSHV infection by microscopy using a specific anti-NFAT antibody. Interestingly, NFAT translocates to the nucleus early after KSHV infection (Fig. 6G) and this nuclear translocation was strongly inhibited in cells treated with CsA prior to KSHV infection (data not shown).
The significance of the Cn/NFAT pathway was further assessed at the COX-2 gene expression and PGE2 secretion levels. KSHV infection-induced COX-2 gene expression appeared to be Cn signaling dependent, as it was suppressed by CsA treatment prior to infection (Fig. 6H) at all of the time points tested. Similar to COX-2 gene expression, PGE2 secretion was also reduced in the cells treated with CsA prior to KSHV infection (Fig. 6I). Data presented in Fig. 6 strongly indicate that NFAT activation increases COX-2 expression and PGE2 synthesis, and inactivation of NFAT by CsA significantly diminished them, suggesting that KSHV-induced COX-2 transcription may involve Ca2+ signaling and Cn-dependent NFAT activation. Although in the present study we have not assessed the role of Ca2+ using any pharmacological inhibitors of Ca2+ (BAPTA-AM and TMB-8), in a separate study (19), we documented the role of PGE2 in Ca2+ signaling, which might also be one of the factors inducing NFAT activity in KSHV-infected cells.
Data presented in Fig. 6E to I clearly show that KSHV infection or PGE2 per se upon interaction with target cells can induce NFAT activation (Fig. 6E and F), COX-2 gene expression (Fig. 6H), and PGE2 secretion (Fig. 6I) via Ca2+/Cn-mediated NFAT dephosphorylation and nuclear translocation (Fig. 6G).
CREB serine 133 phosphorylation is involved in KSHV-induced COX-2 gene expression.
Since COX-2 promoter analysis indicated a role for CRE (Fig. 5C, E, and G), next we assessed whether KSHV infection induces the binding of transcription factor CREB to CRE. CREB is a member of a large family of structurally related transcription factors (AFT1 to -4, c-Fos, c-Jun, c-Myc, and C/EBP) (59) which specifically recognize the CRE promoter site (5′-TGACGTCA-3′) (39). CREB transcriptional activity is controlled by homo- and heterodimers (CREB/ATF-1) of CREB family members binding to the CRE promoter site (30). CREB activity is also controlled by its cofactor, CREB-binding protein (5), and a variety of protein kinases like protein kinase A (PKA), PKC, MAPKs, and Ca2+/calmodulin-dependent protein kinases that phosphorylate CREB at Ser-133 (59), which increases CREB's affinity for its promoter site (59) (Fig. 7D).
FIG. 7.
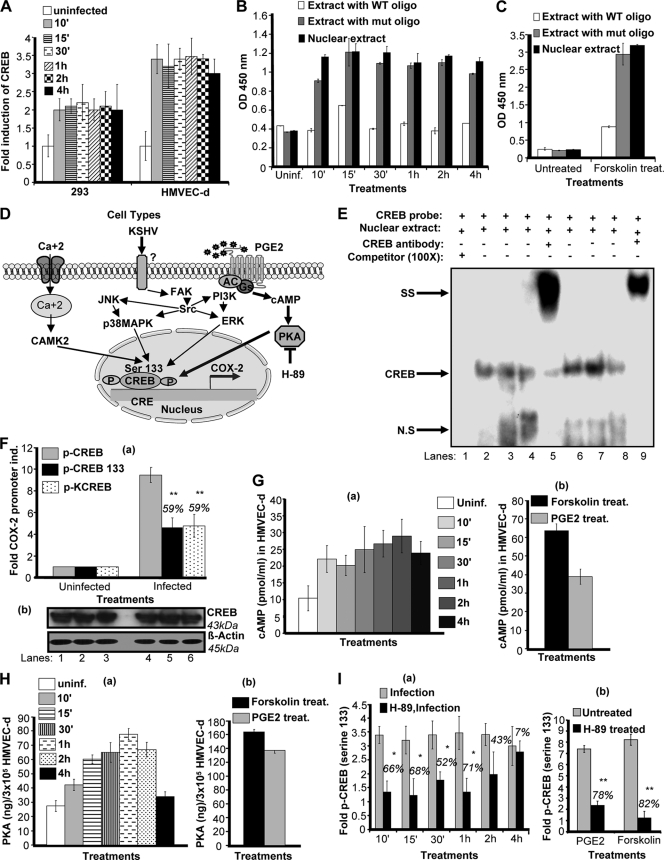
Effect of KSHV de novo infection-activated, cAMP-PKA-mediated CREB serine 133 phosphorylation and COX-2 promoter induction. (A) KSHV de novo infection of HMVEC-d and 293 cells induces CREB activation. Relative activation was calculated by taking activation in uninfected cells as 1-fold. (B) To monitor specificity of CREB activation, nuclear extracts prepared from HMVEC-d cells left uninfected or infected with KSHV for different times were tested for CREB activity by incubating the nuclear extracts with WT consensus oligonucleotides (competitor for CREB binding) or mutant oligonucleotides for 30 min prior to performing ELISA as described for Fig. 6B. (C) Nuclear lysates prepared from forskolin-treated cells were used as a positive control, and the specificity of the assay was verified as shown in Fig. 6B. Data represent the average ± SD of three experiments. (D) Diagrammatic representation of the possible pathways involved in CREB serine 133 phosphorylation and CRE activation. (E) EMSA for nuclear extracts prepared from HMVEC-d or 293 cells using CRE probe. This EMSA experiment shows the binding of nuclear extracts prepared from virus-infected and uninfected HMVEC-d cells with a CRE probe. The specificity of the DNA-protein interaction was assessed by supershift EMSA using anti-CREB-1 antibody. Each EMSA is representative of at least three independent experiments. (F, a) 293 cells cotransfected with a P2-1900 COX-2 promoter full-length construct along with 1 μg of p-CMV-CREB, p-CMV-CREB-133, or p-CMV-KCREB expression plasmid were left uninfected or KSHV infected for 2 h. The promoter induction in uninfected cells was taken as 1-fold. Percent inhibition of COX-2 induction was calculated with reference to the n-fold induction in p-CMV-CREB-transfected and KSHV-infected (2 h) cells, which was taken as 100%. Expression of various CREB constructs was checked by Western blotting (F, b). (G, a and b). Lysates prepared from serum-starved HMVEC-d cells left uninfected or KSHV infected or treated with 1 μΜ PGE2 or forskolin for the indicated times were used to measure intracellular cAMP levels. Data are expressed as mean pmol/ml. (H, a and b). Lysates prepared as described for panel G (a and b) were used to quantitate PKA levels by the methods described previously. Data are expressed as mean ng/3 × 105 HMVEC-d cells. Each reaction was done in triplicate, and each point represents the average ± SD of five experiments. (I, a and b). Serum-starved HMVEC-d cells were left untreated or pretreated with PKA inhibitor H-89 and then KSHV infected for the indicated times, treated with 1 μΜ PGE2 for 2 h, or treated with forskolin for 30 min. Nuclear lysates prepared from these cells were used to quantitate CREB serine 133 phosphorylation levels as described above. Each reaction was done in triplicate, and each point represents the average ± SD of five experiments. Percent inhibition was calculated by taking the CREB serine 133 phosphorylation in non-inhibitor-treated, KSHV-infected (2 h) cells as 100%.
