Summary
Chromatin regulators play fundamental roles in the regulation of gene expression and chromosome maintenance, but the regions of the genome where most of these regulators function has not been established. We explored the genome-wide occupancy of four different chromatin regulators encoded in Saccharomyces cerevisiae. The results reveal that the histone acetyltransferases Gcn5 and Esa1 are both generally recruited to the promoters of active protein-coding genes. In contrast, the histone deacetylases Hst1 and Rpd3 are recruited to specific sets of genes associated with distinct cellular functions. Our results provide new insights into the association of histone acetyltransferases and histone deacetylases with the yeast genome, and together with previous studies, suggest how these chromatin regulators are recruited to specific regions of the genome.
Introduction
Chromatin regulators play important roles in a broad range of biological activities such as transcription, replication, recombination, and repair (reviewed in Felsenfeld and Groudine, 2003). Chromatin regulators fall into two classes. One of these covalently modifies histones by addition or removal of chemical residues on histones (Wu and Grunstein, 2000; Jenuwein and Allis, 2001; Kouzarides, 2002). Members of the other class consist of the ATP-dependent nucleosome-remodeling complexes that noncovalently modify and reposition nucleosomes (Kingston and Narlikar, 1999; Kornberg and Lorch, 1999; Vignali et al., 2000; Urnov and Wolffe, 2001).
Covalent modifications of nucleosomes occur predominantly in the N-terminal tails of histones and include lysine acetylation, methylation, and ubiquitination; arginine methylation; and serine phosphorylation (Berger, 2002). The addition and removal of chemical moieties is a dynamic process that can influence chromatin function by different mechanisms. Histone modifications can generate sites for interaction with additional proteins (Strahl and Allis, 2000; Jenuwein and Allis, 2001). For example, proteins with bromo-, chromo-, and SANT domains interact with modified histones (Bannister et al., 2001; Hassan et al., 2002). Histone modifications may also directly affect the condensation of chromatin (Schwarz et al., 1996; Tse et al., 1998a, 1998b), possibly by modulating exposed charge patches on nucleosome surfaces (Durrin et al., 1991; Dou and Gorovsky, 2000; Ren and Gorovsky, 2001).
Chromatin regulators do not have sequence-specific DNA-recognition properties of their own, but appear to be recruited to specific locations in the genome by interacting with other proteins (reviewed in Cosma et al., 1999; Hampsey and Reinberg, 2003). For example, studies on individual genes suggest that transcriptional activators recruit the histone acetylases Gcn5 and Esa1 and the chromatin-remodeling complex Swi/Snf to the promoters of protein-coding genes (see Peterson and Herskowitz, 1992; Brownell et al., 1996; Smith et al., 1998; Cosma et al., 1999; Reid et al., 2000; Bhaumik and Green, 2001; Larschan and Winston, 2001; Nourani et al., 2001). Transcriptional activators also recruit the general transcription factor TFIID, whose large subunit TAF1 is a histone acetylase (Mizzen et al., 1996). Some activators and repressors can recruit the RSC chromatin-remodeling complex to promoters (Ng et al., 2002). Recruitment of the Set1 histone methyltransferase, which is responsible for histone H3K4 trimethylation at nucleosomes located near the start site of transcription, is dependent on components of the transcription apparatus (Bernstein et al., 2002; Santos-Rosa et al., 2002; Ng et al., 2003; Krogan et al., 2003).
Knowledge of the genome-wide location of a chromatin regulator has the potential to (1) determine whether the regulator is associated with all or a subset of genes transcribed by specific RNA polymerases (Lieb et al., 2001; Wang et al., 2002; Ng et al., 2002, 2003), (2) reveal whether a regulator is associated with the promoter or transcribed region of genes (Wang et al., 2002; Ng et al., 2002, 2003), (3) lay the foundation for studies that reveal the factors responsible for recruiting the regulator to specific regions of the genome (Ng et al., 2002, 2003), and (4) extend to many genes a model for regulator function based on previous studies of one or a few genes (Lieb et al., 2001). Thus far, however, the genome-wide occupancy of only a few chromatin regulators has been determined (Lieb et al., 2001; Wang et al., 2002; Kurdistani et al., 2002; Ng et al., 2002, 2003). We report here genome-wide location results for two HATs and two HDACs in yeast. Our results show that the HATs Gcn5 (SAGA) and Esa1 (NuA4) are generally recruited to the promoters of active protein-coding genes, whereas the HDACs Rpd3/Sin3 and Hst1 are targeted to specific sets of genes associated with distinct cellular functions.
Results
Overview
The histone acetyltransferases (HATs) Gcn5 and Esa1 and the histone deacetylases (HDACs) Hst1 and Rpd3 were epitope tagged at their chromosomal locus (Lee et al., 2002). Genome-wide location analysis experiments were performed in triplicate with yeast cells grown in rich media using DNA arrays representing the yeast genome (Ren et al., 2000; Iyer et al., 2001). In some cases, additional subunits of the protein complex within which the regulators reside were also profiled (see below). Control experiments using an isogenic strain containing no tagged protein were also performed. The results for the four chromatin regulators are summarized in Table 1, together with those for two other regulators we reported previously (Ng et al., 2002, 2003). Complete data sets are available at http://web.wi.mit.edu/young/chromatin_regulators.
Table 1.
Summary of the Chromatin Regulators Analyzed in This Study
| Pol II Genes |
|||||||
|---|---|---|---|---|---|---|---|
| Gene Classa |
|||||||
| Subunit | Complex | Activity | Pol I | Pol II | Pol III | General versus Gene Specificb |
Promoter versus ORFc |
| Gcn5 | SAGA and others | HAT | + | general | promoter | ||
| Esa1 | NuA4 | HAT | + | general | promoter | ||
| Hst1 | Hst1/Sum1 | HDAC | + | specific | promoter | ||
| Rpd3 | HDB | HDAC | + | specific | promoter | ||
| Sth1d | RSC | remodeling | + | + | specific | promoter | |
| Set1e | COMPASS | HMT | + | general | ORF | ||
The predominant class(es) of genes occupied by each chromatin regulator is indicated by a “+.” Some regulators occupy a significant portion of more than one class of gene.
