SUMMARY
When it escapes early detection, malignant melanoma becomes a highly-lethal and treatment-refractory cancer. Melastatin is greatly downregulated in metastatic melanomas and is widely believed to function as a melanoma tumor suppressor. Here we report that tumor suppressive activity is not mediated by melastatin but instead by a microRNA (miR-211) hosted within an intron of melastatin. Increasing expression of miR-211 but not melastatin reduced migration and invasion of malignant and highly invasive human melanomas characterized by low levels of melastatin and miR-211. An unbiased network analysis of melanoma-expressed genes filtered for their roles in metastasis identified three central node genes: IGF2R, TGFBR2, and NFAT5. Expression of these genes was reduced by miR-211 and knockdown of each gene phenocopied the effects of increased miR-211 on melanoma invasiveness. These data implicate miR-211 as a suppressor of melanoma invasion whose expression is silenced (or selected against) via suppression of the entire melastatin locus during human melanoma progression.
INTRODUCTION
A large proportion of advanced melanomas show down-regulation of melastatin, a founding member of the transient receptor potential (TRPM) cation channel family that is highly expressed in melanocytes and retinal pigmented epithelium. Whereas melastatin (TRPM1) is robustly expressed in benign and dysplastic nevi and in melanomas in situ, it is only variably expressed in invasive melanomas, shows widespread down-regulation in melanoma metastases, and is inversely correlated with metastatic potential and prognosis of melanomas (Duncan et al., 2001; Duncan et al., 1998). However, the mechanism(s) by which melastatin might suppress melanoma metastasis remain unknown.
Interestingly, intron 6 of the TRPM1 gene (Hunter et al., 1998) hosts the gene for miR-211, a microRNA (miRNA) whose expression is restricted to the melanocyte lineage (Gaur et al., 2007). Numerous miRNAs have been implicated in tumorigenesis (e.g. miR-17~92 as a oncogenic cluster (He et al., 2005; O’Donnell et al., 2005); let-7 as a tumor suppressor (Takamizawa et al., 2004; Yanaihara et al., 2006); and miR-10b (Ma et al., 2007), miR-373 and miR-520c (Huang et al., 2008), miR-335 (Tavazoie et al., 2008), and miR-9 (Ma et al., 2010) in tumor invasion and metastasis). These observations, together with the lack of mechanistic data linking melastatin to melanoma tumor suppression, led us to hypothesize that miR-211 activity might regulate melanoma invasion and metastasis independently of its host gene function.
Melanoma short-term cultures (MSTCs) represent a physiologically-relevant and an experimentally-tractable cancer model because they have relatively low passage numbers since biopsy, are readily annotated with respect to genetic alterations, and are easily grown in culture. Through genetic and functional studies performed on a collection of MSTCs, we identified two populations of malignant melanomas: one with low miR-211 expression and high invasive activity and the other with high miR-211 expression and low invasive activity. Here we report that manipulation of miR-211 levels, but not melastatin levels, alters the invasive potential of malignant melanomas and we demonstrate a network of miR-211-responsive genes affecting melanoma invasive activity.
RESULTS AND DISCUSSION
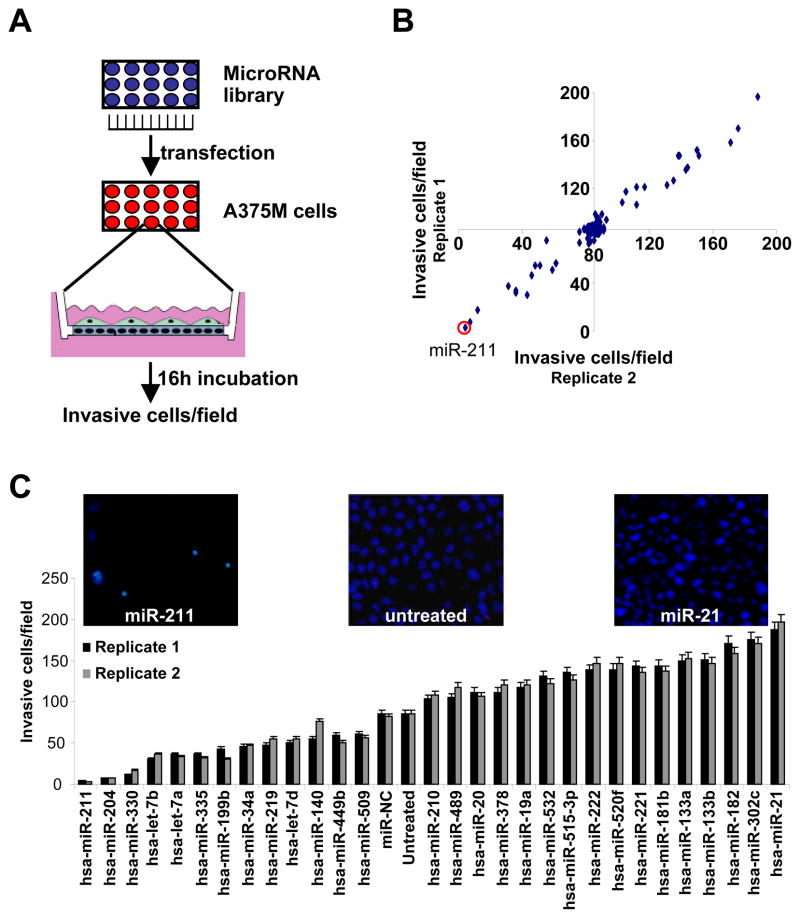
To determine whether miR-211 or other miRNAs can regulate melanoma invasiveness independently from melastatin, we performed an unbiased miRNA library screen in a highly invasive melanoma cell line, A375M (Figure 1A, B and S1). We found that expression of the melanoma-specific miR-211 significantly decreased invasiveness of A375M cells. Interestingly, the second highest hit in this screen was the miR-211 paralog, miR-204, which is not expressed in melanocytes but shares the same seed region as miR-211 and therefore targets similar genes. Each of the miRNAs identified in the primary screen was validated and ranked by invasiveness in a secondary invasive activity screen (Figure 1C). Importantly, increased expression of tumor suppressor miRNAs miR-335 (Tavazoie et al., 2008), let-7a, let-7b, and let-7d (Takamizawa et al., 2004; Yanaihara et al., 2006) led to decreased melanoma invasiveness (Figure 1C). Conversely, increases in the oncogenic miRNAs miR-19a, miR-20 (He et al., 2005; O’Donnell et al., 2005), and miR-21 (Medina et al., 2010) led to increased melanoma invasiveness (Figure 1C).
