Abstract
Autism Speaks’ Autism Genetic Resource Exchange (AGRE) represents the largest private collection of genetic and phenotype data for families with ASD that is made available to qualified researchers worldwide. The availability of large and comprehensive registries that include detailed phenotype and genetic information for individuals affected with an ASD and family members is crucial for the discovery of autism susceptibility genes and the development and application of biologically-based approaches to diagnosis and treatment. The model that AGRE has developed can be applied broadly to other disorders with complex etiologies
Introduction
Autism Spectrum Disorders (ASD) are a group of highly prevalent, neurodevelopmental disorders affecting social, communicative, and behavioral functioning, which pose a substantial public health burden. With an estimated U.S. prevalence of 1 in 110, care for individuals with ASD costs the U.S. approximately $35 billion annually (Ganz, 2007). Although it is well-established that ASD is heritable, the field is only now beginning to identify specific ASD susceptibility genes. The goal is for gene discovery to lead to improved methods for early risk detection and a deeper understanding of the biological mechanisms underlying ASD so that targeted therapeutics can be developed.
The availability of large and comprehensive registries that include detailed phenotype and genetic information for individuals affected with an ASD and family members is crucial for the discovery of autism susceptibility genes and the development and application of biologically-based approaches to diagnosis and treatment of individuals with ASD. Complex neuropsychiatric disorders such as autism are best approached by collaborative research efforts that pool large samples of affected individuals.. Autism Speaks’ Autism Genetic Resource Exchange (AGRE) represents the largest private collection of genetic and phenotype data for families with ASD that is made available to qualified researchers worldwide. The creation of large public resources allows both junior and senior scientists with ambition and ingenuity to accelerate progress. Advocacy organizations such as Autism Speaks have played a crucial role in advancing science by influencing policymakers to recognize the unique challenges faced by their constituents and sharing their sense of urgency and purpose to ensure that policies do not stand in the way of research and treatment.
Build it and they will come
The AGRE program was founded in 1997 by parents, scientists, and clinicians who felt that in order to facilitate more rapid progress in the identification of the genetic underpinnings of ASD, they needed to collect critical phenotypic and genetic information from families with autism and make these data readily available to the scientific community. Jon Shestack and Portia Iversen, founders of the autism advocacy group Cure Autism Now (CAN), emphasized the urgent need for scientific progress and urged parents and scientists alike to be proactive and make it happen. A year earlier, in May 1996, the parents held a think tank where they asked scientists to tell them what they could do to speed progress in autism research. Their answer was to establish a DNA resource that would make biological samples from well-characterized families available to the scientific community at large. This notion of collaboration and data sharing was what piqued the interest of UCLA clinician and researcher, Daniel Geschwind, MD, PhD, scientific founder of the program, who felt that the way to attract researchers to the field was to give them the tools they needed. In return, scientists would give back by returning their research results to be shared with the research community. It was with this vision in mind that the foundation enlisted the help of key scientists and clinicians who also bought into the notion that science could be hurried to develop a resource that would serve that purpose. Although a daunting proposition for parents who knew little about science and researchers who were skeptical about broad data sharing, the parents knew that if they built it and they built it right, researchers would come.
AGRE is the realization of a number of ideals in scientific research. Standardization, collaboration, and data sharing stand out as three key guiding principles that have marked the success of the AGRE program. It is focused on the rapid accumulation of high-quality data from a large number of families, which is critical for molecular genetic research of complex diseases. Both the data and the biomaterials are made available to any and all qualified researchers around the world as soon as the data are processed. The infrastructure for the sharing of data and biomaterials has been carefully developed and has proven to be highly efficient. The program has maintained superb rapport with the families, who appreciate this effort and who then enlist the participation of fellow families, and gladly volunteer their time to re-engage in supplemental phenotyping efforts. The decision-making process for balancing cost with throughput, balancing depth of assessment with sample size, prioritizing the work plan for fine-mapping, resequencing, and refining phenotypic characterization is apolitical and guided by a panel of scientists who welcome input from their peers and who have exhibited long-standing investment to identify the causes of ASD.