KSHV infection-induced CREB activity in 293 (∼2.5-fold) and HMVEC-d (∼3-fold) cells (Fig. 7A) was measured by a transcription factor ELISA with a CREB-specific probe. Using an ELISA-based technique as described in Fig. 6B, nuclear extracts from uninfected and KSHV-infected HMVEC-d and 293 cells were assessed for the ability of host transcription factors to bind to their respective WT DNA sequences. Figure 7B shows a representative example of the CREB activity in HMVEC-d cells infected with KSHV for different times. The specificity of activation was assessed by a reduction of binding in the nuclear extracts incubated with WT CREB consensus sequences (Fig. 7B). Levels of these transcription factors were barely detectable in serum-starved uninfected cells (Fig. 7B). Since cAMP induction is required upstream of CREB serine 133 phosphorylation and CRE binding, we prepared nuclear lysates from forskolin-treated HMVEC-d cells (Fig. 7C). These nuclear lysates showed very high activation of CREB-serine phosphorylation and exhibited reduced binding when preincubated with WT consensus sequences (Fig. 7C).
Multiple scenarios can be hypothesized for KSHV- or PGE2-induced CREB serine 133 phosphorylation (Fig. 7D). KSHV binding initiates various MAPKs (p38, JNK), and FAK/Src kinases can induce production of cAMP, which can activate PKA and mediate phosphorylation of CREB serine 133. PGE2 secreted from infected cells can induce adenylate cyclase activation via binding to its prostanoid receptors (EP receptors) present on the infected cell or the neighboring infected/uninfected cells. These interactions can upregulate cAMP production, PKA activation, and CREB serine phosphorylation. Apart from PGE2/EP receptor signaling, Ca2+ signaling could also regulate CREB activation, as reported in other systems.
To further confirm a functional activation of CREB by KSHV, CREB DNA binding was assessed by EMSAs with an oligonucleotide containing a consensus CRE binding site as a probe (Fig. 7E). Nuclear extracts prepared from HMVEC-d (Fig. 7E, lanes 6 to 9) or 293 (Fig. 7E, lanes 2 to 5) cells left uninfected or KSHV infected for different times were incubated with consensus oligonucleotides for the CRE binding site as labeled probes. Compared with leaving HMVEC-d cells uninfected (Fig. 7E, lane 8), KSHV infection for 1 h (Fig. 7E, lane 7) or 2 h (Fig. 7E, lane 6) markedly induced the formation of a complex with the consensus CRE probe. Compared with leaving 293 cells uninfected (Fig. 7E, lane 4), KSHV infection for 1 h (Fig. 7E, lane 3) or 2 h (Fig. 7E, lane 2) markedly induced the formation of a complex with the consensus CRE probe. Competition assay with an excess of unlabeled probe (COX-2-CRE) completely abolished complex formation with the labeled probe, suggesting that the complex is specific to COX-2-CREB dNFAT (Fig. 7E, lane 1). The specificity of in vitro DNA was checked by supershift EMSA by preincubating nuclear extracts prepared from 293 (Fig. 7E, lane 5) and HMVEC-d (Fig. 7E, lane 9) cells KSHV infected for 2 h with anti-CREB antibody. The strong supershift verified the specific binding.
To verify that CREB binding to the CRE consensus site in the COX-2 promoter is essential for COX-2 promoter activation, 293 cells were cotransfected with P2-1900 (COX-2 full-length promoter construct) along with the p-CMV-CREB, p-CMV-CREB 133, or p-CMV-KCREB vector (Fig. 7F, a). Equal expression of CREB constructs was checked by Western blotting (Fig. 7F, b). At 24 h posttransfection, these cells were either left uninfected or KSHV infected (30 DNA copies per cell) for 2 h. Upon KSHV infection, maximum promoter induction was observed in cells cotransfected with p-CMV-CREB, as it expressed constitutive WT CREB (Fig. 7F). An ∼60% reduction in COX-2 promoter activation was observed in the presence of the p-CMV-CREB 133 vector, suggesting that mutation of serine 133 to alanine in CREB confirms that CREB phosphorylation is essential for KSHV-induced COX-2 transcription (Fig. 7F). Similarly, a 60% reduction in COX-2 promoter activity was observed in the presence of the p-CMV-KCREB vector expressing a mutant variant of human CREB containing mutations in its DNA-binding domain. These results suggest that CREB binding to CRE is critical for KSHV-induced COX-2 promoter activity, as KCREB, which blocks CREB binding to CRE, effectively reduced COX-2 promoter activation (Fig. 7F).
CREB activation has been shown to be dependent on cAMP production via PGE2 in other systems (1); therefore, we checked intracellular cAMP levels in uninfected, KSHV-infected, or PGE2-treated HMVEC-d cells. We observed appreciable levels of cAMP induction in KSHV-infected cells (Fig. 7G, a), but this induction was lower than the levels observed in the presence of exogenous PGE2 (Fig. 7G, b). Forskolin (cAMP) agonist was used as a positive control (Fig. 7G, b).
Higher levels of intracellular cAMP in KSHV-infected or PGE2-treated cells suggested the involvement of PKA, a cAMP-dependent protein kinase (69) which appears to be the principal mediator of cAMP action. The PKA holoenzyme is a heterotetramer composed of a regulatory (R) subunit dimer and two monomeric catalytic (C) subunits. cAMP-dependent PKA is the principal intracellular target for cAMP in mammalian cells, and binding of cAMP to tandem sites in each R subunit causes dissociation of the holoenzyme, which subsequently alleviates an autoinhibitory contact that releases the active C subunit. Active kinase enters the cell nucleus and is then free to phosphorylate substrates on serine or threonine residues present in various transcription factors, including CREB. PKA therefore plays a role in the transcriptional control of genes downstream of CRE (50). We then quantified the levels of PKA activation in uninfected, KSHV-infected (Fig. 7H, a), and PGE2-treated HMVEC-d cells (Fig. 7H, b). We observed higher levels of PKA activity in KSHV-infected cells (Fig. 7H, a), but this induction was lower than that observed in the presence of exogenous PGE2 (Fig. 7H, b). Lower levels of PKA activation were observed in samples KSHV infected for 4 h (Fig. 7H, a), indicating that cAMP and PKA levels might not be stable at later times p.i.
