Abstract
Although marine picophytoplankton are at the base of the global food chain, accounting for half of the planetary primary production, they are outnumbered 10 to 1 and are largely controlled by hugely diverse populations of viruses. Eukaryotic microalgae form a ubiquitous and particularly dynamic fraction of such plankton, with environmental clone libraries from coastal regions sometimes being dominated by one or more of the three genera Bathycoccus, Micromonas, and Ostreococcus (class Prasinophyceae). The complete sequences of two double-stranded (dsDNA) Bathycoccus, one dsDNA Micromonas, and one new dsDNA Ostreococcus virus genomes are described. Genome comparison of these giant viruses revealed a high degree of conservation, both for orthologous genes and for synteny, except for one 36-kb inversion in the Ostreococcus lucimarinus virus and two very large predicted proteins in Bathycoccus prasinos viruses. These viruses encode a gene repertoire of certain amino acid biosynthesis pathways never previously observed in viruses that are likely to have been acquired from lateral gene transfer from their host or from bacteria. Pairwise comparisons of whole genomes using all coding sequences with homologous counterparts, either between viruses or between their corresponding hosts, revealed that the evolutionary divergences between viruses are lower than those between their hosts, suggesting either multiple recent host transfers or lower viral evolution rates.
Phytoplankton is responsible for about half of the photosynthetic activity of the planet (13), with the other half being ensured by terrestrial plants. Phytoplankton is essentially composed of unicellular organisms which have a high turnover rate, and whereas terrestrial plants are renewed on average once every 9 years, the global phytoplankton population is replaced approximately every week (13). Although the ecological importance of viruses has previously been debated, they are now recognized as major players in regulating these highly dynamic phytoplankton populations. Indeed, viruses are the most numerous biological entities in the ocean, infecting all marine organisms from prokaryotes to uni- and multicellular eukaryotes (36). Cell death following viral infection produces particulate and dissolved organic matter that in turn fuels the growth of other phytoplankton. The importance of this viral shunt is not yet well understood although some studies suggest that it constitutes an important flux that must be taken into account in marine trophic transfer models.
Among viruses affecting the eukaryotic phytoplankton, several large double-stranded DNA (dsDNA) viruses have been described, and these viruses have been named phycodnaviruses because they infect algae (12). However, the term “alga” has no evolutionary significance, and phycodnaviruses infect phylogenetically distantly related organisms. Thus, comparisons of dsDNA viruses infecting organisms as diverse as haptophytes, dinoflagellates, and green algae likely span the same order of evolutionary distances as comparisons of viruses of animals with those of plants. In order to understand the evolution of these viruses, comparisons between more closely related host-virus combinations are desirable and are even more valuable if DNA sequence information about their host species' genomes is available. Viruses infecting Chlorophyta, which include most green algae, thus present attractive systems for such analyses. In this phylum, both prasinoviruses and chloroviruses, infecting Prasinophyceae and Trebouxiophyceae, respectively, have been described.
Several dsDNA viruses have been described infecting different Chlorella sp. unicellular green algae (Trebouxiophyceae), which are symbionts of the ciliate Paramecium bursaria (14, 15, 44) or of the heliozoon Acanthocystis turfacea (16). They belong to the nucleocytoplasmic large DNA viruses (NCLDV), indicating that they either replicate exclusively in the cytoplasm of the host cell or start their life cycle in the host nucleus but complete it in the cytoplasm (20, 46). NCLDV can also infect members of the Prasinophyceae, an ecologically important class of microalgae that are found in all oceans (39). Prasinophyceae can dominate the eukaryotic picoplankton fraction in coastal areas, and a high proportion of the DNA sequences in many environmental DNA clone libraries can be attributed to one or more of the three genera Bathycoccus, Micromonas, and Ostreococcus (31, 42). Two dsDNA Ostreococcus viruses have been sequenced (9, 40), but no viruses specific to Bathycoccus have yet been reported (2, 6). Both dsDNA and RNA Micromonas viruses have been described although information about their genomes is not yet available (5, 8). Phylogenetic analyses based on their DNA polymerase or major capsid gene sequences suggest that chloroviruses and prasinoviruses form a monophyletic group (4). Since host genomes of two Chlorella species and three Prasinophyceae genera are available, the possibility of horizontal gene transfer (HGT) between hosts and their viruses can be investigated and might provide key insights into their coevolution. Both chloroviruses and prasinoviruses have a DNA polymerase gene but no DNA-dependent RNA polymerase, in contrast to the Emiliania huxleyi virus EhV-86 (41), which is consistent with a large evolutionary divergence between these viruses.
Here, we describe the complete sequences of two dsDNA Bathycoccus virus genomes, one dsDNA Micromonas virus genome, and one new dsDNA Ostreococcus virus genome. Comparison between them revealed a high degree of conservation, both for orthologous genes and for synteny. Several specific pathways, such as amino acid biosynthesis, are encoded differentially by genes never previously identified before in viruses, and we compared these genomes with those of the six available Chlorella viruses. We propose a new phylogeny to reconcile the wide evolutionary distances between phycodnavirus genomes with those of their hosts.
MATERIALS AND METHODS
Culture of host algal strains and virus OtV5.
The Prasinophyceae host strains Bathycoccus sp. strain RCC1105, Micromonas sp. strain RCC1109, and Ostreococcus lucimarinus CCMP2972 were used to screen for the presence of viruses in seawater by plaque lysis. After two rounds of single-plaque purifications, viruses were subsequently produced as described for OtV5 (9) by infecting 2 liters of liquid culture in exponential phase (e.g., 5 × 107 cells/ml). Briefly, after centrifugation, lysed cultures were passed sequentially through 5-μm- and 0.45-μm-pore-size filters to remove large cellular debris. Virus filtrates were concentrated by ultrafiltration with a 50,000-molecular-weight (MW) cutoff unit (Amicon Ultra; Millipore) to a final volume of 1 ml. Virus concentrates were embedded in agarose for pulsed-field gel electrophoresis (PFGE).
Viral genomes.
