Abstract
Recurrence and reinfection of tuberculosis have quite different implications for prevention. We identified 267 spoligotypes of Mycobacterium tuberculosis from consecutive tuberculosis patients in Acapulco, Mexico, to assess the level of clustering and risk factors for clustered strains. Point cluster analysis examined spatial clustering. Risk analysis relied on the Mantel Haenszel procedure to examine bivariate associations, then to develop risk profiles of combinations of risk factors. Supplementary analysis of the spoligotyping data used SpolTools. Spoligotyping identified 85 types, 50 of them previously unreported. The five most common spoligotypes accounted for 55% of tuberculosis cases. One cluster of 70 patients (26% of the series) produced a single spoligotype from the Manila Family (Clade EAI2). The high proportion (78%) of patients infected with cluster strains is compatible with recent transmission of TB in Acapulco. Geomatic analysis showed no spatial clustering; clustering was associated with a risk profile of uneducated cases who lived in single-room dwellings. The Manila emerging strain accounted for one in every four cases, confirming that one strain can predominate in a hyperendemic area.
1. Introduction
Tuberculosis (TB) remains a global health problem, mainly related to poverty and concomitant diseases [1]. The reported annual incidence of pulmonary TB in Mexico is 14.27 cases per 100,000 people; in Guerrero state, the rate is 34.56 per 100,000 [2]. Acapulco is of particular concern as a centre for internal migration in Guerrero state, in addition to its role in international tourism.
TB recurrence and reinfection have quite different implications for prevention. A key issue among migrants to a city like Acapulco is whether TB infection contracted earlier in life is reactivated, to be detected in the city as clinical cases, or whether these are new infections contracted in the city. One research device to help differentiate between recurrence and reinfection is to establish whether contemporary cases are clustered; unless they all came from the same distant place of original infection, clustering suggests a shared source of infection in the locality of the study.
Molecular typing is a well-recognized tool for identifying clustering. The sequence insertion of IS6110 is often the basis for molecular fingerprinting, as the number of copies of this insertion and its localization in the chromosome varies among different strains [6]. Another sequence targeted for molecular typing is the 36-base pair Direct Repeat (DR) locus. Spoligotyping is a PCR-based typing method that generates distinct patterns based on the hybridization of 43 different oligonucleotides to amplified spacer sequences that lie between DR loci in the M. tuberculosis chromosome [7]. This estimates the frequency of recent infection with M. tuberculosis estimated by identification of clusters of a shared spoligotype.
This paper studies spoligotypes isolates of M. tuberculosis from a series of patients in Acapulco, to determine the level of clustering and to identify risk factors for clustering.
2. Material and Methods
2.1. Study Location and Population
A total of 330 patients with pulmonary TB presented a positive diagnosis between February 2001 and September 2002 in the municipality of Acapulco. Receipt of funding set the start of the series which closed with 330 consecutive participants recruited through a review of recent medical records. The series excluded only 30 who had left the city and who could not be localised. Using addresses from clinical records, researchers approached each participant and explained the study, obtaining written consent prior to administering a pretested questionnaire. The study provided an opportunity to follow patients with a preliminary sputum positive result; the Ministry of Health offered directly observed therapy, short course strategy (DOTS) [8] for all cases.
2.2. Bacterial Strains
The 300 participants contacted yielded 273 isolates of M. tuberculosis from sputum samples collected by the Acapulco General Hospital, the Donato G. Alarcón Hospital, the State Public Health Laboratory and the Clínica Avanzada de Atención Primaria a la Salud. Processing began with the Petroff method for preparation and decontamination of sputum samples, followed by inoculation of 0.5 mL of each sample onto Löwenstein-Jensen media, incubated at 37°C for eight weeks.
2.3. Genotyping
Extraction of DNA required resuspension of M. tuberculosis colonies in 1 mL of sterile distilled water, with samples lysed at 90°C for 10 minutes and frozen at −70°C for 15 minutes. Samples were then thawed and incubated at 90°C for 10 minutes. Centrifugation for three minutes concentrated bacterial contents, and the supernatant was transferred to a new tube. Spoligotyping relied on a commercially available kit (Isogen Bioscience BV Maarssen, The Netherlands) for amplification of DNA from the DR locus, the region with the highest level of polymorphism in the M. tuberculosis chromosome [7]. PCR amplification used 5 uL of extracted DNA from each sample combined with 1.5 uL MgCl2, 2.5 mM of each dNTP, 5 μl 10X buffer, 0.5 U Taq polymerase, and 20 pmol of each primer (DRa: biotin-5′-CCG AGA GGG GAC GGA AAC-3′ and DRb: 5′-GGT TTT GGG TCT GAC GAC-3′) in a final volume of 50 uL. The amplification protocol required 5 minutes at 94°C followed by 30 cycles of 1 minute at 94°C, 1 minute at 55°C, and 30 seconds at 72°C, with a final extension of 10 minutes at 72°C. The assay included two positive controls (chromosomal DNA from M. tuberculosis H37Rv and from M. bovis BCG P3) and a negative control (sterile H2O).
We hybridized the amplified DNA with 43 oligonucleotides covalently linked to a nylon membrane (Isogen Bioscience BV, Maarssen, The Netherlands) at 60°C for 1 hour in a blotter with 45 lines (Miniblotter 45; Immunetics, Cambridge, Mass). Afterwards, hybridized DNA fragments were incubated with streptavidin, conjugated to peroxidase (Boehringer Mannheim), and then analyzed by chemi-luminescence by incubating for 1 minute in 20 mL of ECL detection reagent (Amersham, Buckinghamshire, England) and exposing to X-ray film for 20 minutes.
2.4. Data Capture and Analysis
Data capture from the questionnaires relied on Epi-Info (CDC, version 6.04). We compared spoligotyping results with the SpolDB4 spoligotyping database from the Institute Pasteur de Guadeloupe which, at the time of the analysis, contained 1,939 types from 39,295 strains contributed by 122 countries [3, 9]. The analysis defined a cluster as two or more isolates with identical genetic patterns. We used the preexisting code for those already identified matching patterns. When the spoligotype was not found in SpolDB4, we labeled it as “Mx” with a number (for example, Mx1).