To understand the downstream effect of KSHV infection or exogenous PGE2 in COX-2 promoter induction, we checked the levels of CREB serine 133 phosphorylation in cells left untreated or treated with the PKA inhibitor H-89 and then KSHV infected (Fig. 7I, a) and in PGE2-treated HMVEC-d cells (Fig. 7I, b). Pretreatment of cells with H-89 (50 μM) for 1 h significantly inhibited KSHV infection-induced CREB serine phosphorylation, but this inhibition was lower than that observed in PGE2-stimulated cells (Fig. 7I, a and b). There was no change in total CREB levels. These data clearly suggested a role for COX-2-PGE2-cAMP-PKA-CREB serine 133 phosphorylation in COX-2 promoter induction.
KSHV infection-induced multiple signal cascades regulating CREB activation.
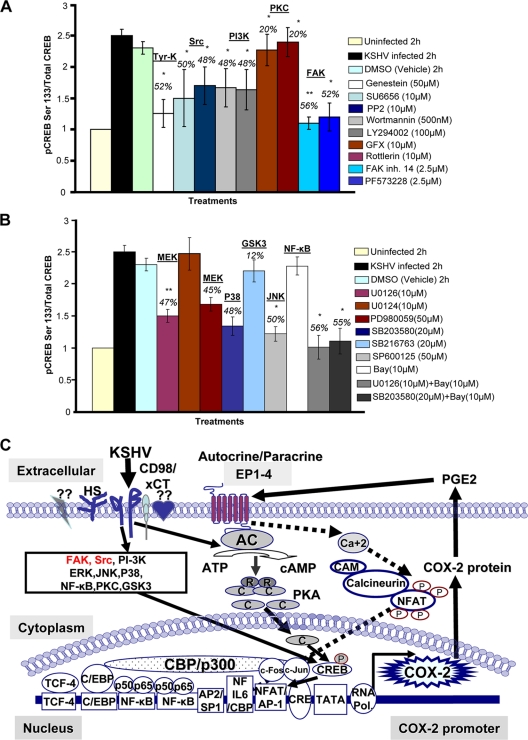
To test whether kinases induced early upon KSHV infection play an important role in CREB serine 133 phosphorylation, nuclear lysates from all of the signaling inhibitor-treated, KSHV-infected, and solvent control-treated cells were prepared (Fig. 8A and B). Of the virus binding- and entry-related kinases, maximum inhibition was obtained in cells treated with genistein, followed by Src and PI3K inhibitors (Fig. 8A). Of the MAPKs tested, a drastic reduction in CREB serine 133 phosphorylation was observed in cells pretreated with the ERK and NF-κB or p38 and NF-κB combination of inhibitors (Fig. 8B). These data suggested a vital link between signaling cascades induced early upon infection and transcription factor activation which eventually regulates the transcription of KSHV-induced master latency and pathogenesis controlling the gene for inflammation-related COX-2 and its metabolite PGE2.
FIG. 8.
Roles of multiple signaling pathways involved in regulating transcription factor (CREB) activation involved in KSHV-induced COX-2. (A and B) Nuclear lysates prepared from HMVEC-d cells were used to measure CREB serine 133 phosphorylation by transcription factor ELISA. Activation in uninfected cells was taken as 1-fold for comparison. Percent inhibition was calculated by taking the CREB activation in KSHV-infected cells as 100%. * and **, statistically significant at P < 0.01 and P < 0.005, respectively. (C) Schematic diagram depicting the multiple signaling pathways and transcription factors involved in the transcriptional regulation of KSHV-induced COX-2. A variety of transcription factors can stimulate COX-2 expression, as there are several binding sites for a number of transcription factors, including NF-κB, NFAT, NF-IL-6/cEBP, AP-1, and CRE, in the 5′ region of the COX-2 gene. Intervention at each step of COX-2 induction can be used as a potential therapeutic target. The role of cell signaling (NF-κB, ERK, p38, JNK, PI3K/AKT, PKC, and Rho-GTPases) in the regulation of COX-2 expression in other systems has been well defined (65, 67, 75). Since KSHV has been shown to induce a variety of overlapping cell signaling cascades (ERK, PI3K, Rho-GTPases, FAK, Src, NF-κB, and PKC-ζ) and transcription factors (c-Fos, c-Jun, c-Myc, and STAT1-α) early during infection (42, 51, 53, 54, 70, 71), here we determined the possible cell signaling events upstream of COX-2 induction upon KSHV infection. Our results suggest that the signal molecules, especially FAK and Src (indicated in red), induced upon virus binding and entry are critical for COX-2 promoter activity, gene expression, protein levels, and PGE2 secretion. Of the MAPKs tested, JNK and p38 appear to control COX-2 promoter activity. The ERK or NF-κ B inhibitor alone is not sufficient to effectively inhibit COX-2 promoter activity, gene expression, protein levels, and PGE2 secretion, compared to the ERK and NF-κB or p38 and NF-κB combination of inhibitors. The Cn/NFAT and CREB transcription factors are exclusively involved in COX-2 promoter activation, gene transcription, and PGE2 secretion. Multiple signaling pathways induced early during KSHV infection are essential for CREB phosphorylation at serine 133 and regulate CRE-mediated COX-2 promoter activity. KSHV- or PGE2-induced cAMP-PKA activation also participates in CREB phosphorylation at serine 133 and enhances COX-2 transcription. Cn/NFAT- and CREB-regulated pathways are represented by dashed and solid black lines, respectively.
DISCUSSION
The novel findings of the present study (Fig. 8C) are as follows. (i) Multiple signaling pathways induced early during KSHV binding and entry events (FAK and Src) are critical for initiation of transcription regulation of the master inflammatory cascade of the COX-2/PGE2 pathway. (ii) NFAT and CREB transcription factors are critical for COX-2 promoter activation, gene transcription, and PGE2 secretion. (iii) KSHV-induced COX-2 promoter activity is exclusively regulated via a distal NFAT site in the COX-2 promoter. (iv) NFAT activation upon KSHV infection is mediated through a Cn pathway. (v) CsA pretreatment could effectively reduce KSHV infection-induced COX-2 gene expression and PGE2 secretion. (vi) Multiple signaling pathways induced early during KSHV infection are essential for CREB phosphorylation at serine 133, which regulates CRE-mediated COX-2 promoter activity. (vii) KSHV infection-induced PGE2 regulates COX-2 promoter induction preferentially via cAMP-mediated PKA activation. (viii) Of the MAPKs tested, GSK3 inactivation via its phosphorylation appears to be required for COX-2 expression.