Genomic DNA for sequence analysis was prepared by embedding viral particles in agarose, lysing the particles using proteinase K, and visualization by PFGE (9). Briefly, a block of agarose containing the viral DNA was then cut out and digested by GELase (Epicentre, Tebu). The DNA was precipitated by adding ethanol. Individual lots of viral nucleic acids were digested overnight at 37°C by DNase, RNase, or different restriction enzymes, and the digested products were analyzed by electrophoresis (on 1% agarose at 100 V/cm for 20 min). Purified viral DNAs were subjected to 454 sequencing and assembly (Cogenics, France) before gap filling by PCR using custom-made primers.
Sequence annotation.
The complete genomic sequence was annotated using the Artemis software, which first considers all open reading frames (ORFs). Putative adjacent coding sequences (CDSs) encoding a minimum of 65 amino acids with appropriate start and stop codons were then chosen and screened against the curated Pfam-A profiles for functional motifs using the Perl script Pfam_scan.pl (Wellcome Trust Sanger Institute, United Kingdom) with default settings. The E-value cutoff of 10−5 was conserved as profiles were mostly defined from distantly related sequences, and many functional motifs with relatively high E values are confirmed by significant BLASTP alignments with proteins having the same putative function.
Whole-proteome divergence comparison.
Host genome annotations were downloaded from the Joint Genome Institute (JGI [http://genome.jgi-psf.org/]) for O. lucimarinus and Micromonas and from the University of Gent (http://bioinformatics.psb.ugent.be/genomes/) for Ostreococcus tauri and Bathycoccus. All orthologous host and virus gene pairs were identified by reciprocal blast best hit (BLASTP E value threshold of 0.001). Each orthologous pair was aligned with the Needleman-Wunsch algorithm (EMBOSS package) with default parameters and processed with codes developed in C language to estimate amino acid identity.
Phylogenetic reconstruction.
Sequences were read and corrected using BioEdit (version 281-7.0.0). BLAST was used to search for similar sequences in public databases, sequences were automatically aligned using MAFFT, version 5, and conserved residues were selected using GBlocks (7). Phylogenetic reconstructions were carried out by using both Bayesian inference (BI) and maximum likelihood (ML). Bayesian analysis was done with MrBayes, version 3.1.2, with four chains of 106 generations; trees were sampled every 100 generations, with the burn-in value set to 20% of the sampled trees. We checked that standard deviations of the split frequencies fell below 0.01 to ensure convergence in tree search. Sequences were analyzed with a mixed amino acid model. Maximum-likelihood reconstructions were carried out using PhyML (19) with an evolutionary model selected via the Akaike information criterion with ProtTest (1) and validated with 100 bootstrap replicates.
Nucleotide sequence accession numbers.
The genome data have been submitted to Genbank under the following accession numbers: for MpV1, HM004429; BpV1, HM004432; BpV2, HM004430; and OlV1, HM004431.
RESULTS
Isolation of viruses and genomic DNA.
Ostreococcus and Micromonas viruses (OlV1 and MpV1, respectively) were isolated from eutrophic northwestern Mediterranean coastal lagoons, whereas the two Bathycoccus viruses (BpV1 and BpV2) were isolated from the open sea close to Banyuls Bay (3) using a plaque lysis method described previously (9). This protocol favors the isolation of lytic viruses. The four new viruses were isolated from the host strains Bathycoccus sp. RCC1105, Micromonas sp. RCC1109, and O. lucimarinus CCMP2972 (Roscoff Culture Collection and Centre for Culture of Marine Phytoplankton).
Global viral genome characteristics.
Genome sizes were first estimated by PFGE, which also showed their linearity, and then confirmed by sequencing. The four new viruses (BpV1, BpV2, MpV1, and OlV1) have similar GC contents of between 37 to 45%, clearly lower than the GC contents of their corresponding hosts (50 to 64%) (10, 33, 43). The six prasinoviruses (including OtV1 and OtV5 already published) have similar genome sizes, ranging from 184 to 198 kb, which is up to 50% smaller than the 288 to 368 kb reported for the six published chlorovirus genomes (Table 1) (14, 15, 16). The smaller genome size of prasinoviruses is partly explained by their smaller CDSs and intergenic regions (IR) (744 ± 27 bp for the CDSs and 40 ± 5 bp for the IRs of prasinoviruses versus 846 ± 34 bp for the CDSs and 90 ± 12 bp for the IRs of the chloroviruses) and partly because they encode fewer CDSs (203 to 251 CDSs for prasinoviruses and 327 to 385 CDSs for chloroviruses). Terminal inverted repeats have been found at the ends of the four new viral genomes and range from 250 bp for BpV2 to 2,150 bp for OlV1.
TABLE 1.
General characteristics of the 12 Chlorophyta viral genomes
| Virusa | Host(s)b | Genome size (bp) | No. of ORFs | Avg ORF length (bp) | Avg intergenic length (bp) | %G+C | No. of tRNAs |
|---|---|---|---|---|---|---|---|
| Prasinoviruses | |||||||
| BpV1 | Bathycoccus sp. RCC1105 | 198,519 | 203 | 793b | 37 | 37 | 4 |
| BpV2 | Bathycoccus sp. RCC1105 | 187,069 | 210 | 746b | 38 | 37 | 4 |
| MpV1 | Micromonas sp. RCC1109 | 184,095 | 244 | 715 | 39 | 41 | 6 |
| OlV1 | O. lucimarinus CCMP2972 | 194,022 | 251 | 730 | 43 | 41 | 5 |
| OtV1 | O. tauri RCC745 | 191,761 | 240 | 750 | 49 | 45 | 4 |
| OtV5 | O. tauri RCC745 | 186,245 | 243 | 732 | 34 | 45 | 5 |
| Chloroviruses | |||||||
| AR158 | Paramecium-Chlorella NC64A | 344,690 | 360 | 862 | 95 | 41 | 6 |
| ATCV-1 | Acanthocystis-Chlorella SAG3.83 | 288,047 | 327 | 809 | 72 | 49 | 11 |
| FR483 | Paramecium-Chlorella Pbi | 321,240 | 331 | 876 | 94 | 45 | 9 |
| MT325 | Paramecium-Chlorella Pbi | 314,335 | 331 | 868 | 82 | 45 | 10 |
| NY-2A | Paramecium-Chlorella NC64A | 368,683 | 385 | 865 | 93 | 41 | 7 |
| PBCV-1 | Paramecium-Chlorella NC64A | 330,743 | 366 | 798 | 106 | 40 | 11 |
For the two Bathycoccus viruses, the two large unknown and specific CDSs, which represent up to 14% of the viral genomes, have been removed for calculation of the average CDS size. The six chloroviruses have been reannotated, using the protocol used for prasinoviruses, from sequences submitted to GenBank.
