Abstract
The roles of conserved nucleotides on the stem-loop (SL) structure in the intergenic region of the hepatitis E virus (HEV) genome in virus replication were determined by using Huh7 cells transfected with HEV SL mutant replicons containing reporter genes. One or two nucleotide mutations of the AGA motif on the loop significantly reduced HEV replication, and three or more nucleotide mutations on the loop abolished HEV replication. Mutations on the stem and of the subgenome start sequence also significantly inhibited HEV replication. The results indicated that both the sequence and the SL structure in the junction region play important roles in HEV replication.
Hepatitis E virus (HEV) is the causative agent of hepatitis E, and at least four major genotypes have been recognized in mammalian species: genotypes 1 and 2 are restricted to humans, whereas genotypes 3 and 4 are zoonotic (1-3, 9, 21, 26-29). The genome is a single-strand, positive-sense RNA molecule (12) consisting of a 5′ noncoding region (NCR), open reading frame 1 (ORF1) encoding the nonstructural proteins, ORF2 encoding the capsid protein, ORF3 encoding a small multifunctional protein (6, 22, 23, 31, 32, 38-40, 42), and a 3′ NCR. ORF2 and ORF3 are translated from a single bicistronic mRNA and overlap each other, but neither overlaps ORF1 (15, 19). The HEV genome contains two cis-reactive elements (CRE): the first CRE overlaps the 3′ end of ORF2 and the 3′ NCR and is essential for virus replication (13), and the second CRE may be the promoter for synthesis of the 2.0-kb subgenomic (SG) mRNA (14, 15). Graff et al. showed that neither ORF2 expression nor ORF3 expression was detectable when 6-nucleotide (nt) or 4-nt mutations were introduced into the junction region of the HEV genome; however, the roles of individual nucleotides in the junction region and its surrounding sequences in virus replication remain unknown (14).
We identified a region within the junction region (Fig. 1) of the HEV genome that shares nucleotide sequence identity with rubella virus and with the conserved alphavirus subgenomic promoter sequence. A highly conserved stem-loop (SL) structure was predicted to occur in the alphavirus junction region with sequences homologous to those of the HEV antigenome RNA in the junction region of the HEV genome (7, 19). The objective of this study was to determine the effect of mutations of the conserved nucleotides in the SL and its RNA structure on HEV replication.
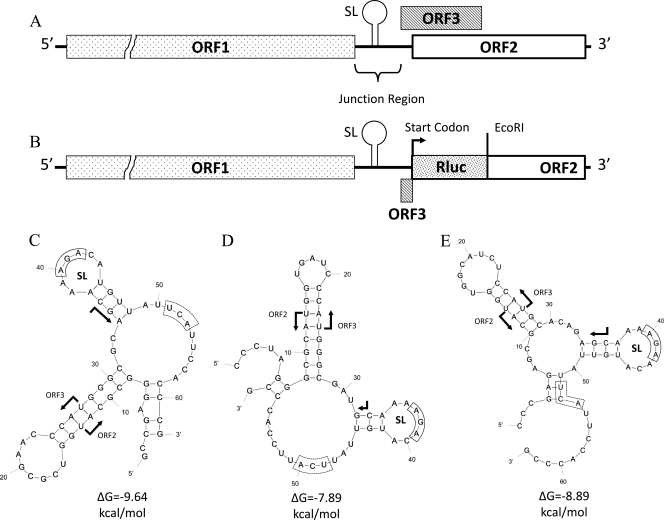
FIG. 1.
(A) Organization of the HEV genome. The position of the predicted RNA stem-loop (SL) structure in the junction region is depicted. (B) Schematic diagram of the HEV Rluc replicon that was used as the backbone for the construction of various mutants. (C) Predicted secondary structure of the negative-polarity complement of the HEV genotype 1 (Sar55 strain) junction region. The sequence shown extends from nt 5096 through 5157. (D) Predicted secondary structure of the negative-polarity complement of the HEV genotype 3 (pSHEV-3 strain) junction region. The sequence shown extends from nt 5142 through 5200. (E) Predicted secondary structure of the negative-polarity complement of the HEV genotype 4 (T1 strain) junction region. The sequence shown extends from nt 5138 through 5200. The conserved AGA triplets in the SG promoters of alphavirus family members are boxed with solid lines. The HEV subgenome start site is indicated with arrows. The start sites of ORF2 and ORF3 are also indicated; the stop codon of ORF1 is boxed with dotted lines.