The chromatin regulators that occupy genes in a manner that correlates with transcription rate (see text) were classified as “general,” and those whose occupancy does not were classified as “gene specific.”
The predominant location, promoter versus open reading frame (ORF), is listed for each regulator as determined by genome-wide location and high-resolution ChIP on target genes.
Data published previously (Ng et al., 2002).
Data published previously (Ng et al., 2003).
Gcn5 and Esa1 Are Generally Recruited to Active Protein-Coding Genes
Gcn5 and Esa1 are the catalytic subunits of the SAGA and NuA4 complexes respectively (Grant et al., 1997; Allard et al., 1999). Both these HATs are involved in transcriptional regulation and have been shown to be recruited to specific genes by DNA binding transcription factors (reviewed in Naar et al., 2001). Genome-wide expression studies with mutants have shown that only a subset of protein-coding genes depend on the function of SAGA or NuA4 for optimal expression (Holstege et al., 1998; Lee et al., 2000; Reid et al., 2004). However, it is not clear whether SAGA and NuA4 are associated with and function at this subset of protein-coding genes alone. It is also possible that they are recruited to and function at essentially all actively transcribed protein-coding genes in wild-type cells, but that the loss of function due to mutation does not fully reveal their roles due to the ability of other chromatin regulators to compensate for the loss of a single regulator.
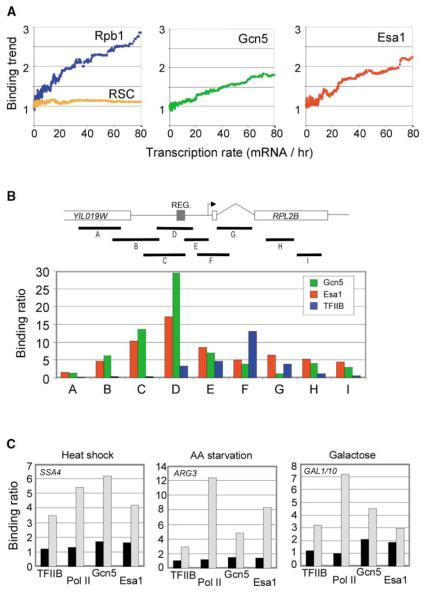
When a regulator is associated with most or all transcriptionally active genes, we expect that its occupancy of these genes will correlate with global transcriptional rates (Lieb et al., 2001; Wang et al., 2002; Kurdistani et al., 2002; Ng et al., 2002, 2003). We found that the genome-wide occupancy of protein-coding genes by both Gcn5 and Esa1 correlates with transcription rates (Figure 1A). When similar DNA material was hybridized on ORF arrays, much smaller enrichment was observed, suggesting Gcn5 and Esa1 are binding to promoter regions rather than transcribed regions (see Supplemental Data at http://www.molecule.org/cgi/content/full/16/2/ 199/DC1/ and Table 1). Independent experiments, performed on a gene-by-gene level, demonstrated that both Gcn5 and Esa1 are located predominantly at the upstream activating sequence (UAS) of the ribosomal protein gene RPL2B and all other active genes we tested (Figure 1B and Supplemental Data). In contrast, the general transcription factor TFIIB is located near the transcriptional start site, as expected. Furthermore, we found that Gcn5 and Esa1 are recruited to otherwise inactive genes upon gene activation (Figure 1C). Collectively, our data indicate that both HATs occupy the promoters of most transcriptionally active protein-coding genes and that recruitment is most likely through the DNA binding transcription factors that bind to UAS elements.
Figure 1.
Recruitment of Histone Acetylases Gcn5 and Esa1 to Promoters of Actively Transcribed Genes
(A) Correlation between chromatin regulator occupancy and transcription rates. The binding trend was calculated by computing the moving median of the binding ratio over a sliding window of 100 genes across all genes ordered by transcription rate as described previously for other regulators such as Set1 (Ng et al., 2002, 2003). The transcription rates for yeast genes and the binding data for RSC were determined previously (Holstege et al., 1998; Ng et al., 2002). Rpb1 occupancy correlates with transcription rate across the genome, whereas RSC occupancy does not, and these proteins served as controls.
(B) Fine mapping of Gcn5 (green), Esa1 (red), and TFIIB (blue) occupancy within the RPL2B locus. The binding ratios from segment-specific chromatin IP experiments are shown for Gcn5, Esa1, and TFIIB for each of the DNA segments A–I. The binding ratios (representing the enrichment generated by the ChIP) were all normalized to the promoter of ARN1, which was set to 1.
(C) The binding ratios for the general transcription factor TFIIB, RNA pol II (8WG16), Gcn5, and Esa1 are shown in uninduced (black) and induced (gray) states for heat shock (25°C versus 15 min at 37°C) at the SSA4 promoter, amino acid starvation (YNB versus 10 min in minimal synthetic media + sulfometuron methyl) at the ARG3 promoter, and galactose (raffinose versus galactose) at the GAL1/10 promoters. Binding ratios were calculated as in (B). The exact growth and inducing conditions can be found in the Supplemental Data. These experiments were repeated multiple times and the variation was never more than 15%.
Hst1 Directly Regulates Midsporulation and Kynureine Pathway Genes
Unlike Gcn5 and Esa1, the genome-wide occupancy of the HDAC Hst1 does not correlate with transcription rate, suggesting that it is not generally recruited to active genes. Hst1 target genes (p < 0.005) were found to be highly associated with MIPS categories containing meiosis and sporulation genes (see Supplemental Data). Conventional ChIP confirmed that Hst1 occupies the promoters of all midsporulation genes that were tested (Figure 2A and Supplemental Data). Hst1 was previously shown to be required for the Sum1-mediated repression of a few midsporulation genes during vegetative growth, but was not shown to be physically associated with these genes (Xie et al., 1999; McCord et al., 2003). Our data demonstrate that Hst1 is directly associated with midsporulation genes, in a Sum1-dependent manner (Figure 2A), during vegetative growth.
Figure 2.
Recruitment of Hst1 to Promoters of Sporulation and Kynureine Genes by the Sum1 Repressor
(A) The binding ratio for Hst1 (blue), Sum1 (red), and Hst1 in sum1Δ cells (green) across the SPS4 and BNA5 loci, as determined by ChIP, is shown. A diagram of the regions amplified by sequence-specific PCR is shown on top.
