Abstract
Genetic variations in cell cycle checkpoints and DNA repair genes are associated with prolonged cell cycle G2 delay following ionizing radiation (IR) treatment and breast cancer risk. However, different studies reported conflicting results examining the association between post-IR cell cycle delay and breast cancer risk utilizing four different parameters: cell cycle G2 delay index, %G2–M, G2/G0–G1, and (G2/G0–G1)/S. Therefore, we evaluated whether different parameters may influence study results using a data set from 118 breast cancer cases and 225 controls as well as lymphoblastoid and breast cancer cell lines with different genetic defects. Our results suggest that cell cycle G2 delay index may serve as the best parameter in assessing breast cancer risk, genetic regulation of IR-sensitivity, and mutations of ataxia telangiectasia mutated (ATM) and TP53. Cell cycle delay in 21 lymphoblastoid cell lines derived from BRCA1 mutation carriers was not different from that in controls. We also showed that IR-induced DNA-damage signaling, as measured by phosphorylation of H2AX on serine 139 (γ-H2AX) was inversely associated with cell cycle G2 delay index. In summary, the cellular responses to IR are extremely complex; mutations or genetic variations in DNA damage signaling, cell cycle checkpoints, and DNA repair contribute to cell cycle G2 delay and breast cancer risk. The cell cycle G2 delay assay characterized in this study may help identify subpopulations with elevated risk of breast cancer or susceptibility to adverse effects in normal tissue following radiotherapy.
Keywords: breast cancer, ionizing radiation sensitivity, cell cycle, G2 delay, radiotherapy
Introduction
Breast cancer is the most common cancer among American women. In 2008, it is estimated that 182,460 new cases would be diagnosed and 40,480 women would die of breast cancer.1 Rare germ line mutations in DNA damage/repair response genes, such as BRCA1, BRCA2, ATM, FANC, and CHEK2, are associated with breast cancer susceptibility and highlight the importance of DNA damage/repair in breast cancer development.2 Prevalent low-penetrance polymorphisms in cell cycle checkpoint and DNA repair genes and their gene–gene and gene–environment interactions may underlie the etiology of breast cancer. Genetic variations in DNA damage signaling, cell cycle checkpoints, and DNA repair pathways may result in downregulation of damage signaling and repair functions which, when combined with environmental exposures, may result in genomic instability that promotes breast cancer. Accordingly, breast cancer risk may be associated with single nucleotide polymorphisms (SNPs) and mutations in cell cycle checkpoint and DNA repair genes, as well as overall cellular response to genotoxic insults, such as ionizing radiation (IR), the repair of which involves the combined action of multiple cell cycle checkpoints and DNA repair pathways.
Cellular exposure to IR induces a myriad of cytotoxic and premutagenic DNA damages, including double-strand breaks and oxidative DNA damage.3,4 Failure of cell cycle checkpoint and repair pathways to correct DNA damages prior to replication may result in the propagation of deleterious chromosomal aberrations and mutations that decrease genomic stability and lead to transformation. Therefore, assessment of defects in cell cycle checkpoints and DNA repair at the cellular level may allow for identification of individuals at high risk for breast cancer and afford opportunities for surveillance and behavioral intervention. IR sensitivity has been correlated with breast cancer risk.5–9 Methodologies for assessing IR sensitivity and breast cancer risk include genetic screening, comet assay, G2-irradiation chromosomal hypersensitivity assay (GICH), G2-micronucleus assay, and analysis of post-IR cell cycle arrest function in peripheral blood lymphocytes (PBLs).8,10–13 GICH and the G2-micronucleus assay, though effective at identifying IR sensitivity, are laborious and frequently utilize subjective analysis. The analysis of cell cycle G2 delay in PBLs following IR exposure has been used in numerous studies as a probe for genetic variations of cell cycle checkpoint and DNA repair genes that are associated with cancer risk.8,9,12,14 Such assays are simple, inexpensive, and utilize automated sample analysis. However, sample size and the use of different methods for analysis of radiation-induced changes in cell cycle distribution, including G2 delay index, %G2–M, G2/G0–G1, and (G2/G0–G1)/S, may have contributed to conflicting results in population-based studies.5,8,15,16
We previously demonstrated an association between breast cancer risk and cell cycle G2 delay in a breast cancer case-control study.9 However, a recent study with smaller sample size found no association between breast cancer risk and IR sensitivity when cell cycle arrest was quantified using the parameter (G2/G0–G1)/S.15 Therefore, we first examined the IR sensitivity data from 118 breast cancer cases and 225 controls reported in a previous study9 and compared the results obtained using four parameters described in previous studies: G2 delay index, %G2–M, G2/G0–G1, and %(G2/G0–G1)/S.5,8,15,16 Furthermore, we compared these four parameters in 54 lymphoblastoid and three breast cancer cell lines with different genetic defects in cell cycle checkpoint and DNA repair. Lastly, we evaluated the association between phosphorylated histone H2AX, a measure of DNA damage signaling17 and cell cycle G2 delay index. This study aims to further characterize the cell cycle G2 delay assay as a simple and reproducible screening tool for assessing IR sensitivity and breast cancer risk.
Materials and methods
Study population
Basal and IR-induced cell cycle distribution data in T lymphocytes were derived from a breast cancer-control study of 118 breast cancer cases and 225 controls as described previously.9 In brief, cancer-free women were recruited from the Comprehensive Breast Center and the Cancer Assessment and Risk Evaluation program at Georgetown University Medical Center from August 1995 to November 1996 as study controls. Breast cancer cases were recruited from the Breast Cancer Section of the Division of Hematology/Oncology. Each woman completed a self-administered questionnaire requesting information about demographics, medical conditions, and family history (FH) of breast cancer in first-degree relatives. A woman with at least one first-degree relative with breast cancer was considered to have a positive FH. The general eligibility criteria were, (i) English-speaking and able to comprehend informed consent, (ii) no personal history of other cancers, and (iii) at least 18 years of age. Blood samples were taken from all study subjects. Subjects received a detailed description of the study protocol and signed informed consent as approved by the Institutional Review Board of the Georgetown University Medical Center.