All of the chloroviruses grow inside green algae of the genus Chlorella, which may in turn be a symbiont of the larger protist Acanthacystis turfacea or Paramecium bursaria, whereas the prasinoviruses grow inside free-living algae of the order Mamiellales (class Prasinophyceae).
The six prasinoviruses, like the six chloroviruses, encode a DNA polymerase but not a DNA-dependent RNA polymerase. These green algal viruses thus use the host RNA polymerase to transcribe their genes and consequently must reach the nucleus to start a lytic cycle.
Genes common to the six prasinoviruses.
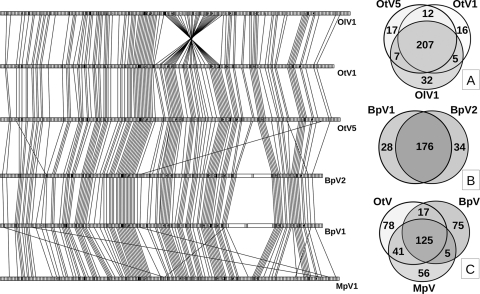
The four new viruses have 125 predicted genes in common with OtV1 and OtV5 (best BLAST hits, E-value cutoff of 10−5). These “core genes” tend to cluster in the central region of the six genomes, whereas specific genes cluster at their extremities (a chi square test on core gene distribution using a 20-gene-long sampling window gave P values varying from <0.0001 to 0.022 for the six prasinoviruses) (see Table S1 in the supplemental material). Among the subset of genes also common to all of the chloroviruses, 20 have potential functions: 6 are involved in DNA replication and recombination, 4 are involved in transcription, 4 are involved in nucleotide metabolism, and 6 are putative capsomers (see Table S2 in the supplemental material). The predicted functions of the remaining six genes include an adenine methyl transferase, a prolyl hydroxylase, a ubiquitin hydrolase like cysteine peptidase, a patatin-like phospholipase, a GDP-d-mannose dehydratase epimerase, and a Ser/Thr protein kinase. Three to five tRNAs are also encoded in prasinoviruses, but Asn-tRNA is the only one conserved in all prasinoviruses and in chloroviruses (Table 2). Seventy-five, 56, and 78 CDSs specific to the phylogenetic genera of their corresponding hosts were identified for Bathycoccus, Micromonas, and Ostreococcus viruses, respectively. Remarkable global colinearity was observed between the six genomes (Fig. 1; see also Fig. S1 in the supplemental material), except at their extremities, i.e., the first 10,000 and the last 10,000 bp, where the terminal inverted repeats are present (see Table S2), and a large 36-kbp inversion was observed in OlV1. Seven or eight CDSs for putative capsid-like proteins (clp) were identified in each of the viral genomes through their similarity to the characterized chloroviruses PBCV-1 major capsid protein (mcp) Vp54 (see Fig. S2 in the supplemental material) (45). Two surprisingly long CDSs (11,202 and 17,067 bp for BpV1 and 9,378 and 11,028 bp for BpV2) represent 10 to 15% of the two Bathycoccus viral genomes (Fig. 1). They have no identifiable Pfam domains and share regions showing similarities ranging from nearly identical to weakly similar. This suggests that these four genes may have a common origin.
TABLE 2.
tRNAs in viruses of Chlorophyta and E. huxleyi
| tRNA speciesa | No. of tRNAs |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prasinovirusb |
Chlorovirus |
E. huxleyi virus |
|||||||||||
| BpV1 | BpV2 | MpV1 | OtV1 | OtV5 | OlV1 | PBCV-1 | NY-2A | AR-158 | MT-325 | FR-483 | ATCV-1 | EhV-86 | |
| Lys-tRNA | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 1 | 1 | 1 | 0 |
| Leu-tRNA | 1(AAG) | 1(AAG) | 1(TAA) | 0 | 0 | 0 | 2 | 2 | 2 | 1 | 1 | 1 | 1 |
| Arg-tRNA | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Val-tRNA | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 0 |
| Ile-tRNA | 1(TAT) | 1(TAT) | 1(TAT) | 1(TAT) | 1(TAT) | 1(TAT) | 1 | 0 | 1 | 1 | 1 | 0 | 1 |
| Gln-tRNA | 0 | 0 | 1(TTG) | 1(TTG) | 1(TTG) | 1(TTG) | 0 | 0 | 0 | 1 | 1 | 0 | 1 |
| Tyr-tRNA | 0 | 1(GTA) | 1(GTA) | 0 | 1(GTA) | 1(GTA) | 1 | 1 | 0 | 1 | 1 | 1 | 0 |
| Asn-tRNA | 1(GTT) | 1(GTT) | 1(GTT) | 1(GTT) | 1(GTT) | 1(GTT) | 2 | 1 | 1 | 2 | 2 | 2 | 1 |
| Thr-tRNA | 0 | 0 | 1(AGT) | 0 | 1(AGT) | 1(AGT) | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| Gly-tRNA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 |
| Phe-tRNA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| Ser-tRNA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Asp-tRNA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
Among the diverse RNA molecules encoded by this group of viruses, only asparagine (Asn) tRNA (in boldface) is present in all of the viruses.
The anticodons encoded by the prasinovirus tRNAs are shown as subscripts.
FIG. 1.
Schematic representation of colinearity of common prasinovirus genes. The six prasinovirus genomes are represented proportionally to their respective sizes, and each CDS is shown as a small open rectangle. Each rectangle has an identical size, unrelated to its real size, except for the two big extra CDSs found in BpV1 and BpV2. The genes common to the six viral genomes are shaded, similarly and joined to their orthologs in neighboring genomes by lines. At right are Venn diagrams showing the numbers of common and specific genes among prasinoviruses: the three Ostreococcus viruses OlV1, OtV1, OtV5 (A), the two Bathycoccus viruses (B), and the six prasinoviruses, showing counts of genes specific to each genus and those common to two or three genera (C).