(B) The log binding ratio (as determined by genome-wide location analysis) of Hst1, Sum1, and Rpd3 (used as a control) is displayed for the promoters of genes bound by Hst1 (p < 0.005). The log binding ratio of Hst1 in sum1 deletion cells (sum1Δ) is also shown.
(C) The level of histone H3 (K9/K14) acetylation and histone H4 (K5, K8, K12, K16) acetylation, as determined by ChIP, is shown in wild-type cells (black) and in cells deleted for HST1 (gray) at the SPS4 and BNA5 promoters. The binding ratios in (A) and (C) were all calculated as in Figure 1B. The binding ratios in hst1Δ in (C) were normalized to wt. These experiments were repeated multiple times and the variation was never more than 15%.
Hst1 has also been shown to be required for repression of the kynureine pathway, which is used to produce de novo NAD+ from tryptophan (Bedalov et al., 2003). Previous attempts to detect a physical association of Hst1 with these genes were unsuccessful, however, so Hst1 might repress these genes indirectly (Bedalov et al., 2003). Our genome-wide data revealed that Hst1 occupies the promoters of two genes from the kynureine pathway, BNA1 and BNA5. Conventional ChIP was used to confirm that Hst1 occupies the regulatory regions of BNA1 and BNA5 (Figure 2A and Supplemental Data). These results support the elegant model proposed by Bedalov et al. (2003) that Hst1 is directly sensing and regulating NAD+ levels through regulation of the kynurenine pathway.
The HDAC Hst1 Is Recruited by a Single Transcription Factor
In order to identify potential DNA binding transcription factors that might recruit Hst1 to its target genes, we compared the genes bound by Hst1 with those bound by more than 100 different DNA binding transcription factors (Lee et al., 2002). A single transcription factor, the repressor of midsporulation genes Sum1, stood out as highly overlapping with Hst1 (see Supplemental Data). We expected to find Sum1 in this search because it was previously shown to be required for the Hst1-mediated repression of midsporulation genes (Xie et al., 1999) and to bind to genes from the kynureine pathway (Bedalov et al., 2003). It was striking, however, that the set of genes occupied by Sum1 and Hst1 were nearly identical (Figure 2B), suggesting they might be functioning exclusively as a pair. In order to test this possibility, we profiled the genome-wide location of Hst1 in a sum1Δ background. Virtually all the genes bound by Hst1 in wild-type cells showed a dramatic decrease in Hst1 binding in the absence of Sum1 (Figure 2B). Together with knowledge that Hst1 and Sum1 physically interact (Rusche and Rine, 2001; Bedalov et al., 2003; McCord et al., 2003), these data suggest that Hst1 is exclusively recruited by Sum1 during vegetative growth. To our knowledge, this is the only known case of a chromatin regulator so specifically dedicated to a single transcription factor.
Hst1 Is a Bona Fide HDAC In Vivo
Hst1 was shown to have NAD+-dependent HDAC activity in vitro (Rusche and Rine, 2001), but it has not been shown to affect histone acetylation in vivo. Previous studies failed to detect changes in acetylation levels for sporulation genes (Rusche and Rine, 2001) or kynureine pathway genes (Bedalov et al., 2003) in Hst1 mutants. It is possible that Hst1 (1) does deacetylate histones at these genes, but the activity is limited to a domain too small to be detected in these experiments; (2) was not detected with the antibodies used; or (3) is difficult to detect in mutants due to redundant activities. To reevaluate the role of Hst1 as a HDAC in vivo, we performed ChIP using antibodies directed against acetylated histone H3 and H4 in wild-type and in hst1Δ cells. Figure 2C shows that acetylation of histone H3 and H4 is modestly, but reproducibly, increased at Hst1 target genes in the hst1Δ stain. Similar results were obtained using a sum1Δ strain (Supplemental Data). The modest effect of HST1 and SUM1 deletion mutants on histone acetylation in vivo might be due to the presence of partially redundant activities. Nevertheless, our data is consistent with Hst1 being a bona fide HDAC in vivo. We conclude that Hst1 recruitment by Sum1 can lead to local deacetylation of histones.
Rpd3 Is Associated with Cell Cycle Genes
The HDAC Rpd3 is part of a large protein complex composed of many different proteins, including Sin3, Sap30, and Sds3 (Kadosh and Struhl, 1997; Kasten et al., 1997; Zhang et al., 1998; Lechner et al., 2000). The Rpd3 complex negatively regulates early meiosis genes during vegetative growth (Kadosh and Struhl, 1997), and deletion of RPD3 leads to defects in sporulation and meiosis (Dora et al., 1999). The Rpd3p complex is recruited to the promoter sequences of some sporulation genes by the transcription factor Ume6 (Kadosh and Struhl, 1997), but there is evidence that Rpd3 regulates other genes independently of Ume6 (Fazzio et al., 2001; Kurdistani et al., 2002; Robyr et al., 2002).
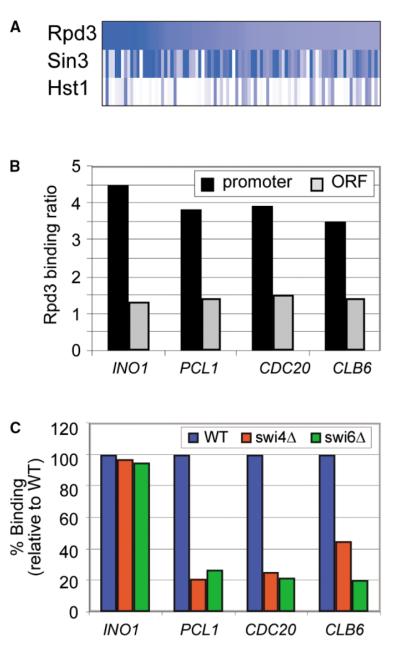
To identify genes occupied by the Rpd3 complex, we performed genome-wide location analysis with both Rpd3 and Sin3. In contrast to a previous report (Kurdistani et al., 2002), our results reveal that the genome-wide occupancy of Rpd3 and Sin3 do not correlate with transcription rate. Rather, Rpd3 and Sin3 occupy approximately 100 genes (p < 0.005). The data show that both Rpd3 and Sin3 are associated with essentially the same genes (Figure 3A), which is consistent with the observation that these proteins can be purified as a complex (Kadosh and Struhl, 1997; Kasten et al., 1997), and indicating that they function together in vivo.