Lymphoblastoid and breast cancer cell lines
Epstein–Barr virus-immortalized human lymphoblastoid cell lines from 21 BRCA1 mutation carriers, 12 controls, and 21 individuals from ATM families were obtained from the Coriell Institute for Medical Research (Camden, NJ). Lymphoblastoid cell lines were maintained in RPMI 1640, 15% fetal bovine serum (FBS), at 37°C, 5% CO2. Human breast cancer cell lines MCF7, BT-20, and HCC1937 were obtained from American Type Culture Collection (Manassas, VA) and maintained in the recommended growth medium at 37°C, 5% CO2.
Mitogen response and cell cycle G2 delay assay
The cell cycle distributions and IR-induced G2 delay data on 118 breast cancer cases and 225 controls were obtained from a previous study.9 In brief, PBLs were stimulated with phytohemagglutinin-P (PHA-P, Sigma-Aldrich, St Louis, MO) for 72 hours prior to irradiation. We reported the cell cycle G2 delay data but not the mitogen response data.9 In the current study, we performed the cell cycle delay assay in 54 lymphoblastoid and three breast cancer cell lines using the method as described previously with minor modifications.9 In brief, lymphoblastoid and breast cancer cell lines were plated 24 hours prior to irradiation at a cell concentration of 0.5 × 106/ml. Irradiation was performed using a Nordion Gammacell 137Cs irradiator at a dose rate of 0.9 Gy/min. Twenty-four hours post-irradiation, cells were harvested and stained with Vindelov’s propidium iodide (PI)18 containing 10 mM Tris-HCl, pH 8.0, 10 mM NaCl, 10 μg/mL RNase A, 0.1% NP-40 (Igepal CA-630; Sigma-Aldrich), and 75 μM PI. Untreated and IR-irradiated cells (3 Gy) were analyzed for cell cycle distribution (10,000 cells per sample) using an LSR dual laser flow cytometer and CellQuest Pro software (BD BioSciences, San Jose, CA).
The cell cycle G2 delay index was calculated as (% irradiated cells in G2–M – % control cells in G2–M)/(% control cells in S phase) × 100.9 Other parameters were calculated as follows: %G2–M (% of irradiated cells in G2–M),16 G2/G0–G1 (% irradiated cells in G2–M)/(% irradiated cells in G0–G1) × 100,5 (G2/G0–G1)/S, (% irradiated cells in G2–M)/(% irradiated cells in G0–G1)/(% controls cells in S) × 100.15 For assay quality control, we performed repeat measures at least twice for 54 lymphoblastoid cell lines until the coefficient of variation (CV) for batch-to-batch variation dropped to less than 30%. For a majority of the samples, the inter-assay CV was under 10% (range 5%–27%). We also established that the intra-individual variation in 23 lymphocyte samples with repeat visits was very low (mean CV at 6%).
Histone H2AX phosphorylation (γ-H2AX) assay
Lymphoblastoid cell lines in culture were centrifuged at 300 × g for 10 min and resuspended in 4°C growth medium (RPMI 1640, 15% FBS, antibiotics) at a concentration of 3.3 × 106 cells/ml. One hundred and fifty microliters (0.5 × 106 cells) of cells was irradiated on ice for 10 min (9 Gy) at a dose rate of 0.9 Gy/min in a 137Cs irradiator. Following irradiation, cells were immediately placed in a 37°C water bath for repair. After repair for up to 4 hours, tubes were placed on ice and 150 μl of cold medium containing 0.2% NP-40 was added to each tube. Tubes were inverted 10x and kept on ice in the dark for one hour to block nonspecific binding sites. 33 μL of a 1:200 solution of γ-H2AX-FITC antibody (16-202A; Millipore, Billerica, MA) in ice cold medium containing 0.1% NP-40 was added to each tube for a final 1:2000 antibody dilution. Tubes were mixed by inversion 10x and placed on ice for 1 hour in the dark. Cells were transferred to flow cytometry tubes and stained with 3.3 μl of PI (50 μg/mL). PI positive cells were analyzed for relative FITC staining (10,000 cells) using an LSR flow cytometer and fluorescence intensity was quantified using CellQuest Pro software.
Statistical analysis
To compare cell cycle distribution in PHA-activated T lymphocytes and cell cycle delay in breast cancer cases and controls, a one-way ANOVA, stratified by breast cancer family history, was performed using SPSS software (version 15.0; SPSS, Inc., Chicago, Illinois). We also performed Bonferroni post hoc pair wise comparisons. The means and standard deviations (SD) were presented. For quality control, CV (SD/mean) was calculated for intra-individual variations of repeat visits in 23 subjects and assay batch-to-batch variations of 54 lymphoblastoid cell lines.
Results
PHA response and IR sensitivity and breast cancer risk
In Table 1, the cellular response to PHA in PBLs from 118 breast cancer cases was significantly lower compared to that in 225 controls based on the percentage of cells in S phase of the cell cycle (24.8% vs 28.5%; p < 0.05). After 3 Gy of IR treatment, breast cancer cases have a significantly higher percentage of cells in G0–G1 phase (63.3% vs 59.0%, p < 0.05) and lower percentage of cells in S phase (17.7% vs 21.2%; p < 0.05). In Table 2, we used four different parameters for measuring IR-induced cell cycle delay: G2 delay index, %G2–M, G2/G0–G1, and (G2/G0–G1)/S. The G2 delay index was the only parameter that showed significant increases in IR-induced cell cycle delay in breast cancer cases compared to controls (36.0 vs 31.4; p < 0.01). The other three parameters did not yield useful findings; the G2/G0–G1 ratio even showed a significantly lower value in breast cancer cases compared to controls (30.8 vs 34.7; p < 0.05).
Table 1.