Figure 1.
A genome-wide miRNA screen identifies reduced miR-211 in melanoma invasive activity. (A) Screening strategy followed to identify miRNAs regulating A375M melanoma cell invasiveness. (B) Number of invasive cells (average of 5 high power fields) after transfection with the miRNA library in A375M cells. The experiment was performed in duplicate (Replicates 1, 2). (C) Number of invasive cells after transfection with positive hits derived from the miRNA library screen experiment. The experiment was performed in duplicate; means ± s.e.m.
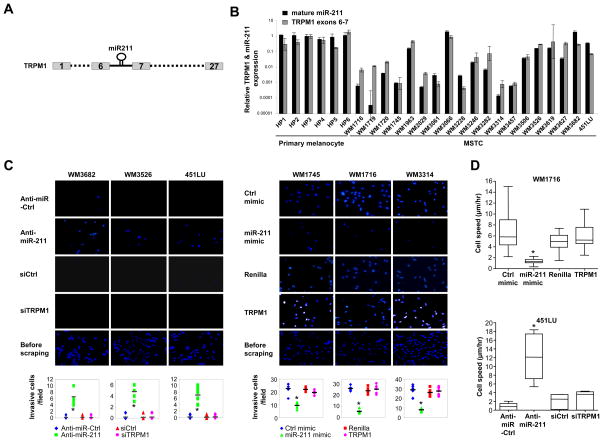
To determine whether the expression levels of miR-211 or its host gene melastatin (Figure 2A) affects invasive activity of human melanomas, we compared mature miR-211 and its completely processed host gene mRNA levels across human melanocytes and malignant melanomas by qRT-PCR (Figure 2B). Consistent with possible tumor suppressor roles in melanomas, both melastatin and miR-211 were reduced in almost all malignant melanomas compared to melanocytes (Figure 2B). Linear regression analysis identified a tight correlation between melastatin mRNA levels and mature miR-211 levels (R2 = 0.73, Figure S2A). Moreover, our data indicate that miR-211 and melastatin share a common promoter (Figure S2B–D) which is consistent with previous observations (Marson et al., 2008; Ozsolak et al., 2008). In addition the expression of both genes is MITF-dependent (Figure S2E–H). Together, these data demonstrate that the levels of melastatin and miR-211 are coordinately increased or decreased in melanomas and indicate coupled transcriptional regulation by MITF.
Figure 2.
Perturbing miR-211 but not melastatin affects melanoma invasion and motility. (A) Schematic presentation of the miR-211 gene in intron 6 of the melastatin (TRPM1) gene. (B) qRT-PCR of TRPM1 (black bars) normalized to actin, and mature miR-211 (gray bars) normalized to RNU48 in multiple human primary melanocytes (HP) and melanoma short term cultures (MSTCs). Y-axis is logarithmic scale. (N=5; means ± s.e.m.). (C) Matrigel assays for WM3682, WM3526, and 451LU (panels at left) transfected with control antagomir (Anti-miR-Ctrl, row 1), a miR-211-specific antagomir (Anti-miR-211, row 2), a control siRNA (siCtrl, row 3), or a melastatin-specific siRNA (siTRPM1, row 4). Membranes prior to scraping non-invasive cells from the top are shown (row 5). Matrigel assays for WM1745, WM1716, or WM3314 (panels at right) transfected with control miRNA mimic (Ctrl mimic, row 1) a miR-211 mimic (miR-211-mimic, row 2), a control Renilla cDNA (Renilla) or TRPM1 cDNA (TRPM1, row 3). The membranes prior to scraping cells from the top of the membrane are shown (row 4). Graphical presentation of the data quantifies the number of invasive cells in 10 high power fields (means ± s.e.m.; *p<0.005). (D) Modulating miR-211 but not melastatin affects melanoma motility. Real-time video microscopy of WM1716 (top panel) expressing Renilla cDNA, melastatin cDNA (TRPM1), a control miRNA mimic (Ctrl mimic), or miR-211 mimic. Real-time video microscopy of 451LU (bottom panel) expressing a control antagomir (Anti-miR-Ctrl), a miR-211 antagomir (Anti-miR-211) a siRNA control (siCtrl), or a melastatin siRNA (siTRPM1). Results quantify cell migration in μm/hr (>25 representative cells analyzed for each category from 2 movies, means ± s.e.m.; *p<0.005). Real-time video images are added as web-browsable links.