Figure 3.
Occupancy and Recruitment of Rpd3 to Promoters of Cell Cycle Genes
(A) Sin3 is associated with virtually all the genes bound by Rpd3. The log binding ratio (as determined by genome-wide location analysis) of Rpd3, Sin3, and Hst1 (used as a control) is displayed for the promoters of genes bound by Rpd3 (p < 0.005).
(B) Rpd3 is associated with the promoters but not the open reading frames (ORFs) of target genes. The Rpd3 binding ratio, as determined by gene-specific ChIP, is shown for the promoter (black) and ORF (gray) of selected Rpd3 target genes.
(C) The association of Rpd3 with promoters of PCL1, CDC20, and CLB6 is dependent on Swi4 and Swi6. The binding ratio of Rpd3 is shown for various Rpd3 target promoters in wild-type cells (blue) and in cells deleted for SWI4 (red) or SWI6 (green). The binding ratios in (B) and (C) were all calculated as in Figure 1B. These experiments were repeated multiple times and the variation was never more than 15%.
Rpd3 target genes (p < 0.005) were found to be highly associated with MIPS categories associated with cell cycle regulation (see Supplemental Data). We used conventional ChIP to confirm that Rpd3 can occupy the promoters of several key cell cycle regulators (Figure 3B). These results are consistent with previous reports that have suggested that Rpd3 might play a role in cell cycle regulation (Fazzio et al., 2001).
In order to identify potential DNA binding transcription factors that might recruit Rpd3, we compared the genes bound by Rpd3 with genome-wide location data for more than 100 transcription factors (Lee et al., 2002). Cell cycle transcription factors (Mbp1, Swi4, Swi6, Fkh1, Fkh2, Mcm1, and Ndd1) were among the most frequent class of factors associated with Rpd3 target genes (see Supplemental Data). To determine whether some of these DNA binding factors are involved in recruitment of Rpd3 to cell cycle genes, we tested whether the heterodimeric transcription factor Swi4/Swi6 is essential for Rpd3 occupancy of three cell cycle regulator genes occupied by both Rpd3 and Swi4/Swi6 (PCL1, CDC20, and CLB6). If Rpd3 is recruited to these genes by Swi4 and Swi6, then Rpd3 should not occupy these genes in SWI4 and SWI6 deletion mutants. As shown in Figure 3C, Rpd3 occupies the promoter of these genes in wild-type cells, but this occupancy is reduced significantly in swi4Δ or swi6Δ cells. This effect is specific to Rpd3 occupancy of the cell cycle regulators’ promoters since Rpd3 occupancy of the INO1 promoter was not affected by the deletion of SWI4 or SWI6 (Figure 3C). Furthermore, when genome-wide location analysis of Rpd3 was carried out with cells lacking Swi4, we found that Rpd3 continued to occupy genes that are not regulated by Swi4 (see Supplemental Data), showing that the absence of Swi4 does not affect Rpd3 occupancy at genes controlled by other regulators. Thus, in contrast to Hst1, which is recruited to the genome by a single DNA binding protein, Rpd3 is recruited to specific sites in the genome via interactions with multiple transcription factors, including Ume6 (Kadosh and Struhl, 1997; Rundlett et al., 1998) and the heterodimeric transcription factor Swi4/Swi6 (this study).
Rpd3 Is Recruited to Ribosomal Protein Genes upon Cold Shock
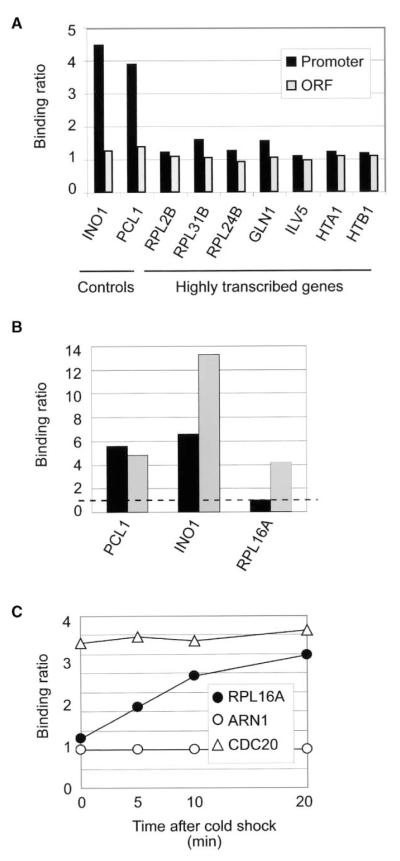
Our interpretation of the genome-wide distribution of Rpd3 differs from a similar study that concluded that Rpd3 is generally associated with genes in a manner that positively correlates with their transcription rate (Kurdistani et al., 2002). We did not find that Rpd3-dependent ChIP signals are enriched significantly relative to controls at highly transcribed genes such as ribosomal protein (RP) genes using our ChIP protocol, whether assayed by genome-wide location analysis (data not shown) or by gene-specific ChIP (Figure 4A). To determine the cause of this discrepancy, we repeated the Rpd3 ChIP experiments using both our protocol and that of Kurdistani et al. (2002). As shown in Figure 4B, both protocols produced significant enrichment with the PCL1 and INO1 promoters, but only the Kurdistani et al. (2002) method led to enrichment of the promoter of the ribosomal promoter gene RPL16A. The two significant differences between the two protocols are the addition of a second crosslinking agent (DMA) and a wash step in ice-cold buffer prior to the addition of the crosslinking agents. The addition of DMA to our protocol did not increase enrichment of ribosomal protein promoters. However, washing in ice-cold buffer prior to crosslinking does have this effect. It appears that this “cold shock” triggers the rapid association of Rpd3 with the RP genes (Figure 4C). This effect is specific since the binding of Rpd3 to the promoters of CDC20 or ARN1 (negative control) is not affected by cold shock (Figure 4C). We therefore propose that Rpd3 can be recruited to RP genes during stress responses when ribosomal protein gene expression is downregulated (Causton et al., 2001; Gasch et al., 2000), and when Rpd3 might thus play a role in transcriptional repression.