Cell cycle distribution in breast cancer cases and controls
|
(A) Without IR treatment |
Control |
Case |
||||||
|---|---|---|---|---|---|---|---|---|
| Family history | (N) | % G0–G1 | % S | % G2–M | (N) | % G0–G1 | % S | % G2–M |
| None | 87 | 60.7 ± 7.1 | 28.4 ± 5.7 | 10.9 ± 2.1 | 61 | 63.8 ± 6.8a | 25.5 ± 5.7a | 10.7 ± 2.4 |
| Mother or sister | 69 | 60.3 ± 5.8 | 29.0 ± 4.7 | 10.7 ± 1.9 | 11 | 65.9 ± 7.5a | 24.1 ± 5.2a | 10.0 ± 2.8 |
| Mother and sister | 13 | 60.6 ± 7.5 | 28.0 ± 5.7 | 11.4 ± 2.9 | 3 | 70.8 ± 4.4a | 21.4 ± 5.9 | 7.8 ± 2.0 |
| Missing | 56 | 60.9 ± 4.8 | 28.0 ± 3.9 | 11.0 ± 1.9 | 43 | 66.0 ± 5.4a | 24.1 ± 4.3a | 9.8 ± 2.0a |
| Total | 225 | 60.6 ± 6.2 | 28.5 ± 5.0 | 10.9 ± 2.1 | 118 | 64.9 ± 6.4a | 24.8 ± 5.2a | 10.3 ± 2.2a |
| (B) With IR treatment | ||||||||
| None | 87 | 59.1 ± 8.0 | 21.1 ± 5.6 | 19.8 ± 4.1 | 61 | 62.4 ± 7.0a | 18.2 ± 5.3a | 19.4 ± 3.3 |
| Mother or sister | 69 | 59.1 ± 7.9 | 21.4 ± 5.4 | 19.5 ± 4.2 | 11 | 62.9 ± 7.3 | 19.5 ± 4.4 | 17.5 ± 4.3 |
| Mother and sister | 13 | 57.0 ± 6.2 | 21.3 ± 5.5 | 21.7 ± 4.3 | 3 | 67.3 ± 2.6a | 13.8 ± 2.2a | 18.9 ± 2.6 |
| Missing | 56 | 59.3 ± 6.1 | 20.9 ± 4.3 | 19.8 ± 3.9 | 43 | 64.4 ± 6.3a | 16.9 ± 4.3a | 18.7 ± 3.4 |
| Total | 225 | 59.0 ± 7.4 | 21.2 ± 5.2 | 19.8 ± 4.1 | 118 | 63.3 ± 6.7a | 17.7 ± 4.9a | 19.0 ± 3.4 |
Notes:
p < 0.05, pair-wise comparison, cases vs controls.
Abbreviation: IR, ionizing radiation.
Table 2.
Ionizing radiation-induced cell cycle delay in breast cancer cases and controls
| Family history |
(N) |
G2 delay index |
%G2–M |
G2/G0–G1 |
(G2/G0–G1)/S |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | Case | Control | Case | Control | Case | Control | Case | Control | Case | |
| None | 87 | 61 | 31.6 ± 11.5 | 35.1 ± 12.8 | 19.8 ± 4.1 | 19.4 ± 3.3 | 34.7 ± 10.9 | 31.9 ± 7.9 | 1.21 ± 0.3 | 1.26 ± 0.2 |
| Mother or sister | 69 | 11 | 30.1 ± 10.7 | 31.8 ± 12.3 | 19.5 ± 4.2 | 17.5 ± 4.3 | 34.3 ± 10.7 | 29.0 ± 10.7 | 1.17 ± 0.3 | 1.18 ± 0.2 |
| Mother and sister | 13 | 3 | 38.3 ± 17.8 | 51.4 ± 12.3 | 21.7 ± 4.3 | 18.9 ± 2.6 | 38.8 ± 9.77 | 28.2 ± 4.7 | 1.40 ± 0.3 | 1.34 ± 0.2 |
| Missing | 56 | 43 | 30.9 ± 10.5 | 37.2 ± 13.4a | 19.8 ± 3.9 | 18.7 ± 3.4 | 34.2 ± 9.72 | 29.8 ± 8.1a | 1.22 ± 0.3 | 1.24 ± 0.3 |
| Total | 225 | 118 | 31.4 ± 11.5 | 36.0 ± 13.1a | 19.8 ± 4.1 | 19.0 ± 3.4 | 34.7 ± 10.5 | 30.8 ± 8.2a | 1.21 ± 0.3 | 1.25 ± 0.2 |
Notes:
p < 0.001, pair-wise comparison, cases vs controls.
Cell cycle distribution and IR sensitivity in lymphoblastoid cell lines
In Table 3A, lymphoblastoid cell lines used in this study had similar cell cycle distributions without IR treatment. With IR treatment, cell lines from ATM mutation carriers, particularly five AT patients, had a lower percentage of cells in the G0–G1 phase (38.7% vs 56.0%, Table 3B) or obligate ATM mutation carriers (38.7% vs 53.5%, Table 3B) and a higher percentage of cells in the G2–M phase compared to controls (55.6% vs 35.2%, Table 3B), BRCA mutation carriers (55.6% vs 37.0%, Table 3B), or obligate ATM mutation carriers (55.6% vs 41.7%, Table 3B). In Table 4, the mean cell cycle delay following IR treatment in 21 heterozygous BRCA1 mutation carriers was not significantly different from that in controls using all four parameters. Although all four parameters were able to detect significantly higher cell cycle G2 delay in AT patients compared to each of the three other groups (p < 0.05), only G2 delay index and (G2/G0–G1)/S detected significant higher cell cycle delay in obligate ATM mutation carriers (Table 4C) compared to controls (Table 4A) or cell lines with BRCA1 mutations (Table 4B).
Table 3.
Cell cycle distribution in lymphoblastoid cell lines
| (A) Without IR treatment | N | %G0–G1 | % Sa | %G2–M |
|---|---|---|---|---|
| Controls | 12 | 60.8 ± 6.3 | 29.6 ± 5.9b | 9.6 ± 1.0 |
| BRCA mutation carriers | 21 | 60.1 ± 5.4 | 29.0 ± 4.9 | 10.8 ± 1.6 |
| ATM mutation carriers | 16 | 63.7 ± 5.2 | 24.8 ± 4.9 | 11.6 ± 5.0 |
| AT patients | 5 | 60.7 ± 2.9 | 26.5 ± 3.0 | 12.8 ± 0.9 |
| (B) With IR treatment | %G0–G1a | % Sa | %G2–Ma | |
| Controls | 12 | 56.0 ± 6.3b | 8.9 ± 1.8b,e | 35.2 ± 5.9b |
| BRCA mutation carriers | 21 | 55.9 ± 8.2c | 7.1 ± 2.1f | 37.0 ± 7.3c |
| ATM mutation carriers | 16 | 53.5 ± 7.9d | 4.8 ± 1.6 | 41.7 ± 7.0d |
| AT patients | 5 | 38.7 ± 5.9 | 5.7 ± 0.3 | 55.6 ± 5.8 |
Notes:
p < 0.05, ANOVA test for differences among four cell lines;
p < 0.05, Bonferroni post hoc pair-wise comparison, cell lines from controls compared to AT patients;
p < 0.05, Bonferroni post hoc pair-wise comparison, cell lines form BRCA1 mutation carriers compared to AT patients;
p < 0.05, Bonferroni post hoc pair-wise comparison, cell lines from ATM mutation carriers compared to AT patients;
p < 0.05, Bonferroni post hoc pair-wise comparison, cell lines from controls compared to ATM mutation carriers;
p < 0.05, Bonferroni post hoc pair-wise comparison, cell lines from BRCA1 mutation carriers compared to ATM mutation carriers.
