Abstract
Nonstructural protein 4B (NS4B) is a key organizer of hepatitis C virus (HCV) replication complex formation. It induces a specific membrane rearrangement, designated membranous web, that serves as a scaffold for the HCV replication complex. However, the mechanisms underlying membranous web formation are poorly understood. Based on fluorescence resonance energy transfer (FRET) and confirmatory coimmunoprecipitation analyses, we provide evidence for an oligomerization of NS4B in the membrane environment of intact cells. Several conserved determinants were found to be involved in NS4B oligomerization, through homotypic and heterotypic interactions. N-terminal amphipathic α-helix AH2, comprising amino acids 42 to 66, was identified as a major determinant for NS4B oligomerization. Mutations that affected the oligomerization of NS4B disrupted membranous web formation and HCV RNA replication, implying that oligomerization of NS4B is required for the creation of a functional replication complex. These findings enhance our understanding of the functional architecture of the HCV replication complex and may provide new angles for therapeutic intervention. At the same time, they expand the list of positive-strand RNA virus replicase components acting as oligomers.
With an estimated 120 to 180 million chronically infected individuals, hepatitis C virus (HCV) is a leading cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma worldwide (26). HCV contains a 9.6-kb positive-strand RNA genome encoding a polyprotein precursor that is co- and posttranslationally processed into 10 structural and nonstructural proteins (28, 36). Like all positive-strand RNA viruses investigated thus far, HCV replicates its genome in a membrane-associated replication complex composed of viral proteins, replicating RNA, rearranged intracellular membranes, and additional host factors (6, 33, 36). Nonstructural protein 4B (NS4B) is the least-characterized HCV protein. However, evidence from biochemical, structural, and genetic studies as well as electron microscopy (EM) indicates that NS4B is a key organizer of HCV replication complex formation (reviewed in reference 16). Indeed, one of the best-documented functions of NS4B is to induce the specific membrane rearrangement, designated membranous web, that serves as a scaffold for the viral replication complex (8, 13). However, the mechanisms underlying membranous web formation are poorly understood.
NS4B is a 27-kDa integral membrane protein comprising an N-terminal part (amino acids 1 to ∼69), a central part harboring four predicted transmembrane segments (amino acids ∼70 to ∼190), and a C-terminal part (amino acids ∼191 to 261). The N-terminal part comprises a predicted and a structurally resolved amphipathic α-helix, designated AH1 and AH2, respectively. AH2 comprises amino acids 42 to 66 and has been shown to play an important role in HCV RNA replication (14). Intriguingly, it has the potential to traverse the phospholipid bilayer as a transmembrane segment, likely upon oligomerization (14).
Oligomerization of membrane proteins represents a potential mechanism to induce membrane curvature and vesicle formation (44, 50). A previous study involving chemical cross-linking provided evidence for the oligomerization of NS4B (49). However, interactions of membrane proteins are inherently difficult to study. Therefore, we aimed to validate and extend these observations by using a different experimental strategy.
In this study, we explored fluorescence resonance energy transfer (FRET) to investigate the determinants for oligomerization of NS4B. FRET is based on the transfer of energy from a fluorescent donor protein (e.g., cyan fluorescent protein [CFP]) to an acceptor protein (e.g., yellow fluorescent protein [YFP]). It has been employed successfully to investigate interactions of membrane proteins, e.g., the G-protein-coupled receptors (5) and nodavirus replicase protein A (7). In acceptor photobleaching FRET, photobleaching of the acceptor results in increased donor emission when the distance between the two is <10 nm, i.e., when the two proteins or protein segments fused to the fluorophores physically interact (5). Thus, acceptor photobleaching FRET offers the unique opportunity to investigate protein-protein interactions at a defined subcellular location within the membrane environment of intact cells.
By the use of FRET and confirmatory coimmunoprecipitation analyses, we found that HCV NS4B oligomerizes through several conserved determinants involving homotypic and heterotypic interactions. Amphipathic α-helix AH2 was identified as a major determinant for the oligomerization of NS4B. Furthermore, mutations in NS4B that affected oligomerization disrupted membranous web formation and HCV RNA replication, implying that oligomerization of NS4B is required for the creation of a functional replication complex.
MATERIALS AND METHODS
Cell lines and reagents.
U-2 OS human osteosarcoma (40) and Huh-7 human hepatocellular carcinoma (38) cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum. The U-2 OS-derived, tetracycline-regulated cell lines UHCVcon-57.3 and UHCVcon-AH2mut, expressing the entire polyprotein derived from the HCV H77 consensus clone and harboring wild-type NS4B and NS4B with alanine substitutions of the 6 fully conserved aromatic residues in AH2 (AH2mut), respectively, have been described previously (14, 42). Transfections were performed by calcium phosphate precipitation (3). Polyclonal anti-NS4B antibody 86 was kindly provided by Ralf Bartenschlager (University of Heidelberg, Germany). Polyclonal anti-hemagglutinin (anti-HA) and monoclonal anti-FLAG antibodies were from Sigma-Aldrich (St. Louis, MO). Alexa 594-conjugated goat anti-rabbit antibody was from Molecular Probes (Eugene, OR).
Plasmids.
HCV NS4B fragments were derived either from the H77 consensus clone present in pBRTM/HCV1-3011con (23) (kindly provided by Charles M. Rice, The Rockefeller University, New York, NY) or from the JFH-1 clone present in pSRG-JFH1 (22) (kindly provided by Takaji Wakita, Tokyo University, Japan). The BamHI restriction site present in the NS4B sequence of the JFH-1 strain was inactivated by site-directed mutagenesis with primers JFH4B-G174mut-fd and JFH4B-G174mut-rv (see Table S1 in the supplemental material), resulting in a G-to-A substitution at nucleotide position 1450.
Green fluorescent protein (GFP) fusion constructs harboring full-length NS4B or NS4B fragments 1-130, 1-116, 40-130, 61-130, and 130-261 (derived from the HCV H77 consensus clone), designated pCMVNS4B-GFP, pCMVNS4B1-130-GFP, pCMVNS4B1-116-GFP, pCMVNS4B40-130-GFP, pCMVNS4B61-130-GFP, and pCMVNS4B130-261-GFP, respectively, were obtained by PCR, using pBRTM/HCV1-3011con as a template and the primers listed in Table S1 in the supplemental material, followed by cloning into the EcoRI-BamHI sites of pCMVKEB-EGFP (2).
Optimized Cerulean CFP (41) (kindly provided by David W. Piston, Vanderbilt University, Nashville, TN) and Venus YFP (37) (kindly provided by Atsushi Miyawaki, RIKEN Brain Science Institute, Saitama, Japan) sequences (referred to as CFP and YFP, respectively, in the following) were used to prepare pcDNA3.1(+) (Invitrogen, La Jolla, CA)-based founder constructs pCMVCFP-X-HA, pCMVYFP-X-FLAG, pCMVHA-X-CFP, and pCMVFLAG-X-YFP, allowing N- or C-terminal fusion of CFP or YFP to a protein of interest (X), with or without an HA or FLAG tag (1). Subcloning through BspEI or BamHI sites yielded short SG or GS linkers, respectively (pCMVCFP-X-HA, KpnI-YFP-SG[BspEI]-X-GS[BamHI]-FLAG-ApaI; pCMVYFP-X-FLAG, KpnI-CFP-SG[BspEI]-X-GS[BamHI]-HA-ApaI; pCMVHA-X-CFP, KpnI-FLAG-SG[BspEI]-X-GS[BamHI]-YFP-ApaI; pCMVFLAG-X-YFP, KpnI-HA-SG[BspEI]-X-GS[BamHI]-CFP-ApaI).