Next, we examined invasive activity and proliferation rates as a function of melastatin/miR-211 expression across several representative melanomas with mildly (~10-fold) and greatly (~1,000-fold) reduced miR-211 levels (Figure 2B). More than 20-fold higher invasive activity was observed in melanomas with greatly reduced miR-211 levels (WM1716, WM1745, and WM3314) compared to melanomas with mildly reduced miR-211 (WM3682, WM3526, and 451LU; Figure 2C top panel and Figure S2I). Consistent with reduced melastatin expression in slowly proliferating but highly invasive melanomas (Hoek et al., 2006), the doubling times of melanomas with higher melastatin/miR-211 (1.2 days and 2 days for 451LU and WM3526, respectively) were considerably shorter than the doubling times of melanomas with lower melastatin/miR-211 (more than 8 days for WM1745; Figure S2J).
To distinguish effects of miR-211 from its host gene, melanomas with no reduction (WM3682) or with 10-fold reduction (WM3526 and 451LU) in melastatin and miR-211 relative to human primary melanocytes were transfected with miR-211-specific antagomirs (antisense oligonucleotides that inhibit miRNAs in a sequence specific fashion (Krutzfeldt et al., 2005)) and the effect on melanoma invasiveness was assessed in matrigel assays (Figure 2C and Figure S2K). Inhibition of miR-211 increased melanoma invasiveness by at least 10-fold compared to a control antagomir. Importantly, knockdown of melastatin had no effect on melanoma invasiveness (Figure 2C and Figure S2K). These data indicate that melanoma invasiveness is suppressed by miR-211 but not melastatin.
The effects of increasing the expression of miR-211 were also examined. Melanomas with ~1,000-fold reductions in miR-211 (WM1716, WM1745, and WM3314) relative to human primary melanocytes were transfected with a plasmid expressing a miR-211 precursor (Figure S2N). Increasing miR-211 levels in these melanomas had no effect on growth rates (Figure S2O) but reduced the invasive potential by two- to three-fold compared to scrambled controls (Figure 2C). In contrast, modulation of melastatin expression in these melanomas had no effect on melanoma invasiveness (Figure 2C and Figure S2L-N). Modulating miR-211 had no effect on melastatin expression (Figure S2M). Similarly, modulating melastatin had no effect on miR-211 expression (Figure S2M). Modulating MITF levels, however, had similar affects on melanoma invasion likely due to its affect on the expression of miR-211 (Figure S2G, H, P). We conclude that changes in expression of miR-211 affect melanoma invasiveness independent of melastatin and do not affect melanoma proliferation rates.
To determine the effect of miR-211 expression levels on melanoma cell migration, we performed real-time video microscopy (Figure 2D and Supplementary Movies). WM1716 cells, expressing reduced miR-211 (Figure S2N), exhibited spindle-like morphology when cultured on a deformable collagen-matrigel matrix. When transfected with a plasmid expressing melastatin cDNA or controls (renilla luciferase cDNA or a scrambled miRNA), WM1716 demonstrated an average motility speed of approximately 5 μm/hour and maintained the spindle-like morphology. Strikingly, expression of miR-211 significantly reduced cell migration to only 1.5 μm/hour and induced rounded cell morphology. The effects of reducing miR-211 or melastatin on cell migration were also examined in 451LU cells expressing elevated miR-211. In agreement with the results in WM1716 cells, reduction of miR-211 but not melastatin increased cell motility (Figure 2D). Thus miR-211, but not melastatin, affects melanoma cell motility and invasiveness.