Analyses of the RNA SL structure and its surrounding sequences in the junction region of the HEV genome.
RNA secondary structures often play important roles in viral replication, SG RNA synthesis, and translation efficiency (24, 36). It is believed that the complementary negative strand of the HEV SG promoter is recognized by RdRp or another viral component or host factor that interacts with RdRp, which then initiates the SG RNA synthesis in a primer-independent fashion at the SG start site (30). Using the mfold program (43), we identified two highly conserved SL structures in the intergenic region between the end of ORF1 and the start of ORF2 (Fig. 1C to E). The first SL is beyond the SG sequence (20) and thus may function as an SG promoter, and the SL also overlaps with the CRE region (14). Sequence analyses revealed an AGA triplet (in negative polarity) at nt −4 to −6 in the junction region (Fig. 1), positions similar to that of the AGA triplet in the rubella virus SG promoter (nt −8 to −10). The AGA triplet is also conserved in the SG promoters of alphavirus family members. Therefore, it is important to determine the function of the HEV AGA triplet and its surrounding nucleotides.
Both the sequence and structure of the SL in the HEV junction region are important for HEV replication.
Although HEV infectious clones are available (13, 18, 35), the lack of an efficient cell culture for HEV prevents us from directly testing the replication of mutant viruses in vitro (34). It has been shown that a green fluorescence protein (GFP) HEV replicon is a good system to study HEV replication in vitro (10, 37). Therefore, in this study, we first constructed a genotype 1 HEV enhanced GFP (EGFP) replicon system (Y.-W. Huang and X. J. Meng, unpublished data) by using the Sar55 infectious clone (a gift from Sue Emerson, NIH, Bethesda, MD). We then constructed eight EGFP replicon-based HEV mutants and tested for their effects on HEV replication. Unfortunately, the sensitivity of the EGFP HEV replicon system was low (data not shown).
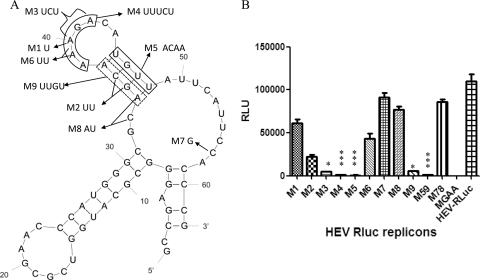
To definitively assess the roles of the nucleotides of the junction region in HEV replication, we subsequently constructed a novel HEV replicon system by replacing nt 5148 to 5816 of the infectious clone pSK-HEV-2 with the Renilla luciferase (Rluc) gene (Fig. 1B). By utilizing the start codon of HEV ORF2, the Rluc HEV replicon expresses Renilla luciferase (Rluc) (Fig. 1B), which was used as a reporter for quantifying HEV replication. By using the Rluc HEV replicon as the backbone, we constructed 11 SL mutants, designated as follows (Fig. 2A; Table 1) (sequences shown as the negative polarity complement of the HEV genome): M1 (A5118→U); M2 (C5122→U, G5123→U); M3 (AGA5116 to -5118→UCU); M4 (AAAGA5116 to -5120→UUUCU); M5 (UGUU5110 to -5113→ACAA), which contains mutations on one leg of the SL stem; M6 (AA5119 to -5120→UU); M7 (C5101→G); M8 (CA5124 to -5125→AU); M9 (AGCA5121 to -5124→UUGU), which contains mutations on another leg of the stem; M59 (UGUU5110 to -5113→ACAA, AGCA5121 to -5124→UUGU), with mutations on both legs of the stem; and M78 (C5101→G, CA5124 to -5125→AU), which is a combination of M7 and M8. In addition, the HEV Rluc replicon mutant with a GDD→GAA mutation on RdRp (MGAA) was constructed and used as a negative control. Capped RNA transcripts from each of the 11 mutant replicons along with the MGAA and wild-type replicon were synthesized in vitro with an mMessage mMachine T7 kit (Ambion) (17, 19, 37). The capped RNA transcripts of each mutant and control were transfected into the Huh7-S10-3 liver cell line (a gift from Sue Emerson, NIH, Bethesda, MD) (11, 15) with 1,2-dimyristyl Rosenthal inhibitor ether (DMRIE-C) reagent (Invitrogen). The luciferase activities were measured with a dual luciferase reporter assay system (Promega) at 5 days posttransfection. Firefly luciferase RNA was cotransfected with HEV Rluc replicon RNAs to normalize the Renilla luciferase signal.