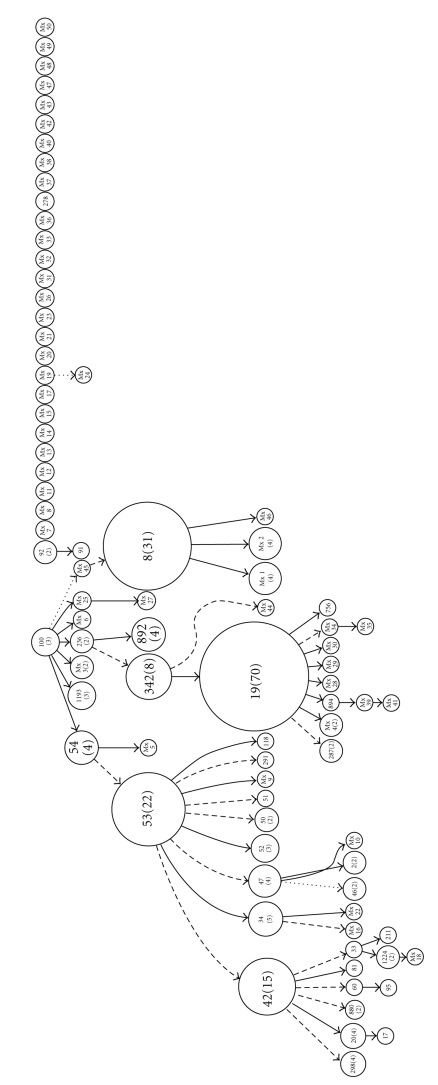
We estimated the Recent Transmission Index (RTI) using the formula of RTIn − 1 [10] and RTIn, which takes into account the number of patterns with unique genotype (singletons) described by Luciani et al. [11]. Supplementary analysis of the spoligotyping data used SpolTools [12]: DESTUS (Detecting Emerging Strains of Tuberculosis Using Spoligotypes) focused on the detection of rapidly propagating strains; spoligoforests allowed visualization of probable relations between the spoligotypes based on a plausible history of mutation events. The clusters of strains sharing the same spoligotype in the diagram are nodes, labeled with shared type (ST) numbers in SpolDB4 [3].
2.5. Risk and Point Cluster Analysis
Potential risk factors for clustering included age (less than or older than 30 years), sex, marital status, education, employment, ethnicity, area of residence (urban or rural), duration of residence, number of people in the dwelling, use of alcohol, concomitant illness, or spouse with TB. When simple bivariate risk analysis revealed no statistically significant contrasts, we derived risk profiles of factor combinations associated with clustering. We examined these risk profiles in a multivariate analysis, beginning with a saturated model and eliminating the weakest association stepwise until only significant associations remained in the final model. We report this as an adjusted odds ratio and 95% confidence interval.
For the spatial analysis, we geoindexed cases on a map of Acapulco at a topographic chart scale of 1 : 50,000 in DXF vector data format (Instituto Nacional de Estadística, Geografía e Informática; 2001). Point cluster analysis of quadrants relied on QUADRAT (IDRISI 32, Clark Labs, Worcester, MA), which determines the total number, mean number or density, variance and variance/mean ratio of points in cases where each grid cell measures the total point count found in that cell. The variance/mean ratio describes the pattern of a point set, with values close to 1.0 suggesting a random point pattern, values significantly smaller than 1.0 suggesting a regularly distributed pattern, and values greater than 1.0 suggesting a clustered pattern. The location of each case was compared with the locations of the reported TB cases. Final display relied on ArcView GIS software (ArcView GIS 3.2, Environmental Systems Research Institute Inc., Redlands, CA).
This study was approved by the Committee of Research Ethics at the Centro de Investigación de Enfermedades Tropicales of the Universidad Autónoma de Guerrero.
3. Results
3.1. Spoligotyping Patterns of M. tuberculosis Isolates
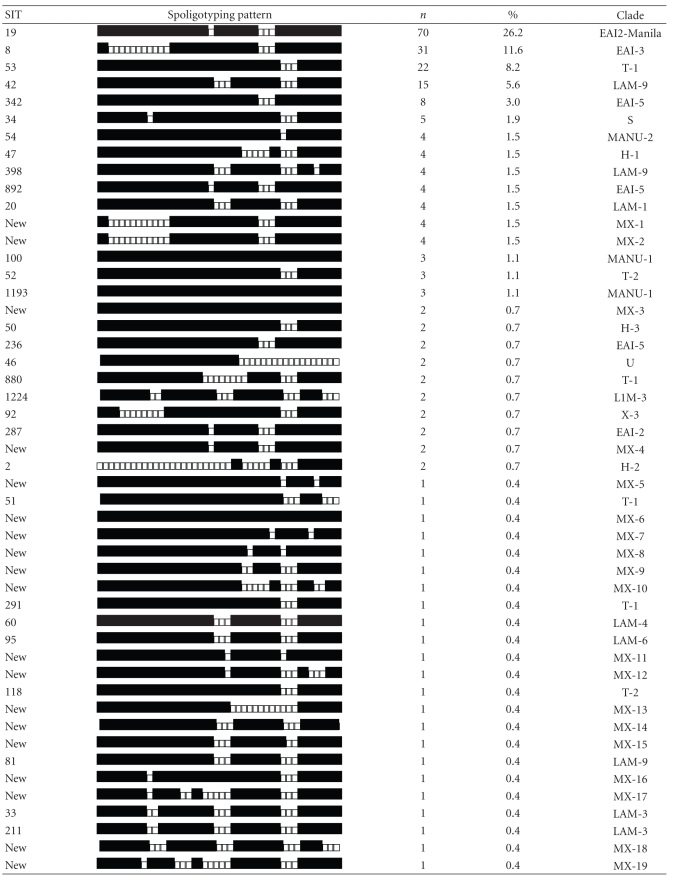
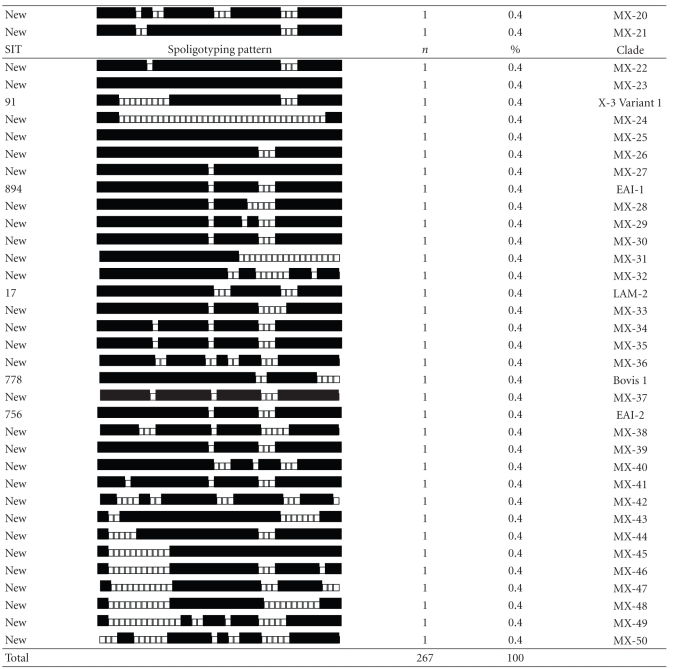
Spoligotyping the 267 isolates produced 85 distinct genotypes, 59 of them with a unique pattern. The 208 (77.9%) remaining isolates were grouped into 26 clusters that were shared with at least one patient. The cluster size varied from 2 to 70 patients, however, most (21/26) included 2 to 5 patients. The Recent Transmission Index, or RTIn, taking into account the number of cases with unique genotypes (singletons), was 0.78, and the RTIn − 1 was 0.68. Table 1 presents a comparison of the Acapulco spoligotypes with data from the global spoligotype database of the cluster genotypes; four were new and 22 had been previously identified by this database.
Table 1.
Acapulco patterns of Mycobacterium tuberculosis spoligotypes.
|