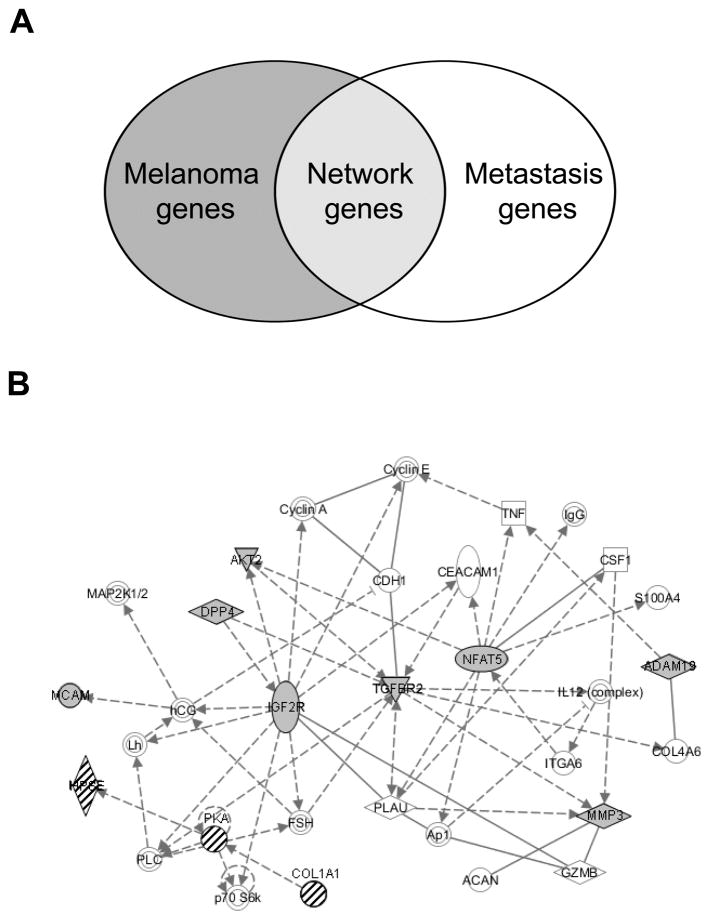
To discover the genetic network mediating the effects of miR-211 on melanoma invasiveness, we interrogated a literature-based set of genes restricted for melanoma expression and filtered for annotated functions in metastasis (Figure 3A and Table S1). A computationally-predicted, melanoma-specific metastasis network was generated using the IPA gene network software (described in Methods). This analysis revealed two statistically-significant (p<10−5) gene networks, with the most statistically-significant network (p=10−37) containing three central nodes (defined in Methods): IGF2R, TGFBR2, and NFAT5 (Figure 3B and Figure S3).
Figure 3.
Gene network analysis identifies three central nodes regulating melanoma metastasis potentially targeted by miR-211. (A) Literature-based melanoma and metastasis gene lists as demonstrated in the Venn diagram were used to construct the melanoma metastasis gene network. (B) Bioinformatically-predicted melanoma metastasis gene network (p=10−37). Solid lines indicate direct interactions and dashed lines indicate indirect interactions. Arrows indicate activation. Lines ending in short perpendicular lines indicate repression. Targetscan-predicted miR-211 targets genes are shaded in grey (IGFR= −0.42, TGFBR2 = −0.49, NFAT= −0.06, DPP4= 0.14, −0.08, AKT2 = −0.01, MMP3= −0.09, ADAM19 = −0.11, −0.08, MCAM= −0.12). Knockdown of each central nodes genes is predicted to inhibit the entire metastasis network except for the genes indicated by hatch marks.
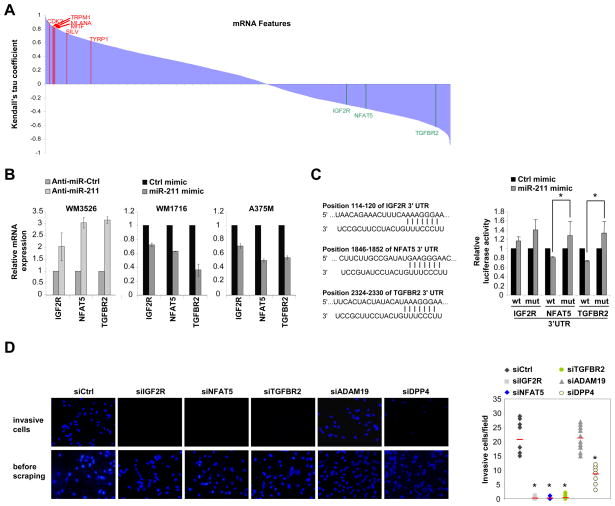
To identify any function of miR-211 in the bioinformatically-predicted melanoma metastasis networks, miR-211 expression data was compared to sample-matched mRNA expression data across a large collection of melanomas (Lin et al., 2008). Translational repression mediated by miRNAs is usually accompanied by reduction of target mRNA levels (Filipowicz et al., 2008). Thus, to a first approximation, potential target mRNAs are expected to demonstrate reciprocal expression relative to miR-211. Kendall’s tau (τ) rank coefficient was calculated for all mRNA/miR-211 pairs to discern concentration-dependent relationships between miR-211 and mRNAs expressed in melanomas (Figure 4A). As an internal measure of the accuracy of this analysis, a strong correlation was observed between the abundance of miR-211 and important target genes of MITF, including TYRP1, SILV, MLANA, and CDK2 (Levy et al., 2006). A similar ranked list of genes correlated with miR-211 expression was obtained using class-dependent identification of marker genes with the ComparativeMarkerSelection module in GenePattern (data not shown). The mRNAs with significant Kendall’s tau coefficients were compared with predicted targets of miR-211 using TargetScan. Possible amplification effects through genomic amplification were eliminated by removing mRNAs that showed increased gene copies in our cell lines based on matching single nucleotide polymorphism data for those genes (Lin et al., 2008) (Table S2). Six genes were found to be represented in both the list of plausible miR-211 targets as well as the independently-generated melanoma metastasis network, including FBXW7, ANGPT1, IGF2R (Targetscan context score = −0.42), NFAT5 (Targetscan context score = −0.06), TGFBR2 (Targetscan context score = −0.49), and VHL. Interestingly, several genes demonstrating inversely correlated expression with miR-211 levels and with lowest–rank coefficients had established roles in melanomagenesis including the central nodes IGF2R, TGFBR2, and NFAT5.