A positive-control construct for FRET in which CFP and YFP were fused through a 5-amino-acid (SGGGG) linker sequence (25) was kindly provided by Roland Nitschke (University of Freiburg, Germany). Cotransfection of unfused CFP and YFP served as a negative control.
Fusion constructs harboring N-terminal CFP or YFP and full-length NS4B derived from the HCV H77 consensus clone were obtained by PCR, using pBRTM/HCV1-3011con as the template and primers NS4B-1-Bsp-fd and NS4B-261-Apa-rv (see Table S1 in the supplemental material), followed by cloning into the BspEI-ApaI sites of pCMVCFP-X-HA and pCMVYFP-Y-FLAG to yield constructs pCMVCFP-NS4B and pCMVYFP-NS4B, respectively. Note that cloning through the BspEI-ApaI sites removed the HA and FLAG tags present in the founder constructs (see above).
Fusion constructs harboring full-length NS4B, NS4B fragment 40-130 or 130-261 (derived from the HCV H77 consensus), or JFH-1 clones and C-terminal CFP or YFP were obtained by PCR, using pBRTM/HCV1-3011con or pSRG-JFH1 as the template and using the primers listed in Table S1 in the supplemental material. Amplification products were cloned into the KpnI-BamHI sites of pCMVHA-X-CFP and pCMVFLAG-X-YFP to yield the H77-derived constructs pCMVNS4B-CFP, pCMVNS4B-YFP, pCMVNS4B40-130-CFP, pCMVNS4B40-130-YFP, pCMVNS4B130-261-CFP, and pCMVNS4B130-261-YFP and the JFH-1-derived constructs pCMVJFH4B-CFP, pCMVJFH4B-YFP, pCMVJFH4B40-130-CFP, pCMVJFH4B40-130-YFP, pCMVJFH4B130-261-CFP, and pCMVJFH4B130-261-YFP. Note that cloning through the KpnI-BamHI sites removed the HA and FLAG tags present in the founder constructs (see above).
Fusion proteins harboring the dengue virus (DV) 2K-NS4B sequence and C-terminal CFP or YFP were obtained by PCR, using pTM1.4-DV-2K(1-249)GFP (34) (kindly provided by Ralf Bartenschlager) as the template and primers DV2K4B-Kpn-fd and DV2K4B-Bam-rv (see Table S1 in the supplemental material), followed by cloning into the KpnI-BamHI sites of pCMVHA-X-CFP and pCMVFLAG-X-YFP to yield constructs pCMVDV4B-CFP and pCMVDV4B-YFP, respectively.
Fusion constructs harboring full-length NS4B or fragment 40-130 with alanine substitutions of the 6 fully conserved aromatic residues in AH2 (AH2mut) and C-terminal CFP or YFP were obtained by PCR, using pUHDHCV(H)conAH2mut (14) as the template and using primer pair NS4B-1-Kpn-fd-NS4B-261-Bam-rv or NS4B-40-Kpn-fd-NS4B-130-Bam-rv (see Table S1 in the supplemental material), respectively, followed by cloning into the KpnI-BamHI sites of pCMVHA-X-CFP or pCMVFLAG-X-YFP to yield constructs pCMVNS4BAH2mut-CFP, pCMVNS4BAH2mut-YFP, pCMVNS4B40-130AH2mut-CFP, and pCMVNS4B40-130AH2mut-YFP.
Constructs with a C-terminal HA tag were prepared by subcloning of the KpnI-BamHI fragments from pCMVNS4B-CFP, pCMVNS4B40-130-CFP, and pCMVNS4B130-261-CFP into the KpnI-BamHI sites of pCMVCFP-X-HA to yield constructs pCMVNS4B-HA, pCMVNS4B40-130-HA, and pCMVNS4B130-261-HA, respectively. Subcloning of the same fragments into the KpnI-BamHI sites of pCMVYFP-X-FLAG yielded the FLAG-tagged constructs pCMVNS4B-FLAG, pCMVNS4B40-130-FLAG, and pCMVNS4B130-261-FLAG, respectively. Finally, DV NS4B constructs with a C-terminal HA or FLAG tag were prepared by subcloning of the KpnI-BamHI fragment from pCMVDV4B-CFP into the KpnI-BamHI sites of pCMVCFP-X-HA and pCMVYFP-X-FLAG to yield constructs pCMVDV4B-HA and pCMVDV4B-FLAG, respectively. Note that this subcloning strategy removed the CFP and YFP sequences present in the founder constructs (see above).
Fluorescence resonance energy transfer.
U-2 OS or Huh-7 cells cultured on glass coverslips were transfected with constructs expressing CFP- and YFP-tagged proteins and fixed at 24 h posttransfection with 2% paraformaldehyde for 5 min. The coverslips were mounted on glass slides with SlowFade reagent (Molecular Probes). Acceptor photobleaching FRET was performed using an SP5 AOBS confocal laser scanning microscope (Leica, Wetzlar, Germany). CFP was excited with a 405-nm blue diode laser and detected at 445 to 485 nm. YFP was excited with the 514-nm line of an argon laser (458, 476, 488, and 514 nm) and detected at 525 to 575 nm. Cells were examined with a 63× oil immersion objective following well-defined rules (see Results). Two regions of interest (ROI) per cell were photobleached in the YFP channel, using the 514-nm argon laser line at 100% intensity. Ten different healthy-looking and intact cells were analyzed, yielding 20 measurements for each combination. CFP images were collected pre- and postphotobleaching to measure changes in donor fluorescence. FRET efficiency (FRETeff) was expressed as the percentage of CFP fluorescence gain after YFP photobleaching. To calculate the apparent FRETeff in the ROI, the Leica software uses the formula FRETeff = [(EDpost − EDpre)/EDpost] × 100, where ED represents the emitted donor fluorescence before (EDpre) and after (EDpost) photobleaching of the acceptor. Statistical analyses were performed by using GraphPad Prism software (GraphPad Software, La Jolla, CA). Unpaired t tests were applied to assess the significance of differences in mean FRETeff values. A P value of <0.05 was considered to indicate a significant difference between two groups.
Coimmunoprecipitation.
U-2 OS cells expressing HA- and FLAG-tagged proteins were harvested at 48 h posttransfection, followed by lysis in a buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and a protease inhibitor cocktail (Roche, Basel, Switzerland). Cell lysates were incubated for 12 to 16 h at 4°C with anti-FLAG M2 agarose slurry (Sigma-Aldrich) on a rotating wheel. After repeated washing, agarose beads were eluted with 3×FLAG peptide solution (150 ng/μl) (Sigma-Aldrich). Eluates were subjected to conventional SDS-PAGE, followed by immunoblotting.
Electron microscopy.
UHCVcon-57.3 and UHCVcon-NS4BAH2mut cells were cultured for 48 h in the presence or absence of tetracycline. Cells were fixed and processed for EM as described previously (11). Ultrathin sections were examined with a JEOL 1230 transmission electron microscope (Tokyo, Japan) connected to a Gatan digital camera driven by Digital Micrograph software (Gatan, Pleasanton, CA) for image acquisition and analysis.