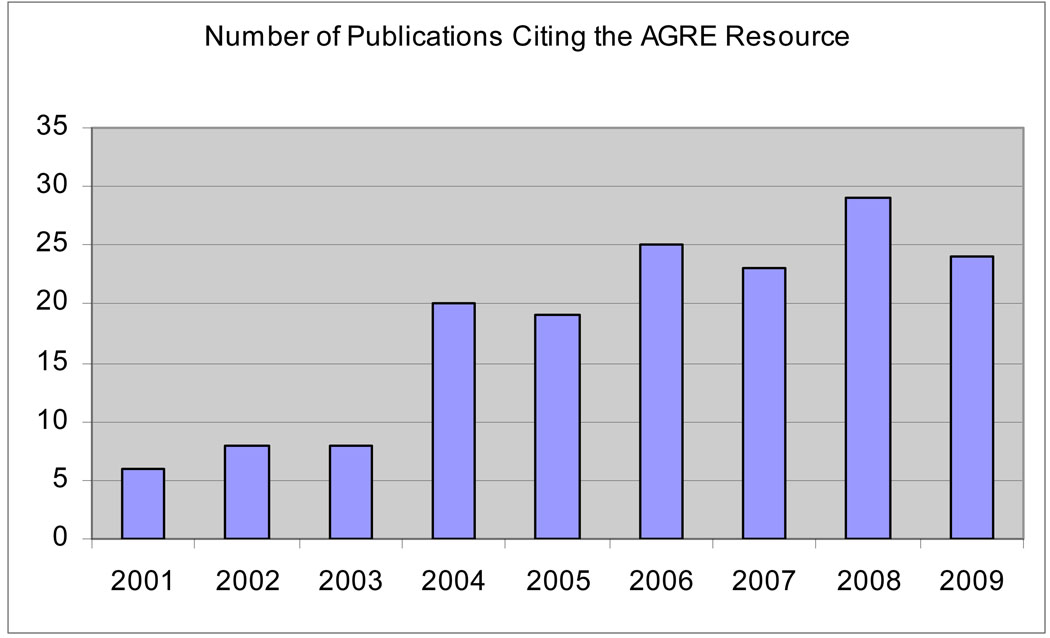
To date, there are 335 active researchers from 21 countries who have used the AGRE resource to publish 169 scientific papers, making it the most widely accessed resource for genetic studies of any mental disorder. Figure 1 illustrates the increase in the number of peer-reviewed publications that have resulted from analyses of AGRE data since the first paper in 2001. The emphasis on collaboration and data sharing created a paradigm shift among scientists who weren’t always motivated to share their data or their findings and influenced larger collaborative programs such as Autism Speaks’ Autism Genome Project (AGP), an Autism Speaks-support international consortium of over 120 scientists from 50 institutions worldwide that pools resources for their genetic analyses. Over the last 2 years, the AGP and its members have been involved in the majority of scientific discoveries in autism genetics. As one of many contributors to the AGP, AGRE continues to sit on the cutting edge of research.
Figure 1.
Increase in number of publications generated by the AGRE resource.
Sample Characteristics
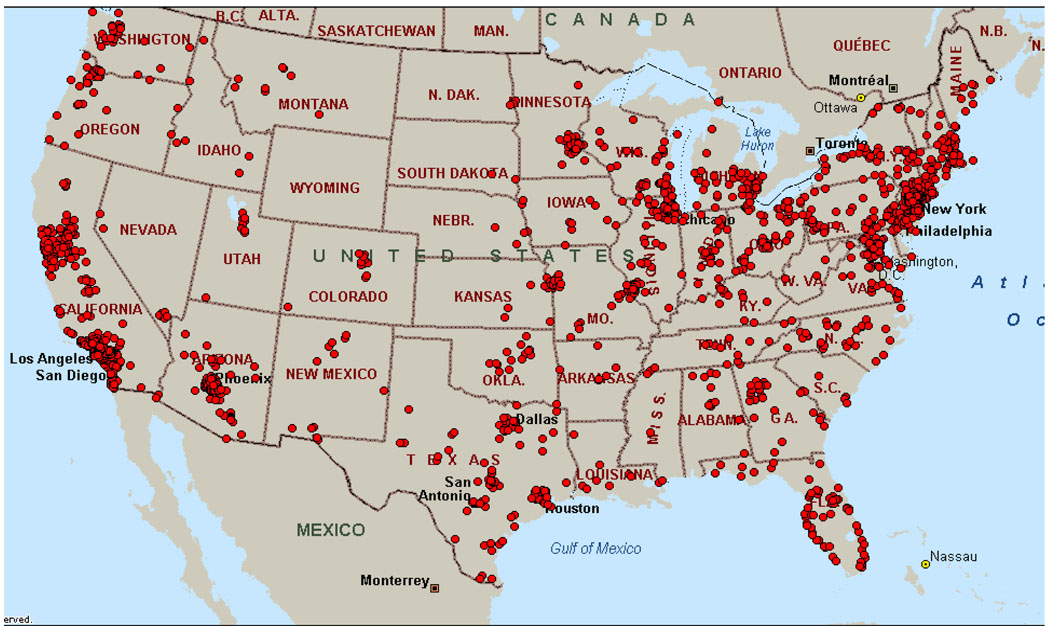
The success of AGRE lies in the unrivaled community support provided by advocacy groups such as CAN and now Autism Speaks. Families understand that the solution to complex disorders such as autism lies in the partnership between parents and researchers and through the AGRE program families are able to become part of the solution. Figure 2 illustrates the nationwide reach of the AGRE program. The majority of the participating families are recruited from areas where Autism Speaks has strong chapter representation reflecting the power of community support. Information about AGRE is provided to families during community events, through the Autism Speaks website, and through Autism Speaks’ national walk program. With the advent of social networking, AGRE has been able to take advantage of sites such as Facebook, Twitter, and Ning to get the word out about the program and alert families about opportunities for research participation. This movement towards web-based recruitment has significantly improved AGRE’s ability to reach out to families more broadly.