FIG. 2.
Mutational analyses of the predicted stem-loop (SL) structure in the junction region of the HEV genome. (A) Mutations were introduced into the stems and loop sequences of the SL structure in mutants M1 to M9, M59, and M78 for the HEV Rluc replicon. (B) Relative luciferase activities in Huh7 S10-3 cells transfected with HEV Rluc mutants M1 to M9, M59 (i.e., M5 plus M9), M78 (i.e., M7 plus M8), HEV Rluc MGAA (negative control), and the wild-type Rluc replicon (HEV-RLuc). The relative luciferase activities are shown at 5 days posttransfection and normalized with cotransfected firefly luciferase RNA. Data are from an average of eight separate replicate experiments, and the error bars indicate standard deviations (SD). The differences in signal produced by HEV Rluc mutants and the wild-type Rluc replicon were compared by one-way analysis of variance (ANOVA) using the Kruskal-Wallis test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; RLU, relative light units.
TABLE 1.
Primers used in the generation of HEV mutants and LNAs used for inhibition of HEV replicon replication
| Oligonucleotide | Polaritya | nt positionb | Sequencec |
|---|---|---|---|
| Mutagenic primers | |||
| HEV2m1R | − | 5090-5139 | 5′-CGAACCCATGGGCGCAGCAAATGACATGTTATTCATTCCACCCGACACAG-3′ |
| HEV2m1F | + | 5′-CTGTGTCGGGTGGAATGAATAACATGTCATTTGCTGCGCCCATGGGTTCG-3′ | |
| HEV2m2R | − | 5097-5141 | 5′-CGCGAACCCATGGGCGCATTAAAAGACATGTTATTCATTCCACCC-3′ |
| HEV2m2F | + | 5′-GGGTGGAATGAATAACATGTCTTTTAATGCGCCCATGGGTTCGCG-3′ | |
| HEV2m3R | − | 5094-5140 | 5′-GCGAACCCATGGGCGCAGCAAATCTCATGTTATTCATTCCACCCGAC-3′ |
| HEV2m3F | + | 5′-GTCGGGTGGAATGAATAACATGAGATTTGCTGCGCCCATGGGTTCGC-3′ | |
| HEV2m4R | − | 5094-5140 | 5′-GCGAACCCATGGGCGCAGCATTTCTCATGTTATTCATTCCACCCGAC-3′ |
| HEV2m4F | + | 5′-GTCGGGTGGAATGAATAACATGAGAAATGCTGCGCCCATGGGTTCGC-3′ | |
| HEV2m5R | − | 5085-5134 | 5′-CCATGGGCGCAGCAAAAGACAACAAATTCATTCCACCCGACACAGAATTG-3′ |
| HEV2m5F | + | 5′-CAATTCTGTGTCGGGTGGAATGAATTTGTTGTCTTTTGCTGCGCCCATGG-3′ | |
| HEV2m6R | − | 5090-5139 | 5′-CGAACCCATGGGCGCAGCAATTGACATGTTATTCATTCCACCCGACACAG-3′ |
| HEV2m6F | + | 5′-CTGTGTCGGGTGGAATGAATAACATGTCAATTGCTGCGCCCATGGGTTCG-3′ | |
| HEV2m7R | − | 5079-5125 | 5′-CAGCAAAAGACATGTTATTCATTCGACCCGACACAGAATTGAATTTG-3′ |
| HEV2m7F | + | 5′-CAAATTCAATTCTGTGTCGGGTCGAATGAATAACATGTCTTTTGCTG-3′ | |
| HEV2m8R | − | 5101-5144 | 5′-GGTCGCGAACCCATGGGCGATGCAAAAGACATGTTATTCATTCC-3′ |
| HEV2m8F | + | 5′-GGAATGAATAACATGTCTTTTGCATCGCCCATGGGTTCGCGACC-3′ | |
| HEV2m9L | − | 5090-5143 | 5′-GGTCGCGAACCCATGGGCGCTTGTAAAGACATGTTATTCATTCCACCCGACACAG-3′ |
| HEV2m9U | + | 5′-CTGTGTCGGGTGGAATGAATAACATGTCTTTACAAGCGCCCATGGGTTCGCGAC-3′ | |
| HEV2m59L | − | 5090-5144 | 5′-GGTCGCGAACCCATGGGCGCTTGTAAAGACAACAAATTCATTCCACCCGACACAG-3′ |
| HEV2m59U | + | 5′-CTGTGTCGGGTGGAATGAATTTGTTGTCTTTACAAGCGCCCATGGGTTCGCGACC-3′ | |
| HEV2mGAAU | + | 4655-4705 | 5′-CAGGTGGCTGCCTTTAAAGGTGCCGCCTCGATAGTGCTTTGCAGTGAGTAC-3′ |
| HEV2mGAAL | − | 5′-GTACTCACTGCAAAGCACTATCGAGGCGGCACCTTTAAAGGCAGCCACCTG-3′ | |
| LNAs | |||
| p19 antisense | − | 5105-5126 | 5′-G+CAG+CA+AA+AG+AC+ATGTT+ATT+CA-3′ |
| p19 sense | + | 5′-T+GAA+TAACA+TGT+CTTT+TG+CT+GC-3′ |
Polarity of primers or LNAs on the HEV genome. +, forward; −, reverse.