RESULTS
FRET analyses reveal oligomerization of HCV NS4B.
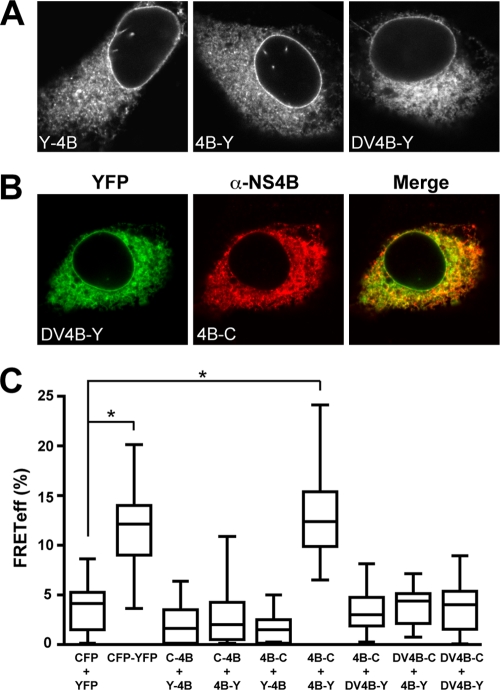
Optimized Venus YFP and Cerulean CFP (referred to in the following as YFP and CFP, respectively) were fused to either the N or the C terminus of NS4B derived from the HCV H77 consensus clone in order to investigate interactions between NS4B molecules by acceptor photobleaching FRET. As shown in Fig. 1A, fusion constructs harboring YFP at the N or the C terminus of NS4B showed a fluorescence pattern that included the nuclear membrane, was strongest in the perinuclear region, and extended in a reticular as well as dot-like fashion throughout the cytoplasm. Analogous results were obtained for the corresponding CFP fusion constructs (data not illustrated). This fluorescence pattern was previously observed for NS4B alone or for NS4B-GFP fusion constructs and has been shown to correspond to endoplasmic reticulum (ER) and seemingly ER-derived modified membranes (9, 14, 15, 17, 19, 30, 31).
FIG. 1.
FRET analyses reveal oligomerization of HCV NS4B. (A) Subcellular localization of HCV NS4B and DV NS4B fusion proteins. Constructs pCMVYFP-NS4B (Y-4B), pCMVNS4B-YFP (4B-Y), and pCMVDV4B-YFP (DV4B-Y) were transfected into U-2 OS cells, followed by fixation with 2% paraformaldehyde and confocal laser scanning microscopy, as described in Materials and Methods. (B) Colocalization of HCV NS4B and DV NS4B. Constructs pCMVDV4B-YFP (DV4B-Y) and pCMVNS4B-CFP (4B-C) were cotransfected into U-2 OS cells, followed by fixation with 2% paraformaldehyde. Since the CFP emission was not sufficiently bright for optimal colocalization studies, HCV NS4B in construct 4B-C was detected with polyclonal antibody 86 and Alexa 594-conjugated secondary antibody, as described in Materials and Methods. Confocal laser scanning microscopy revealed colocalization of DV4B-Y and 4B-C. (C) FRET analyses. Constructs pCMVCFP-NS4B (C-4B), pCMVYFP-NS4B (Y-4B), pCMVNS4B-YFP (4B-Y), pCMVNS4B- CFP (4B-C), pCMVDV4B-YFP (DV4B-Y), and pCMVDV4B-CFP (DV4B-C) were transfected into U-2 OS cells in different combinations, as indicated, followed by acceptor photobleaching FRET analyses as described in Materials and Methods. The CFP-YFP fusion protein and cotransfection of unfused CFP and YFP served as positive and negative controls for FRET, respectively. Box-and-whisker plots represent the median FRETeff values of 20 measurements (middle line), the FRETeff values from the lower to the upper quartile (central box), and the minimum and maximum values (vertical line). The significance of the observed differences was assessed as described in Materials and Methods (*, P < 0.0001).
NS4B from the related virus DV was used as a specificity control, given its similarities in molecular mass, multispanning membrane topology, and subcellular localization (34). As expected, a fluorescence pattern virtually identical to that of HCV NS4B was observed for constructs harboring the DV 2K-NS4B sequence fused to YFP (Fig. 1A) or CFP (data not illustrated). Consistent with this observation, DV NS4B was found to broadly colocalize with HCV NS4B, as shown in Fig. 1B. Hence, DV NS4B and HCV NS4B share similar membrane topologies and broadly colocalize on ER and ER-derived modified membranes but are not expected to interact.
FRET analyses were performed with cells cotransfected with different combinations of YFP and CFP fusion constructs according to the acceptor photobleaching protocol. With this technique, photobleaching of the acceptor (YFP) results in increased donor (CFP) emission when the distance between the two molecules is <10 nm, i.e., when the two proteins or protein segments of interest fused to the fluorophores interact. This protocol allowed us to investigate interactions in selected cells and at defined subcellular locations. Therefore, the following rules were observed throughout this work: (i) only healthy-looking and intact cells were examined, (ii) ROIs in the cytoplasm were carefully selected based on the expected reticular and dot-like fluorescence pattern, (iii) two ROIs per cell were always assayed simultaneously, and (iv) 10 different cells were analyzed for each protein combination. Thus, in each box-and-whisker plot shown in Fig. 1C and for subsequent experiments, the middle line represents the median of 20 measurements, the central box represents the FRETeff values from the lower to the upper quartile, and the vertical line extends from the minimum to the maximum value. Representative results from at least two independent experiments are shown throughout this work. In addition, experiments were performed in U-2 OS and Huh-7 cells, with analogous results (data not illustrated).
As shown in Fig. 1C, FRET was observed when both fluorescent proteins were fused to the C terminus of HCV NS4B (mean FRETeff ± standard deviation [SD], 13.4% ± 1.1%). This result was comparable to that obtained with the CFP-YFP fusion protein, which served as a positive control (FRETeff, 12.5% ± 0.9%). Thus, FRET analyses revealed a self-interaction of HCV NS4B in the membrane environment of intact cells. No FRET was observed when one or both partners harbored the fluorescent protein at the N terminus (FRETeff, <10% [comparable to the values obtained after cotransfection of unfused CFP and YFP, i.e., the negative control]). Thus, although an interaction may have occurred between the two NS4B moieties, the fluorophores may be positioned improperly for FRET when fused to the N terminus of NS4B. This observation underscores the stringency of the FRET approach and demonstrates that colocalization to the ER or ER-derived modified membranes is not sufficient to produce FRET. Accordingly, no FRET was observed between HCV NS4B and DV NS4B, despite their colocalization, thus corroborating the specificity of the result observed for HCV NS4B. Interestingly, no FRET was observed between DV NS4B molecules in this experimental setting. This may be related to the observation that DV NS4B does not induce membrane rearrangements and may have functions distinct from those of HCV NS4B in the DV life cycle (34). In DV, NS4A is the key protein responsible for the induction of membrane rearrangements (32).
In conclusion, FRET analyses revealed a specific oligomerization of HCV NS4B in the membrane environment of intact cells.
Multiple determinants contribute to the oligomerization of HCV NS4B.