Abbreviation: IR, ionizing radiation.
Table 4.
Ionizing radiation-induced cell cycle delay in lymphoblastoid cell lines
| Repository | G2 delay | %G2–M | G2/G1 | (G2/G1)/S |
|---|---|---|---|---|
| (A) Controls (n = 12) | ||||
| GM14807 | 100 ± 5 | 41 ± 5 | 81 ± 20 | 2.5 ± 0.3 |
| GM14548 | 96 ± 15 | 37 ± 8 | 69 ± 24 | 2.4 ± 0.5 |
| GM14453 | 95 ± 11 | 35 ± 7 | 64 ± 18 | 2.4 ± 0.5 |
| GM01990 | 95 ± 11 | 34 ± 6 | 61 ± 19 | 2.3 ± 0.5 |
| GM10924 | 88 ± 10 | 40 ± 5 | 81 ± 15 | 2.3 ± 0.2 |
| GM14448 | 87 ± 16 | 39 ± 4 | 74 ± 16 | 2.2 ± 0.2 |
| GM14476 | 85 ± 9 | 31 ± 8 | 53 ± 19 | 1.9 ± 0.4 |
| GM14452 | 84 ± 16 | 40 ± 5 | 84 ± 20 | 2.4 ± 0.4 |
| AG09387 | 84 ± 11 | 43 ± 4 | 93 ± 14 | 2.4 ± 0.5 |
| GM14673 | 83 ± 23 | 27 ± 11 | 47 ± 24 | 2.0 ± 0.5 |
| GM01814 | 75 ± 15 | 31 ± 4 | 53 ± 11 | 1.8 ± 0.1 |
| GM13079 | 72 ± 12 | 24 ± 5 | 36 ± 12 | 2.0 ± 0.5 |
| Mean ± SD | 87.0 ± 8.5a,b | 35.2 ± 5.9b | 66.2 ± 17.1b | 2.2 ± 0.2a,b |
| (B) BRCA1 mutation carriers (n = 21) | ||||
| GM13713 | 120 ± 17 | 45 ± 3 | 89 ± 13 | 3.3 ± 0.2 |
| GM15993 | 116 ± 9 | 52 ± 7 | 135 ± 42 | 3.8 ± 0.8 |
| GM14637 | 103 ± 10 | 35 ± 3 | 58 ± 8 | 2.3 ± 0.2 |
| GM13715 | 100 ± 23 | 42 ± 5 | 86 ± 17 | 2.8 ± 0.8 |
| GM13712 | 99 ± 6 | 45 ± 5 | 100 ± 21 | 3.2 ± 0.4 |
| GM13714 | 97 ± 25 | 35 ± 6 | 64 ± 17 | 2.4 ± 0.6 |
| GM13707 | 95 ± 15 | 40 ± 5 | 71 ± 15 | 2.5 ± 0.7 |
| GM13710 | 95 ± 13 | 48 ± 6 | 112 ± 27 | 3.1 ± 0.6 |
| GM14636 | 92 ± 15 | 34 ± 5 | 59 ± 13 | 2.1 ± 0.3 |
| GM15232 | 90 ± 6 | 47 ± 5 | 113± 32 | 2.7 ± 0.4 |
| GM14093 | 89 ± 15 | 32 ± 5 | 52 ± 14 | 2.0 ± 0.5 |
| GM16105 | 89 ± 12 | 38 ± 7 | 72 ± 22 | 2.3 ± 0.5 |
| GM14097 | 88 ± 18 | 34 ± 6 | 61 ± 17 | 2.2 ± 0.4 |
| GM14638 | 88 ± 12 | 35 ± 6 | 63 ± 18 | 2.2 ± 0.3 |
| GM14092 | 82 ± 9 | 34 ± 1 | 56 ± 3 | 1.9 ± 0.2 |
| GM14094 | 80 ± 10 | 29 ± 8 | 45 ± 18 | 1.9 ± 0.3 |
| GM14091 | 80 ± 9 | 27 ± 2 | 40 ± 5 | 2.1 ± 0.4 |
| GM13705 | 78 ± 19 | 36 ± 6 | 68 ± 18 | 2.2 ± 0.5 |
| GM14096 | 78 ± 9 | 36 ± 4 | 70 ± 15 | 2.0 ± 0.3 |
| GM13709 | 71 ± 13 | 28 ± 4 | 43 ± 8 | 1.7 ± 0.3 |
| GM14090 | 61 ± 10 | 25 ± 5 | 37 ± 10 | 1.6 ± 0.3 |
| Mean ± SD | 90.0 ± 13.9c,d | 37.0 ± 7.3d | 71.1 ± 26.2d | 2.4 ± 0.6c,d |
| (C) ATM mutation obligate carriers (n =16) | ||||
| GM09579 | 162 ± 32 | 49 ± 4 | 105 ± 16 | 4.4 ± 1.0 |
| GM00736 | 160 ± 15 | 48 ± 12 | 106 ± 54 | 4.5 ± 1.6 |
| GM02781 | 155 ± 20 | 32 ± 1 | 49 ± 2 | 3.5 ± 0.3 |
| GM02782 | 134 ± 12 | 46 ± 25 | 129 ± 122 | 4.6 ± 2.3 |
| GM09583 | 130 ± 23 | 45 ± 3 | 94 ± 12 | 3.4 ± 0.7 |
| GM03383 | 127 ± 11 | 39 ± 4 | 66 ± 13 | 2.8 ± 0.1 |
| GM03188 | 125 ± 34 | 46 ± 5 | 97 ± 22 | 3.6 ± 1.7 |
| GM09580 | 124 ± 16 | 47 ± 2 | 103 ± 11 | 3.3 ± 0.4 |
| GM02783 | 122 ± 70 | 46 ± 7 | 105 ± 36 | 4.3 ± 0.9 |
| GM03325 | 118 ± 10 | 48 ± 1 | 104 ± 4 | 3.2 ± 0.1 |
| GM03187 | 116 ± 14 | 40 ± 1 | 71 ± 3 | 2.7 ± 0.3 |
| GM03324 | 115 ± 16 | 40 ± 5 | 73 ± 17 | 2.8 ± 0.5 |
| GM03323 | 107 ± 17 | 40 ± 3 | 75 ± 10 | 2.6 ± 0.4 |
| GM03334 | 97 ± 12 | 30 ± 4 | 46 ± 10 | 2.2 ± 0.5 |
| GM03333 | 84 ± 19 | 44 ± 9 | 90 ± 40 | 6.8 ± 5.6 |
| GM03380 | 77 ± 16 | 26 ±1 | 37 ± 3 | 1.6 ± 0.4 |
| Mean ± SD | 122.1 ± 24.2e | 41.7 ± 7.0e | 84.4 ± 25.8e | 3.5 ± 1.2e |
| (D) AT patients (n = 5) | ||||
| GM09581 | 200 ± 10 | 61 ± 2 | 179 ± 7 | 7.7 ± 0.5 |
| GM09582 | 164 ± 14 | 60 ± 0.2 | 173 ± 3 | 5.9 ± 0.4 |
| GM00719 | 159 ± 27 | 59 ± 8 | 178 ± 66 | 5.9 ± 0.6 |
| GM03332 | 148 ± 17 | 48 ± 23 | 136 ± 122 | 5.1 ± 2.9 |