Positions of primers or LNAs on the HEV genome.
Sequences of primers or LNAs. The mutated nucleotides are underlined and in boldface. The modified nucleotides in LNAs are indicated with +.
The results showed that the Rluc signal is lower in cells transfected with RNA of mutant M1, M2, M3, M4, M5, M6, M9, or M59 than that in cells transfected with RNA of the wild-type HEV Rluc replicon (Fig. 2B), with statistically significant differences for mutants M3, M4, M5, M9, and M59. The mutant M3, which changed only the AGA motif, abolished HEV replication as efficiently as did mutant M4 that contained two additional adenosine nucleotide changes compared to M3. Even a single nucleotide mutation of the AGA motif on the loop (M1) inhibited HEV replication, suggesting that the nucleotides on the loop of SL are important for HEV replication and that the AGA motif is critical for HEV replication. The mutation on either leg of the stem (M5 and M9) also significantly inhibits HEV replication. The mutation that broke one base pair (U-G) on one leg of the stem (M2) also inhibited HEV replication, indicating that the structure of SL is also important for HEV replication. Although the HEV EGFP replicon system is not as sensitive as the Rluc system, the results with the EGFP replicon-based SL mutants are qualitatively similar to those with the Rluc-based mutants (data not shown).
It is noteworthy that the conserved AAUAAC sequence in the sense genome of the junction region, which was identified as an important motif for HEV replication in vivo (19), has 3 nt overlapped with one leg of the SL stem. The mutations of AAUAAC to AACAUG that resulted in less-efficient replication (19) actually broke two base pairs on the SL stem and thus may change the SL structure and inhibit HEV replication. However, we failed to rescue HEV replication by replacing the stem with a mutated complement sequence (M59) on the stem of SL (Fig. 2B), suggesting that both the sequence and structure of the SL play an important role in HEV replication. Elimination of the predicted JC virus (JCV) repeated sequence (25) and enhancer core motif (41) (M7) has no significant effect on HEV replication. Furthermore, mutations in the metal response element (MRE) motif CS2 (8) (M8) reduced HEV replication, and the combination of M7 and M8 mutations (M78) has a similar effect on HEV replication compared to that of the single mutation (M7 or M8), suggesting that these motifs may regulate but are not important for HEV replication.
LNAs targeting the SL structure inhibit HEV replication.