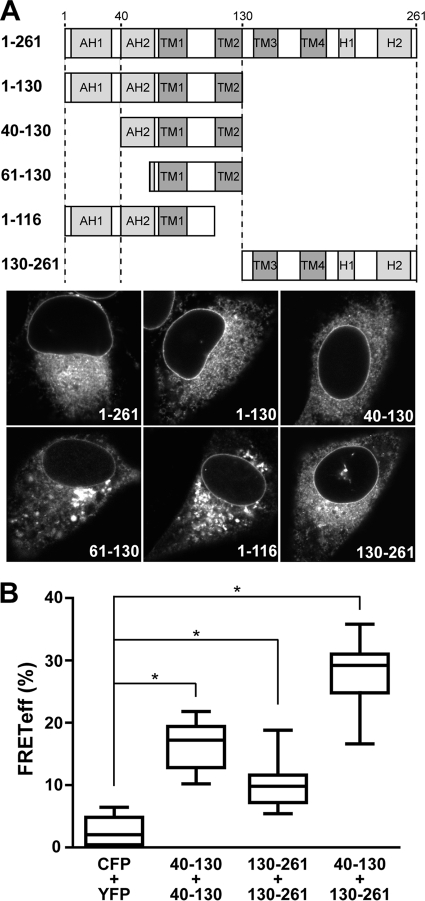
NS4B is a 261-amino-acid integral membrane protein comprising an N-terminal part with a predicted and a structurally resolved amphipathic α-helix (AH1 and AH2, respectively), a central part harboring four predicted transmembrane segments, and a C-terminal part comprising a predicted and a structurally resolved α-helix (H1 and H2, respectively) (Fig. 2A; see Fig. S1 in the supplemental material) (16). To identify regions involved in the oligomerization of NS4B, we designed fragments based on these structural elements and fused GFP to their C termini to investigate their subcellular localization. A fragment comprising the N-terminal half of NS4B (amino acids 1 to 130) displayed the same subcellular localization as full-length NS4B (Fig. 2A and data not shown). Localization on ER and ER-derived modified membranes was preserved after deletion of AH1 (segment 40-130). However, further truncation at the N terminus (segment 61-130) or C terminus (segment 1-116) abrogated the typical fluorescence pattern and produced coarse cytoplasmic aggregates (Fig. 2A). As shown in Fig. 2A, a fragment comprising the C-terminal half of NS4B (amino acids 130 to 261) also displayed the same subcellular localization as full-length NS4B, while it was shown previously that N-terminal truncation of this fragment alters the subcellular localization (15).
FIG. 2.
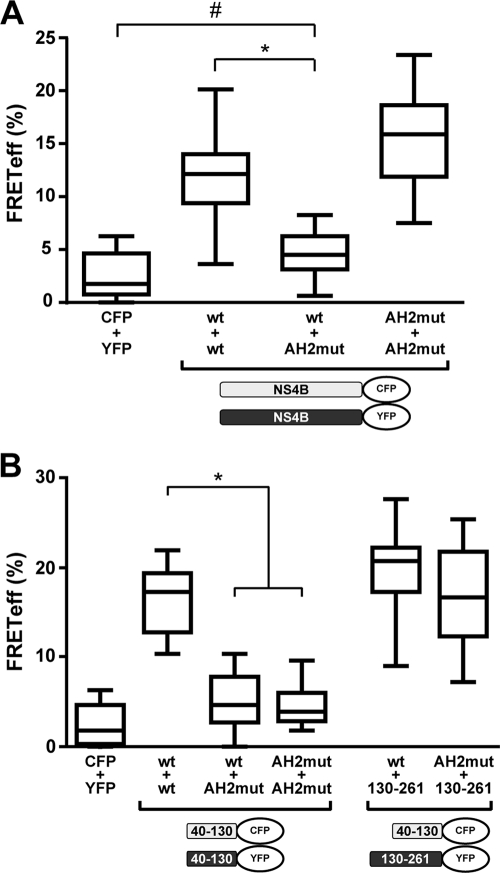
Several determinants contribute to the oligomerization of HCV NS4B. (A) Subcellular localization of NS4B fragments. A schematic representation of NS4B's known and predicted structural elements as well as the different fragments analyzed is depicted at the top. AH, amphipathic α-helix; TM, transmembrane segment; H, α-helix. Constructs pCMVNS4B-GFP (1-261), pCMVNS4B1-130-GFP (1-130), pCMVNS4B40-130-GFP (40-130), pCMVNS4B61-130-GFP (61-130), pCMVNS4B1-116-GFP (1-116), and pCMVNS4B130-261-GFP (130-261) were transfected into U-2 OS cells, followed by fixation with 2% paraformaldehyde and confocal laser scanning microscopy. (B) FRET analyses. Constructs pCMVNS4B40-130-CFP and pCMVNS4B40-130-YFP (40-130 + 40-130), pCMVNS4B130-261-YFP and pCMVNS4B130-261-CFP (130-261 + 130-261), and pCMVNS4B40-130-CFP and pCMVNS4B130-261-YFP (40-130 + 130-261) were cotransfected into U-2 OS cells, followed by acceptor photobleaching FRET analyses. Cotransfection of unfused CFP and YFP (CFP + YFP) served as a negative control. The significance of the observed differences was assessed as described in Materials and Methods (*, P < 0.0001).
Based on these preliminary experiments, interactions of segments 40-130 and 130-261 fused to CFP or YFP were investigated by FRET. As shown in Fig. 2B, these N- and C-terminal fragments were found to interact both with themselves and with each other. These results demonstrate that several determinants contribute to the oligomerization of NS4B, through homotypic (i.e., occurring between the same protein segment) and heterotypic (i.e., occurring between different protein segments) interactions. Whether the heterotypic interaction between the N- and C-terminal fragments occurs as an intermolecular or intramolecular association in the context of the full-length protein remains unknown.
Confirmation of FRET results by coimmunoprecipitation.
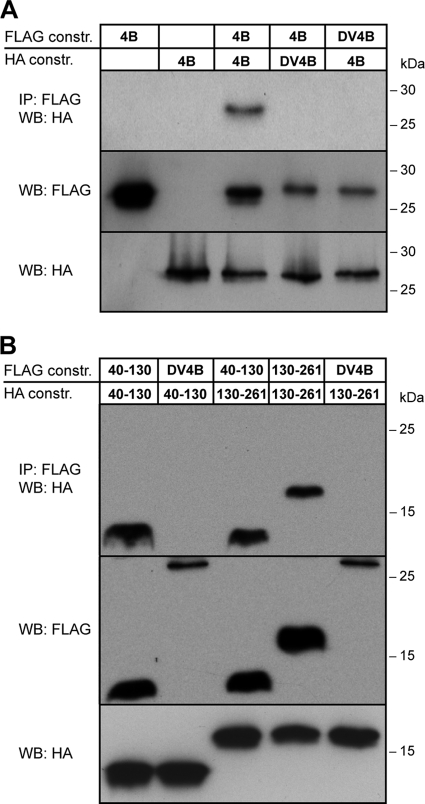
To corroborate the results obtained by FRET, we performed coimmunoprecipitation analyses. To this end, the HA or FLAG tag was fused to the C terminus of full-length HCV NS4B or DV NS4B, as well as to fragments 40-130 and 130-261 from HCV NS4B. Lysates from cells coexpressing different combinations of these constructs were subjected to immunoprecipitation and Western blot analyses using anti-HA and anti-FLAG antibodies. As shown in Fig. 3A, these analyses revealed a specific self-interaction of HCV NS4B, while as expected, no interaction was observed between the NS4B proteins from HCV and DV. In addition, coimmunoprecipitation analyses confirmed the specificity of the homotypic and heterotypic interactions observed between the N- and C-terminal NS4B fragments 40-130 and 130-261 (Fig. 3B). Taken together, the results from the coimmunoprecipitation analyses confirm the oligomerization of HCV NS4B through several determinants.