COX-2 expression is low and undetectable in normal cells but is highly elevated in inflammation-associated malignancies (10, 11). COX-2 expression can be readily induced by growth factors, cytokines, and tumor promoters (10, 11). COX-2 is well established as an important regulator of KSHV latency and its pathogenesis (19, 55, 56). Blockade of COX-2 enzymatic activity by either COXIB (NS-398) or COX-2 silencing effectively downregulated viral pathogenesis-related events like cell invasion, inflammation, angiogenesis, and cell survival (55). COX-2 transcription mechanisms vary according to the type of stimulus and the target cell type. A variety of transcription factors play roles in COX-2 induction, synthesis, and maintenance. COX-2 levels are tightly controlled, and regulation occurs at both the transcriptional and posttranscriptional levels (63).
Since KS is an endothelial cell vascular neoplasm, we chose to work with endothelial (HMVEC-d) and HFF cells, which makes this study highly significant and biologically relevant. KSHV infection of in vitro target (HMVEC-d, HFF) cells has been shown to be associated with latency-associated nuclear antigen 1 (LANA-1), and the absence of progeny virus production and this latent infection can be switched into the lytic cycle by viral lytic cycle ORF50 protein. Depending on the viral lytic and latent gene expression pattern, in vitro-infected endothelial and fibroblast cells display two phases in their life cycle, i.e., early lytic (peaks at 2 h p.i. and dips at 24 h p.i.) and latent (is low at 2 h p.i., increases subsequently, is maintained at a steady state, and declines by 5 days). This study represents only early (2 and 4 h p.i.) events of host factor modulation by KSHV binding and entry and limited early lytic genes (K2, K4, K5, K6, K7, and vIRF2) (31). Our results clearly show that COX-2 promoter activity is markedly induced early after KSHV de novo infection. Conversely, COX-1 promoter-driven luciferase activity was not augmented. Induction of COX-2/PGE2 early during KSHV infection is regulated at the transcriptional level, as the induction of both COX-2 and PGE2 was significantly inhibited in the presence of the transcriptional inhibitor actinomycin D.
PGE2 secreted upon KSHV infection could not sustain COX-2 gene expression by itself for the long term, suggesting the involvement of other secreted proinflammatory and angiogenic factors, including VEGF, ILs, and HIF-1α, which are known to prolong sustained induction of signal cascades and COX-2 levels (21, 68). Another reason for the short-lived activity of COX-2 upon PGE2 stimulation might be the short half-life of PGE2 (14).
Given the findings of EMSAs, mutation/deletion construct reporter gene transfection assays, and the transcription factor assessment experiments presented herein, it is evident that binding of NFAT and binding of CRE are necessary steps involved in the transcriptional activation of COX-2. Selective mutation of the dNFAT site diminished (more than 75%) KSHV infection-induced COX-2 promoter activity. It is well established that NFAT activity is tightly regulated by the Ca2+/calmodulin-dependent phosphatase Cn, which regulates NFAT protein translocation from the cytoplasm to the nucleus via dephosphorylation in response to increased levels of intracellular calcium. Our studies indicated that KSHV infection activates NFAT and CREB in biologically relevant (HMVEC-d) cells, as quantified by transcription factor ELISAs and gel shift assays. Activation of NFAT by KSHV was inhibited by CsA, an inhibitor of the NFAT-activating protein Cn. Inhibition of NFAT by CsA led to a significant reduction in COX-2 gene expression and PGE2 secretion, strengthening the involvement of the Cn/NFAT pathway in KSHV-induced COX-2 transcription. Although the details of the mechanism of specific NFAT activation by KSHV infection in HMVEC-d cells are unclear, the best explanation is probable cross talk among the various KSHV-induced signaling pathways and increased levels of intracellular Ca2+. Increased intracellular Ca2+ levels might either be due to viral glycoprotein interactions with their specific receptors on target endothelial cells or occur through PGE2-mediated pathways, as indicated in a separate study (19) where a role for PGE2-mediated Ca2+ in KSHV latency was indicated. NFAT activation may be important for KSHV lytic replication too, since Cn-dependent signaling has been shown to be essential for calcium-dependent reactivation of KSHV (79).
Cn/NFAT signaling has been implicated in many broad-spectrum pathological conditions (47) but never studied in detail for the early events of KSHV infection. The role of Cn/NFAT signaling involved in the transcription of COX-2, a factor involved in maintenance of viral latency, is a novel finding. Although CsA (a Cn/NFAT inhibitor) has revolutionized transplant biology by inducing immunosuppression via NFAT inactivation in T cells, its potential application in cancer therapy remains untested. Newer advances with novel analogues of CsA and small peptides blocking Cn/NFAT interaction without inhibiting Cn phosphatase activity have exhibited efficacy without significant toxicity. Data indicating inhibition of COX-2 gene expression and PGE2 secretion upon CsA pretreatment in infected endothelial cells further suggest that NFAT merits additional investigation as a transcription factor in KSHV biology. The potential to exploit NFAT signaling for therapeutic benefit with COX-2 inhibitors appears valuable and clearly warrants future investigation.
Another interesting observation of our study is the transcriptional regulation of COX-2 promoter activity via NF-κB. Removal of one NF-κB binding site decreased COX-2 promoter activity, while elimination of both NF-κB binding sites relieved this inhibition, indicating the interplay of epigenetic control to tightly regulate COX-2 levels. The NF-κB and COX-2 connection in the KSHV infection scenario seems context dependent.
Our study indicates that of the MAPK inhibitors tested, the GSK3 inhibitor was the only agent that did not reduce COX-2 promoter activity. Rather, it increased COX-2 gene expression and maintained PGE2 secretion, suggesting that GSK3 inactivation has a potential role in controlling COX-2 induced by KSHV infection. GSK3 has been shown to be a key regulator in several physiological processes, such as cell cycle, oncogenesis, Wnt and Hedgehog signaling, transcription, differentiation, Wnt-associated developmental patterning and β-catenin-driven tumorigenesis, response to DNA damage, cell death, cell survival, and apoptosis in metabolic syndromes and many cancers, including KSHV-associated malignancies (17). KSHV latent protein LANA-1 has been reported to interact with GSK3β and regulate its activity, leading to inhibition of GSK3-dependent β-catenin degradation, an important transcriptional coactivator of T-cell factor/LEF transcription factors required for infected-cell proliferation. There is compelling evidence that GSK3 regulates NFAT activity by phosphorylation; its nuclear translocation and its inhibition by potent inhibitors increase the levels of nuclear NFAT, which prolongs the synthesis of ILs (40). It has been shown in other systems that PKA can either phosphorylate serine 21 or 9 of GSK3α and GSK3β, inactivating GSK3, or indirectly inhibit AKT while H-89 treatment can inhibit GSK3α phosphorylation (12). In other words, our study suggests that higher time-dependent GSK3 phosphorylation could potentially be dependent on KSHV-induced PKA and cAMP levels.