All of the CDSs identified in the six viral genomes were classified in one of three categories: (i) coding sequences showing similarity to known genes in databases and referred to as putative identified genes (PIG); (ii) putative unknown genes (PUG) representing genes having similarity to sequences of unknown functions; and (iii) sequences showing no hits and being putative orphan genes (POG). The distribution of genes among these three categories differs sharply between core and specific genes. The proportion of PIG in core genes is 35% compared to 8, 11, or 13% in specific genes for BpV1, MpV1, or OtV5, respectively (see Fig. S3 in the supplemental material), which is consistent with the higher proportion of orphans in the host's specific genes (22).
Phylogeny of green algal viruses.
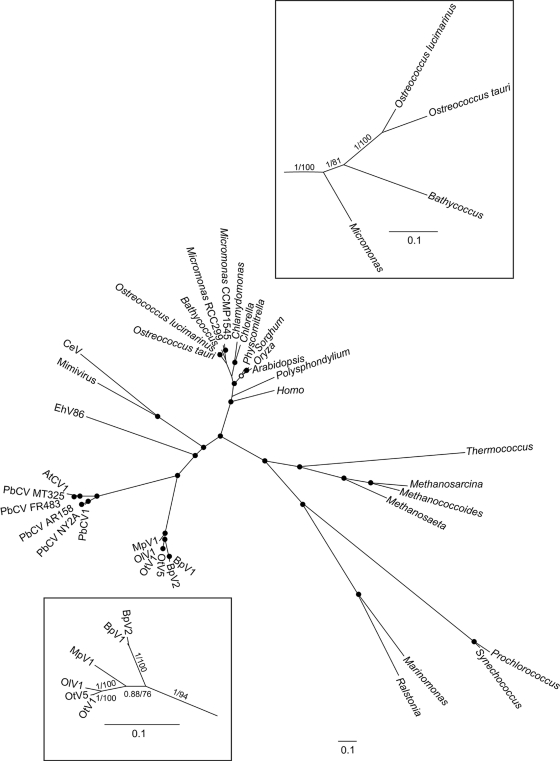
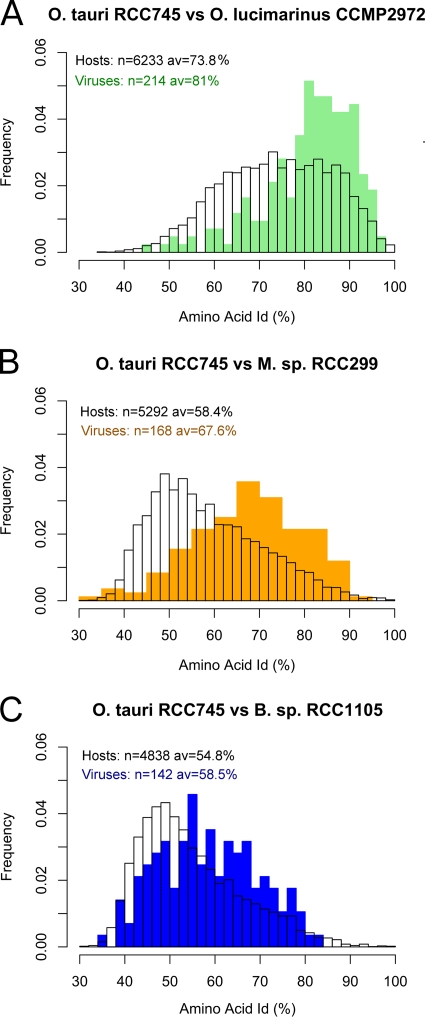
Based on the DNA polymerase gene, the six Prasinophyceae viruses are monophyletic, and the six chloroviruses are also grouped in one clade emerging beside prasinoviruses (Fig. 2). These two clades are grouped in one superclade that can be named Chlorophytaviruses or green algal viruses. Other NCLDV infecting different eukaryotic lineages appeared to be distantly related, and no closer relationship was observed between Chlorophytaviruses and another phycodnavirus like the Emiliania virus than with other NCLDV infecting other eukaryote lineages such as metazoans (Fig. 2). Host cellular polymerase genes form a distinct clade, completely independent from the viral DNA polymerases, and reflect the known phylogenetic relationships between these groups. Average amino acid identity among all orthologous genes between all pairs of these six prasinoviruses varied between 58% and 98% for OtV1 and BpV2 and for OtV1 and OtV5, respectively (see Fig. S4 in the supplemental material). Surprisingly, identity was lower between the host's orthologous proteins than between the orthologous proteins of their virus, whatever host/virus pair was considered (Fig. 3), suggesting greater evolutionary distance between hosts than between viruses. Consistent with this, the percentage of common genes is lower between host pairs than between their viruses (e.g., O. tauri has 81% common genes with O. lucimarinus, whereas OtV5 has 88% common genes with OlV1; Micromonas sp. RCC299 has 55% common genes with O. lucimarinus, whereas MpV has 72% common genes with OlV).
FIG. 2.
Phylogeny of NCLDV, including diverse eukaryote and prokaryote sequences, based on Bayesian inference (BI) and maximum likelihood (ML) analyses. Black dots indicate posterior probabilities (pp; BI) of >0.9 and bootstrap proportions (bp; ML) of >70%, and white dots indicate pp of >0.9 or bp of >70%. The overall phylogeny of diverse lineages was accomplished using the full-length DNA polymerase B, predicted amino acid sequence, which is a gene conserved in all NCLDV and all prokaryotes and eukaryotes. To resolve the branches concerning prasinoviruses and their hosts more clearly (boxes), the amino acid sequences of five conserved genes common to both hosts and viruses (DNA polymerase B, proliferating cell nuclear antigen, ribonucleotide reductase large and small subunits, and thymidine synthase) were concatenated, permitting comparison of evolutionary distances. Numbers on branches are maximum likelihood/Bayesian inference support values.
FIG. 3.