SIT, designation of the spoligotype in the international database. Spoligotyping patterns, binary description: (■) hybridization and (□) no hybridization. n, number of strains; clade, defined mainly as described [4, 5]: EAI-2/Manila: East African Indian 2/Manila, T (ill-defined T cluster), LAM: Latin American and Mediterranean, S: S clade, MANU: Manu family, H: Haarlem, U: Undesignated, X: X cluster, Bovis. New clusters designated as MX (Mexican).
Approximately, 59% of the 85 Acapulco spoligotypes, involving 58 (21.7%) of the 267 M. tuberculosis isolates, were previously unreported. The main SITs (Spoligo International Type Number)—19, 8, 53, 42, and 342—permitted the classification of isolates in representative patterns described as families or groups [3, 13, 14]. The largest single cluster of 70 patients included 26.2% (70/267) of M. tuberculosis isolates of Acapulco; the spoligotype pattern was the East African Indian group (EAI2), previously reported as M. tuberculosis Manila family [15]. Approximately 44.6% (119/267) of the isolates descended from East African Indian family (EAI) subgroups 1, 2, 3, and 5. Another 11.6% (31/267) came from Latin American and Mediterranean families (LAM) subgroups 1, 2, 3, 4, 6, and 9. A further 11.2% (30/267) belonged to the “T” family (strain of modern TB) subgroups 1 and 2. Another 3.7% (10/267) were of the “Manu” family, subgroups 1 and 2, and 3.0% (8/267) consisted of the Haarlem family, subgroups 1, 2, and 3. A further 1.9% (5/267) were group “S”, and 1.1% (3/267) were “X” family subgroup 3 and 3 variant 1. Two cases (0.8%) were not classified (“U”), and 0.4% (1/267) pertained to the Bovis group, subgroup 1. We identified no isolate of the Beijing genotype.
3.2. Detection of an Emerging Strain and Visualization of the Relations between Spoligotypes
The S19 strain demonstrates an elevated rate of transmission, independent of the sampling fraction f (Table 2), behaving as an emerging strain according to the method proposed by Tanaka and Francis [16]. According to the SpolDB4 database, S19 corresponds to the EAI2-Manila strain.
Table 2.
Emerging strains detected in the pulmonary TB by using spoligotypes in Acapulco.
| f | |||||||
|---|---|---|---|---|---|---|---|
| 0.95 | 0.5 | 0.1 | 0.01 | ||||
| Strain | q value | Strain | q value | Strain | q value | Strain | q value |
|
| |||||||
| S19 | 0.00000000 | S19 | 0.00000006 | S19 | 0.00000477 | S19 | 0.00001087 |
| S8 | 0.03205437 | S8 | 0.08223924 | S8 | 0.17184278 | S8 | 0.20014382 |
f = sampling fraction.
Figure 1 shows the Spoligoforest hierarchical layout, with the lines between the nodes denoting the weights calculated using this model. The spoligoforest contains three trees with connected components and 26 unconnected nodes. The biggest tree has the ST 100 strain as a root, suggesting that it is the oldest spoligotype in this tree. Seven spoligotypes descend from ST 100, four of which are in small clusters (ST 54, ST 1193, ST 236, and Mx 3, with 4, 3, 2, and 2 isolates, resp.). The ST 54 and ST 236 spoligotypes form two lineages distinct from ST 100. Comparing these spoligotypes with the families in the SpolDB4 database, these lineages are classified as Manu Family and EAI (East African Indian). The Mx 45 genotype, not registered in the SpolDB4 database, also descending from ST 100, originates another lineage. ST 53 and its descendents ST 42, ST 34, ST 47, ST 52, ST 50, ST 51, ST 291, and ST 118 belong to the LAM (Latin American Mediterranean), Haarlem, cluster T and Clade S family of strains. ST 342 and its descendents ST 19 and Mx 44, ST 19 belongs to the Manila family (EAI2-Manila) and is the biggest node, representing 70 isolates and Mx 44 are not registered in SpolDB4. Finally, ST 8 is from the EAI (East African Indian) family, and its descendents Mx 1, Mx 2, and Mx 46 are not registered in SpolDB4.