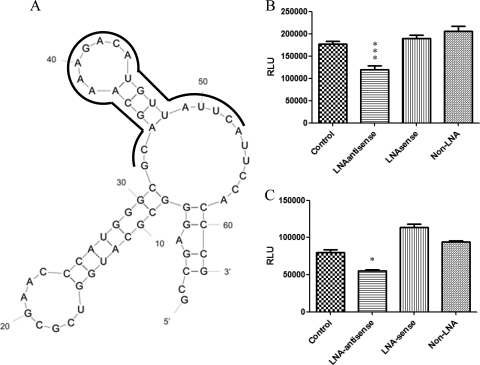
Locked nucleic acid (LNA) bases contain a bridging methylene carbon between the 2′ and 4′ position of the ribose ring (4), and this constraint preorganizes the oligonucleotide backbone and can increase melting temperature (Tm) values by as much as 10°C per LNA substitution. Chimeric LNAs have been demonstrated not only to be active antisense agents (5, 16) but also to block the internal ribosomal entry site and inhibit translation (33). Thus, to further verify the importance of the SL on HEV replication, we designed and synthesized two LNAs that specifically target both the sense and the antisense sequences of the SL structure (Fig. 3A; Table 1). An oligonucleotide unrelated to HEV sequence, the M13 forward primer, was used as the non-LNA control. After cotransfecting each LNA with HEV Rluc replicon RNA at the same time into Huh7 cells, we measured the Rluc signal at 5 days posttransfection. The results showed that the antisense LNA inhibited Rluc signals by 42% (Fig. 3B), whereas the sense LNA has no effect on HEV replication. When the LNAs were transfected into cells at 48 h after the transfection of HEV Rluc replicon RNAs, similar inhibition results were observed with the antisense LNA (Fig. 3C). The inhibition of antisense LNA on HEV replication may function through blocking the negative RNA from binding to the RdRp and/or another factor(s) that initiates the replication of subgenomic RNA. The signal from Huh7 cells transfected with HEV Rluc replicon and LNA sense is higher than that from cells transfected with M13 forward primer, but the difference is not statistically significant. This could be due to the variation of nonspecific inhibition of LNA sense and M13 forward primer on the replication of HEV Rluc RNA. The LNA results further confirmed that the SL sequence is important for HEV replication and that the conserved SL structure in the junction region could be a potential target for HEV drug development.
FIG. 3.
Inhibition of HEV Rluc replicon replication by LNAs. (A) The position of LNA on the stem-loop structure in the junction region of the HEV genome (targeting nt 5105 through 5126) is indicated with a boldface solid line. (B) Relative luciferase activity in Huh7 S10-3 cells cotransfected at the same time with HEV Rluc replicon and LNA sense, LNA antisense, or control oligonucleotide (100 pmol per well in 24-well plate), respectively. (C) Relative luciferase activity in Huh7 S10-3 cells transfected with LNA sense, LNA antisense, or control oligonucleotide at 48 h after the transfection of the HEV Rluc replicon RNA. Data are from an average of eight separate replicate experiments, and the error bars indicate SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In summary, we identified the nucleotides on the SL structure of the junction region in the HEV genome that are important for HEV replication and demonstrated that both the sequence and the structure of the SL are critical for HEV replication.
Acknowledgments
We thank Barbara Dryman for technical assistance.
This study is supported by grants from the National Institutes of Health (R01 AI074667 and R01 AI050611).
Footnotes
Published ahead of print on 13 October 2010.
REFERENCES
- 1.Aikawa, T., M. Kojima, M. Takahashi, T. Nishizawa, and H. Okamoto. 2002. Identification of indigenous hepatitis E virus from a Japanese patient who contracted sporadic acute hepatitis in 1982. J. Infect. Dis. 186:1535-1536. (Author reply, 186:1536-1537.) [DOI] [PubMed] [Google Scholar]
- 2.Arankalle, V. A., L. P. Chobe, M. V. Joshi, M. S. Chadha, B. Kundu, and A. M. Walimbe. 2002. Human and swine hepatitis E viruses from Western India belong to different genotypes. J. Hepatol. 36:417-425. [DOI] [PubMed] [Google Scholar]
- 3.Arankalle, V. A., S. Paranjape, S. U. Emerson, R. H. Purcell, and A. M. Walimbe. 1999. Phylogenetic analysis of hepatitis E virus isolates from India (1976-1993). J. Gen. Virol. 80:1691-1700. [DOI] [PubMed] [Google Scholar]
- 4.Braasch, D. A., and D. R. Corey. 2001. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. 8:1-7. [DOI] [PubMed] [Google Scholar]
- 5.Braasch, D. A., Y. Liu, and D. R. Corey. 2002. Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Res. 30:5160-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandra, V., M. Kalia, K. Hajela, and S. Jameel. 2010. The ORF3 protein of hepatitis E virus delays degradation of activated growth factor receptors by interacting with CIN85 and blocking formation of the Cbl-CIN85 complex. J. Virol. 84:3857-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandra, V., S. Taneja, M. Kalia, and S. Jameel. 2008. Molecular biology and pathogenesis of hepatitis E virus. J. Biosci. 33:451-464. [DOI] [PubMed] [Google Scholar]
- 8.Culotta, V. C., and D. H. Hamer. 1989. Fine mapping of a mouse metallothionein gene metal response element. Mol. Cell. Biol. 9:1376-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emerson, S. U., D. Anderson, A. Arankalle, X. J. Meng, M. Purdy, G. G. Schlauder, and S. A. Tsarev. 2004. Hepevirus, p. 851-855. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: VIIIth report of the ICTV. Elsevier/Academic Press, London, United Kingdom.