FIG. 3.
Confirmation of FRET results by coimmunoprecipitation. (A) Constructs pCMVNS4B-HA, pCMVNS4B-FLAG, pCMVDV4B-HA, and pCMVDV4B-FLAG were transfected into U-2 OS cells as indicated, followed by immunoprecipitation (IP) of cell lysates with anti-FLAG M2 agarose and Western blotting (WB) with an anti-HA antibody, as described in Materials and Methods. Direct Western blotting of the cell lysates by use of anti-FLAG and anti-HA antibodies is shown in the lower panels. Molecular mass markers are indicated on the right. (B) Constructs pCMVNS4B40-130-HA, pCMVNS4B40-130-FLAG, pCMVNS4B130-261-HA, pCMVNS4B130-261-FLAG, and pCMVDV4B-FLAG were transfected into U-2 OS cells as indicated, followed by immunoprecipitation and Western analyses as described for panel A. Molecular mass markers are indicated on the right.
Oligomerization of NS4B proteins from different HCV genotypes.
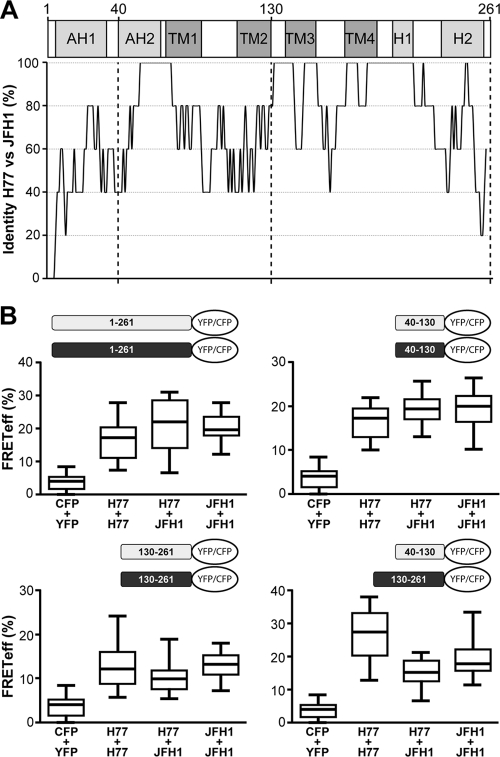
In order to assess whether our observations can be extended to other HCV strains and to gain insights into the genotype specificity of the identified interactions, we investigated FRET between NS4B sequences derived from the HCV H77 consensus (genotype 1a) and JFH-1 (genotype 2a) clones. The overall amino acid sequence identity between these two NS4B sequences is 72%, while the 40-130 and 130-261 fragments display 70% and 82% identity, respectively (see Fig. S1 in the supplemental material). A schematic representation of the percent amino acid identity along the NS4B sequence, derived from the sequence alignment shown in Fig. S1, highlights the regions that are conserved between these relatively distant HCV strains (Fig. 4A).
FIG. 4.
Oligomerization of NS4B proteins from different HCV genotypes. (A) Amino acid sequence conservation between NS4B proteins from the HCV H77 consensus (genotype 1a; GenBank accession number AF009606) and JFH-1 (genotype 2a; GenBank accession number AB047639) clones. After Clustal W alignment of the two sequences, a score of 1 was attributed to a conserved position and a score of 0 to a divergent one. The mean score of overlapping 7-amino-acid windows was calculated along the sequence and illustrated as the percent identity for each central position of the window. (B) FRET analyses. Constructs comprising H77- and JFH-1-derived NS4B sequences fused to CFP or YFP were transfected in different combinations, as illustrated. See Materials and Methods for individual plasmid denominations. Cotransfection of unfused CFP and YFP (CFP + YFP) served as a negative control. All combinations tested were significantly different from the negative control (P < 0.0001).
As shown in Fig. 4B, the results obtained for the H77 strain can be extended to the JFH-1 strain. Indeed, FRET was observed for full-length NS4B as well as the different combinations of fragments 40-130 and 130-261 derived from the JFH-1 strain. More interestingly, interactions were observed between full-length NS4B and the different fragments derived from the H77 and JFH-1 strains (Fig. 4B). Therefore, oligomerization is a general feature of HCV NS4B and appears to involve conserved determinants.
Role of amphipathic α-helix AH2 in NS4B oligomerization.
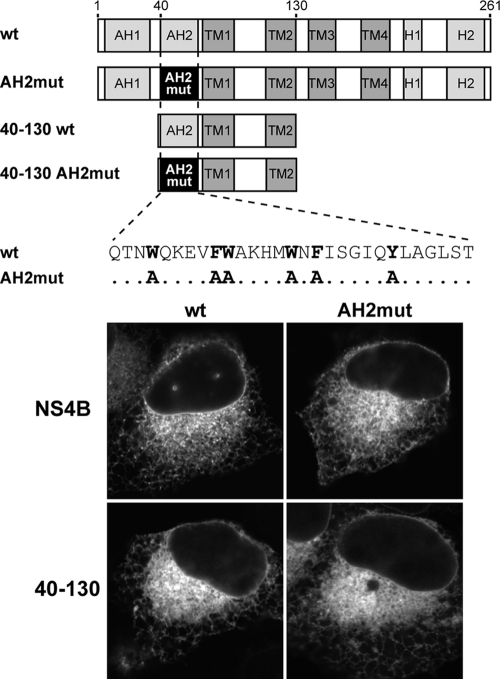
One of the most conserved regions in NS4B maps to membrane-associated amphipathic α-helix AH2. Interestingly, AH2 can adopt a transmembrane orientation, likely upon oligomerization (14). We previously reported that the replacement of 6 fully conserved aromatic residues on the hydrophobic side of AH2 with alanine (mutant AH2mut) preserves the α-helical fold but disrupts the membrane association and transmembrane orientation of AH2, as well as RNA replication, when introduced into the context of a subgenomic HCV replicon (14). In order to examine the role of AH2 in the oligomerization of NS4B, we introduced the AH2mut substitutions into constructs comprising full-length NS4B or fragment 40-130 fused to CFP or YFP (Fig. 5). In accordance with and in extension of our previous observations (14), these substitutions did not interfere with the membrane association of full-length NS4B or fragment 40-130 fused to YFP (Fig. 5) or CFP (data not illustrated). Indeed, the constructs harboring the AH2mut substitutions displayed the same fluorescence pattern as the wild-type constructs, indicating that one or more additional internal determinants in NS4B can ensure membrane association in the context of the full-length protein or the 40-130 segment.
FIG. 5.
Subcellular localization of NS4B and NS4B segment 40-130 harboring the AH2mut mutations. Constructs pCMVNS4B-YFP (wt), pCMVNS4BAH2mut-YFP (AH2mut), pCMVNS4B40-130-YFP (40-130 wt), and pCMVNS4B40-130AH2mut-YFP (40-130 AH2mut), illustrated schematically at the top, were transfected into U-2 OS cells, followed by fixation with 2% paraformaldehyde and confocal laser scanning microscopy as described in Materials and Methods.