Figure 2.
Nationwide distribution of AGRE families
Despite the geographic distribution of AGRE families, the sample is not population-based. Given that AGRE was conceived as a neurogenetic resource, there is a sampling bias towards the recruitment of families with more than one individual affected with an ASD (multiplex) since these families would have an increased genetic load. .In addition to the statistical benefits of family-based cohorts, several studies have reported differences between multiplex and simplex families in rates of copy number variations and autism related traits in otherwise unaffected family members suggesting that the mechanisms of genetic transmission could have a differential effect on outcome (Constantino et al, 2010; Sebat et al, 2007; Campbell et al., 2006). The uniqueness of this collection makes it a very sought after cohort for family-based studies and other novel approaches for gene discovery.
Participants are recruited based on autism diagnosis and all individuals are included as long as they meet diagnostic criteria and have an English-speaking parent. Recruitment priority is given to families with two or more immediate family members affected with an ASD. Because it is not known which types of families will become valuable as new genetic research technologies unfold, AGRE’s philosophy is to include a broad range of ASD’s, including autistic disorder, pervasive developmental disorder not otherwise specified (PDD-NOS), and Asperger's disorder.
A SWAT Team Approach
The phenotypic characterization of individuals with ASD is incredibly important given the behavioral and genetic complexity of the disorder. In order to recruit the large sample sizes required to identify rare and common genetic variants, some researchers have challenged the notion of “deep phenotyping” in favor of faster, more-scalable mechanisms for data collection including those that are internet-based (Lee et al., 2010). While these “lighter” phenotyping efforts may ultimately prove successful for some studies, the strategy taken at AGRE to meet the emerging needs of science is to consider that the breadth of the data made available to researchers is as important as the sample size. The availability of a more comprehensive dataset will ultimately allow researchers to develop phenotype selection algorithms that will help stratify the sample by symptom clusters or subtypes.. The clinical assessment battery is extremely comprehensive and includes an ever expanding library of diagnostic and cognitive measures (www.agre.org) as well as questionnaires about environmental exposures, medical issues, social communication, sleep habits, and quantitative information about parents and unaffected siblings. Using an expanded phenotype battery, AGRE hopes to meet the emerging needs of science by offering more information on unaffected family members (parents and siblings) to better understand the broader autism phenotype. AGRE’s responsibility is to provide the highest quality data to the research community and by taking family recruitment and data collection out of the hands of the researchers, the onus is on AGRE to balance cost with efficiency and maintain the highest levels of quality control.
The AGRE steering committee felt strongly that the best way to maximize family participation and maintain the integrity of the data was to perform all the clinical evaluations and blood draws in the home. This SWAT team approach requires clinical evaluation teams to target families in regional clusters around the country. Funds from Autism Speaks and the NIMH support a phlebotomist and a full-time clinical staff that includes four psychometricians who are all trained and research-reliable on the state of the art autism diagnostic and cognitive evaluations and a full time clinical psychologist who ensures the quality of these evaluations. These staff members are highly sensitive and compassionate about the needs and burdens experienced by families with children with autism. The goal of these evaluations is to confirm the diagnosis of ASD and provide the data to researchers in real-time. Interestingly, less than 7% of individuals with a clinician-reported diagnosis of autism are found to be elsewhere on the autism spectrum (e.g. Pervasive Developmental Disorder-Not Otherwise Specified) whereas 76% of individuals with a reported diagnosis of PDD-NOS are found to meet criteria for full autism. Similarly, 82% of individuals with reported diagnoses of Aspergers also meet full criteria for autism. These latter findings likely reflect trends among community providers to under diagnose autism because of a lack of knowledge about the disorder or an avoidance of the “autism” label.