- 10.Emerson, S. U., H. Nguyen, J. Graff, D. A. Stephany, A. Brockington, and R. H. Purcell. 2004. In vitro replication of hepatitis E virus (HEV) genomes and of an HEV replicon expressing green fluorescent protein. J. Virol. 78:4838-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emerson, S. U., H. Nguyen, U. Torian, and R. H. Purcell. 2006. ORF3 protein of hepatitis E virus is not required for replication, virion assembly, or infection of hepatoma cells in vitro. J. Virol. 80:10457-10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emerson, S. U., and R. H. Purcell. 2007. Hepatitis E virus, p. 3048-3059. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 13.Emerson, S. U., M. Zhang, X. J. Meng, H. Nguyen, M. St. Claire, S. Govindarajan, Y. K. Huang, and R. H. Purcell. 2001. Recombinant hepatitis E virus genomes infectious for primates: importance of capping and discovery of a cis-reactive element. Proc. Natl. Acad. Sci. U. S. A. 98:15270-15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graff, J., H. Nguyen, C. Yu, W. R. Elkins, M. St. Claire, R. H. Purcell, and S. U. Emerson. 2005. The open reading frame 3 gene of hepatitis E virus contains a cis-reactive element and encodes a protein required for infection of macaques. J. Virol. 79:6680-6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graff, J., U. Torian, H. Nguyen, and S. U. Emerson. 2006. A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J. Virol. 80:5919-5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grunweller, A., E. Wyszko, B. Bieber, R. Jahnel, V. A. Erdmann, and J. Kurreck. 2003. Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2′-O-methyl RNA, phosphorothioates and small interfering RNA. Nucleic Acids Res. 31:3185-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, F. F., F. W. Pierson, T. E. Toth, and X. J. Meng. 2005. Construction and characterization of infectious cDNA clones of a chicken strain of hepatitis E virus (HEV), avian HEV. J. Gen. Virol. 86:2585-2593. [DOI] [PubMed] [Google Scholar]
- 18.Huang, Y. W., G. Haqshenas, C. Kasorndorkbua, P. G. Halbur, S. U. Emerson, and X. J. Meng. 2005. Capped RNA transcripts of full-length cDNA clones of swine hepatitis E virus are replication competent when transfected into Huh7 cells and infectious when intrahepatically inoculated into pigs. J. Virol. 79:1552-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, Y. W., T. Opriessnig, P. G. Halbur, and X. J. Meng. 2007. Initiation at the third in-frame AUG codon of open reading frame 3 of the hepatitis E virus is essential for viral infectivity in vivo. J. Virol. 81:3018-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ichiyama, K., K. Yamada, T. Tanaka, S. Nagashima, Jirintai, M. Takahashi, and H. Okamoto. 2009. Determination of the 5′-terminal sequence of subgenomic RNA of hepatitis E virus strains in cultured cells. Arch. Virol. 154:1945-1951. [DOI] [PubMed] [Google Scholar]
- 21.Jameel, S. 1999. Molecular biology and pathogenesis of hepatitis E virus. Expert Rev. Mol. Med. 1999:1-16. [DOI] [PubMed] [Google Scholar]
- 22.Kar-Roy, A., H. Korkaya, R. Oberoi, S. K. Lal, and S. Jameel. 2004. The hepatitis E virus open reading frame 3 protein activates ERK through binding and inhibition of the MAPK phosphatase. J. Biol. Chem. 279:28345-28357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, F., J. Torresi, S. A. Locarnini, H. Zhuang, W. Zhu, X. Guo, and D. A. Anderson. 1997. Amino-terminal epitopes are exposed when full-length open reading frame 2 of hepatitis E virus is expressed in Escherichia coli, but carboxy-terminal epitopes are masked. J. Med. Virol. 52:289-300. [DOI] [PubMed] [Google Scholar]
- 24.Maia, I. G., K. Seron, A. L. Haenni, and F. Bernardi. 1996. Gene expression from viral RNA genomes. Plant Mol. Biol. 32:367-391. [DOI] [PubMed] [Google Scholar]