Figure 4.
Rpd3 Is Recruited to Ribosomal Protein Genes upon Cold Shock
(A) Rpd3 is not generally associated with highly transcribed protein-coding genes in rich media. Gene-specific ChIPs were performed with an Rpd3-tagged strain at the promoter (black) and ORF (gray) of the indicated highly transcribed genes. INO1 and PCL1 were included as positive controls.
(B) Different protocols give different enrichment at ribosomal protein genes. Rpd3 ChIP experiments were performed using our standard protocol (black) and the Kurdistani et al. (2002) protocol (gray). The binding ratio of Rpd3 is shown at the INO1, PCL1, and RPL16A promoters. These experiments were repeated multiple times and the variation was never more than 15%.
(C) Cold shock treatment induces the binding of Rpd3 to ribosomal protein genes. Myc-Rpd3 cells were grown until mid-log phase and the culture was transferred to an ice-cold water bath. 50 ml samples were collected and subject to ChIP prior to, as well as 5, 10, and 20 min after transferring to cold water bath. The binding of Rpd3 is shown at the CDC20, ARN1 (a negative control), and RPL16A promoters. The binding ratios were all calculated as in Figure 1B. These experiments were repeated multiple times and the variation was never more than 15%.
Discussion
We have investigated how two histone acetyltransferases, Gcn5 and Esa1, and two histone deacetylases, Rpd3 and Hst1, occupy the yeast genome. Our results show that the HATs Gcn5 (SAGA) and Esa1 (NuA4) are generally recruited to the promoters of active protein-coding genes, whereas the HDACs Rpd3/Sin3 and Hst1 are targeted to specific sets of genes associated with distinct cellular functions. Although previous studies established that these chromatin regulators make important contributions to gene regulation at some well-studied genes, these new results demonstrate that Gcn5- and Esa1-containing complexes are generally recruited to protein-coding genes and help explain the long-standing observation that histone acetylation correlates with gene expression (Allfrey et al., 1964). These results also revise and improve our understanding of the genome-wide targets of the HDACs Rpd3 and Hst1.
Recruitment of Gcn5 and Esa1 to Transcriptionally Active Protein-Coding Genes
Previous studies showed that Gcn5 occupies the promoters of specific genes in yeast and that occupancy is influenced by specific DNA binding transcription factors. For example, Gcn5 is recruited to the HO promoter and this recruitment is dependent on the heterodimeric transcription factor SBF (Cosma et al., 1999). Other transcription factors have been shown to recruit Gcn5 to different genes (see Natarajan et al., 1998; Massari et al., 1999; Vignali et al., 2000; Kuo et al., 2000; Bhaumik and Green, 2001; Larschan and Winston, 2001; Proft and Struhl, 2002). These studies have not, however, demonstrated that Gcn5 is generally recruited to protein-coding genes. We found that Gcn5 occupancy of protein-coding genes correlates with the transcription rates of these genes. In addition, we found that Gcn5 was recruited to the promoter of every inducible gene tested (n = 9). These results indicate that Gcn5 is recruited to most transcriptionally active protein-coding genes. The occupancy of these chromatin regulators was highest at the UAS and was optimal during gene activation, suggesting that recruitment of the Gcn5-containing complex most likely occurs through interactions with the transcription factors that bind to UAS elements.
Esa1 has also previously been shown to be recruited to specific genes through specific DNA binding transcription factors (see Reid et al., 2000; Vignali et al., 2000; Nourani et al., 2001; Brown et al., 2001; Baek et al., 2002; Boudreault et al., 2003; Frank et al., 2003; Nourani et al., 2004; Taubert et al., 2004). A previous genome-wide location study led to the conclusion that Esa1 is associated with ribosomal protein gene promoters, where it is recruited by the DNA binding transcription factor Rap1, but the signal was inadequate to ascertain whether Esa1 is associated with a substantial number of other genes (Reid et al., 2000). For these reasons, it might be assumed that Esa1 is targeted to specific subsets of genes. Our data, however, show that Esa1 occupancy of protein-coding genes correlates with transcription rates of these genes and that Esa1 can be recruited to the promoter of all the inducible genes tested (n = 9). In agreement with previous studies, we found that Esa1 occupancy is maximal at the UAS, suggesting it is recruited through DNA binding transcription factors. We conclude that Esa1, like Gcn5, is recruited to most protein-coding gene promoters by DNA binding transcription factors that recognize UAS elements.
The observation that Gcn5- and Esa1-containing complexes are both recruited to the promoters of most protein-coding genes makes it interesting to consider how transcription factors manage to recruit both complexes to promoters. It would require, in fact, that the number of transcription factors able to interact with those two complexes be large and that complexes have the property of being able to interact with a large number of different transcription factors. One possibility is that DNA binding regulators interact with the Tra1 subunit shared by SAGA and NuA4 (Brown et al., 2001). Tra1 is a large (~400 kDa) protein that has been shown to interact with many different transcription factors (Brown et al., 2001; Bhaumik et al., 2004). It is also possible that the activating domains of transcription activators are capable of interacting with multiple components of these chromatin-regulating complexes.
Although the genome-wide location data and conventional chromatin IP data indicate that Gcn5 and Esa1 are recruited to most transcriptionally active protein-coding genes, the expression of some genes is more sensitive to loss-of-function mutations in these chromatin regulators than others (Holstege et al., 1998; Lee et al., 2000; Huisinga and Pugh, 2004). There are at least three explanations for these observations. First, the Gcn5, Esa1, Sas2, Sas3, TFIID, and Elp3 HATs may be somewhat functionally redundant at certain genes (Wittschieben et al., 2000), thus leading to different levels of dependence on one or more HATs. Second, it is not possible to ascertain what effects of mutations are direct or indirect with expression profiling. Finally, expression profiling is generally performed with strains lacking only one subunit of a given HAT complex, which could lead to only a partial loss of function of that complex. Some HAT complexes have been shown to have multiple functions, some of which are independent of their HAT activity (Horiuchi et al., 1997; Dudley et al., 1999a, 1999b; Sterner et al., 1999). Expression profiling experiments performed using an spt20 allele known to disrupt the integrity of the SAGA complex lead to a much wider effect on gene expression that experiments performed using gcn5 alleles (Lee et al., 2000). This result is consistent with SAGA playing a wide role in gene expression regulation, with Gcn5’s function being only part of its function.