Figure 1.
Spoligoforest of tuberculosis cases in Acapulco, Mexico. Nodes are labeled with the ST identifier as indicated in SoplDB4 [3], with the cluster size enclosed in parentheses. Spoligotype not in SpolDB4 was labeled as “Mx” with a number. The size of each node increases as a function of the number of isolates (size of the cluster); the lines among the nodes reflect the evolutionary relationships among spoligotypes with arrows that denote the descendent. If the weight of the line was equal to 1, we drew a solid line; if the weight was greater than or equal to 0.5 but less than 1, we used a dashed line; if the weight was less than 0.5, we used a dotted line.
3.3. Epidemiological and Clinical Characteristics of the Tuberculosis Cases
Of 330 diagnosed TB cases in the series, 30 (9.1%) could not be found; 273 (91.0%) of the 300 contacted yielded a positive culture of M. tuberculosis: 190 (63.3%) of them were male, and the average age was 41.0 years (standard deviation: 15.5, range: 15–86 years). Most (90.7%, 272/300) were from Guerrero state, and 55.1% (150/272) of these were from the municipality of Acapulco (Table 3). Some 59% (172/293) lived less than 16 years in their community of origin. The average duration of residence in Acapulco was 2.6 years (SD: 1.66, range: 1–10 years). Household had an average of 5.1 occupants (SD: 2.83, range: 1–20 people).
Table 3.
Characteristics of the pulmonary TB case series in Acapulco.
| Variable | n = 300 | % |
|---|---|---|
| Sex* | ||
| Male | 190 | 63.3 |
| Female | 110 | 36.7 |
| Age (in years)** | ||
| 15–24 | 51 | 17 |
| 26–34 | 72 | 24 |
| 35–44 | 58 | 19.3 |
| 45–54 | 55 | 18.3 |
| 55–64 | 40 | 13.3 |
| 65 + | 24 | 8 |
| Signs and symptoms | ||
| Duration of the cough >3 months** | 192 | 64 |
| Lost weight** | 271 | 90.3 |
| Fever** | 227 | 75.7 |
| Hemoptysis** | 151 | 50.3 |
| Associated illness** | 132 | 44 |
| Type of associated illness** (n = 132) | ||
| Diabetes | 77 | 58.3 |
| Drug user | 20 | 15.2 |
| AIDS | 12 | 9.1 |
| Alcoholism | 7 | 5.3 |
| Undernutrition | 3 | 2.3 |
| Others | 13 | 9.8 |
| Prior TB** | 26 | 8.7 |
| Husband/wife with prior TB** (n = 190) | 22 | 11.6 |
| Recent contact with someone with TB** | ||
| Yes | 112 | 37.3 |
| No | 150 | 50 |
| Do not know | 37 | 12.3 |
| Indigenous** | 27 | 9 |
| Indigenous group** (n = 27) | ||
| Náhuatl | 16 | 59.3 |
| Mixteco | 9 | 33.3 |
| Amuzgo | 2 | 7.4 |
| State of birth** | ||
| Guerrero | 272 | 90.7 |
| Oaxaca | 11 | 3.7 |
| Estado de México | 4 | 1.3 |
| Michoacán | 3 | 1 |
| Other state | 10 | 3.3 |
| Region of birth** (n = 272) | ||
| Acapulco | 150 | 55.1 |
| Zone Centro | 25 | 9.2 |
| La Montaña | 7 | 2.6 |
| Costa Chica | 46 | 16.9 |
| Costa Grande | 21 | 7.7 |
| Zone Norte | 8 | 2.9 |
| Tierra Caliente | 12 | 4.4 |
*P value =.05,
**P value <.05.
Data on the start date of symptoms permitted identification of the “first case” (index case) in the largest clusters. The “first case” of the Manila cluster, for example, was a 35-year-old adult addicted to drugs and alcohol who had symptoms for 10 years and, on examination, had advanced pulmonary TB (bacilloscopy positive, grade 3). Thirty of the 70 cases in this cluster reported a concomitant disease, diabetes being the most common (17/70).
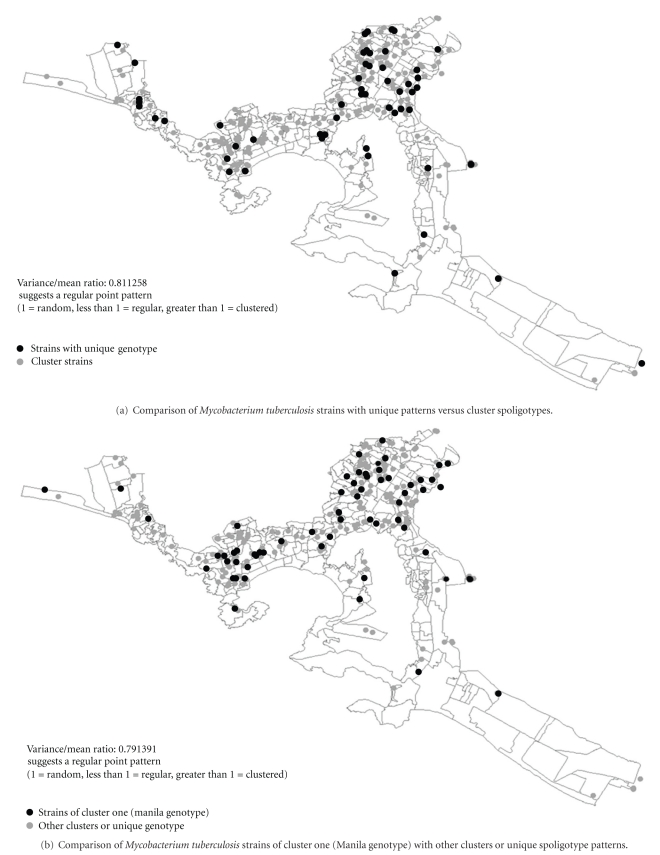
The largest five clusters included 70, 31, 22, 15, and 8 cases, together contributing 70% (146/208) of the case series (Table 4). Figure 2 shows a random distribution pattern of spatial variances. There was no evidence of spatial grouping, consistent with recent infection associated with factors other than residence.