| GM03189 | 146 ± 30 | 50 ± 3 | 114 ± 9 | 4.6 ± 1.7 |
| Mean ± SD | 163.3 ± 21.9c | 55.6 ± 5.8c | 155.9 ± 29.4c | 5.8 ± 1.2c |
Notes:
p < 0.05, controls vs ATM mutation carriers;
p < 0.05, controls vs AT patients;
p < 0.05, BRCA1 vs ATM mutation carriers;
p < 0.05, BRCA1 mutation carriers vs AT patients;
p < 0.05, ATM mutation carriers vs AT patients.
Cell cycle distribution and IR sensitivity in breast cancer cell lines
Three breast cancer cell lines with different genetic backgrounds were evaluated for their response to IR. As shown in Table 5, without IR treatment, a higher percentage of cells from BT-20 and HCC1937 cell lines were present in the G2–M phase of the cell cycle compared to MCF7 cell line (31.9% and 32.1% vs 16.6%). Post-IR treatment, these two TP53-mutant lines, BT-20 and HCC1937, showed significantly higher cell cycle delay compared to that of MCF7 using all four parameters. The results suggest that TP53 mutations may influence IR-induced cell cycle delay (Table 5). However, G2 delay index showed the maximal 20.7-fold difference in cell cycle delay between MCF7 and HCC1937 cell lines compared to 3.2-, 10.0-, and 8.9-fold differences using %G2–M, G2/G0–G1, and (G2/G0–G1)/S, respectively.
Table 5.
Cell cycle distribution and mitotic delay in breast cancer cell lines
| Cell line |
Without IR Treatment |
With IR treatment |
G2delay indexa | %G2–Ma | G2/G0–G1a | (G2/G0–G1)/Sa | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| %G0–G1a | % Sa | %G2–Ma | %G0–G1a | % Sa | %G2–Ma | |||||
| MCF7b | 63.4 ± 5.6f | 18.8 ± 2.7e | 16.6 ± 2.1e,f | 68.2 ± 3.2e,f | 11.2 ± 1.1f | 17.5 ± 1.4e,f | 5.6 ± 10.8e,f | 17.5 ± 1.4e,f | 25.8 ± 2.7e,f | 1.4 ± 0.1e,f |
| BT-20c | 52.2 ± 15.6 | 13.9 ± 1.4g | 31.9 ± 14.6 | 41.6 ± 15.3g | 11.8 ± 1.0g | 45.2 ± 15.6 | 95.6 ± 14.0g | 45.2 ± 15.6 | 146.7 ± 117.4g | 10.3 ± 7.8 |
| HCC1937d | 45.4 ± 6.1 | 20.7 ± 1.6 | 32.1 ± 4.0 | 22.3 ± 3.2 | 18.6 ± 1.3 | 56.0 ± 1.9 | 116.2 ± 18.2 | 56.0 ± 1.9 0 | 257.4 ± 49.70 | 12.4 ± 1.9 |
| Fold difference | 20.7 | 3.2 | 10.0 | 8.9 | ||||||
Notes:
p<0.05, ANOVA test for differences among three cell lines;
HCC1937 cells with mutant BRCA1 and mutant p53; 44
p < 0.05, Bonferroni post hoc pair-wise comparison, MCF7 compared to BT-20 cell line;
p < 0.05, Bonferroni post hoc pair-wise comparison, MCF7 compared to HCC1937 cell line;
p < 0.05, Bonferroni post hoc pair-wise comparison, BT20 compared to HCC1937 cell line.
Abbreviation: IR, ionizing radiation.
γ-H2AX induction in irradiated lymphoblastoid cell lines
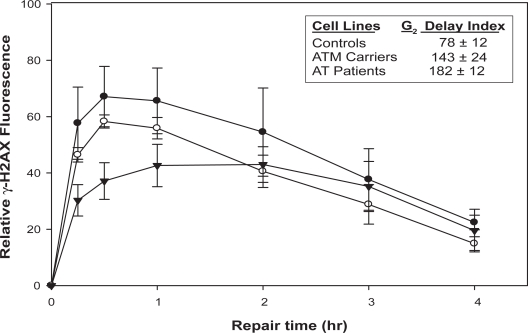
H2AX phosphorylation kinetics differed significantly in irradiated lymphoblastoid cell lines from controls, obligate ATM mutation carriers, and AT patients (Figure 1). Control cells had the most rapid and largest increase in γ-H2AX level following IR exposure. Cells from controls and obligate ATM mutation carriers reached maximal γ-H2AX induction within 30 min post-irradiation. However, cells from AT patients had a slower and lower γ-H2AX induction (p < 0.05 at 15, 30, and 60 min) which peaked two hours after irradiation. γ-H2AX induction in cells from obligate ATM mutation carriers followed kinetics similar to controls, but the maximum γ-H2AX induction level was intermediate between that observed in controls and AT patients. At four hours post-IR treatment, all three groups of cells showed similar levels of γ-H2AX.
Figure 1.