As shown in Fig. 6A, introduction of the AH2mut substitutions into one of the two full-length NS4B partners resulted in a significant reduction of the FRET signal. The residual FRET signal remained statistically different from that of the negative control (P = 0.0036). Interestingly, FRET could be recovered fully when both partners harbored the AH2mut substitutions. A possible explanation for this observation may involve a preferential self-association and sequestration of wild-type NS4B in the combination where only one partner carries the mutations. In this scenario, determinants for oligomerization other than AH2 may result in strong FRET only when both partners carry the mutations. To further explore this possibility, interactions between N- and C-terminal NS4B fragments were analyzed. As shown in Fig. 6B, the 40-130 segment harboring the AH2mut substitutions interacted with neither the wild-type 40-130 fragment nor itself, indicating that oligomerization of the N-terminal segment of NS4B relies on AH2. In contrast, FRET was observed between the 40-130 segment harboring the AH2mut substitutions and the C-terminal segment 130-261, indicating that the interaction between N- and C-terminal NS4B fragments may involve distinct, likely weaker determinants. These findings are in line with the report by Yu et al., who provided evidence for NS4B oligomerization through multiple determinants based on chemical cross-linking studies (49). Interestingly, they mapped a major determinant for NS4B oligomerization to the N-terminal 70 amino acids, which comprise amphipathic α-helix AH2.
FIG. 6.
Role of amphipathic α-helix AH2 in NS4B oligomerization. (A) Constructs pCMVNS4B-CFP and pCMVNS4B-YFP (both designated wt) as well as pCMVNS4BAH2mut-YFP and pCMVNS4BAH2mut-CFP (both designated AH2mut) were transfected into U-2 OS cells in different combinations, as indicated. (B) Constructs pCMVNS4B40-130-CFP and pCMVNS4B40-130-YFP (both designated wt), pCMVNS4B40-130AH2mut-YFP and pCMVNS4B40-130AH2mut-CFP (both designated AH2mut), and pCMVNS4B130-261-YFP (130-261) were transfected into U-2 OS cells in different combinations, as indicated. The significance of the observed differences was assessed as described in Materials and Methods (*, P < 0.0001; #, P = 0.0036).
In conclusion, the results above indicate that amphipathic α-helix AH2 is a major determinant for the oligomerization of NS4B, while additional determinants contribute to the interaction.
NS4B oligomerization correlates with membranous web formation.
We hypothesized that oligomerization may be a mechanism by which NS4B induces formation of the membranous web as a scaffold for the HCV replication complex. To further explore this hypothesis, we investigated the effect of the AH2mut alanine substitutions in NS4B on membranous web formation. To this end, we used EM to examine a cell line, designated UHCVcon-AH2mut (14), which can be induced to express an HCV H77-derived polyprotein harboring the AH2mut mutations in NS4B. A well-characterized cell line that can be induced to express the wild-type HCV H77 polyprotein served as a control (42). We have previously shown that the two cell lines express the HCV structural and nonstructural proteins at comparable levels and that polyprotein processing is not affected by the AH2mut mutations (14).
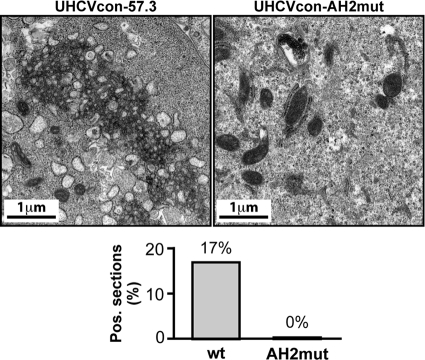
As shown in Fig. 7 and in accordance with the initial report by Egger et al. (8), specific membrane rearrangements composed of tightly organized vesicles embedded in a membranous matrix could be detected readily in UHCVcon-57.3 cells expressing the wild-type HCV polyprotein. These membrane rearrangements correspond to typical membranous webs. In fact, 17 of 100 cell sections analyzed showed a membranous web. In contrast, no membranous web was observed in 200 sections from UHCVcon-AH2mut cells expressing the polyprotein carrying the AH2mut mutations in NS4B. As expected, no membranous web was observed in UHCVcon-57.3 and UHCVcon-AH2mut cells cultured in the presence of tetracycline, i.e., in the absence of HCV polyprotein expression (data not shown). This observation likely explains the previously reported replication defect of subgenomic replicons harboring the AH2mut substitutions (14).
FIG. 7.
AH2mut mutations disrupt membranous web formation. Ultrathin sections from UHCVcon-57.3 and UHCVcon-AH2mut cells cultured for 48 h in the absence of tetracycline were examined by electron microscopy. While typical membranous webs were found in 17 of 100 sections from UHCVcon-57.3 cells expressing the wild-type (wt) HCV H77 polyprotein, no membranous web was found in 200 sections from UHCVcon-AH2mut cells expressing the H77 polyprotein harboring the AH2mut alanine substitutions in NS4B.
In conclusion, mutations that affect NS4B oligomerization disrupt membranous web formation and, as shown previously (14), HCV RNA replication. Although we cannot formally exclude the possibility that interactions with other replicase components are affected by the AH2mut substitutions, these findings are consistent with an important role for NS4B oligomerization in membranous web formation and in the assembly of a functional replication complex.
DISCUSSION
Based on carefully controlled FRET and confirmatory coimmunoprecipitation analyses, we provide evidence for oligomerization of HCV NS4B in the membrane environment of intact cells. Several conserved determinants were found to contribute to the oligomerization of NS4B, through homotypic and heterotypic interactions. Amphipathic α-helix AH2 was identified as a major determinant for the oligomerization of NS4B. Furthermore, mutations in NS4B that affected oligomerization disrupted membranous web formation and HCV RNA replication, implying that oligomerization of NS4B is essential for at least one of its functions in the viral life cycle.
Acceptor photobleaching FRET offers the unique opportunity to investigate protein-protein interactions at a defined subcellular location within the membrane environment of intact cells (5). Analyses were performed following carefully established criteria and involved a large number of measurements for each protein combination. In particular, analyses were restricted to NS4B fragments with a subcellular localization corresponding to that of the full-length protein, i.e., the ER or ER-derived modified membranes. Constructs displaying aberrant subcellular localization or protein aggregation as well as cells that appeared to suffer as a result of the transfection procedure or membrane protein overexpression were excluded from our analyses. Under these conditions, we found that not only full-length NS4B but also N- and C-terminal fragments comprising amino acids 40 to 130 and 130 to 261, respectively, interacted both with themselves and with each other. In our experience, FRET did not allow us to further dissect the different determinants involved in NS4B self-interaction. Indeed, shorter segments were often aberrantly localized or showed aggregation, precluding meaningful interaction studies by FRET.