Figure 4.
miR-211 targets multiple genes regulating melanoma metastasis. (A) Statistical association analysis of previously published mRNA profiling data (13,211 transcripts) versus miR-211 expression using Kendall’s tau. mRNA names are sorted by Kendall’s tau coefficient and periodically listed on the horizontal axis. Magnitude of coefficient measures the degree of association, and a negative coefficient denotes an inverse association. Representative melanoma-relevant genes directly correlated with miR-211 are highlighted in red: MLANA (τ = 0.831), MITF (τ = 0.821), CDK2 (τ = 0.776), SILV (τ = 0.738), and TYRP1 (τ = 0.627). Predicted miR-211 target genes are inversely correlated with miR-211 and are highlighted in green: IGF2R (τ = −0.297), NFAT5 (τ = −0.357), and TGFB2R (τ = −0.620). (B) miR-211 represses endogenous IGF2R, NFAT and TGFBR2 expression in melanoma cells. Transfection of a miR-211 antagomir into WM3526 de-represses IGF2R, NFAT5, and TGFBR2 expression compared to a control antagomir (Anti-miR-Ctrl). Transfection of a miR-211 mimic into WM1716 or A375M represses IGF2R, NFAT5, and TGFBR2 expression compared to a control miRNA mimic (Ctrl mimic). N=3; means ± s.e.m. (C) NFAT5 and TGFBR2 but not IGF2R are direct miR-211 target genes. Schematic presentation of predicted miR-211 target sites identified in the IGF2R, NFAT5 or TGFBR2. Numbers indicate positions of miR-211 binding sites on target mRNA 3′UTRs. HeLa cells were transfected with luciferase constructs possessing wildtype (wt) or miR-211 binding site mutant (mut) 3′UTRs of IGF2R, NFAT5 or TGFBR2 with a miR-211 mimic or a control miRNA mimic (ctrl Mimic). N=5; means ± s.e.m. (D) Knockdown of IGF2R, NFAT5, or TGFBR2 phenocopies the effect of increasing miR-211 on melanoma invasive activity. WM1716 cells were transfected with a control siRNA (siCtrl), or siRNAs against indicated genes and subjected to matrigel assays. Invasive cells were counted (top panels and are quantitated (graphs, right). The bottom panels demonstrate the membranes prior to scraping (N=3; means ± s.e.m.).
To determine whether these central node genes are biological targets of miR-211, the levels of miR-211 were perturbed in melanomas and IGF2R, TGFBR2, and NFAT5 expression levels were examined (Figure 4B and Figure S4A, B). Reducing miR-211 in WM3526 cells increased the expression of all three mRNAs. Conversely, increasing miR-211 in WM1716 and in A375M cells reduced levels of all three mRNAs, indicating that endogenous IGF2R, TGFBR2, and NFAT5 are biological targets of miR-211 in melanomas. To determine whether these genes are direct targets of miR-211, we transfected luciferase-expressing constructs containing the wildtype and predicted miR-211 binding site mutant 3′ untranslated regions (3′ UTRs) of these genes into HeLa cells (which lack endogenous miR-211) with or without a miR-211 mimic. Unexpectedly, TGFBR2 and NFAT5, but not IGF2R, were shown to be direct targets of miR-211 (Figure 4C).
Because miR-211 affects endogenous expression of the central node genes, we tested the effect of knockdown of these genes on melanoma invasive activity. Interestingly, knockdown of any central node gene phenocopied the effect of increasing miR-211 levels in melanomas with high levels of IGF2R, TGFBR2, and NFAT5 (Figure 4D). Consistent with a lack of direct interaction between these genes as predicted bioinformatically (Figure 3B), knockdown of each central node gene did not affect the expression of the other central node genes (Figure S4C). However, we noted that one direct input to IGF2R is DPP4, which has two predicted miR-211 binding sites (Targetscan context score = 0.14 and −0.08). Knockdown of the peripheral node gene DPP4, but not ADAM19 (another predicted miR-211 target gene), phenocopied the effect of increased miR-211 on melanoma invasiveness (Figure 4D and Figure S4D, E). Interestingly, knockdown of DPP4 also led to reduced expression of IGF2R, TGFBR2 and NFAT5 (Figure S4D). Together, these data demonstrate the high-predictive index of the melanoma metastasis network and provides a mechanistic basis for reduced IGF2R expression upon increased miR-211 expression. Taken together, our data indicates that increasing miR-211 directly represses NFAT5 and TGFBR2, and indirectly represses IGF2R, leading to the reduction of the invasive potential of malignant melanomas.
Here we report an intronic miRNA that assumes a tumor suppressive function previously ascribed to its host gene. Modulation of miR-211 levels, but not melastatin levels, is sufficient to convert a non-invasive melanoma to invasive melanoma and vice versa. Previous observations comparing primary melanomas identified reduced expression of melastatin in distant metastasis (Duncan et al., 2001; Duncan et al., 1998) and an inverse correlation between melastatin expression and tumor thickness (Deeds et al., 2000). Our data raise the possibility, however, that these observations may have been due to altered expression levels of miR-211. Because miRNA genes are frequently hosted by protein coding genes, phenotypes attributed to genetic deletion of protein-coding genes may actually be attributable to abrogated expression of the hosted miRNAs (Moffett and Novina, 2007).