PGE2 might be amplifying COX-2 levels via PGE2 receptor-linked Gs activation-induced adenylyl cyclase/cAMP/PKA signaling-mediated inactivation of GSK3. Studies involving COX-2 stability and its connection with signal cascades in KSHV-infected cells are in progress. Though we have not assessed the role of GSK3 inhibition in latently infected cells, we found that KSHV de novo infection of HMVEC-d cells increased GSK3 phosphorylation (serine 9) in a time-dependent manner (see Fig. S6A in the supplemental material), which was followed by decreased β-catenin degradation and nuclear translocation of stabilized β-catenin (Fig. S6B), similar to what has been established for KSHV-linked B-cell lymphomas (17). GSK3β inhibition is known to increase β-catenin activity (24) and is similar to what we observed upon KSHV infection. Our study also indicates that KSHV efficiently utilizes and tightly controls host signal pathways and ensures its survival in the host cell.
The human COX-2 3′ untranslated region (UTR) possesses the AU motif that contains 22 repeats of the sequence AUUUA which is also described as a mediator of mRNA degradation (57) and is associated with instability of the COX-2 transcript (8). COX-2 mRNA stability has also been shown in other systems to be regulated at the posttranscriptional level via various cytokines (41, 48, 49). KSHV infection induces the secretion of a variety of cytokines, which could modify some signal transduction pathways, eventually regulating (at least in part) the degradation of COX-2 mRNA, and needs further investigation. Although COX-2 transcription during KSHV de novo infection was completely repressed by actinomycin D pretreatment, these results should be interpreted with caution as this regulatory mechanism might not be similar in latently infected cells, where abundant levels of COX-2 exist (unpublished results). Aberrant COX-2 expression in latently KSHV-infected endothelial cells (55) and PEL cells (60) indicates that a critical role for posttranscriptional, translational, or posttranslational mechanisms of regulation might be more prevalent than transcriptional mechanisms. Therefore, it becomes relevant to study the role of the 3′ UTR in the regulation of COX-2 in latently infected cells. As the COX-2 3′ UTR possesses several prospective polyadenylation sites, involvement of RNA-binding proteins, KSHV microRNAs, in the recruitment and stabilization of core adenylation factors on the COX-2 mRNAs in latently infected cells cannot be ruled out. Studies involving posttranscriptional regulation of COX-2 via its 3′ UTR affecting COX-2 RNA stability and translational efficiency are in progress.
In summary, we provide compelling evidence that KSHV infection efficiently controls COX-2 promoter activity at multiple levels (Fig. 8C). COX-2 transcriptional activation upon KSHV infection appears to be the overall coordinated effect of many factors, including multiple signaling pathways and various growth factors/cytokines which are known to upregulate COX-2/PGE2 in other systems (10, 11). Our study clearly shows that transcriptional activation at the 5′ UTR of the COX-2 gene involves cross talk between multiple signaling pathways induced upon KSHV binding and entry into the target cells, which exclusively regulates the activities of transcription factors like NFAT and CREB present on the COX-2 promoter. We demonstrated that as a consequence of NFAT inhibition, PGE2 secretion in cells pretreated with CsA and then KSHV infected is significantly impaired. Studies to identify viral factors involved in the prolonged induction and huge levels of COX-2 in lymphoma cells are currently in progress.
As the expectation of developing a “magic bullet” for curing KSHV-associated hematologic and endothelial malignancies is low, our study predicts that a combination of the signaling inhibitors controlling inflammatory molecules like COX-2, along with COXIBs, seems to be very promising. Intervention in COX-2 at multiple levels would provide an effective therapeutic approach for KS-associated malignancies.
Supplementary Material
Acknowledgments
We gratefully acknowledge M. A. Iñiguez of the Universidad Autónoma de Madrid, Spain, for providing all of the COX-2 and COX-1 promoter constructs. We gratefully acknowledge Karsten Schroer of the Institut für Pharmakologie und Klinische Pharmakologie for providing COX-2-CRE-WT and COX-2-CRE-Mut promoter constructs. We thank Keith Philibert for critically reading the manuscript.
This study was supported in part by Public Health Service grants CA128560 (N.S.-W.) and CA 099925 and the Rosalind Franklin University of Medicine and Science H. M. Bligh Cancer Research Fund (B.C.).
Footnotes
Published ahead of print on 13 October 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ansari, K. M., Y. M. Sung, G. He, and S. M. Fischer. 2007. Prostaglandin receptor EP2 is responsible for cyclooxygenase-2 induction by prostaglandin E2 in mouse skin. Carcinogenesis 28:2063-2068. [DOI] [PubMed] [Google Scholar]
- 2.Aramburu, J., M. B. Yaffe, C. Lopez-Rodriguez, L. C. Cantley, P. G. Hogan, and A. Rao. 1999. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science 285:2129-2133. [DOI] [PubMed] [Google Scholar]
- 3.Bradbury, D., D. Clarke, C. Seedhouse, L. Corbett, J. Stocks, and A. Knox. 2005. Vascular endothelial growth factor induction by prostaglandin E2 in human airway smooth muscle cells is mediated by E prostanoid EP2/EP4 receptors and SP-1 transcription factor binding sites. J. Biol. Chem. 280:29993-30000. [DOI] [PubMed] [Google Scholar]
- 4.Buchanan, F. G., and R. N. DuBois. 2006. Connecting COX-2 and Wnt in cancer. Cancer Cell 9:6-8. [DOI] [PubMed] [Google Scholar]
- 5.Chrivia, J. C., R. P. Kwok, N. Lamb, M. Hagiwara, M. R. Montminy, and R. H. Goodman. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365:855-859. [DOI] [PubMed] [Google Scholar]
- 6.Chuang, P. C., H. S. Sun, T. M. Chen, and S. J. Tsai. 2006. Prostaglandin E2 induces fibroblast growth factor 9 via EP3-dependent protein kinase Cdelta and Elk-1 signaling. Mol. Cell. Biol. 26:8281-8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clipstone, N. A., and G. R. Crabtree. 1992. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature 357:695-697. [DOI] [PubMed] [Google Scholar]