Other HCV nonstructural proteins have been shown to form dimers, notably NS2 (29) and NS5A (46). In addition, NS3 and NS5B have been proposed to form higher-order oligomeric complexes (20, 48). Our assays do not allow us to distinguish between dimer formation and the formation of higher-order oligomeric complexes or arrays of NS4B. However, based on the role of NS4B in the induction of vesicular membrane rearrangements, the involvement of homotypic and heterotypic interactions, and the chemical cross-linking studies reported by Yu et al. (49), we favor the latter scenario. Indeed, a number of viral and cellular proteins induce and maintain membrane curvature and vesicle induction by oligomerization (44, 50). Of particular importance, key organizers of the replication complexes of other positive-strand RNA viruses have been shown to exert their function as multimeric complexes. For example, replicase protein A from flock house virus (FHV), an alphanodavirus, has been shown to self-interact through multiple determinants (7) and to induce invaginations of the outer mitochondrial membrane, designated spherules (24). These FHV-induced replication vesicles are believed to be lined by a continuous shell of protein A (6). In addition, brome mosaic virus (BMV), a member of the alphavirus-like superfamily, encodes a membrane-associated multifunctional protein, designated 1a, that self-interacts through multiple determinants and induces ER-derived replication vesicles (6, 43).
A major mechanism to generate membrane curvature involves hydrophobic insertion or wedging (44). In this mechanism, proteins insert hydrophobic domains, such as the hydrophobic side of an amphipathic α-helix, into one leaflet of the membrane bilayer, thereby generating local membrane curvature. Alternatively, integral membrane proteins that occupy more space in one leaflet of the membrane than the other could also generate membrane curvature. A second major mechanism is termed scaffolding (44). In this case, a protein or an oligomeric protein complex with a curved surface interacts with a membrane to induce membrane curvature. Importantly, most membrane curvature-inducing proteins use a combination of hydrophobic insertion and scaffolding mechanisms. For example, N-BAR domain-containing proteins, such as amphiphysin, insert an N-terminal amphipathic α-helix into the lipid bilayer and use their banana shape to scaffold the curved membrane (39). COPII vesicle formation also involves both mechanisms. For this process, the small GTP-binding protein Sar1 induces membrane curvature by the insertion of an N-terminal amphipathic α-helix, while oligomeric complexes of COPII coat proteins serve as a scaffold (10, 27). Finally, reticulons and DP1/Yop1p are believed to shape ER tubules by hydrophobic insertion and scaffolding (18, 45, 47). Clearly, both mechanisms may also be operative in the case of HCV NS4B, where the amphipathic α-helices, notably AH2, the central transmembrane segments, and C-terminal membrane-associated α-helix H2 (15) may induce local membrane curvature that is scaffolded by NS4B oligomerization. Thus, cooperative mechanisms may be involved in the formation of the membranous web by NS4B.
The observation that N- and C-terminal determinants contribute to NS4B oligomerization through homotypic and heterotypic interactions raises several and potentially dynamic models for the formation of NS4B oligomeric structures. As evoked previously for FHV protein A (7), it is possible that these contacts form simultaneously such that two NS4B molecules in a dimer contact one another at multiple interfaces. If NS4B forms higher-order oligomers, then multiple NS4B molecules could interact such that each molecule contacts another by using a different interaction domain. Also, certain interactions and multimeric states may be specific to distinct NS4B functions. Indeed, recent evidence indicates that NS4B may not only be a key organizer of replication complex formation but also have a role in virus assembly and release (21; J. Gouttenoire and D. Moradpour, unpublished data). Since the N-terminal part of NS4B can acquire a dual topology through the posttranslational membrane insertion of AH2 (14, 30, 31), one could also speculate that the membrane topology (in-plane versus transmembrane) of AH2 may determine distinct NS4B oligomerization states and, thereby, different functions. Moreover, additional complex interactions of NS4B with other replicase components may influence its functions during different stages of the viral life cycle. Clearly, further studies will be necessary to explore these exciting possibilities. Obtaining a three-dimensional structure of full-length NS4B will be key to such studies.
A recent report showed that a synthetic peptide corresponding to amphipathic α-helix AH2 triggered liposome aggregation in vitro (4). Interestingly, a chemical compound blocking this activity inhibited HCV RNA replication, presumably by interfering with AH2-mediated oligomerization of NS4B. Hence, interfering with the oligomerization of NS4B may provide new opportunities for antiviral intervention. The observation that oligomerization can occur between NS4B proteins from different HCV genotypes through conserved determinants is particularly attractive in this regard. Notably, unprecedented antiviral activity against different HCV genotypes was recently achieved with a small molecule targeting NS5A at a site that may be involved in the formation of oligomeric NS5A structures (12, 35).
In conclusion, we provide evidence for oligomerization of HCV NS4B in the membrane environment of intact cells. Several conserved determinants were found to contribute to oligomerization, through homotypic and heterotypic interactions. Amphipathic α-helix AH2 was identified as a major determinant for the oligomerization of NS4B. Furthermore, mutations that affected NS4B oligomerization disrupted membranous web formation, implying that NS4B functions as an oligomer at least during a key step in the viral life cycle, i.e., the formation of the membranous web. These findings enhance our understanding of the functional architecture of the HCV replication complex and may provide new angles for therapeutic intervention. At the same time, they expand the list of positive-strand RNA virus replicase components acting as oligomers.
Supplementary Material
Acknowledgments
We gratefully acknowledge Jan Martin Berke, Volker Brass, and Naveen Arora for early contributions to this study, Audrey Kennel and Juliette Rousseau for expert technical assistance, Jean-Yves Chatton, Jérôme N. Feige, and Laurent Gelman for advice on FRET, and Atsushi Miyawaki, David W. Piston, Roland Nitschke, and Ralf Bartenschlager for reagents. FRET analyses were performed at the Cellular Imaging Facility of the University of Lausanne. EM analyses were performed at the RIO Electron Microscopy Facility of the University of Tours.
This work was supported by the Swiss National Science Foundation (grant 3100A0-122447), the Novartis Foundation (grant 09C53), the Centre National de la Recherche Scientifique (CNRS), and the Agence Nationale pour la Recherche sur le SIDA et les Hépatites Virales (ANRS).
Footnotes
Published ahead of print on 6 October 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Berke, J. M. 2007. Determinants for membrane association of the hepatitis C virus NS3-4A complex. Ph.D. thesis. University of Freiburg, Freiburg, Germany.