Table 4.
Characteristics of the largest clusters detected by spoligotyping in Acapulco.
| No. Cluster |
Cluster size (Number of male) |
Age (range) |
Place of birth (n) |
Indigenous group (n) |
Associated illness % (n) |
Immune- compromised illness (n) |
Previous TB % (n) |
Contact/ TB case % (n) |
Husband/ wife with TB % (n) |
BAAR diagnostic % |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 70 (45) | 41.6 (15–86) |
Guerrero (62) Oaxaca (4) Mexico (1) DF (1) Gto (1) Tabasco (1) |
Nah (4) | 43% (30) | Diabetes (17) AIDS (2) Alcoholism (2) Drug user (2) undernourished (2) |
10% (7) | 39% (27) | 8% (5) | + 47 ++ 23 +++ 30 |
|
| ||||||||||
| 2 | 31 (21) | 41.2 (21–73) |
Guerrero (28) Oaxaca (1) EdoMex (1) Veracruz (1) |
Nah (2) | 45% (14) | Diabetes (10) AIDS (1) |
13% (4) | 26% (8) | 17% (5) | + 36 ++ 19 +++ 45 |
|
| ||||||||||
| 3 | 22 (19) | 44.0 (22–76) |
Guerrero (20) Oaxaca (1) EdoMex (1) |
Nah (3) Mix (1) |
27% (6) | Diabetes (4) Alcoholism (1) Drug user (1) |
14% (3) | 41% (9) | 0 | ++ 36 +++ 64 |
|
| ||||||||||
| 4 | 15 (7) | 45.4 (19–75) |
Guerrero (13) Oaxaca (1) Michoacán (1) |
Nah (1) Mix (2) |
60% (9) | Diabetes (3) AIDS (1) Alcoholism (1) Drug user (3) |
0 | 53% (8) | 0 | + 33 ++ 27 +++ 40 |
|
| ||||||||||
| 5 | 8 (4) | 43.6 (23–68) |
Guerrero (8) | Mix (2) | 50% (4) | Diabetes (1) Drug user (2) |
0 | 63% (5) | 0 | + 12.5 +++ 88 |
Place of birth: DF= Federal district, Mexico; Gto= Guanajuato state, EdoMex= State of México. Indigenous group: Nah= Náhuatl, Mix= Mixteco.
Figure 2.
Geographical distribution of tuberculosis cases in Acapulco, Mexico. Mapping and georeferencing of tuberculosis cases used a scale of 1 : 50000. Point cluster analysis relied on the IDRISI QUADRAT module. The variance/mean ratio significantly smaller than 1.0 suggests a regular distribution, while values greater than 1.0 suggest spatial clustering.
Table 5 shows the list of conventional TB risk factors, none of which on their own showed a statistically significant association with clustering on univariate analysis. Seven risk profiles, combining two risk factors, did show significant differences between clustered and nonclustered TB cases (Table 6): males who consumed alcohol, unmarried men under the age of 30 years, unemployed single cases, indigenous cases without remunerated employment, young people in urban areas, and uneducated people living in one-roomed dwellings. The size of the study did not permit combining all of these profiles in a single multivariate model. Each was therefore tested to see if it was explained by each other profile in turn. Only one risk profile remained that could not be explained by any others—uneducated cases living in single-room dwellings.
Table 5.
Bivariate analysis of conventional TB risk factors, contrasting clustered and nonclustered cases.
| Variable | Clustered | Not clustered | OR | 95% CI | |||
|---|---|---|---|---|---|---|---|
| N | % | n | % | ||||
| Sex | Male | 135 | 80.6 | 32 | 19.4 | 1.60 | 0.87–2.2 |
| Female | 72 | 72.7 | 27 | 27.3 | 1 | ||
| Age | <30 years | 55 | 76.4 | 17 | 23.6 | 0.91 | 0.48–1.72 |
| ≥30 years | 150 | 78.1 | 42 | 21.9 | 1 | ||
| Marital status | Single | 65 | 83.3 | 13 | 16.4 | 1.64 | 0.83–3.2 |
| Married | 140 | 75.3 | 46 | 24.7 | 1 | ||
| Education | No formal studies | 39 | 72.2 | 15 | 27.8 | 0.67 | 0.34–1.33 |
| Formal study | 166 | 79.4 | 43 | 20.6 | 1 | ||
| Occupation | Unemployed | 70 | 74.5 | 24 | 25.5 | 0.76 | 0.40–1.43 |
| Employed | 135 | 79.4 | 35 | 20.6 | 1 | ||
| Ethnicity | Indigenous | 19 | 73.1 | 7 | 26.9 | 0.75 | 0.28–2.08 |
| Mestizo | 189 | 78.4 | 52 | 21.6 | 1 | ||
| Migration | Yes | 99 | 76.2 | 31 | 23.8 | 0.82 | 0.45–1.47 |
| No | 105 | 79.5 | 27 | 20.5 | 1 | ||
| Area of residence | Urban | 153 | 78.5 | 42 | 21.5 | 1.19 | 0.59–2.37 |
| Rural | 52 | 75.4 | 17 | 24.6 | 1 | ||
| Duration of residence | ≥17 years | 123 | 79.4 | 32 | 20.6 | 1.09 | 0.59–2.0 |
| <17 years | 74 | 77.9 | 21 | 22.1 | 1 | ||
| Number of residents per household | ≥6 persons | 70 | 72.9 | 26 | 27.1 | 0.66 | 0.37–1.19 |
| <6 persons | 135 | 80.4 | 33 | 16.6 | 1 | ||
| Use of alcohol | Yes | 67 | 75.3 | 22 | 24.7 | 0.82 | 0.45–1.50 |
| No | 137 | 78.7 | 37 | 21.3 | 1 | ||
| Concomitant illness | Yes | 94 | 77.7 | 27 | 23.3 | 1.02 | 0.55–1.91 |
| No | 111 | 77.6 | 32 | 23.4 | 1 | ||
| Spouse with TB | Yes | 15 | 71.4 | 6 | 28.6 | 0.66 | 0.24–1.85 |
| No | 113 | 79.0 | 30 | 21.0 | 1 | ||
Table 6.