TGFB has been implicated in the metastatic spread of melanoma and other cancers (Giampieri et al., 2009; Pinner et al., 2009; Van Belle et al., 1996). TGFB signaling has also been shown to promote hypopigmentation (Martinez-Esparza et al., 1997; Pinner et al., 2009), downregulation of MITF (which controls the melastatin/miR-211 locus (Miller et al., 2004)), disruption of cell-cell adhesion, and single cell invasion to escape the primary tumor and invasion of blood vessels (Giampieri et al., 2009). Therefore, increased TGFB signaling through increased TGFBR2 expression might partially explain increased melanoma invasion and motility when miR-211 levels are low. To our knowledge, NFAT5 and IGF2R have not been previously linked to melanoma metastasis. Several genes represented in the melanoma metastasis network are predicted targets of miR-211 and demonstrate reduced expression upon increased miR-211 (Figure 4 and data not shown), a phenomenon which will be valuable to examine using melanoma tissue microarrays.
Complex phenotypes such as changes in cell morphology, motility, and invasiveness are rarely regulated by a single gene. Because a single miRNA may target thousands of genes, it is possible that altered expression of a single miRNA can regulate complex phenotypes. It has been suggested that therapeutic manipulation of a single miRNA may result in the regulation of many genes and may effect the “normalization” of a diseased phenotype (Wurdinger and Costa, 2007). Our data suggest that modulating miR-211 or its target gene levels may pave the way towards new therapies for metastatic melanoma.
EXPERIMENTAL PROCEDURES
Cell culture
HeLa cells and A375M cells were cultured in DMEM medium supplemented with 10% FBS and 1% penicillin-streptomycin.
Melanoma short term cultures (MSTCs) were selected from cryopreserved collections at the Wistar Institute (62 lines), the University Hospital of Zurich (24 lines), and the Dana-Farber Cancer Institute (DFCI, 37 lines). All MSTC were cultured in RPMI medium (MediaTech) supplemented with 10% fetal bovine serum (MediaTech) and 1% Penicillin-Streptomycin-Glutamine (Invitrogen), except for DFCI MSTCs, which were cultivated in DMEM (MediaTech) supplemented with 10% serum. For some Wistar lines, to enhance cell adherence, tissue culture dishes were first coated with 1% porcine gel solution (Sigma). Primary human melanocytes were isolated and grown from neotal foreskins as described (McGill et al., 2002). Primary melanocytes between passages 2 and 5 were stimulated with 20 uM forskolin (Sigma Aldrich) for indicated times after an overnight starvation in F10 medium supplemented only with penicillin/streptomycin/glutamine. For microarrays primary melanocyte were stimulated for 16 hours with forskolin. 501mel human melanoma cells (gift from Dr. Ruth Halaban, Yale University) maintained in Ham’s F-10 media (Mediatech) supplemented with 10% fetal bovine serum (FBS, Sigma Aldrich) and 1% Penicillin-Streptomycin-Glutamine (PSQ). UACC62 human melanoma cell line was obtained from NCI and maintained in RPMI-1640 media (Mediatech) supplemented with 10% fetal bovine serum (FBS) (Sigma Aldrich) and 1% Penicillin-Streptomycin-Glutamine (PSQ).
Genome-wide miRNA screen
A miRNA library of 300 precursor miRNAs (Ambion Inc) was transfected in triplicate (100 nM) using siPORT NeoFX into A375M cells, seeded in 24-well plates. SiPORT NeoFX is a lipid transfection agent consisting of a mixture of lipids that spontaneously complex small interfering RNAs and facilitates its transfer to the cells. After 24h, cells were serum-starved over-night. 1×105 cells were added to invasion chambers coated with Matrigel (BD Biosciences), cells were allowed to invade for 16 hours towards media containing 10% FBS. Cells remaining on the top side of the membrane were removed using a cotton swab and invading cells were counted in five fields per insert. The library experiment was performed in duplicate. No cell toxicity from the transfection agent was detected. Transfection conditions were optimized with a miRNA negative control (AM17110, Ambion Inc) and miR-182 as a positive control. For validation, the 29 precursor miRNAs identified from the primary screen were transfected into A375M cells, seeded in 24-well plates and the invasion assay was repeated. Each miRNA was transfected in triplicate.
RNA purification and qRT-PCR
Total RNA was purified using Trizol reagent (Invitrogen) according to the manufacturer’s instructions, followed by treatment with RNase-free DNase (Qiagen). Dried RNA pellets were re-suspended in appropriate volumes of DEPC/ddH2O. RNA was quantified by measuring OD260/280. For qRT-PCR analysis of mature miR-211 or miR-107, 10 ng total RNA was used in a TaqMan microRNA assay according to manufacturer’s protocols (Applied Biosystems). Mature miR-211 expression was normalized to expression of RNU48, also detected by TaqMan assay (Applied Biosystems). For qRT-PCR analysis of TRPM1, NFAT5, IGF2R, TGFBR2 and other genes, RNA was subjected to one-step qRT-PCR using a QuantiTect RT-PCR kit (Qiagen) and iQ SYBRgreen Supermix (Biorad). Primer sequences and manufacturers are listed in Table S3. All experiments are n 3. Standard error of the mean (s.e.m.) is presented.