General Model for Recruitment of Chromatin Regulators at Protein-Coding Genes
Studies in yeast with a few genes have indicated that multiple chromatin regulators are recruited in a temporal order (for reviews, see Cosma, 2002; Hampsey and Reinberg, 2003). The results described here suggest that it is the case that DNA binding transcription factors generally recruit the Gcn5 and Esa1 complexes to nucleosomes located near UAS elements in protein-coding genes upon activation. This accounts, at least in part, for the association between gene activity and histone acetylation (Allfrey et al., 1964). Once the transcription initiation apparatus is recruited and initiates transcript elongation at protein-coding genes, it is also generally the case that the Set1 histone methyl transferase is recruited to the beginning of the ORFs through interactions with the elongating RNA polymerase II (reviewed by Hampsey and Reinberg, 2003). This leads to trimethylation of lysine 4 of histone H3 in nucleosomes located near the start site. It will be interesting to determine whether additional regulatory events noted for chromatin regulators at specific genes also occur generally at protein-coding genes (see Hampsey and Reinberg, 2003).
Recruitment of HDACs to Specific Sets of Genes
Our results indicate that the HDACs Hst1 and Rpd3 are not generally associated with actively transcribed protein-coding genes; rather, they occupy specific sets of genes associated with distinct cellular functions. While the genome-wide binding of Hst1 has not been studied previously, our data on Rpd3 is in striking contrast with a previous study of the genome-wide location of this HDAC (Kurdistani et al., 2002).
Hst1 was previously shown to be involved in the regulation of several sporulation genes (Xie et al., 1999; McCord et al., 2003) as well as a number of genes from the kynureine pathway (Bedalov et al., 2003). The interaction of Hst1 with these genes, however, was never directly observed and its HDAC activity was not demonstrated to occur in vivo. Our results show that Hst1 is physically recruited to sporulation and kynureine pathway genes by the Sum1 DNA binding transcriptional repressor, leading to a decrease in histone acetylation at those genes. We conclude that Hst1 directly regulates the expression of genes involved in sporulation and the kynureine pathway in vivo and does so through histone deacetylation. We also found that Sum1 is required for virtually all the binding activity of Hst1, which indicates that Hst1 is recruited to all its genomic sites by Sum1 under the growth conditions studied here. To our knowledge, this is the only case of a chromatin regulator whose association with the genome is dependent on a single DNA binding transcription factor.
Initial studies on Rpd3 focused on its role in the regulation of sporulation-specific genes through the DNA binding transcription factor Ume6 (see Kadosh and Struhl, 1997, 1998a, 1998b; Rundlett et al., 1998; Burgess et al., 1999; Fazzio et al., 2001). Recent studies suggest that Rpd3 regulates additional cellular functions (see De Rubertis et al., 1996; Dora et al., 1999; Arevalo-Rodriguez et al., 2000; Vannier et al., 2001; Kurdistani et al., 2002; Sandmeier et al., 2002), and the genome-wide targets we find occupied by Rpd3 are consistent with these studies. One interesting set of genes occupied by Rpd3 encodes important cell cycle regulators including cyclins and cyclin-dependent kinases. This may explain, at least in part, the importance of Rpd3 in meiosis (Vannier et al., 2001, and references therein). In addition, our results are consistent with previous reports that have suggested that Rpd3 might play a role in cell cycle regulation. The expression of cyclin genes is upregulated by rpd3 mutations (Fazzio et al., 2001). Loss of RPD3 function causes hyperacetylation of the chromatin surrounding those same genes (Robyr et al., 2002). An rpd3 deletion is synthetically lethal with deletions of either swi4 or swi6 (Vannier et al., 2001), which encode cell cycle transcription factors that regulate G1/S gene expression. Overexpression of Rpd3p can lead to cell cycle arrest in some genetic backgrounds (Arevalo-Rodriguez et al., 2000). Taken together, the global binding data and the genetic evidence argue that Rpd3p plays an important role in regulation of the cell cycle gene expression program.
Our data indicate that the genome-wide occupancy of Rpd3 does not correlate with transcription rates and that Rpd3 is recruited to specific genes by specific DNA binding regulators. This result is inconsistent with the view that Rpd3 is globally recruited to active protein-coding genes (Kurdistani et al., 2002). Much of the discrepancy between the genome-wide data we report and that of Kurdistani et al. (2002) involves the ribosomal protein (RP) genes, which are reported as occupied by Rpd3 in the latter study. We found that Rpd3 is rapidly recruited to RP genes upon cold shock, and we show that the use of ice-cold buffer to wash cells before the addition of crosslinking agents (Kurdistani et al., 2002) can explain the discrepancy in genome-wide location data. We therefore propose that Rpd3 is recruited to the promoters of RP genes, but only in stress conditions like cold shock. In these conditions, it is likely that Rpd3 acts as a repressor of RP genes, since the expression of these genes is generally downregulated upon exposure of yeast cells to stress conditions (Causton et al., 2001; Gasch et al., 2000).
The HATs and HDACs studied here are highly conserved in eukaryotes, so several of our observations have implications for studies in higher eukaryotes. It will be interesting to determine whether Gcn5 and Esa1 homologs are generally recruited to the promoters of protein-coding genes in higher eukaryotes. It will also be important to determine the identity of specific HDACs in higher eukaryotes that may be devoted to particular cellular functions. It might then be possible to modify specific cellular functions through chemical compounds that target the enzymatic activities of specific chromatin regulators (Melnick and Licht, 2002).