γ-H2AX staining in irradiated lymphoblastoid cell lines. Induced γ-H2AX fluoresence in lymphoblastoid cell lines stained with γ-H2AX-FITC antibody at the indicated time interval following exposure to 9 Gy IR. Closed circles, average γ-H2AX fluorescence of three control lymphoblastoid cell lines GM10924, GM01814, and GM13079. Open circles, average γ-H2AX fluorescence of two ATM carrier lymphoblastoid cell lines GM09579 and GM09580. Closed triangles, average γ-H2AX fluorescence of two AT patient lymphoblastoid cell lines GM09581 and GM09582. Inset: G2 delay index of corresponding lymphoblast groups assayed for γ-H2AX. All values are means of three independent experiments with standard deviation.
Discussion
Using data derived from a relatively large breast cancer case-control study and cell lines with different genetic defects, our current results support previous findings that prolonged cell cycle G2 delay in response to IR may serve as a sensitive biomarker for assessing IR sensitivity and breast cancer predisposition. Some of the conflicting results in the literature may be related to: (1) study sample size and related statistical power, (2) PHA-induced PBLs vs lymphoblastoid cell lines, and (3) parameters used for interpreting the study results.5,9,15,16
Another interesting finding in this study is that response of PBLs to PHA is lower in breast cancer cases compared to that in controls. Several earlier studies reported that PBLs from cancer patients are less responsive to mitogenic stimulation by PHA and other plant lectins compared to healthy controls.19–20 Our current data clearly demonstrate that PHA-stimulated PBLs from breast cancer cases had a significantly higher percentage of cells in the G0–G1 and a lower percentage of cells in the S phase compared to controls. PHA-induced proliferation of PBLs derived from patients with breast cancer may reflect tumor load and be a good clinical predictor for the further course of the disease.21 Antitumor T lymphocytes play a pivotal role in immune surveillance of cancer cells. Therefore, PHA-stimulated T lymphocyte cell cycle distribution may also serve as an immune function marker for cancer risk. Our observation is also consistent with the results from a recent study showing that genetic polymorphisms in cytotoxic T-lymphocyte antigen 4 influenced T-cell activation and modified the susceptibility to breast and other cancers.22
Breast cancer case-control differences in PBL response to PHA have an impact on studies of IR-induced cell cycle delay. Since only cycling cells are arrested in G2–M following irradiation, PBLs with lower percentages of cycling cells would produce a lower percentage of G2-arrested cells, subsequently influencing study results. Therefore, we introduced the G2 delay index to normalize the results with the percentage of cells in S phase without IR treatment.8,9 The importance of adjusting for PHA response is clearly demonstrated in Table 2, where significant prolonged cell cycle delay in breast cancer cases was observed only when G2 delay index was used. Neither %G2–M nor (G2/G0–G1)/S showed a case-control difference in cell cycle delay. When G2/G0–G1 was used, we even observed a lower cell cycle delay response in breast cancer cases compared to that in controls.
PBLs and lymphoblastoid cell lines from BRCA1 and ATM mutation carriers have previously been analyzed for associations between IR-induced cell cycle delay and mutation status.12,15,16,23 Mutations in BRCA1 or ATM are risk factors for breast and ovarian cancers.2,24 The results from recent studies suggest that BRCA1 plays critical roles in DNA repair, recombination, checkpoint control of the cell cycle, transcription, and S- and G2-phase checkpoints in response to IR.25–27 We compared the IR-induced G2 delay in lymphoblastoid cell lines from controls and BRCA1 carriers using G2 delay index and three other parameters (Table 4). None of the methods could distinguish a difference in BRCA1 heterozygote mutation carriers compared to controls. Our findings are consistent with a recent study that showed normal post-IR cell cycle kinetics in BRCA1 mutation carriers23 and suggest that one functional BRCA1 allele may be sufficient for its normal post-IR checkpoint function.
An association between mutations in the ATM gene and IR hypersensitivity has been demonstrated in numerous studies as summarized in a recent review.28 Early studies identified that AT cells are defective in immediate cell cycle checkpoints in response to IR, but a later ATM-independent response causes these irradiated cells to accumulate in G2 relative to normal cells.12,29 Therefore, we also evaluated the four parameters in lymphoblast cell lines derived from AT patients and obligate ATM mutation carriers. All four parameters showed significantly higher cell cycle delay in cell from AT patients. In contrast to a previous study,15 we also showed significantly higher cell cycle delay in cells from obligate ATM mutation carriers compared to that in controls using G2 delay index and (G2/G0–G1)/S. The difference in study results may be related to a larger sample size of our study with 16 ATM mutation carriers and 12 controls compared to the previous study with four ATM mutation carriers and four controls.15
We further investigated the association between G2 delay index and IR-induced H2AX phosphorylation in seven lymphoblastoid cell lines (Figure 1). In response to DNA double-strand strand breaks induced by IR or other genotoxic agents, H2AX is rapidly phosphorylated at Ser-139 (γ-H2AX) by ATM and other kinases, including ATR and DNA PK.30 γ-H2AX is immediately localized to DSB sites where it is believed to recruit additional factors required for completion of DNA repair.31 The formation and disappearance of γ-H2AX foci in a nucleus are proportional to the induction and repair of double-strand breaks, respectively.32 The result of DNA double-strand breaks measured by γ-H2AX levels is in agreement with data obtained using the comet assay.33 Thus, the kinetics of γ-H2AX formation may be a useful surrogate for DNA double-strand break induction and repair. The established role of ATM kinase in post-IR double-strand break repair and checkpoint function underlies the DNA repair deficient- and IR-sensitive phenotype observed in AT patients.34 Following IR exposure, cells from AT patients had slower and lower levels of γ-H2AX induction than those in obligate ATM carriers and controls (Figure 1). In addition to genetic variations in DNA repair, the results from the current study also demonstrate that defective DNA-damage signaling is associated with prolonged cell cycle G2 delay.
Analysis of IR-induced cell cycle delay in breast cancer cell lines utilizing the four parameters further supports the advantage of using the G2 delay index parameter. In three breast cancer cell lines, the G2 delay index was able to detect the largest fold difference between the MCF7 and the HCC1937 cell lines (Table 5). Although both MCF7 and BT-20 cells express wild-type BRCA1,35 they exhibit very different IR responses. The difference may be related to TP53 mutation (AAG→ CAG; K132Q) in BT-20 cells36 since p53 plays critical role in checkpoint regulation.37 In HCC1937 cells, the presence of mutations in both BRCA1 (5382C) and TP53 (CGA→TGA; R306X)36 genes may contribute to its having the highest cell cycle delay among the three breast cancer cell lines.