- 8.Crofford, L. J., R. L. Wilder, A. P. Ristimaki, H. Sano, E. F. Remmers, H. R. Epps, and T. Hla. 1994. Cyclooxygenase-1 and -2 expression in rheumatoid synovial tissues. Effects of interleukin-1 beta, phorbol ester, and corticosteroids. J. Clin. Invest. 93:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degousee, N., J. Martindale, E. Stefanski, M. Cieslak, T. F. Lindsay, J. E. Fish, P. A. Marsden, D. J. Thuerauf, C. C. Glembotski, and B. B. Rubin. 2003. MAP kinase kinase 6-p38 MAP kinase signaling cascade regulates cyclooxygenase-2 expression in cardiac myocytes in vitro and in vivo. Circ. Res. 92:757-764. [DOI] [PubMed] [Google Scholar]
- 10.Dubois, R. N. 2000. Review article: cyclooxygenase—a target for colon cancer prevention. Aliment. Pharmacol. Ther. 14(Suppl. 1):64-67. [DOI] [PubMed] [Google Scholar]
- 11.Dubois, R. N., S. B. Abramson, L. Crofford, R. A. Gupta, L. S. Simon, L. B. Van De Putte, and P. E. Lipsky. 1998. Cyclooxygenase in biology and disease. FASEB J. 12:1063-1073. [PubMed] [Google Scholar]
- 12.Fang, X., S. X. Yu, Y. Lu, R. C. Bast, Jr., J. R. Woodgett, and G. B. Mills. 2000. Phosphorylation and inactivation of glycogen synthase kinase 3 by protein kinase A. Proc. Natl. Acad. Sci. U. S. A. 97:11960-11965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feske, S., H. Okamura, P. G. Hogan, and A. Rao. 2003. Ca2+/calcineurin signalling in cells of the immune system. Biochem. Biophys. Res. Commun. 311:1117-1132. [DOI] [PubMed] [Google Scholar]
- 14.Fitzpatrick, F. A., R. Aguirre, J. E. Pike, and F. H. Lincoln. 1980. The stability of 13,14-dihydro-15 keto-PGE2. Prostaglandins 19:917-931. [DOI] [PubMed] [Google Scholar]
- 15.Fletcher, B. S., D. A. Kujubu, D. M. Perrin, and H. R. Herschman. 1992. Structure of the mitogen-inducible TIS10 gene and demonstration that the TIS10-encoded protein is a functional prostaglandin G/H synthase. J. Biol. Chem. 267:4338-4344. [PubMed] [Google Scholar]
- 16.Frias, M. A., M. C. Rebsamen, C. Gerber-Wicht, and U. Lang. 2007. Prostaglandin E2 activates Stat3 in neonatal rat ventricular cardiomyocytes: a role in cardiac hypertrophy. Cardiovasc. Res. 73:57-65. [DOI] [PubMed] [Google Scholar]
- 17.Fujimuro, M., F. Y. Wu, C. ApRhys, H. Kajumbula, D. B. Young, G. S. Hayward, and S. D. Hayward. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300-306. [DOI] [PubMed] [Google Scholar]
- 18.George, R. J., M. A. Sturmoski, S. Anant, and C. W. Houchen. 2007. EP4 mediates PGE2 dependent cell survival through the PI3 kinase/AKT pathway. Prostaglandins Other Lipid Mediat. 83:112-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.George Paul, A., N. Sharma-Walia, N. Kerur, C. White, and B. Chandran. 2010. Piracy of prostaglandin E2/EP receptor-mediated signaling by Kaposi's sarcoma-associated herpes virus (HHV-8) for latency gene expression: strategy of a successful pathogen. Cancer Res. 70:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harper, K. A., and A. J. Tyson-Capper. 2008. Complexity of COX-2 gene regulation. Biochem. Soc Trans. 36:543-545. [DOI] [PubMed] [Google Scholar]
- 21.Hierholzer, C., B. G. Harbrecht, T. R. Billiar, and D. J. Tweardy. 2001. Hypoxia-inducible factor-1 activation and cyclo-oxygenase-2 induction are early reperfusion-independent inflammatory events in hemorrhagic shock. Arch. Orthop. Trauma Surg. 121:219-222. [DOI] [PubMed] [Google Scholar]
- 22.Hla, T., and K. Neilson. 1992. Human cyclooxygenase-2 cDNA. Proc. Natl. Acad. Sci. U. S. A. 89:7384-7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogan, P. G., L. Chen, J. Nardone, and A. Rao. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17:2205-2232. [DOI] [PubMed] [Google Scholar]
- 24.Howe, L. R., H. C. Crawford, K. Subbaramaiah, J. A. Hassell, A. J. Dannenberg, and A. M. Brown. 2001. PEA3 is up-regulated in response to Wnt1 and activates the expression of cyclooxygenase-2. J. Biol. Chem. 276:20108-20115. [DOI] [PubMed] [Google Scholar]
- 25.Iñiguez, M. A., S. Martinez-Martinez, C. Punzon, J. M. Redondo, and M. Fresno. 2000. An essential role of the nuclear factor of activated T cells in the regulation of the expression of the cyclooxygenase-2 gene in human T lymphocytes. J. Biol. Chem. 275:23627-23635. [DOI] [PubMed] [Google Scholar]
- 26.Iñiguez, M. A., C. Punzon, and M. Fresno. 1999. Induction of cyclooxygenase-2 on activated T lymphocytes: regulation of T cell activation by cyclooxygenase-2 inhibitors. J. Immunol. 163:111-119. [PubMed] [Google Scholar]
- 27.Ito, H., M. Duxbury, E. Benoit, T. E. Clancy, M. J. Zinner, S. W. Ashley, and E. E. Whang. 2004. Prostaglandin E2 enhances pancreatic cancer invasiveness through an Ets-1-dependent induction of matrix metalloproteinase-2. Cancer Res. 64:7439-7446. [DOI] [PubMed] [Google Scholar]
- 28.Jain, J., P. G. McCaffrey, Z. Miner, T. K. Kerppola, J. N. Lambert, G. L. Verdine, T. Curran, and A. Rao. 1993. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature 365:352-355. [DOI] [PubMed] [Google Scholar]
- 29.Jung, Y. J., J. S. Isaacs, S. Lee, J. Trepel, and L. Neckers. 2003. IL-1beta-mediated up-regulation of HIF-1alpha via an NFkappaB/COX-2 pathway identifies HIF-1 as a critical link between inflammation and oncogenesis. FASEB J. 17:2115-2117. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi, M., and K. Kawakami. 1995. ATF-1CREB heterodimer is involved in constitutive expression of the housekeeping Na,K-ATPase alpha 1 subunit gene. Nucleic Acids Res. 23:2848-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnan, H. H., P. P. Naranatt, M. S. Smith, L. Zeng, C. Bloomer, and B. Chandran. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J. Virol. 78:3601-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krysan, K., K. L. Reckamp, H. Dalwadi, S. Sharma, E. Rozengurt, M. Dohadwala, and S. M. Dubinett. 2005. Prostaglandin E2 activates mitogen-activated protein kinase/Erk pathway signaling and cell proliferation in non-small cell lung cancer cells in an epidermal growth factor receptor-independent manner. Cancer Res. 65:6275-6281. [DOI] [PubMed] [Google Scholar]