Experimental Procedures
Epitope Tagging of Strains
Chromatin regulators were tagged at the C terminus by inserting multiple copies of the Myc epitope coding sequence into the normal chomosomal loci of these genes. The plasmids pWZV88 and p3747 were used as a template to generate PCR products containing the Myc epitope coding sequence and a selectable marker (TRP1 or URA3, respectively) flanked by homologous regions designed to recombine at the 3′ end of the targeted transcriptional regulator. In most cases the PCR products were transformed into the W303 strain Z1256 or one of its derivatives. Clones were selected for growth on TRP- or URA-selective plates. Insertion of the epitope coding sequence was confirmed by PCR and expression of the epitope-tagged protein was confirmed by Western blotting using an anti-Myc antibody. The complete list of the tagged strains used in this study can be found in the Supplemental Data.
Gene Deletions
The plasmid pRS306 was used to amplify PCR products containing an URA marker flanked by homologous regions designed to recombine on either side of the gene to be deleted. The PCR products were transformed into the appropriate yeast strains and the stable integration of the marker was selected on URA-selective plates. The appropriate gene replacement was confirmed by PCR. The complete list of the deletion strains used in this study can be found in the Supplemental Data.
Chromatin Immunoprecipitation and Genome-Wide Location Analysis
Chromatin immunoprecipitation and genome-wide location analysis were performed as described previously (Ren et al., 2000) except that the crosslinking time was reduced to 30 min at room temperature. A detailed protocol can be found at http://web.wi.mit.edu/young/chromatin_regulators. Each tagged strain was grown in three independent cultures in rich medium. Genome-wide location data were subjected to quality control filters, normalized, and the ratio of immunoprecipitated to control DNA was determined for each spot. A confidence value (p value) was calculated for each spot from each array using an error model, and the data for each of the three samples comprising an experiment were combined using a weighted average method (Ren et al., 2000).
Among the control experiments, we included a control immunoprecipitation (with myc antibody) of lysates from a strain lacking the epitope tag. This data is available at http://web.wi.mit.edu/young/chromatin_regulators.
Induction of specific classes of genes including galactose, heat shock, and animo acid starvation genes was performed as follows. For galactose induction experiments, galactose (2% final concentration) was added for 45 min to cell cultures grown at mid-log phase in YEP medium containing 2% raffinose at 30°C. For heat shock induction experiments, cell cultures grown at mid-log phase in YPD medium at 25°C were transferred to prewarmed flasks in 37°C thermostat and equal volume of YPD medium, prewarmed to 50°C was added. Cultures were then incubated for another 15 min at 37°C. For animo acid starvation experiments, cells were grown in YNB supplemented with all amino acids and 2% glucose at 30°C until reaching mid-log phase (OD600 0.6–0.8). Then, cells were spun down, washed twice with YNB meduim supplemented with required amino acids and 2% glucose, and incubated at 30°C for additional 10 min in this same meduim with 0.2 μg/ml sulfometuron methyl.
Antibodies
The antibodies used in this study are anti-Myc (9E11), anti-Pol II CTD (8WG16), anti-histone H3 (Abcam), anti-acetylated histone H3 (K9,K14) (Upstate Biotech), and anti-acetylated H4 (K5, K8, K12, K16) (Upstate Biotech).
Analysis of Genome-Wide Location Data
The intensity of the signal obtained for spots containing no DNA was measured, averaged, and subtracted from the intensity of all DNA containing spots prior to calculation. Next, we calculated the log of the ratio of intensity in the IP-enriched channel to intensity in the genomic DNA channel for each intergenic region across a large set of hybridization experiments (the set contains many hundreds unrelated hybridization experiments including the ones analyzed here). To account for systematic biases introduced by the immunoprecipitation, all of the log ratios for a specific intergenic region were then normalized by subtracting the average log ratio for that intergenic region. Adjusted intensity values for the IP-enriched channel were calculated from these normalized ratios. The data were then analyzed using an error model and a weighted averaged method as described by Ren et al. (2000). A detail description of this error model can be found at http://web.wi.mit.edu/young/regulatory_network/(follow links to Analysis Methods).
Supplementary Material
Acknowledgments
We thank members of our laboratories, as well as Kevin Struhl, Huck Hui Ng, and Sudhanshu Dole for helpful discussion. This work was supported by grants from the NIH.
Footnotes
Accession Numbers The data sets desribed in this paper have been deposited in the ArrayExpress database under the accession number E-WMIT-2.
References
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo-Rodriguez M, Cardenas ME, Wu X, Hanes SD, Heitman J. Cyclophilin A and Ess1 interact with and regulate silencing by the Sin3-Rpd3 histone deacetylase. EMBO J. 2000;19:3739–3749. doi: 10.1093/emboj/19.14.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bedalov A, Hirao M, Posakony J, Nelson M, Simon JA. NAD+-dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:7044–7054. doi: 10.1128/MCB.23.19.7044-7054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik SR, Green MR. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Cote J. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 2003;17:1415–1428. doi: 10.1101/gad.1056603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Ajimura M, Kleckner N. GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth, in budding yeast. Proc. Natl. Acad. Sci. USA. 1999;96:6835–6840. doi: 10.1073/pnas.96.12.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma MP. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell. 2002;10:227–236. doi: 10.1016/s1097-2765(02)00604-4. [DOI] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- De Rubertis F, Kadosh D, Henchoz S, Pauli D, Reuter G, Struhl K, Spierer P. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature. 1996;384:589–591. doi: 10.1038/384589a0. [DOI] [PubMed] [Google Scholar]
- Dora EG, Rudin N, Martell JR, Esposito MS, Ramirez RM. RPD3 (REC3) mutations affect mitotic recombination in Saccharomyces cerevisiae. Curr. Genet. 1999;35:68–76. doi: 10.1007/s002940050434. [DOI] [PubMed] [Google Scholar]
- Dou Y, Gorovsky MA. Phosphorylation of linker histone H1 regulates gene expression in vivo by creating a charge patch. Mol. Cell. 2000;6:225–231. doi: 10.1016/s1097-2765(00)00024-1. [DOI] [PubMed] [Google Scholar]