Oligonucleotide transfection
MiRNA mimics, antagomirs or siRNAs were transfected into MSTCs using HiPerFect according to manufacturer’s protocols (Qiagen). The sequences and manufacturers of the miR-211 mimic, miR-211 antagomir, control miRNA mimic, control antagomir, TRPM1, NFAT5, IGF2R, TGFBR2, DPP4 and ADAM19 siRNA, and control siRNA are listed in Table S4. Cells were transfected twice with 100 pmol of oligonucleotide per well (0.5 × 106 cells) at 24 hour intervals. Transfected MSTCs were assayed 48 hours after the second transfection.
Invasion and migration assays
MSTCs were transfected with oligonucleotides as described above. Forty-eight hours after the second transfection, cells were serum-starved over-night. Fifty thousand cells were added in duplicate to invasion chambers coated with Matrigel (BD Biosciences). Cells were allowed to invade for 9 hours towards media containing 10% FBS. Cells remaining on the top side of the membrane were removed using a cotton swab and invading cells were fixed and stained with 4′-6-Diamidino-2-phenylindole (DAPI, Vector Laboratories Inc.). In all assays, ten fields per insert were photographed and scored s.e.m. was measured.
Cell growth and metabolic rate assay
Cells (104/well) were plated in multiple 24 well plates. The following day was considered day 0. For analysis of cell growth/number, cells were fixed using 10% ethanol/10% acetic acid, and stained with 0.2% crystal violet fixative. Following washes with water, the plates were dried. Each 24 well plate was quantified by crystal violet staining. The crystal violet dye was redissolved in fixation solution. One hundred μl of the re-dissolved dye were used for color measurement, in duplicate. Color intensity was measured at OD595 in a 96-well plate reader, and normalized to day 0 as 100%. For metabolic rate test, 5×104 cell were cultured at 96 well, 48h after siRNA, miRNA or control nucleic acid transfection, cells were subjected to cell proliferation assay. The Cell Proliferation Reagent WST-1 (Roche) was added and cell proliferation was measured according to manufacture instructions (Roche). All experiments are n 3 and s.e.m. is presented.
Microscopy real-time cell mobility
A ZeissLSM510META confocal microscope was used. Analysis of cell migration speed was performed as described (Pinner et al., 2009) by tracking phase-contrast movies of primary melanoma cell lines plated on collagen-matrigel gels (Hooper et al., 2006) spread on 24-well glass-bottomed plates (MatTek corporation). Multi-position imaging was conducted over 10 hrs. and images were acquired at 10 min. intervals. Mobility of Cherry/GFP positive cells (>25 cells) was measured from each movie. Tracks were calculated using ImageJ and Excel software.
Gene Network Analysis
Gene networks were constructed and identified important hubs using Ingenuity Gene Network Analysis. Specifically, we created a melanoma-metastasis gene list including common genes derived from a literature-based melanoma gene list and a literature-based metastasis gene list. These literature-based gene lists were created using IPA software analysis (Ingenuity Inc). Using this melanoma metastasis gene list we performed gene network analysis. Pathways of highly interconnected genes were identified by statistical likelihood using the following equation:
Where N is the number of genes in the network of which G are central node genes, for a pathway of s genes of which f are central node genes. C (n, k) is the binomial coefficient. Central nodes are considered genes with very high flux capacity (total number of incoming and outgoing links higher than 9). The flux capacity score for TGFBR2 is 10, for NFAT5 is 9, and for IGF2R is 12. The nodes with flux capacity score lower than 9 were considered peripheral nodes. We considered statistically significant gene networks those with a p value <10−10. Grey color indicates genes included in the melanoma-metastasis gene list. Solid lines indicate direct gene interactions while dashed lines indicate indirect gene interactions.
Hierarchical clustering of genomic profiles
An input matrix was generated by calculating the base-2 logorithm of normalized mRNA expression profiles. The input matrix was visualized in TIGR MeV v4.1 (http://www.tm4.org/mev.html). Both the gene tree and the sample tree were generated using Euclidean distance and average linkage.
Unsupervised rank correlation of microarrays
To identify concentration-dependent relationships involving miR-211, rigid non-parametric association statistical analyses were performed. Computations were done using scripts written in Python (http://www.python.org) and the SciPy module (http://www.scipy.org/). A matrix was generated with each column of the input representing mRNA expression values from a single culture and each row of the input representing expression values for each gene. A copy of the miR-211 expression array was then paired with each row of the input matrix, and the association between each pair was evaluated by calculating the Kendall’s tau (|) rank coefficient.
Class-dependent identification of marker genes
MSTCs were separated into three classes based on miR-211 expression level: high, medium, and low. Only the “high” and “low” classes are used in the actual calculation. For each gene, the degree of difference in mRNA expression between the “high” and “low” classes were measured using pair-wise, two-sided t-test and median values in each class. The calculations were done with 1000 iterations and smooth p-values through the ComparativeMarkerSelection module in the GenePattern software suite (http://genepattern.broad.mit.edu/; version 3.1.1). Genes were ranked by t-test score. A negative score denotes an inverse relationship between a gene’s mRNA expression and miR-211 expression, whereas a positive score denotes a direct relationship.