Bivariate analysis of risk profiles associated with clustering.
| Variable | Cluster | No cluster | OR | 95% CI | |||
|---|---|---|---|---|---|---|---|
| N | % | n | % | ||||
| Among cases who used alcohol | Male | 71 | 85.5 | 12 | 14.5 | 2.1 | 1.05–4.03 |
| Female | 68 | 73.1 | 25 | 26.9 | 1 | ||
| Among cases over the age of 29 years | Single | 36 | 92.3 | 3 | 7.7 | 4.1 | 1.29–13.1 |
| Married or cohabiting | 114 | 74.5 | 39 | 23.5 | 1 | ||
| Among cases single/unmarried cases | Unemployed | 7 | 87.5 | 1 | 12.5 | 13.7 | 1.81–104 |
| Employed | 8 | 40.0 | 12 | 60.0 | 1 | ||
| Among cases with remunerated employment | Mestizo | 132 | 81.0 | 31 | 19.0 | 3.2 | 1.07–9.1 |
| Indigenous | 8 | 57.1 | 6 | 42.9 | 1 | ||
| Among cases who live in one place for less than 15 years | Urban | 66 | 78.6 | 18 | 21.4 | 3.0 | 1.22–10.0 |
| Rural | 11 | 55.0 | 9 | 45.0 | 1 | ||
| Cases younger than 30 years of age | Urban | 43 | 82.7 | 9 | 17.3 | 3.2 | 1.03–9.82 |
| Rural | 12 | 60.0 | 8 | 40.0 | 1 | ||
| Among cases without formal education | 1 room | 22 | 91.7 | 2 | 8.3 | 8.9 | 2.0–39.1 |
| 2 or more rooms | 16 | 55.2 | 13 | 44.8 | 1 | ||
OR= Odds Ratio, CI= Confidence Interval.
4. Discussion
Reflecting the still very partial documentation of spoligotypes worldwide, 58% of our spoligotypes were unique to Acapulco; 21.7% of cases were of a type not previously registered in the global database.
Our five most frequent spoligotypes included more than one half (54.7%) of the cases; these types are commonly recognized worldwide [3, 13, 14]. The T1 Spoligotype (shared type, 53), EAI (shared type 19, 8, and 342), and LAM9 (shared type 42) currently represent 28.75% of M. tuberculosis isolates in the global spoligotype database, being prevalent in Europe, Africa, India, and other countries [3].
The largest cluster, including 33.6% (70/208) of the isolates, was the Manila family. This demonstrates the public health impact of this strain in Acapulco. This relates to Mexico's historic ties with Asia, as the Manila strain is found throughout South East Asia, particularly in the Philippines (73%), Myanmar and Malaysia (53%), and Vietnam and Thailand (32%) [3, 15, 17]. In a study in the Philippines, 90% (43/48) of isolates were of the Manila genotype [15]. Phylogenetic studies of related spoligotype strains demonstrated that the EAI (East African Indian) genotype has shared ancestral relations [17]. This group includes the Manila or EAI2-Manila strain [3].
Internationally, certain M. tuberculosis strains are linked with a large proportion of recent infection, suggesting these strains might have greater transmissibility or higher probability to cause disease soon after transmission. These strains are associated with families or groups of related isolates, such as the genotypes of the Beijing family strain W, Haarlem, and Africa [3, 18, 19]. These “cluster strains” come from the Latin American Mediterranean families (LAM), Haarlem (H), and M. Bovis [20]. The Manila strain, responsible for a third of our cluster strain cases, might share this greater transmission or progress to active disease—at least in Acapulco.
The method used for detection of rapidly propagating strains [16] allowed identification of an emerging strain (strain S19), previously classified as EAI2-Manila [3]. This result coincides with the spoligoforest result, where node S19 (n = 70) has the most rapid transmission. Spoligoforest demonstrates all of the possible relations between spoligotypes under the assumption of spoligotype mutation [12]. In our data set, the largest root of the tree was the ST 100 strain, which corresponds to the MANU family. Among the principal descendants, we identified strains ST 54, ST 236, and ST 1193. However, the majority of the 26 unconnected nodes had not been previously registered in the SpolDB4 database.
Our proportion of clustered strains (77.9%) is higher than in other studies. A six-year study in two urban communities in South Africa identified 72% of the cases as belonging to clusters [21]. Similar results came from the Grand Canary Islands of Spain [4]. Other studies in South Africa and in Equatorial Guinea identified a level of grouping of 67 and 61.6%, respectively [5, 22]. A range of clustering rates (31 to 67.7%) have been reported in cities from industrial countries such as Spain, Italy, Holland, Denmark, Slovenia, Canada, and United States [23–29], while France, London, and Switzerland reported a minor portion of 18 to 27.6% cluster strains [30–32].
Considering the probable existence of an index case in each cluster, we estimated that 68% (208–26) of our 267 cases could have been due to recent infection [10]. Taking into account the number of singletons, the estimate of RTI was greater (78%). Migration, duration of the study [33], predominance of a local strain, simultaneous reactivation of an infection acquired remotely, and laboratory error can all influence the reliability of this inference [34]. In our study, other factors support a high proportion of recent transmission, including the high prevalence rate, the mobility of population [29], the bacillary load of the cases, and the duration of their symptoms.