- Dudley AM, Gansheroff LJ, Winston F. Specific components of the SAGA complex are required for Gcn4- and Gcr1-mediated activation of the his4-912delta promoter in Saccharomyces cerevisiae. Genetics. 1999a;151:1365–1378. doi: 10.1093/genetics/151.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AM, Rougeulle C, Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 1999b;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrin LK, Mann RK, Kayne PS, Grunstein M. Yeast histone H4 N-terminal sequence is required for promoter activation in vivo. Cell. 1991;65:1023–1031. doi: 10.1016/0092-8674(91)90554-c. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Kooperberg C, Goldmark JP, Neal C, Basom R, Delrow J, Tsukiyama T. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 2001;21:6450–6460. doi: 10.1128/MCB.21.19.6450-6460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D. Tails of intrigue. Phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Horiuchi J, Silverman N, Pina B, Marcus GA, Guarente L. ADA1, a novel component of the ADA/GCN5 complex, has broader effects than GCN5, ADA2, or ADA3. Mol. Cell. Biol. 1997;17:3220–3228. doi: 10.1128/mcb.17.6.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998a;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 1998b;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasten MM, Dorland S, Stillman DJ. A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Mol. Cell. Biol. 1997;17:4852–4858. doi: 10.1128/mcb.17.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Chromatin-modifying and -remodeling complexes. Curr. Opin. Genet. Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Kuo MH, vom Baur E, Struhl K, Allis CD. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell. 2000;6:1309–1320. doi: 10.1016/s1097-2765(00)00129-5. [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Robyr D, Tavazoie S, Grunstein M. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 2002;31:248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner T, Carrozza MJ, Yu Y, Grant PA, Eberharter A, Vannier D, Brosch G, Stillman DJ, Shore D, Workman JL. Sds3 (suppressor of defective silencing 3) is an integral component of the yeast Sin3[middle dot]Rpd3 histone deacetylase complex and is required for histone deacetylase activity. J. Biol. Chem. 2000;275:40961–40966. doi: 10.1074/jbc.M005730200. [DOI] [PubMed] [Google Scholar]
- Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Massari ME, Grant PA, Pray-Grant MG, Berger SL, Workman JL, Murre C. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol. Cell. 1999;4:63–73. doi: 10.1016/s1097-2765(00)80188-4. [DOI] [PubMed] [Google Scholar]
- McCord R, Pierce M, Xie J, Wonkatal S, Mickel C, Vershon AK. Rfm1, a novel tethering factor required to recruit the Hst1 histone deacetylase for repression of middle sporulation genes. Mol. Cell. Biol. 2003;23:2009–2016. doi: 10.1128/MCB.23.6.2009-2016.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick A, Licht JD. Histone deacetylases as therapeutic targets in hematologic malignancies. Curr. Opin. Hematol. 2002;9:322–332. doi: 10.1097/00062752-200207000-00010. [DOI] [PubMed] [Google Scholar]
- Mizzen CA, Yang XJ, Kokubo T, Brownell JE, Bannister AJ, Owen-Hughes T, Workman J, Wang L, Berger SL, Kouzarides T, et al. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- Naar AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annu. Rev. Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- Natarajan K, Jackson BM, Rhee E, Hinnebusch AG. yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol. Cell. 1998;2:683–692. doi: 10.1016/s1097-2765(00)80166-5. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Nourani A, Doyon Y, Utley RT, Allard S, Lane WS, Cote J. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol. 2001;21:7629–7640. doi: 10.1128/MCB.21.22.7629-7640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourani A, Utley RT, Allard S, Cote J. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J. 2004;23:2597–2607. doi: 10.1038/sj.emboj.7600230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- Proft M, Struhl K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell. 2002;9:1307–1317. doi: 10.1016/s1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- Reid JL, Iyer VR, Brown PO, Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell. 2000;6:1297–1307. doi: 10.1016/s1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- Reid JL, Moqtaderi Z, Struhl K. Eaf3 regulates the global pattern of histone acetylation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:757–764. doi: 10.1128/MCB.24.2.757-764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q, Gorovsky MA. Histone H2A.Z acetylation modulates an essential charge patch. Mol. Cell. 2001;7:1329–1335. doi: 10.1016/s1097-2765(01)00269-6. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell. 2002;109:437–446. doi: 10.1016/s0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Suka N, Turner BM, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- Rusche LN, Rine J. Conversion of a gene-specific repressor to a regional silencer. Genes Dev. 2001;15:955–967. doi: 10.1101/gad.873601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier JJ, French S, Osheim Y, Cheung WL, Gallo CM, Beyer AL, Smith JS. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 2002;21:4959–4968. doi: 10.1093/emboj/cdf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schwarz PM, Felthauser A, Fletcher TM, Hansen JC. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry. 1996;35:4009–4015. doi: 10.1021/bi9525684. [DOI] [PubMed] [Google Scholar]
- Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook RG, Lucchesi JC, Allis CD. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. USA. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, Livingston DM, Amati B. E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol. Cell. Biol. 2004;24:4546–4556. doi: 10.1128/MCB.24.10.4546-4556.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse C, Fletcher TM, Hansen JC. Enhanced transcription factor access to arrays of histone H3/H4 tetramer.DNA complexes in vitro: implications for replication and transcription. Proc. Natl. Acad. Sci. USA. 1998a;95:12169–12173. doi: 10.1073/pnas.95.21.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 1998b;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Wolffe AP. Above and within the genome: epigenetics past and present. J. Mammary Gland Biol. Neoplasia. 2001;6:153–167. doi: 10.1023/a:1011304606604. [DOI] [PubMed] [Google Scholar]
- Vannier D, Damay P, Shore D. A role for Sds3p, a component of the Rpd3p/Sin3p deacetylase complex, in maintaining cellular integrity in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2001;265:560–568. doi: 10.1007/s004380100447. [DOI] [PubMed] [Google Scholar]
- Vignali M, Steger DJ, Neely KE, Workman JL. Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J. 2000;19:2629–2640. doi: 10.1093/emboj/19.11.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Kurdistani SK, Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science. 2002;298:1412–1414. doi: 10.1126/science.1077790. [DOI] [PubMed] [Google Scholar]
- Wittschieben BO, Fellows J, Du W, Stillman DJ, Svejstrup JQ. Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo. EMBO J. 2000;19:3060–3068. doi: 10.1093/emboj/19.12.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Grunstein M. 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- Xie J, Pierce M, Gailus-Durner V, Wagner M, Winter E, Vershon AK. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 1999;18:6448–6454. doi: 10.1093/emboj/18.22.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sun ZW, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.