- 33.Lim, K., C. Han, L. Xu, K. Isse, A. J. Demetris, and T. Wu. 2008. Cyclooxygenase-2-derived prostaglandin E2 activates beta-catenin in human cholangiocarcinoma cells: evidence for inhibition of these signaling pathways by omega 3 polyunsaturated fatty acids. Cancer Res. 68:553-560. [DOI] [PubMed] [Google Scholar]
- 34.Lin, D. T., K. Subbaramaiah, J. P. Shah, A. J. Dannenberg, and J. O. Boyle. 2002. Cyclooxygenase-2: a novel molecular target for the prevention and treatment of head and neck cancer. Head Neck 24:792-799. [DOI] [PubMed] [Google Scholar]
- 35.Liu, X. H., A. Kirschenbaum, M. Lu, S. Yao, A. Dosoretz, J. F. Holland, and A. C. Levine. 2002. Prostaglandin E2 induces hypoxia-inducible factor-1alpha stabilization and nuclear localization in a human prostate cancer cell line. J. Biol. Chem. 277:50081-50086. [DOI] [PubMed] [Google Scholar]
- 36.Loh, C., K. T. Shaw, J. Carew, J. P. Viola, C. Luo, B. A. Perrino, and A. Rao. 1996. Calcineurin binds the transcription factor NFAT1 and reversibly regulates its activity. J. Biol. Chem. 271:10884-10891. [DOI] [PubMed] [Google Scholar]
- 37.Micali, A., N. Medici, A. Sottile, M. Venza, I. Venza, V. Nigro, G. A. Puca, and D. Teti. 1996. Prostaglandin E2 induction of binding activity to CRE and AP-2 elements in human T lymphocytes. Cell. Immunol. 174:99-105. [DOI] [PubMed] [Google Scholar]
- 38.Mochizuki-Oda, N., K. Mori, M. Negishi, and S. Ito. 1991. Prostaglandin E2 activates Ca2+ channels in bovine adrenal chromaffin cells. J. Neurochem. 56:541-547. [DOI] [PubMed] [Google Scholar]
- 39.Montminy, M., P. Brindle, J. Arias, K. Ferreri, and R. Armstrong. 1996. Regulation of somatostatin gene transcription by cyclic adenosine monophosphate. Metabolism 45(8 Suppl. 1):4-7. [DOI] [PubMed] [Google Scholar]
- 40.Murphy, P. M. 1997. Pirated genes in Kaposi's sarcoma. Nature 385:296-297, 299. [DOI] [PubMed] [Google Scholar]
- 41.Nabors, L. B., G. Y. Gillespie, L. Harkins, and P. H. King. 2001. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res. 61:2154-2161. [PubMed] [Google Scholar]
- 42.Naranatt, P. P., S. M. Akula, C. A. Zien, H. H. Krishnan, and B. Chandran. 2003. Kaposi's sarcoma-associated herpesvirus induces the phosphatidylinositol 3-kinase-PKC-zeta-MEK-ERK signaling pathway in target cells early during infection: implications for infectivity. J. Virol. 77:1524-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naranatt, P. P., H. H. Krishnan, S. R. Svojanovsky, C. Bloomer, S. Mathur, and B. Chandran. 2004. Host gene induction and transcriptional reprogramming in Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8)-infected endothelial, fibroblast, and B cells: insights into modulation events early during infection. Cancer Res. 64:72-84. [DOI] [PubMed] [Google Scholar]
- 44.Okamura, H., J. Aramburu, C. Garcia-Rodriguez, J. P. Viola, A. Raghavan, M. Tahiliani, X. Zhang, J. Qin, P. G. Hogan, and A. Rao. 2000. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol. Cell 6:539-550. [DOI] [PubMed] [Google Scholar]
- 45.Park, J., N. R. Yaseen, P. G. Hogan, A. Rao, and S. Sharma. 1995. Phosphorylation of the transcription factor NFATp inhibits its DNA binding activity in cyclosporin A-treated human B and T cells. J. Biol. Chem. 270:20653-20659. [DOI] [PubMed] [Google Scholar]
- 46.Pham, L., O. Bezouglaia, P. M. Camargo, J. M. Nervina, and S. Tetradis. 2007. Prostanoids induce egr1 gene expression in cementoblastic OCCM cells. J. Periodontal Res. 42:486-493. [DOI] [PubMed] [Google Scholar]
- 47.Rao, A., C. Luo, and P. G. Hogan. 1997. Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 15:707-747. [DOI] [PubMed] [Google Scholar]
- 48.Ridley, S. H., J. L. Dean, S. J. Sarsfield, M. Brook, A. R. Clark, and J. Saklatvala. 1998. A p38 MAP kinase inhibitor regulates stability of interleukin-1-induced cyclooxygenase-2 mRNA. FEBS Lett. 439:75-80. [DOI] [PubMed] [Google Scholar]
- 49.Ristimaki, A., S. Garfinkel, J. Wessendorf, T. Maciag, and T. Hla. 1994. Induction of cyclooxygenase-2 by interleukin-1 alpha. Evidence for post-transcriptional regulation. J. Biol. Chem. 269:11769-11775. [PubMed] [Google Scholar]
- 50.Rosenberg, D., L. Groussin, E. Jullian, K. Perlemoine, X. Bertagna, and J. Bertherat. 2002. Role of the PKA-regulated transcription factor CREB in development and tumorigenesis of endocrine tissues. Ann. N. Y. Acad. Sci. 968:65-74. [DOI] [PubMed] [Google Scholar]
- 51.Sadagopan, S., N. Sharma-Walia, M. V. Veettil, H. Raghu, R. Sivakumar, V. Bottero, and B. Chandran. 2007. Kaposi's sarcoma-associated herpesvirus induces sustained NF-kappaB activation during de novo infection of primary human dermal microvascular endothelial cells that is essential for viral gene expression. J. Virol. 81:3949-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sawasdikosol, S., S. Pyarajan, S. Alzabin, G. Matejovic, and S. J. Burakoff. 2007. Prostaglandin E2 activates HPK1 kinase activity via a PKA-dependent pathway. J. Biol. Chem. 282:34693-34699. [DOI] [PubMed] [Google Scholar]
- 53.Sharma-Walia, N., H. H. Krishnan, P. P. Naranatt, L. Zeng, M. S. Smith, and B. Chandran. 2005. ERK1/2 and MEK1/2 induced by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) early during infection of target cells are essential for expression of viral genes and for establishment of infection. J. Virol. 79:10308-10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma-Walia, N., P. P. Naranatt, H. H. Krishnan, L. Zeng, and B. Chandran. 2004. Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 envelope glycoprotein gB induces the integrin-dependent focal adhesion kinase-Src-phosphatidylinositol 3-kinase-rho GTPase signal pathways and cytoskeletal rearrangements. J. Virol. 78:4207-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharma-Walia, N., A. G. Paul, V. Bottero, S. Sadagopan, M. V. Veettil, N. Kerur, and B. Chandran. 2010. Kaposi's sarcoma associated herpes virus (KSHV) induced COX-2: a key factor in latency, inflammation, angiogenesis, cell survival and invasion. PLoS Pathog. 6:e1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma-Walia, N., H. Raghu, S. Sadagopan, R. Sivakumar, M. V. Veettil, P. P. Naranatt, M. M. Smith, and B. Chandran. 2006. Cyclooxygenase 2 induced by Kaposi's sarcoma-associated herpesvirus early during in vitro infection of target cells plays a role in the maintenance of latent viral gene expression. J. Virol. 80:6534-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaw, G., and R. Kamen. 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46:659-667. [DOI] [PubMed] [Google Scholar]