- 2.Brass, V., J. M. Berke, R. Montserret, H. E. Blum, F. Penin, and D. Moradpour. 2008. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proc. Natl. Acad. Sci. U. S. A. 105:14545-14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho, N. J., H. Dvory-Sobol, C. Lee, S. J. Cho, P. Bryson, M. Masek, M. Elazar, C. W. Frank, and J. S. Glenn. 2010. Identification of a class of HCV inhibitors directed against the nonstructural protein NS4B. Sci. Transl. Med. 2:15ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciruela, F., J. P. Vilardaga, and V. Fernández-Dueñas. 2010. Lighting up multiprotein complexes: lessons from GPCR oligomerization. Trends Biotechnol. 28:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.den Boon, J. A., A. Diaz, and P. Ahlquist. 2010. Cytoplasmic viral replication complexes. Cell Host Microbe 8:77-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dye, B. T., D. J. Miller, and P. Ahlquist. 2005. In vivo self-interaction of nodavirus RNA replicase protein A revealed by fluorescence resonance energy transfer. J. Virol. 79:8909-8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egger, D., B. Wölk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elazar, M., P. Liu, C. M. Rice, and J. S. Glenn. 2004. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J. Virol. 78:11393-11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fath, S., J. D. Mancias, X. Bi, and J. Goldberg. 2007. Structure and organization of coat proteins in the COPII cage. Cell 129:1325-1336. [DOI] [PubMed] [Google Scholar]
- 11.Ferraris, P., E. Blanchard, and P. Roingeard. 2010. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J. Gen. Virol. 91:2230-2237. [DOI] [PubMed] [Google Scholar]
- 12.Gao, M., R. E. Nettles, M. Belema, L. B. Snyder, V. N. Nguyen, R. A. Fridell, M. H. Serrano-Wu, D. R. Langley, J. H. Sun, D. R. O'Boyle II, J. A. Lemm, C. Wang, J. O. Knipe, C. Chien, R. J. Colonno, D. M. Grasela, N. A. Meanwell, and L. G. Hamann. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gouttenoire, J., V. Castet, R. Montserret, N. Arora, V. Raussens, J. M. Ruysschaert, E. Diesis, H. E. Blum, F. Penin, and D. Moradpour. 2009. Identification of a novel determinant for membrane association in hepatitis C virus nonstructural protein 4B. J. Virol. 83:6257-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gouttenoire, J., R. Montserret, A. Kennel, F. Penin, and D. Moradpour. 2009. An amphipathic α-helix at the C terminus of NS4B mediates membrane association. J. Virol. 51:11378-11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouttenoire, J., F. Penin, and D. Moradpour. 2010. Hepatitis C virus nonstructural protein 4B: a journey into unexplored territory. Rev. Med. Virol. 20:117-129. [DOI] [PubMed] [Google Scholar]
- 17.Gretton, S. N., A. I. Taylor, and J. McLauchlan. 2005. Mobility of the hepatitis C virus NS4B protein on the endoplasmic reticulum membrane and membrane-associated foci. J. Gen. Virol. 86:1415-1421. [DOI] [PubMed] [Google Scholar]
- 18.Hu, J., Y. Shibata, C. Voss, T. Shemesh, Z. Li, M. Coughlin, M. M. Kozlov, T. A. Rapoport, and W. A. Prinz. 2008. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science 319:1247-1250. [DOI] [PubMed] [Google Scholar]
- 19.Hügle, T., F. Fehrmann, E. Bieck, M. Kohara, H.-G. Kräusslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 20.Jennings, T. A., S. G. Mackintosh, M. K. Harrison, D. Sikora, B. Sikora, B. Dave, A. J. Tackett, C. E. Cameron, and K. D. Raney. 2009. NS3 helicase from the hepatitis C virus can function as a monomer or oligomer depending on enzyme and substrate concentrations. J. Biol. Chem. 284:4806-4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, D. M., A. H. Patel, P. Targett-Adams, and J. McLauchlan. 2009. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J. Virol. 83:2163-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 23.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 24.Kopek, B. G., G. Perkins, D. J. Miller, M. H. Ellisman, and P. Ahlquist. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuppig, S., and R. Nitschke. 2006. A fusion tag enabling optical marking and tracking of proteins and cells by FRET-acceptor photobleaching. J. Microsc. 222:15-21. [DOI] [PubMed] [Google Scholar]
- 26.Lavanchy, D. 2009. The global burden of hepatitis C. Liver Int. 29(Suppl. 1):74-81. [DOI] [PubMed] [Google Scholar]
- 27.Lee, M. C., L. Orci, S. Hamamoto, E. Futai, M. Ravazzola, and R. Schekman. 2005. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell 122:605-617. [DOI] [PubMed] [Google Scholar]
- 28.Lindenbach, B. D., H. J. Thiel, and C. M. Rice. 2007. Flaviviridae: the viruses and their replication, p. 1101-1152. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott-Raven, Philadelphia, PA.
- 29.Lorenz, I. C., J. Marcotrigiano, T. G. Dentzer, and C. M. Rice. 2006. Structure of the catalytic domain of the hepatitis C virus NS2-3 protease. Nature 442:831-835. [DOI] [PubMed] [Google Scholar]
- 30.Lundin, M., H. Lindstrom, C. Gronwall, and M. A. Persson. 2006. Dual topology of the processed hepatitis C virus protein NS4B is influenced by the NS5A protein. J. Gen. Virol. 87:3263-3272. [DOI] [PubMed] [Google Scholar]
- 31.Lundin, M., M. Monne, A. Widell, G. Von Heijne, and M. A. Persson. 2003. Topology of the membrane-associated hepatitis C virus protein NS4B. J. Virol. 77:5428-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, S., S. Kastner, J. Krijnse-Locker, S. Bühler, and R. Bartenschlager. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 282:8873-8882. [DOI] [PubMed] [Google Scholar]
- 33.Miller, S., and J. Krijnse-Locker. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, S., S. Sparacio, and R. Bartenschlager. 2006. Subcellular localization and membrane topology of the dengue virus type 2 non-structural protein 4B. J. Biol. Chem. 281:8854-8863. [DOI] [PubMed] [Google Scholar]
- 35.Moradpour, D., V. Brass, and F. Penin. 2005. Function follows form: the structure of the N-terminal domain of HCV NS5A. Hepatology 42:732-735. [DOI] [PubMed] [Google Scholar]
- 36.Moradpour, D., F. Penin, and C. M. Rice. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453-463. [DOI] [PubMed] [Google Scholar]
- 37.Nagai, T., K. Ibata, E. S. Park, M. Kubota, K. Mikoshiba, and A. Miyawaki. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87-90. [DOI] [PubMed] [Google Scholar]
- 38.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 39.Peter, B. J., H. M. Kent, I. G. Mills, Y. Vallis, P. J. Butler, P. R. Evans, and H. T. McMahon. 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303:495-499. [DOI] [PubMed] [Google Scholar]
- 40.Ponten, J., and E. Saksela. 1967. Two established in vitro cell lines from human mesenchymal tumours. Int. J. Cancer 2:434-447. [DOI] [PubMed] [Google Scholar]
- 41.Rizzo, M. A., G. H. Springer, B. Granada, and D. W. Piston. 2004. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 22:445-449. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt-Mende, J., E. Bieck, T. Hügle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 43.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 44.Shibata, Y., J. Hu, M. M. Kozlov, and T. A. Rapoport. 2009. Mechanisms shaping the membranes of cellular organelles. Annu. Rev. Cell Dev. Biol. 25:329-354. [DOI] [PubMed] [Google Scholar]
- 45.Shibata, Y., C. Voss, J. M. Rist, J. Hu, T. A. Rapoport, W. A. Prinz, and G. K. Voeltz. 2008. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 283:18892-18904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tellinghuisen, T. L., J. Marcotrigiano, and C. M. Rice. 2005. Structure of the zinc-binding domain of an essential replicase component of hepatitis C virus reveals a novel fold. Nature 435:375-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voeltz, G. K., W. A. Prinz, Y. Shibata, J. M. Rist, and T. A. Rapoport. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124:573-586. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Q. M., M. A. Hockman, K. Staschke, R. B. Johnson, K. A. Case, J. Lu, S. Parsons, F. Zhang, R. Rathnachalam, K. Kirkegaard, and J. M. Colacino. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, G. Y., K. J. Lee, L. Gao, and M. M. Lai. 2006. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J. Virol. 80:6013-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zimmerberg, J., and M. M. Kozlov. 2006. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell. Biol. 7:9-19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.