In several studies, the association between IR sensitivity in patient PBLs and acute radiotherapy (RT) toxicity has been inconclusive.6,7,16,38 Some breast cancer patients experience RT-induced acute adverse skin reactions of varying severity.39 In addition, RT-treated patients may also develop telangiectasia and fibrosis as late effects.40 Accordingly, there is increasing interest in the development of predictive tests for RT-induced adverse reactions. However, several studies have reported conflicting results utilizing chromosomal aberration analysis in predicting acute RT reactions.37,41,42 IR-induced DNA damages activate checkpoints that delay cell cycle progression to facilitate DNA repair. However, continued proliferation after DNA damage in IR-irradiated cells has been well documented. The results from a recent study estimated that cells with about 10 to 20 double-strand breaks are released from a G2 check-point delay and enter mitosis.43 Although these cells continue to divide for one to two cell cycles; the unrepaired damaged DNA eventually results in rapid apoptosis, senescence, a permanent cell cycle arrest, or mitotic catastrophe.44 Therefore, our validation of the cell cycle G2 delay assay suggests that it may serve as a simple screening tool to probe for combined genetic defects and variations in cell cycle checkpoint regulation (eg, ATM and TP53 mutations) and DNA repair (eg, BRCA1 mutations and DNA repair SNPs) in assessing radio-sensitivity and cancer risk.
The major strengths of our study are: (1) large sample size of the case-control study and cell lines, (2) stringent laboratory assay quality control with adequate batch-to-batch assay variations, (3) validation of the G2 delay index as the best parameter for testing IR sensitivity in a case-control study and among cell lines with different genetic defects, and (4) availability of genomic DNA for future testing of genotype–phenotype association studies. However, our study has several limitations. First, the current study uses a dataset from a previous study. Future studies with a larger sample size and a different case-control population will be required to confirm our study findings and to evaluate the genetic regulation of cell cycle delay in IR sensitivity and breast cancer risk. Second, viable cells are required for performing the cell cycle assay, which may limit its application in population-based studies of breast-cancer risk assessment and tumor response. Therefore, we are currently evaluating whether the newly developed antibody-based γ-H2AX assay may overcome this limitation.
In summary, the cellular responses to IR-induced DNA damage are complex. The cell cycle G2 delay assay may serve as a probe for genetic defects/variations in cell cycle checkpoint regulation and DNA repair in assessing IR sensitivity and breast cancer risk. However, there is a need to evaluate future genotype-phenotype relationships in cellular IR responses and compare results from other functional DNA damage and repair phenotype assays in order to establish the utility of the cell cycle G2 delay assay in the assessment of breast-cancer risk and prediction of RT response.
Acknowledgments
We are grateful to the participants and investigators of the original breast cancer case-control study. This research was supported by National Institutes of Health grant RO1 CA73629 (J.J. Hu). The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Turnbull C, Rahman N. Genetic predisposition to breast cancer: past, present, and future. Annu Rev Genomics Hum Genet. 2008;9:321–345. doi: 10.1146/annurev.genom.9.081307.164339. [DOI] [PubMed] [Google Scholar]
- 3.Ayene IS, Koch CJ, Krisch RE. DNA strand breakage by bivalent metal ions and ionizing radiation. Int J Radiat Biol. 2007;83:195–210. doi: 10.1080/09553000601146956. [DOI] [PubMed] [Google Scholar]
- 4.Stepán V, Davídková M. Significance of 8-oxoG in the spectrum of DNA damages caused by ionising radiation of different quality. Radiat Prot Dosimetry. 2006;122:113–115. doi: 10.1093/rpd/ncl418. [DOI] [PubMed] [Google Scholar]
- 5.Lavin MF, Bennett I, Ramsay J, et al. Identification of a potentially radiosensitive subgroup among patients with breast cancer. J Natl Cancer Inst. 1994;86:1627–1634. doi: 10.1093/jnci/86.21.1627. [DOI] [PubMed] [Google Scholar]
- 6.Scott D, Barber JB, Spreadborough AR, Burrill W, Roberts SA. Increased chromosomal radiosensitivity in breast cancer patients: a comparison of two assays. Int J Radiat Biol. 1999;75:1–10. doi: 10.1080/095530099140744. [DOI] [PubMed] [Google Scholar]
- 7.Baeyens A, Thierens H, Claes K, et al. Chromosomal radiosensitivity in breast cancer patients with a known or putative genetic predisposition. Br J Cancer. 2002;87:1379–1385. doi: 10.1038/sj.bjc.6600628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu JJ, Smith TR, Miller MS, Mohrenweiser HW, Golden A, Case LD. Amino acid substitution variants of APE1 and XRCC1 genes associated with ionizing radiation sensitivity. Carcinogenesis. 2001;22:917–922. doi: 10.1093/carcin/22.6.917. [DOI] [PubMed] [Google Scholar]
- 9.Hu JJ, Smith TR, Miller MS, Lohman K, Case LD. Genetic regulation of ionizing radiation sensitivity and breast cancer risk. Environ Mol Mutagen. 2002;39:208–215. doi: 10.1002/em.10058. [DOI] [PubMed] [Google Scholar]
- 10.Djuzenova CS, Mühl B, Fehn M, Oppitz U, Müller B, Flentje M. Radio-sensitivity in breast cancer assessed by the Comet and micronucleus assays. Br J Cancer. 2006;94:1194–1203. doi: 10.1038/sj.bjc.6603005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott D, Spreadborough AR, Jones LA, Roberts SA, Moore CJ. Chromosomal radiosensitivity in G2-phase lymphocytes as an indicator of cancer predisposition. Radiat Res. 1996;145:3–16. [PubMed] [Google Scholar]
- 12.Lavin MF, Le Poidevin P, Bates P. Enhanced levels of radiation-induced G2 phase delay in ataxia telangiectasia heterozygotes. Cancer Genet Cytogenet. 1992;60:183–187. doi: 10.1016/0165-4608(92)90014-y. [DOI] [PubMed] [Google Scholar]