Although one cluster included 70 cases, most clusters had five or fewer cases. Similar results come from the Grand Canary Islands [4]. A study in San Francisco in 1996-97 found 73 of 221 (33%) cases in multiple clusters, implying multiple points of infection in the community.
Analysis of why certain strains of M. tuberculosis propagated effectively points mostly to delayed diagnosis [35]. The index cases in five of our clusters were young adult males with at least one risk factor, such as drug use or alcohol addiction [4, 10, 25, 36]. Cases with an associated pathology have a higher tendency to acquire a new infection of M. tuberculosis [37] or to develop tuberculosis [38].
Our analysis of potential risk factors for clustering did not produce clearly actionable results. The notable absence of spatial clustering implies that place of residence is not a useful risk indicator. Type of residence (single room) combined with lack of formal education, on the other hand, was the single enduring risk profile.
SpolTools permitted the analysis of M. tuberculosis spoligotyping data to identify emerging strains and visualization of the probable evolutionary relationships between the spoligotypes in our series. Considering that a single emerging strain, the EAI2-Manila genotype, can account for so many cases, an evolution of spoligotyping could conceivably be used to evaluate the impact of TB prevention and early diagnosis efforts. Further studies of the virulence and drug sensitivity of the Manila genotype may be warranted.
Conflicts of Interests
The authors do not have funding, commercial or other associations that might pose a conflict of interest.
Acknowledgments
The authors thank Lee Riley, Division of Infectious Diseases, School of Public Health, University of California, Berkeley, for revising the paper. Mauricio Castañón Arreola, Departamento de Microbiología y Parasitología, Universidad Nacional Autónoma de México provided technical assistance with the spoligotyping technique. Also, they acknowledge the cooperation of the State of Guerrero Ministry of Health, the laboratories of the Acapulco General Hospital, the Donato G. Alarcón Hospital, the Clínica Avanzada de Atención Primaria a la Salud, and the State Public Health “Galo Soberón y Parra” for providing the sputum samples and facilities for preparing culture medium. This work was supported by Fondo del Sistema de Investigación Benito Juárez from Consejo Nacional de Ciencia y Tecnología (CONACYT) Grant 20000502018 and the Project SDEI.PTID.05.04.
References
- 1.World Health Organization. Tech. Rep. WHO/HTM/TB/2006.362. Geneva, Switzerland: World Health Organization,; 2006. Global tuberculosis control: surveillance, planning, financing. [Google Scholar]
- 2.Secretaría de Salud. Boletines Epidemiológicos Anuales. Mexico City, México: Dirección General de Epidemiología, Secretaría de Salud; 1990–2005. [Google Scholar]
- 3.Brudey K, Driscoll JR, Rigouts L, et al. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiology. 2006;6, article 23 doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pena MJ, Caminero JA, Campos-Herrero MI, et al. Epidemiology of tuberculosis on Gran Canaria: a 4 year population study using traditional and molecular approaches. Thorax. 2003;58(7):618–622. doi: 10.1136/thorax.58.7.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godfrey-Faussett P, Sonnenberg P, Shearer SC, et al. Tuberculosis control and molecular epidemiology in a South African gold-mining community. The Lancet. 2000;356(9235):1066–1071. doi: 10.1016/s0140-6736(00)02730-6. [DOI] [PubMed] [Google Scholar]
- 6.van Soolingen D, Hermans PWM, de Haas PEW, Soll DR, van Embden JDA. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. Journal of Clinical Microbiology. 1991;29(11):2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamerbeek J, Schouls L, Kolk A, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. Journal of Clinical Microbiology. 1997;35(4):907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Secretaría de Salud. Norma Oficial Mexicana NOM-006-SSA2-1993, para la prevención y control de la tuberculosis en la atención primaria a la salud. Diario Oficial de la Federación. 1995:1–12. [Google Scholar]
- 9.Spol DB4 database. http://www.pasteur-guadeloupe.fr/tb/bd_myco.html.
- 10.Small PM, Hopewell PC, Singh SP, et al. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. The New England Journal of Medicine. 1994;330(24):1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 11.Luciani F, Francis AR, Tanaka MM. Interpreting genotype cluster sizes of Mycobacterium tuberculosis isolates typed with IS6110 and spoligotyping. Infection, Genetics and Evolution. 2008;8(2):182–190. doi: 10.1016/j.meegid.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Tang C, Reyes JF, Luciani F, Francis AR, Tanaka MM. SpolTools: online utilities for analyzing spoligotypes of the Mycobacterium tuberculosis complex. Bioinformatics. 2008;24(20):2414–2415. doi: 10.1093/bioinformatics/btn434. [DOI] [PubMed] [Google Scholar]