Distribution of amino acid identities of three host proteomes versus their respective viruses. (A) O. tauri RCC745 versus O. lucimarinus CCMP2972 with OtV5 versus OlV1. (B) O. tauri RCC745 versus Micromonas (M) sp. RCC299 with OtV5 versus MpV1. (C) O. tauri RCC745 versus Bathycoccus sp. RCC1105 with OtV5 versus BpV1. n, number of orthologous genes analyzed; av, average amino acid identity. All 15 possible host-virus pairwise comparisons showed the same trends.
Prasinovirus-specific metabolic processes.
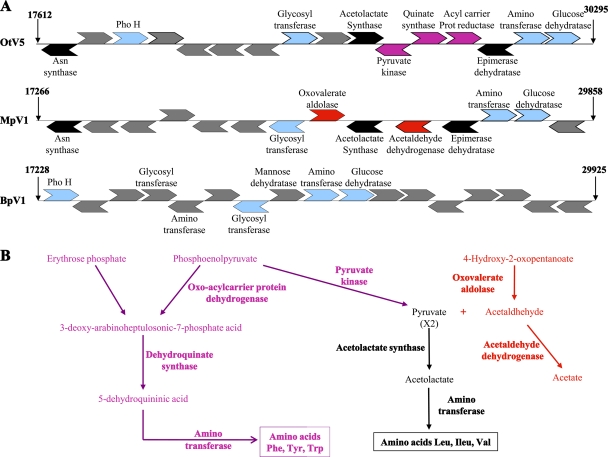
Some predicted virus-specific functionalities are remarkable. Different gene sets encoding enzymes involved in several amino acid biosynthesis pathways are found in Ostreococcus and Micromonas viral genomes. Together, these genes probably provide alternative pathways for the biosynthesis of several amino acids and are grouped in a cluster which is absent in the two Bathycoccus viruses (Fig. 4). The acetolactate synthase (ALS) gene, previously described in OtV1 (40) and OtV5 (9), which is known to be involved in synthesis of nonpolar lateral chain amino acids such as leucine, isoleucine, and valine, is present in the Micromonas and the three Ostreococcus viral genomes but not in those of the Bathycoccus viruses. In contrast, the 3-dehydroquinate synthase gene, involved in synthesis of the aromatic amino acids phenylalanine, tyrosine, and tryptophan is present in the three Ostreococcus viral genomes but not in Micromonas or in Bathycoccus. Finally, oxovalerate aldolase, which catalyzes the transformation of 4-hydroxypentanoate to pyruvate and acetaldehyde, and acetaldehyde dehydrogenase, which transforms the highly toxic acetaldehyde to nontoxic acetate, are encoded only in the Micromonas viral genome. Genes encoding enzymes involved in these interrelated metabolic pathways are all clustered close to the 5′ extremities of the genomes (between bp 17000 and 30000 bp) (Fig. 4A) in both Ostreococcus and Micromonas virus genomes. A predicted asparagine synthase gene is again found only in Micromonas and Ostreococcus viruses but is absent in Bathycoccus. This gene is located at the 5′ extremity of the cluster (Fig. 4A) and has also been described in the mimiviruses (34). To our knowledge, no acetolactate synthase, dehydroquinate synthase, oxovalerate aldolase, or acetaldehyde dehydrogenase genes have been described previously in viral genomes, and this is the first report of these alternative biochemical pathways in viruses. All of these genes give highest BLAST scores with bacterial genes, and phylogenetic analyses also suggest a bacterial origin, except for the asparagine synthase, which is most likely derived from the “green lineage” (see Fig. S5 in the supplemental material).
FIG. 4.
(A) Gene clusters encoding amino acid synthesis pathways. Representation of CDS positions located between bp 17000 and about bp 30000 for BpV1, MpV1, and OtV5. (B) Amino acid biosynthesis pathways encoded by Micromonas or Ostreococcus viruses: biosynthesis pathways for Leu, Ile, Val, Tyr, Trp, and Phe. Text written in black corresponds to reactions catalyzed by enzymes encoded by both Micromonas and Ostreococcus viruses, text in red represents reactions with enzymes found only in the Micromonas genome, and text in pink represents reactions specific to the Ostreococcus genomes. Note that none of these enzymes is present in the two Bathycoccus viral genomes.
Several other genes are predicted to fulfill functions important for host-virus interactions (see Fig S6 in the supplemental material for more details). For example, the heat shock gene hsp70 has been recruited, most likely by HGT, from its host (see Fig. S6A), and numerous independent acquisitions of this gene by taxonomically distant viruses (reviewed in reference 28) highlight the importance of this functionality. Replication of double-stranded DNA viruses requires large amounts of deoxynucleotides, including dTTP, and degraded recycled host DNA may be either insufficient or unavailable when viral genome replication starts. In OtV5 at least, new viral genomes are observed about 4 h after infection, and, at that time, there is no degradation of host chromosomes, which remain intact until the end of the lytic cycle (9). For the huge deoxynucleotide biosynthesis necessary for viral replication, many DNA viruses encode enzymes necessary for deoxynucleotide synthesis, as previously reported for PBCV-1 (47). The enzyme dCMP deaminase encoded by 52 dsDNA viruses (Interpro [http://www.ebi.ac.uk/interpro/IEntry?ac=IPR002125]) is a key enzyme for one of the three synthetic routes for dTTP via dUMP and is also found in the genomes of OlV1 and MpV1, providing an additional route to the ribonucleotide reductase pathway (see Fig. S6B in the supplemental material). MpV1, like several large-animal viruses, encodes a Cu-Zn superoxide dismutase that probably plays a role in the regulation of cellular apoptosis, possibly acting as a decoy for its cellular counterpart by binding a cellular copper chaperone (26, 37, 38) and a mannitol dehydrogenase that may similarly be involved either in controlling cellular superoxide or in the metabolism of mannitol as an energy source (21). Lipases, such as that predicted in OlV1, are known, for example, to manipulate host lipid metabolism during infection of humans (human cytomegalovirus [HCMV]) and are required for virulence in several viruses (23).