- 58.Shaw, K. T., A. M. Ho, A. Raghavan, J. Kim, J. Jain, J. Park, S. Sharma, A. Rao, and P. G. Hogan. 1995. Immunosuppressive drugs prevent a rapid dephosphorylation of transcription factor NFAT1 in stimulated immune cells. Proc. Natl. Acad. Sci. U. S. A. 92:11205-11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaywitz, A. J., and M. E. Greenberg. 1999. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 68:821-861. [DOI] [PubMed] [Google Scholar]
- 60.Shelby, B. D., A. Nelson, and C. Morris. 2005. Gamma-herpesvirus neoplasia: a growing role for COX-2. Microsc. Res. Tech. 68:120-129. [DOI] [PubMed] [Google Scholar]
- 61.Shibasaki, F., E. R. Price, D. Milan, and F. McKeon. 1996. Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature 382:370-373. [DOI] [PubMed] [Google Scholar]
- 62.Sirois, J., and J. S. Richards. 1993. Transcriptional regulation of the rat prostaglandin endoperoxide synthase 2 gene in granulosa cells. Evidence for the role of a cis-acting C/EBP beta promoter element. J. Biol. Chem. 268:21931-21938. [PubMed] [Google Scholar]
- 63.Smith, W. L., D. L. DeWitt, and R. M. Garavito. 2000. Cyclooxygenases: structural, cellular, and molecular biology. Annu. Rev. Biochem. 69:145-182. [DOI] [PubMed] [Google Scholar]
- 64.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steer, S. A., J. M. Moran, B. S. Christmann, L. B. Maggi, Jr., and J. A. Corbett. 2006. Role of MAPK in the regulation of double-stranded RNA- and encephalomyocarditis virus-induced cyclooxygenase-2 expression by macrophages. J. Immunol. 177:3413-3420. [DOI] [PubMed] [Google Scholar]
- 66.Streicher, J. M., S. Ren, H. Herschman, and Y. Wang. 2010. MAPK-activated protein kinase-2 in cardiac hypertrophy and cyclooxygenase-2 regulation in heart. Circ. Res. 106:1434-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Subbaramaiah, K., J. C. Hart, L. Norton, and A. J. Dannenberg. 2000. Microtubule-interfering agents stimulate the transcription of cyclooxygenase-2. Evidence for involvement of ERK1/2 AND p38 mitogen-activated protein kinase pathways. J. Biol. Chem. 275:14838-14845. [DOI] [PubMed] [Google Scholar]
- 68.Tamura, M., S. Sebastian, B. Gurates, S. Yang, Z. Fang, and S. E. Bulun. 2002. Vascular endothelial growth factor up-regulates cyclooxygenase-2 expression in human endothelial cells. J. Clin. Endocrinol. Metab. 87:3504-3507. [DOI] [PubMed] [Google Scholar]
- 69.Taylor, S. S., C. Kim, C. Y. Cheng, S. H. Brown, J. Wu, and N. Kannan. 2008. Signaling through cAMP and cAMP-dependent protein kinase: diverse strategies for drug design. Biochim. Biophys. Acta 1784:16-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Veettil, M. V., S. Sadagopan, N. Sharma-Walia, F. Z. Wang, H. Raghu, L. Varga, and B. Chandran. 2008. Kaposi's sarcoma-associated herpesvirus forms a multimolecular complex of integrins (alphaVbeta5, alphaVbeta3, and alpha3beta1) and CD98-xCT during infection of human dermal microvascular endothelial cells, and CD98-xCT is essential for the postentry stage of infection. J. Virol. 82:12126-12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Veettil, M. V., N. Sharma-Walia, S. Sadagopan, H. Raghu, R. Sivakumar, P. P. Naranatt, and B. Chandran. 2006. RhoA-GTPase facilitates entry of Kaposi's sarcoma-associated herpesvirus into adherent target cells in a Src-dependent manner. J. Virol. 80:11432-11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vieira, J., M. L. Huang, D. M. Koelle, and L. Corey. 1997. Transmissible Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in saliva of men with a history of Kaposi's sarcoma. J. Virol. 71:7083-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang, X., and R. D. Klein. 2007. Prostaglandin E2 induces vascular endothelial growth factor secretion in prostate cancer cells through EP2 receptor-mediated cAMP pathway. Mol. Carcinog. 46:912-923. [DOI] [PubMed] [Google Scholar]
- 74.Wu, G., A. P. Mannam, J. Wu, S. Kirbis, J. L. Shie, C. Chen, R. J. Laham, F. W. Sellke, and J. Li. 2003. Hypoxia induces myocyte-dependent COX-2 regulation in endothelial cells: role of VEGF. Am. J. Physiol. Heart Circ. Physiol. 285:H2420-H2429. [DOI] [PubMed] [Google Scholar]
- 75.Wu, W., R. A. Silbajoris, Y. E. Whang, L. M. Graves, P. A. Bromberg, and J. M. Samet. 2005. p38 and EGF receptor kinase-mediated activation of the phosphatidylinositol 3-kinase/Akt pathway is required for Zn2+-induced cyclooxygenase-2 expression. Am. J. Physiol. Lung Cell. Mol. Physiol. 289:L883-L889. [DOI] [PubMed] [Google Scholar]
- 76.Xie, W., J. R. Merrill, W. S. Bradshaw, and D. L. Simmons. 1993. Structural determination and promoter analysis of the chicken mitogen-inducible prostaglandin G/H synthase gene and genetic mapping of the murine homolog. Arch. Biochem. Biophys. 300:247-252. [DOI] [PubMed] [Google Scholar]
- 77.Yamaki, T., K. Endoh, M. Miyahara, I. Nagamine, N. Thi Thu Huong, H. Sakurai, J. Pokorny, and T. Yano. 2004. Prostaglandin E2 activates Src signaling in lung adenocarcinoma cell via EP3. Cancer Lett. 214:115-120. [DOI] [PubMed] [Google Scholar]
- 78.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. U. S. A. 93:6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zoeteweij, J. P., A. V. Moses, A. S. Rinderknecht, D. A. Davis, W. W. Overwijk, R. Yarchoan, J. M. Orenstein, and A. Blauvelt. 2001. Targeted inhibition of calcineurin signaling blocks calcium-dependent reactivation of Kaposi sarcoma-associated herpesvirus. Blood 97:2374-2380. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.