- 13.Sola C, Filliol I, Gutierrez MC, Mokrousov I, Vincent V, Rastogi N. Spoligotype database of Mycobacterium tuberculosis: biogeographic distribution of shared types and epidemiologic and phylogenetic perspectives. Emerging Infectious Diseases. 2001;7(3):390–396. doi: 10.3201/eid0703.010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filliol I, Driscoll JR, van Soolingen D, et al. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. Journal of Clinical Microbiology. 2003;41(5):1963–1970. doi: 10.1128/JCM.41.5.1963-1970.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douglas JT, Qian L, Montoya JC, et al. Characterization of the Manila family of Mycobacterium tuberculosis . Journal of Clinical Microbiology. 2003;41(6):2723–2726. doi: 10.1128/JCM.41.6.2723-2726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka MM, Francis AR. Detecting emerging strains of tuberculosis by using spoligotypes. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(41):15266–15271. doi: 10.1073/pnas.0603130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filliol I, Motiwala AS, Cavatore M, et al. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. Journal of Bacteriology. 2006;188(2):759–772. doi: 10.1128/JB.188.2.759-772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glynn JR, Whiteley J, Bifani PJ, Kremer K, van Soolingen D. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerging Infectious Diseases. 2002;8(8):843–849. doi: 10.3201/eid0808.020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Soolingen D, Qian L, de Haas PEW, et al. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. Journal of Clinical Microbiology. 1995;33(12):3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aristimuño L, Armengol R, Cebollada A, et al. Molecular characterisation of Mycobacterium tuberculosis isolates in the first national survey of anti-tuberculosis drug resistance from Venezuela. BMC Microbiology. 2006;6, article 90 doi: 10.1186/1471-2180-6-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verver S, Warren RM, Munch Z, et al. Transmission of tuberculosis in a high incidence urban community in South Africa. International Journal of Epidemiology. 2004;33(2):351–357. doi: 10.1093/ije/dyh021. [DOI] [PubMed] [Google Scholar]
- 22.Tudó G, González-Martín J, Obama R, et al. Molecular epidemiology of tuberculosis in the Bata and Malabo districts of Equatorial Guinea. International Journal of Tuberculosis and Lung Disease. 2004;8(12):1458–1463. [PubMed] [Google Scholar]
- 23.Cacho Calvo J, Astray Mochales J, Pérez Meixeira A, et al. Ten-year population-based molecular epidemiological study of tuberculosis transmission in the metropolitan area of Madrid, Spain. International Journal of Tuberculosis and Lung Disease. 2005;9(11):1236–1241. [PubMed] [Google Scholar]
- 24.Moro M, Salamina G, Gori A, et al. Two-year population-based molecular epidemiological study of tuberculosis transmission in the Metropolitan area of Milan, Italy. European Journal of Clinical Microbiology and Infectious Diseases. 2002;21(2):114–122. doi: 10.1007/s10096-001-0664-2. [DOI] [PubMed] [Google Scholar]
- 25.van Soolingen D, Borgdorff MW, de Haas PEW, et al. Molecular epidemiology of tuberculosis in the Netherlands: a nationwide study from 1993 through 1997. Journal of Infectious Diseases. 1999;180(3):726–736. doi: 10.1086/314930. [DOI] [PubMed] [Google Scholar]
- 26.Yang ZH, de Haas PEW, Wachmann CH, van Soolingen D, van Embden JDA, Andersen AB. Molecular epidemiology of tuberculosis in Denmark in 1992. Journal of Clinical Microbiology. 1995;33(8):2077–2081. doi: 10.1128/jcm.33.8.2077-2081.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Žolnir-Dovč M, Poljak M, Eržen D, Şorli J. Molecular epidemiology of tuberculosis in Slovenia: results of a one-year (2001) nation-wide study. Scandinavian Journal of Infectious Diseases. 2003;35(11-12):863–868. doi: 10.1080/00365540310017221. [DOI] [PubMed] [Google Scholar]
- 28.Blackwood KS, Al-Azem A, Elliott LJ, Hershfield ES, Kabani AM. Conventional and molecular epidemiology of Tuberculosis in Manitoba. BMC Infectious Diseases. 2003;3, article 18 doi: 10.1186/1471-2334-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnes PF, Yang Z, Preston-Martin S, et al. Patterns of tuberculosis transmission in Central Los Angeles. Journal of the American Medical Association. 1997;278(14):1159–1163. [PubMed] [Google Scholar]
- 30.Vachée A, Vincent P, Savage C, et al. Molecular epidemiology of tuberculosis in the Nord department of France during 1995. Tubercle and Lung Disease. 1999;79(6):361–366. doi: 10.1054/tuld.1999.0224. [DOI] [PubMed] [Google Scholar]
- 31.Maguire H, Dale JW, McHugh TD, et al. Molecular epidemiology of tuberculosis in London 1995-7 showing low rate of active transmission. Thorax. 2002;57(7):617–622. doi: 10.1136/thorax.57.7.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Genewein A, Telenti A, Bernasconi C, et al. Molecular approach to identifying route of transmission of tuberculosis in the community. The Lancet. 1993;342(8875):841–844. doi: 10.1016/0140-6736(93)92698-s. [DOI] [PubMed] [Google Scholar]
- 33.van Soolingen D. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. Journal of Internal Medicine. 2001;249(1):1–26. doi: 10.1046/j.1365-2796.2001.00772.x. [DOI] [PubMed] [Google Scholar]
- 34.Glynn JR, Bauer J, de Boer AS, et al. Interpreting DNA fingerprint clusters of Mycobacterium tuberculosis. European concerted action on molecular epidemiology and control of tuberculosis. International Journal of Tuberculosis and Lung Disease. 1999;3(12):1055–1060. [PubMed] [Google Scholar]
- 35.Chin DP, Crane CM, Diul MY, et al. Spread of Mycobacterium tuberculosis in a community implementing recommended elements of tuberculosis control. Journal of the American Medical Association. 2000;283(22):2968–2974. doi: 10.1001/jama.283.22.2968. [DOI] [PubMed] [Google Scholar]
- 36.Caminero JA, Pena MJ, Campos-Herrero MI, et al. Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. American Journal of Respiratory and Critical Care Medicine. 2001;164(7):1165–1170. doi: 10.1164/ajrccm.164.7.2101031. [DOI] [PubMed] [Google Scholar]
- 37.Daley CL, Small PM, Schecter GF, et al. An outbreak of tuberculosis with accelerated progression among persons infected with the human immunodeficiency virus: an analysis using restriction-fragment-length polymorphisms. The New England Journal of Medicine. 1991;326(4):231–235. doi: 10.1056/NEJM199201233260404. [DOI] [PubMed] [Google Scholar]
- 38.Pérez A, Brown HS, III, Restrepo BI. Association between tuberculosis and diabetes in the Mexican border and non-border regions of Texas. American Journal of Tropical Medicine and Hygiene. 2006;74(4):604–611. [PMC free article] [PubMed] [Google Scholar]