Accelerating the Pace of Autism Research
Since its inception, the program has enrolled 2880 families between the ages of 2 and 51. There are currently 1308 families with clinical data and banked biospecimens that are available for research user. The median age at time of testing is 7.2 years old and the ratio of boys to girls is 3.8:1, which is consistent with other genetic studies. Prior to 2007, physicians performed medical and neurological evaluations in the home but given the high volume of families, AGRE captures medical and developmental history data using OSCR, AGRE’s online system for clinical research. AGRE has performed fragile-X testing on 99% of the sample and 94% has been genotyped through a partnership with Dr. Daniel Geschwind’s NIH-funded ACE network, which relies on AGRE for access to biological samples and phenotype data for their investigations. In 2010, AGRE reached a new milestone with the collection of its 10,000th DNA sample. Over the last 13 years, cell lines and DNA have been established for 10,236 individuals in this collection and as a result, AGRE has become the most productive study site for the NIMH Genetics Initiative.
The Value of Public-Private Partnerships
Public-private partnerships offer a unique opportunity for advocacy organizations to advance the shared goals of their stakeholders and to leverage the knowledge, skills and resources offered by patient groups, industry, academia, and the federal government to advance science and facilitate translational research. AGRE has a long history of collaboration with the National Institutes of Health (NIH) and other government agencies that recognized the importance of maintaining AGRE as a viable resource for scientific discovery. Support for AGRE is provided in large part by Autism Speaks, who along with its predecessor, CAN, has invested over $15 million dollars through private donations to establish, support, and maintain the resource. These funds are currently supplemented by seven federally-funded grants that have allowed AGRE to play a major role in key autism research efforts. Over the last 7 years, NIH, specifically the NIMH, has invested close to $10 million dollars to increase the depth and breadth of the AGRE collection, including support for the phenotype data collection, biospecimen collection and the bioinformatic infrastructure that researchers use to access clinical and genetic data. In 2007, the NIH made an investment of close to $8M to establish the Center for Genomic and Phenomic Studies of Autism at the University of Southern California, a virtual center designed to promote collaborative autism genetic research. AGRE is the data collection engine for the Center and over the course of 5 years, will be doubling the size of the resource. The Center also supports proof of concept pilot studies on air pollution, environmental exposure, and craniofacial dysmorphology and through a partnership with colleagues at the MIND Institute at the University of California, Davis, neuroimaging and studies of immune function. The NIH’s investment represents $14M of leveraged funding from Autism Speaks and its predecessors over the last 7 years. These long-term partnerships have fostered substantially the pace and quality of ASD research. They have also optimally leveraged the investments made by NIH and Autism Speaks in infrastructure and research and continue to inspire new and more sophisticated means for data analysis. This goal supports the joint mission to accelerate the progress of autism research.
The Importance of Bioinformatics
Over the years, researchers with a willingness to share data have identified significant barriers that have made data sharing difficult and cumbersome. First, access to many of the large-scale resources is limited to a select number of qualified researchers and each of these resources has its own data sharing policies. More clearly defined policies for data and information sharing, such as requiring that funded researchers provide public access to publications through PubMed, will help ensure that researchers make their findings available and also have access to other data that they can use as replication samples or for meta-analyses. In 2008, Autism Speaks pioneered an open-access policy that requires its grantees to make papers resulting from that funding freely accessible in a public database no later than 12 months after publication.
Second, researchers find it difficult to make cross-study comparisons given that few studies adhere to an agreed upon set of common behavioral phenotype measures across a variety of cognitive and behavioral domains. Unfortunately, a lack of uniformity across studies makes it difficult to merge datasets, ensure the comparability of measures and interpret findings across studies and research groups. Therefore, efforts to identify common core phenotype measures and standards for data collection could serve as guidelines for autism researchers worldwide.
The management, availability and use of data are also of critical concern. Most researchers understand that the management of the information is just as critical to success as the data itself. Data systems that are flexible and sustainable are critical tools that researchers need to manage the complexity of the information. As the field expands, the informatics plan has to be able to grow in size and scope to ultimately serve as a knowledge sphere to pull in high quality information from other external databases to help identify research gaps in the autism field and point to areas and connections from which the field has benefited.