Combination analysis of correlation with miRNA and target prediction
Computationally-predicted mRNA targets for Homo sapiens miR-211 (hsa-miR-211) were identified using a database from Memorial Sloan-Kettering Cancer Center (http://www.microrna.org) as the first input for this analysis. The second input comprised a list of genes whose differential mRNA expressions had strong negative correlation with miR-211 expression from unsupervised rank correlation analysis. The two inputs were combined to identify overlaps.
Gel Electrophoresis and Immunoblotting
MSTCs were cultured in 6-well dishes and manipulated as indicated in the figure legend. Subsequently, the cells were lysed in buffer containing 50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 10 μM leupeptin, 1 mM AEBSF, 100 units/ml aprotinin, 10 mM NaF, and 1 mM μg) were resolved by 7% SDS-PAGE, transferred to nitrocellulose Na3VO4. Samples (30 membranes, and then exposed to the appropriate antibodies: anti-MITF (C5), NFAT5 (Abcam), TGFBR2 (Cell Signaling Technologies), anti-tubulin (Sigma Aldrich). Proteins were visualized with the ECL system from Perkin Elmer Life Sciences using horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibody.
The band intensity were quantified usind ImageJ (NIH).
Plasmid construction and stable cell lines establishment
PLKO.1 vector was modified by replacing the puromycin resistance casset with a casset coding for luciferase, mCherry and puromycin using BamHI and KpnI restriction sites. This vector was used for miRNA overexpression. For cDNA overexpression, the U6 promoter of the vector described above was replaced by a HPGK promoter using SanDI and AgeI.
For establishement of stable cell lines, MSTC’s were seeded in 6 well dish and tansfected the modified PLKO.1 vectors using FugeneHD (roche). After 2 days, the cells were grown with 1μg/ml of puromicyn. After 4 to 6 weeks, the cells were used for experiments.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Deeds J, Cronin F, Duncan LM. Patterns of melastatin mRNA expression in melanocytic tumors. Hum Pathol. 2000;31:1346–1356. [PubMed] [Google Scholar]
- Duncan LM, Deeds J, Cronin FE, Donovan M, Sober AJ, Kauffman M, McCarthy JJ. Melastatin expression and prognosis in cutaneous malignant melanoma. J Clin Oncol. 2001;19:568–576. doi: 10.1200/JCO.2001.19.2.568. [DOI] [PubMed] [Google Scholar]
- Duncan LM, Deeds J, Hunter J, Shao J, Holmgren LM, Woolf EA, Tepper RI, Shyjan AW. Down-regulation of the novel gene melastatin correlates with potential for melanoma metastasis. Cancer Res. 1998;58:1515–1520. [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol. 2009;11:1287–1296. doi: 10.1038/ncb1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, Weber BL, Nathanson KL, Phillips DJ, Herlyn M, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- Hooper S, Marshall JF, Sahai E. Tumor cell migration in three dimensions. Methods Enzymol. 2006;406:625–643. doi: 10.1016/S0076-6879(06)06049-6. [DOI] [PubMed] [Google Scholar]
- Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- Hunter JJ, Shao J, Smutko JS, Dussault BJ, Nagle DL, Woolf EA, Holmgren LM, Moore KJ, Shyjan AW. Chromosomal localization and genomic characterization of the mouse melastatin gene (Mlsn1) Genomics. 1998;54:116–123. doi: 10.1006/geno.1998.5549. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, Laine E, Greulich H, Tseng H, Gates C, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Esparza M, Jimenez-Cervantes C, Beermann F, Aparicio P, Lozano JA, Garcia-Borron JC. Transforming growth factor-beta1 inhibits basal melanogenesis in B16/F10 mouse melanoma cells by increasing the rate of degradation of tyrosinase and tyrosinase-related protein-1. J Biol Chem. 1997;272:3967–3972. doi: 10.1074/jbc.272.7.3967. [DOI] [PubMed] [Google Scholar]
- McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK, Lin YL, Ramaswamy S, Avery W, Ding HF, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- Miller AJ, Du J, Rowan S, Hershey CL, Widlund HR, Fisher DE. Transcriptional regulation of the melanoma prognostic marker melastatin (TRPM1) by MITF in melanocytes and melanoma. Cancer Res. 2004;64:509–516. doi: 10.1158/0008-5472.can-03-2440. [DOI] [PubMed] [Google Scholar]
- Moffett HF, Novina CD. A small RNA makes a Bic difference. Genome Biol. 2007;8:221. doi: 10.1186/gb-2007-8-7-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinner S, Jordan P, Sharrock K, Bazley L, Collinson L, Marais R, Bonvin E, Goding C, Sahai E. Intravital imaging reveals transient changes in pigment production and Brn2 expression during metastatic melanoma dissemination. Cancer Res. 2009;69:7969–7977. doi: 10.1158/0008-5472.CAN-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belle P, Rodeck U, Nuamah I, Halpern AC, Elder DE. Melanoma-associated expression of transforming growth factor-beta isoforms. Am J Pathol. 1996;148:1887–1894. [PMC free article] [PubMed] [Google Scholar]
- Wurdinger T, Costa FF. Molecular therapy in the microRNA era. Pharmacogenomics J. 2007;7:297–304. doi: 10.1038/sj.tpj.6500429. [DOI] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.