- 25.Martin, J. D., D. M. King, J. M. Slauch, and R. J. Frisque. 1985. Differences in regulatory sequences of naturally occurring JC virus variants. J. Virol. 53:306-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng, X. J. 2010. Hepatitis E virus: animal reservoirs and zoonotic risk. Vet. Microbiol. 140:256-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng, X. J. 2010. Recent advances in hepatitis E virus. J. Viral Hepat. 17:153-161. [DOI] [PubMed] [Google Scholar]
- 28.Meng, X. J., P. G. Halbur, M. S. Shapiro, S. Govindarajan, J. D. Bruna, I. K. Mushahwar, R. H. Purcell, and S. U. Emerson. 1998. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J. Virol. 72:9714-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng, X. J., R. H. Purcell, P. G. Halbur, J. R. Lehman, D. M. Webb, T. S. Tsareva, J. S. Haynes, B. J. Thacker, and S. U. Emerson. 1997. A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. U. S. A. 94:9860-9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 31.Moin, S. M., V. Chandra, R. Arya, and S. Jameel. 2009. The hepatitis E virus ORF3 protein stabilizes HIF-1alpha and enhances HIF-1-mediated transcriptional activity through p300/CBP. Cell. Microbiol. 11:1409-1421. [DOI] [PubMed] [Google Scholar]
- 32.Moin, S. M., M. Panteva, and S. Jameel. 2007. The hepatitis E virus Orf3 protein protects cells from mitochondrial depolarization and death. J. Biol. Chem. 282:21124-21133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nulf, C. J., and D. Corey. 2004. Intracellular inhibition of hepatitis C virus (HCV) internal ribosomal entry site (IRES)-dependent translation by peptide nucleic acids (PNAs) and locked nucleic acids (LNAs). Nucleic Acids Res. 32:3792-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okamoto, H. 2009. Recent advances in hepatitis E: including the development and application of a robust cell culture system for HEV. Nippon Shokakibyo Gakkai Zasshi 106:177-187. (In Japanese.) [PubMed] [Google Scholar]
- 35.Panda, S. K., I. H. Ansari, H. Durgapal, S. Agrawal, and S. Jameel. 2000. The in vitro-synthesized RNA from a cDNA clone of hepatitis E virus is infectious. J. Virol. 74:2430-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pappas, C. L., W. P. Tzeng, and T. K. Frey. 2006. Evaluation of cis-acting elements in the rubella virus subgenomic RNA that play a role in its translation. Arch. Virol. 151:327-346. [DOI] [PubMed] [Google Scholar]
- 37.Pudupakam, R. S., Y. W. Huang, T. Opriessnig, P. G. Halbur, F. W. Pierson, and X. J. Meng. 2009. Deletions of the hypervariable region (HVR) in open reading frame 1 of hepatitis E virus do not abolish virus infectivity: evidence for attenuation of HVR deletion mutants in vivo. J. Virol. 83:384-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riddell, M. A., F. Li, and D. A. Anderson. 2000. Identification of immunodominant and conformational epitopes in the capsid protein of hepatitis E virus by using monoclonal antibodies. J. Virol. 74:8011-8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sehgal, D., S. Thomas, M. Chakraborty, and S. Jameel. 2006. Expression and processing of the hepatitis E virus ORF1 nonstructural polyprotein. Virol. J. 3:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surjit, M., S. Jameel, and S. K. Lal. 2007. Cytoplasmic localization of the ORF2 protein of hepatitis E virus is dependent on its ability to undergo retrotranslocation from the endoplasmic reticulum. J. Virol. 81:3339-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiher, H., M. Konig, and P. Gruss. 1983. Multiple point mutations affecting the simian virus 40 enhancer. Science 219:626-631. [DOI] [PubMed] [Google Scholar]
- 42.Yamada, K., M. Takahashi, Y. Hoshino, H. Takahashi, K. Ichiyama, S. Nagashima, T. Tanaka, and H. Okamoto. 2009. ORF3 protein of hepatitis E virus is essential for virion release from infected cells. J. Gen. Virol. 90:1880-1891. [DOI] [PubMed] [Google Scholar]
- 43.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]