Because of the large amount of information that is being generated daily and because some sources of readily-accessible information are not credible (e.g., some internet sites), mechanisms that serve as clearinghouses of information for key stakeholders (families, clinicians, researchers) are becoming increasingly important. Advances in gene-chip technology, proteomics, neuroimaging, and metabolomics require more sophisticated integrated databases that can support the vast scientific needs of the research community. Due to the high costs of creating and sustaining large-scale data bases, leveraging internal and external resources through public-private partnerships is essential. Autism Speaks recently received funding from the NIH through the American Reinvestment and Recovery Act to expand the bioinformatic infrastructure of AGRE so that it can serve as a data hub for the NIH National Database for Autism Research (NDAR). This project will facilitate collaboration and data sharing on an even broader scale by making AGRE’s clinical phenotype data, biological data, and existing genetic information available to an even broader group of researchers through the NDAR portal. This will greatly enhance, in both quantity and scope, the knowledge base available to NDAR users, allowing them to advance their analyses and investigations of the underlying causes of autism.
Impact on the Field
Researchers have used the AGRE resource to identify some of the most important findings in autism research. In 2008, a collaborative of researchers (Weiss et al., 2008) identified several families in the AGRE sample set with copy number variations at 16p11.2, implicating this region as important in the development of autism in these families and sparking great interest in copy number variations and de novo mutations in autism. In another study, serum samples collected by AGRE were instrumental in the finding of stereotypic behavior and hyperactivity in rhesus monkeys exposed to immunoglobulins from mothers of children with autism (Martin et al, 2008). More recently, AGRE has been cited in two Nature papers as part of the AGP that identified new autism susceptibility genes including SHANK2, SYNGAP1, DLGAP2 and the X-linked DDX53–PTCHD1 locus. Some of these genes belong to synapse-related pathways, while others are involved in cellular proliferation, projection and motility, and intracellular signaling, functional targets that may lead to the development of new treatment approaches.
Thus, the pioneering vision of a small group of parents and scientists around a kitchen table in 1996 has revolutionized the way people conduct autism science in the 21st century. The influence of advocacy groups has forced a cultural shift among key thought leaders both public and private, which has paved the way for open access to data and fostered a more collaborative environment for innovation. Thinking more strategically rather than reinventing the wheel has leveled the playing field for both young investigators and experienced researchers by creating equal opportunities to test novel ideas within a collaborative environment. The AGRE model has stimulated much interest among other genetic disease and orphan disorder groups who are focused on providing researchers with the biological specimens that will help them fast-track therapeutics through the development pipeline. The model that AGRE has developed has broad application to other complex disorders and because of the tenacity, generosity, and foresight of families affected by these disorders, we now can envision a a better future for their children and those of future generations..
Acknowledgments
We gratefully acknowledge the resources provided by the Autism Genetic Resource Exchange (AGRE) Consortium and the participating AGRE families. The Autism Genetic Resource Exchange is a program of Autism Speaks and is supported, in part, by grant 1U24MH081810 from the National Institute of Mental Health to Clara M. Lajonchere (PI).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Campbell DB, Sutcliffe JS, Ebert PJ, Militerni R, Bravaccio C, Trillo S, Elia M, Schneider C, Melmed R, Sacco R, Persico AM, Levitt P. A genetic variant that disrupts MET transcription is associated with autism. Proc Natl Acad Sci. 2006;103(45):16621–16622. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino J, Zhang Y, Frazier T, Abbacchi AM, Law P. Sibling Recurrence and the Genetic Epidemiology of Autism. Am J Psychiatry. 2010;AiA:1–8. doi: 10.1176/appi.ajp.2010.09101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz ML. The lifetime distribution of the incremental societal costs of autism. Arch Pediatr Adolesc Med. 2007;161(4):343–349. doi: 10.1001/archpedi.161.4.343. [DOI] [PubMed] [Google Scholar]
- Lee H, Marvin AR, Watson T, Piggot J, Law JK, Law PA, Constantino JN, Nelson SF. Accuracy of phenotyping of autistic children based on Internet implemented parent report. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(6):1119–1126. doi: 10.1002/ajmg.b.31103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimäki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M. Strong association of de novo copy number mutations with autism. Science. 2007;316(5823):445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ Autism Consortium. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358(7):667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]