DISCUSSION
Among Prasinophyceae, the three genera Bathycoccus, Micromonas, and Ostreococcus are widespread and often dominate the eukaryotic picophytoplankton fraction, at least in coastal waters. Under appropriate conditions these green microalgae grow rapidly (18), but prasinoviruses are also present everywhere in marine waters, sometimes at high densities (3), and probably play a key role in the regulation of such populations by host cell lysis. However, despite their ecological importance, relatively little is known about these viruses. We show that their genome content and structure are surprisingly well conserved, with similar sizes and many common genes (50 to 60% of all CDSs), although their hosts are rather more divergent, as illustrated by comparing the two most closely related hosts, O. lucimarinus and O. tauri, whose proteome divergence is equivalent to that separating the human from chicken proteomes (22, 33).
Like other members of the Phycodnaviridae (3), some of the putative genes described here were probably recruited from HGT, and we note the importance of these in certain amino acid biosynthesis pathways in specific prasinoviruses. The intriguing cluster of genes involved in the biosynthesis of hydrophobic and aromatic amino acids is one example. However, although the capsomers are probably among the most highly expressed viral proteins, their amino acid contents did not show any overrepresentation of these amino acids in Ostreococcus and Micromonas viruses compared to Bathycoccus viruses, where these genes are absent (this is also true for the complete viral and host proteomes). These pathways might thus not be directly involved in building proteins but, rather, are used to produce metabolites, energy-yielding substrates, nutrients (a nitrogen source for all proteins), or even signaling molecules (17), which might be rate-limiting steps in the viral life cycle. Furthermore, these pathways occur in the chloroplast (35), an organelle that remains intact and active throughout the lytic cycle, at least in Micromonas (5). Conclusive evidence for recent HGT events is rare, being limited to only two CDSs, the hsp70 of Bathycoccus viruses and the proline dehydrogenase of Ostreococcus viruses (9). Taken together, our data are best explained by the classical hypothesis that dsDNA viruses originated by escape of a cellular replicon (reviewed in reference 29), followed initially by vertical coevolution of the viruses before the speciation of diverse hosts. Host speciation would then lead to the development of host specificity by natural selection of viral functions best adapted to that host. This, in turn, would reduce the frequency of host switching, with viruses then evolving with rather closely defined sets of functions within the different domains of known life.
Phylogenetic analyses using the complete polymerase B gene common to both algal hosts and their viruses show them to form two well-supported monophyletic clusters within a diverse range of taxa from different kingdoms and suggest a global pattern of coevolution although the precise phylogenetic relationships between the Prasinophyceae hosts and their viruses are not well resolved (Fig. 2). This is coherent with the proposition that NCLDV had a common ancestor predating the radiation of eukaryotes, encoding both a DNA polymerase and an RNA DNA-directed polymerase, and that they coevolved with their respective hosts, evolving into viruses that kept both enzymes in some lineages, whereas in others (such as in green algae) they kept only the DNA polymerase gene (30). This hypothesis of coevolution from a common ancestor is consistent with the observation that there is little evidence for frequent switching of viruses between hosts and is reminiscent of that proposed recently for picornaviruses, i.e., a common ancestor which coevolved with hosts in different lineages (25). However, at present we have insufficient data to assess a cophylogenetic scenario in the earlier evolution of the Prasinophyceae-virus interactions, and this remains a possibility (in Fig. 2, the boxed phylogenies based on five concatenated genes common to both hosts and viruses are not completely congruent). Given the astounding conclusion that the viral genomes are more conserved than those of their hosts and assuming coevolution, the host genomes might, indeed, be evolving faster than their corresponding viral genomes. This lower substitution rate of virus genomes per unit time could be the consequence of lower mutation rates per generation, longer generation times, or a higher proportion of sites under strong purifying selection in the prasinovirus genomes. There is evidence that the mutation rates per generation of dsDNA viruses are close to those estimated in multicellular species (11), and the first possibility seems unlikely although it has been shown that coevolution with viruses may speed up the mutation rate of the hosts in bacterial populations (32). Alternatively, in contrast to metazoans, prasinovirus hosts are fast-growing unicellular organisms that can divide more than once per day under favorable conditions (18), whereas the replication rate of viruses in the environment is unknown. Indeed, viral particles are stable in seawater in the laboratory over long periods (at least several months at 20°C and over 4 years at 4°C). Furthermore, well-supported data report that some lytic viruses may survive for long periods once buried in sediments, suggesting that viral particles might remain dormant in the environment (27), encountering their specific compatible host genotype infrequently, in contrast to their host cells, which continue to divide every day. Similar mutation rates per generation and different generation times in viruses and hosts may thus lead to an unusual virus-host equilibrium of slowly evolving viruses due to low replication rates, compared to faster-dividing, and consequently faster-evolving, hosts, like white pawns chasing the red queen. Recent theoretical studies suggest that species with more at stake kept ahead in the coevolutionary arms race, so that mutation rates may rise to higher levels in the host (29).
Prasinoviruses are grouped phylogenetically in one well-supported clade, with chloroviruses emerging closely, together forming a “supergroup” which can be named the Chlorophyta or green algal viruses. These green algal viruses are of particular interest for future host-virus interaction investigations as more potential hosts and viruses with completely analyzed genomes are available (see the JGI and Broad Institute websites).
Supplementary Material
Acknowledgments
We are very grateful to Laure Bellec for isolation of some viruses, Michèle Laudié for technical help in DNA sequencing, and Lucie Subirana for technical assistance in the production of host and viral cultures. We thank Florence Le Gall for sending us the RCC827 strain and Yannis Michalakis for stimulating discussions.
Funding was provided by the CNRS and an ANR grant BLAN07-1_200218 PICOVIR (coordinated by N.G.).
Footnotes
Published ahead of print on 22 September 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abascal, F., R. Zardoya, and D. Posada. 2005. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 21:2104-2105. [DOI] [PubMed] [Google Scholar]
- 2.Attoui, H., F. M. Jaafar, M. Belhouchet, P. de Micco, X. de Lamballerie, and C. P. Brussaard. 2006. Micromonas pusilla reovirus: a new member of the family Reoviridae assigned to a novel proposed genus (Mimoreovirus). J. Gen. Virol. 87:1375-1383. [DOI] [PubMed] [Google Scholar]
- 3.Bellec, L., N. Grimsley, E. Derelle, H. Moreau, and Y. Desdevises. 2010. Abundance, spatial distribution and genetic diversity of Ostreococcus tauri viruses in two different environments. Environ. Microbiol. Rep. 2:313-321. [DOI] [PubMed] [Google Scholar]
- 4.Bellec, L., N. Grimsley, H. Moreau, and Y. Desdevises. 2009. Phylogenetic analysis of new prasinoviruses (Phycodnaviridae) that infect the green unicellular algae Ostreococcus, Bathycoccus and Micromonas. Environ. Microbiol. Rep. 1:114-123. [DOI] [PubMed] [Google Scholar]
- 5.Brown, C. M., D. A. Campbell, and J. E. Lawrence. 2007. Resource dynamics during infection of Micromonas pusilla by virus MpV-Sp1. Environ. Microbiol. 9:2720-2727. [DOI] [PubMed] [Google Scholar]
- 6.Brussaard, C. P. D., A. A. M. Noordeloos, R. A. Sandaa, M. Heldal, and G. Bratbak. 2004. Discovery of a dsRNA virus infecting the marine photosynthetic protist Micromonas pusilla. Virology 319:280-291. [DOI] [PubMed] [Google Scholar]
- 7.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540-552. [DOI] [PubMed] [Google Scholar]
- 8.Cottrell, M. T., and C. A. Suttle. 1995. Dynamics of a lytic virus infecting the photosynthetic marine picoflagellate Micromonas pusilla. Limnol. Oceanogr. 40:730-739. [Google Scholar]
- 9.Derelle, E., C. Ferraz, M. L. Escande, S. Eychenié, R. Cooke, G. Piganeau, Y. Desdevises, L. Bellec, H. Moreau, and N. Grimsley. 2008. Life-cycle and genome of OtV5, a large DNA virus of the widespread pelagic marine unicellular green alga Ostreococcus tauri. PLoS One 3:e2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derelle, E., C. Ferraz, S. Rombauts, P. Rouzé, A. Z. Worden, F. Partensky, S. Robbens, S. Degroeve, S. Echeynié, R. Cooke, Y. Saeys, J. Wuyts, K. Jabbari, C. Bowler, S. Ball, J. P. Ral, F. Y. Bouget, G. Piganeau, B. De Baets, A. Picard, M. Delseny, J. Demaille, Y. Van de Peer, and H. Moreau. 2006. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. U. S. A. 103:11647-11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duffy, S., L. A. Shackelton, and E. C. Holmes. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9:267-276. [DOI] [PubMed] [Google Scholar]
- 12.Dunigan, D. D., L. A. Fitzgerald, and J. L. Van Etten. 2006. Phycodnaviruses: a peek at genetic diversity. Virus Res. 117:119-132. [DOI] [PubMed] [Google Scholar]
- 13.Field, B. C., M. J. Behrenfeld, J. T. Randerson, and P. Falkowski. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281:237-240. [DOI] [PubMed] [Google Scholar]
- 14.Fitzgerald, L. A., M. V. Graves, X. Li, T. Feldblyum, J. Hartigan, and J. L. Van Etten. 2007. Sequence and annotation of the 314-kb MT325 and the 321-kb FR483 viruses that infect Chlorella Pbi. Virology 358:459-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzgerald, L. A., M. V. Graves, X. Li, T. Feldblyum, W. C. Nierman, and J. L. Van Etten. 2007. Sequence and annotation of the 369-kb NY-2A and the 345-kb AR158 viruses that infect Chlorella NC64A. Virology 358:472-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fitzgerald, L. A., M. V. Graves, X. Li, J. Hartigan, A. J. Pfitzner, E. Hoffart, and J. L. Van Etten. 2007. Sequence and annotation of the 288-kb ATCV-1 virus that infects an endosymbiotic chlorella strain of the heliozoon Acanthocystis turfacea. Virology 362:350-361. (Erratum, 366:226). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forde, B. G., and P. J. Lea. 2007. Glutamate in plants: metabolism, regulation, and signalling. J. Exp. Bot. 58:2339-2358. [DOI] [PubMed] [Google Scholar]
- 18.Fouilland, E., C. Descolas-Gros, C. Courties, Y. Collos, A. Vaquer, and A. Gasc. 2004. Productivity and growth of a natural population of the smallest free-living eukaryote under nitrogen deficiency and sufficiency. Microb. Ecol. 48:103-110. [DOI] [PubMed] [Google Scholar]
- 19.Guindon, S., F. Lethiec, P. Duroux, and O. Gascuel. 2005. PHYML online: a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acid Res. 33:W557-W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iyer, L. M., S. Balaji, E. V. Koonin, and L. Aravind. 2006. Evolutionary genomics of nucleocytoplasmic large DNA viruses. Virus Res. 117:156-184. [DOI] [PubMed] [Google Scholar]
- 21.Iwamoto, K., and Y. Shiraiwa. 2005. Salt-regulated mannitol metabolism in algae. Mar. Biotechnol. (NY) 5:407-415. [DOI] [PubMed] [Google Scholar]
- 22.Jancek, S., S. Gourbière, H. Moreau, and G. Piganeau. 2008. Genetic basis of adaptation in the proteomes of two Ostreococcus ecotypes (Chlorophyta, Prasinophyceae). Mol. Biol. Evol. 25:2293-2300. [DOI] [PubMed] [Google Scholar]
- 23.Kamil, J. P., B. K. Tischer, S. Trapp, V. K. Nair, N. Osterrieder, and H. J. Kung. 2005. vLIP, a viral lipase homologue, is a virulence factor of Marek's disease virus. J. Virol. 79:6984-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reference deleted.
- 25.Koonin, E. V., Y. I. Wolf, K. Nagasaki, and V. V. Dolja. 2008. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 6:925-939. [DOI] [PubMed] [Google Scholar]
- 26.Lane, N. 2008. Marine microbiology: origins of death. Nature 453:583-585. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence, J. E., A. M. Chan, and C. A. Suttle. 2002. Viruses causing lysis of the toxic bloom-forming alga Heterosigma akashiwo (Raphidophyceae) are widespread in coastal sediments of British Columbia, Canada. Limnol. Oceanogr. 47:545-550. [Google Scholar]
- 28.Mayer, M. P. 2005. Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 153:1-46. [DOI] [PubMed] [Google Scholar]
- 29.M'Gonigle, L. K., J. J. Shen, and S. P. Otto. 2009. Mutating away from your enemies: the evolution of mutation rate in a host-parasite system. Theor. Popul. Biol. 75:301-311. [DOI] [PubMed] [Google Scholar]
- 30.Moreira, D., and C. Brochier-Armanet. 2008. Giant viruses, giant chimeras: the multiple evolutionary histories of Mimivirus genes. BMC Evol. Biol. 8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Not, F., M. Latasa, D. Marie, T. Cariou, D. Vaulot, and N. Simon. 2004. A single species Micromonas pusilla (Prasinophyceae) dominates the eukaryotic picoplankton in the western English Channel. Appl. Environ. Microbiol. 70:4064-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pal, C., M. D. Maciá, A. Oliver, I. Schachar, and A. Buckling. 2007. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature 450:1079-1081. [DOI] [PubMed] [Google Scholar]
- 33.Palenik, B., J. Grimwood, A. Aerts, P. Rouze, A. Salamov, N. Putnam, C. Dupont, R. Jorgensen, E. Derelle, S. Rombauts, K. Zhou, R. Otillar, S. S. Merchant, S. Podell, T. Gaasterland, C. Napoli, K. Gendler, A. Manuell, V. Tai, O. Vallon, G. Piganeau, S. Jancek, M. Heijde, K. Jabbari, C. Bowler, M. Lohr, S. Robbens, G. Werner, I. Dubchak, G. J. Pazour, Q. Ren, I. Paulsen, C. Delwiche, J. Schmutz, D. Rokhsar, Y. Van de Peer, H. Moreau, and I. Grigoriev. 2007. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. U. S. A. 104:7705-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raoult, D., S. Audic, C. Robert, C. Abergel, P. Renesto, H. Ogata, B. La Scola, M. Suzan, and J. M. Claverie. 2004. The 1.2-megabase genome sequence of Mimivirus. Science 306:1344-1350. [DOI] [PubMed] [Google Scholar]
- 35.Rippert, P., J. Puyaubert, D. Grisollet, L. Derrier, and M. Matringe. 2009. Tyrosine and phenylalanine are synthesized within the plastids in Arabidopsis. Plant Physiol. 149:1251-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suttle, C. A. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5:801-812. [DOI] [PubMed] [Google Scholar]
- 37.Teoh, M. L. T., P. J. Walasek, and D. H. Evans. 2003. Leporipoxvirus Cu,Zn-superoxide dismutase (SOD) homologs are catalytically inert decoy proteins that bind copper chaperone for SOD. J. Biol. Chem. 278:33175-33184. [DOI] [PubMed] [Google Scholar]
- 38.Vardi, A., B. A. Van Mooy, H. F. Fredricks, K. J. Popendorf, J. E. Ossolinski, L. Haramaty, and K. D. Bidle. 2009. Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton. Science 326:861-865. [DOI] [PubMed] [Google Scholar]
- 39.Vaulot, D., W. Eikrem, M. Viprey, and H. Moreau. 2008. The diversity of eukaryotic marine picophytoplankton. FEMS Microbiol. Rev. 32:795-820. [DOI] [PubMed] [Google Scholar]
- 40.Weynberg, K. D., M. J. Allen, K. Ashelford, D. J. Scanlan, and W. H. Wilson. 2009. From small hosts come big viruses: the complete genome of a second Ostreococcus tauri virus, OtV-1. Environ. Microbiol. 11:2821-2839. [DOI] [PubMed] [Google Scholar]
- 41.Wilson, W. H., D. C. Schroeder, M. J. Allen, M. T. Holden, J. Parkhill, B. G. Barrell, C. Churcher, N. Hamlin, K. Mungall, H. Norbertczak, M. A. Quail, C. Price, E. Rabbinowitsch, D. Walker, M. Craigon, D. Roy, and P. Ghazal. 2005. Complete genome sequence and lytic phase transcription profile of a coccolithovirus. Science 309:1090-1092. [DOI] [PubMed] [Google Scholar]
- 42.Worden, A. Z. 2006. Picoeukaryote diversity in coastal waters of the Pacific Ocean. Aquat. Microb. Ecol. 43:165-175. [Google Scholar]
- 43.Worden, A. Z., J. H. Lee, T. Mock, P. Rouzé, M. P. Simmons, A. L. Aerts, A. E. Allen, M. L. Cuvelier, E. Derelle, M. V. Everett, E. Foulon, J. Grimwood, H. Gundlach, B. Henrissat, C. Napoli, S. M. McDonald, M. S. Parker, S. Rombauts, A. Salamov, P. Von Dassow, J. H. Badger, P. M. Coutinho, E. Demir, I. Dubchak, C. Gentemann, W. Eikrem, J. E. Gready, U. John, W. Lanier, E. A. Lindquist, S. Lucas, K. F. Mayer, H. Moreau, F. Not, R. Otillar, O. Panaud, J. Pangilinan, I. Paulsen, B. Piegu, A. Poliakov, S. Robbens, J. Schmutz, E. Toulza, T. Wyss, A. Zelensky, K. Zhou, V. Armbrust, D. Bhattacharya, U. W. Goodenough, Y. Van de Peer, and I. V. Grigoriev. 2009. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324:268-272. [DOI] [PubMed] [Google Scholar]
- 44.Yamada, T., H. Onimatsu, and J.-L. Van Etten. 2006. Chlorella viruses. Adv. Virus Res. 66:293-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan, X., Z. Yu, P. Zhang, A. J. Battisti, H. A. Holdaway, P. R. Chipman, C. Bajaj, M. Bergoin, M. G. Rossmann, and T. S. Baker. 2009. The capsid proteins of a large, icosahedral dsDNA virus. J. Mol. Biol. 385:1287-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yutin, N., Y. I. Wolf, D. Raoult, and E. V. Koonin. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol. J. 6:223-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Y., F. Maley, G. F. Maley, G. Duncan, D. D. Dunigan, and J. L. Van Etten. 2007. Chloroviruses, encode a bifunctional dCMP-dCTP deaminase that produces two key intermediates in dTTP formation. J. Virol. 81:7662-7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.