- 13.Smith TR, Miller MS, Lohman KK, Case LD, Hu JJ. DNA damage and breast cancer risk. Carcinogenesis. 2003;5:883–889. doi: 10.1093/carcin/bgg037. [DOI] [PubMed] [Google Scholar]
- 14.Löbrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat Rev Cancer. 2007;7:861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- 15.Kaufmann WK, Filatov L, Oglesbee SE, et al. Radiation clastogenesis and cell cycle checkpoint function as functional markers of breast cancer risk. Carcinogenesis. 2006;27:2519–2527. doi: 10.1093/carcin/bgl103. [DOI] [PubMed] [Google Scholar]
- 16.Perez A, Grabenbauer GG, Sprung CN, Sauer R, Distel LV. Potential for the G2/M arrest assay to predict patient susceptibility to severe reactions following radiotherapy. Strahlenther Onkol. 2007;183:99–106. doi: 10.1007/s00066-007-1565-9. [DOI] [PubMed] [Google Scholar]
- 17.Rothkamm K, Löbrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci U S A. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vindelov LL. Flow microfluorometric analysis of nuclear DNA in cells from solid tumors and cell suspensions. A new method for rapid isolation and straining of nuclei. Virchows Arch B Cell Pathol. 1977;24:227–242. [PubMed] [Google Scholar]
- 19.Silverman NA, Alexander JC, Jr, Potvin C, Chretien PB. In vitro lymphocyte reactivity and T cell levels in patients with melanoma: correlations with clinical and pathological stage. Surgery. 1976;79:332–339. [PubMed] [Google Scholar]
- 20.Gilboa-Garber N, Avichezer D, Rozenszajn LR. Stimulation of peripheral lymphocytes from cancer patients and healthy subjects by Pseudomonas aeruginosa lectin. FEMS Microbiol Lett. 1986;34:237–240. [Google Scholar]
- 21.Wiltschke C, Krainer M, Budinsky AC, et al. Reduced mitogenic stimulation of peripheral blood mononuclear cells as a prognostic parameter for the course of breast cancer: a prospective longitudinal study. Br J Cancer. 1995;71:1292–1296. doi: 10.1038/bjc.1995.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun T, Zhou Y, Yang M, et al. Functional genetic variations in cytotoxic T-lymphocyte antigen 4 and susceptibility to multiple types of cancer. Cancer Res. 2008;68:7025–7034. doi: 10.1158/0008-5472.CAN-08-0806. [DOI] [PubMed] [Google Scholar]
- 23.Barwell J, Pangon L, Georgiou A, et al. Lymphocyte radiosensitivity in BRCA1 and BRCA2 mutation carriers and implications for breast cancer susceptibility. Int J Cancer. 2007;121:1631–1636. doi: 10.1002/ijc.22915. [DOI] [PubMed] [Google Scholar]
- 24.Thorstenson YR, Roxas A, Kroiss R, et al. Contributions of ATM mutations to familial breast and ovarian cancer. Cancer Res. 2003;63:3325–3333. [PubMed] [Google Scholar]
- 25.Venkitaraman A. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 26.Xu B, Kim ST, Kastan MB. Involvement of BRCA1 in S-phase and G2-phase checkpoints after ionizing radiation. Mol Cell Bio. 2001;21:3445–3450. doi: 10.1128/MCB.21.10.3445-3450.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 28.Mavrou A, Tsangaris GT, Roma E, Kolialexi A. The ATM gene and ataxia telangiectasia. Anticancer Res. 2008;28:401–405. [PubMed] [Google Scholar]
- 29.Iliakis G, Wang Y, Guan J, Wang H. DNA damage checkpoint control in cells exposed to ionizing radiation. Oncogene. 2003;22:5834–5847. doi: 10.1038/sj.onc.1206682. [DOI] [PubMed] [Google Scholar]
- 30.Foster ER, Downs JA. Histone H2A phosphorylation in DNA double-strand break repair. FEBS J. 2005;272:3231–3240. doi: 10.1111/j.1742-4658.2005.04741.x. [DOI] [PubMed] [Google Scholar]
- 31.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 32.Rothkamm K, Löbrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci U S A. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ismail IH, Wadhra TI, Hammarsten O. An optimized method for detecting gamma-H2AX in blood cells reveals a significant interindividual variation in the gamma-H2AX response among humans. Nucleic Acids Res. 2007;35:e36. doi: 10.1093/nar/gkl1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lavin MF, Kozlov S. ATM activation and DNA damage response. Cell Cycle. 2007;6:931–942. doi: 10.4161/cc.6.8.4180. [DOI] [PubMed] [Google Scholar]
- 35.Elstrodt F, Hollestelle A, Nagel JH, et al. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 2006;66:41–45. doi: 10.1158/0008-5472.CAN-05-2853. [DOI] [PubMed] [Google Scholar]
- 36.Wasielewski M, Elstrodt F, Klijn JG, et al. Thirteen new p53 gene mutants identified among 41 human breast cancer cell lines. Breast Cancer Res Treat. 2006;99:97–101. doi: 10.1007/s10549-006-9186-z. [DOI] [PubMed] [Google Scholar]
- 37.Di Leonardo A, Linke SP, Clarkin K, Wahl GM. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 38.Barber JB, Burrill W, Spreadborough AR, et al. Relationship between in vitro chromosomal radiosensitivity of peripheral blood lymphocytes and the expression of normal tissue damage following radiotherapy for breast cancer. Radiother Oncol. 2000;55:179–186. doi: 10.1016/s0167-8140(99)00158-9. [DOI] [PubMed] [Google Scholar]
- 39.Fisher J, Scott C, Stevens R, et al. Randomized phase III study comparing best supportive care to Biafine as a prophylactic agent for radiation-induced skin toxicity for women undergoing breast irradiation: Radiation Therapy Oncology Group (RTOG) 97–13. Int J Radiat Oncol Biol Phys. 2000;48:1307–1310. doi: 10.1016/s0360-3016(00)00782-3. [DOI] [PubMed] [Google Scholar]
- 40.Lilla C, Ambrosone CB, Kropp S, et al. Predictive factors for late normal tissue complications following radiotherapy for breast cancer. Breast Cancer Res Treat. 2007;106:143–150. doi: 10.1007/s10549-006-9480-9. [DOI] [PubMed] [Google Scholar]
- 41.Dikomey E, Borgmann K, Peacock J, Jung H. Why recent studies relating normal tissue response to individual radiosensitivity might have failed and how new studies should be performed. Int J Radiat Oncol Biol Phys. 2003;56:1194–1200. doi: 10.1016/s0360-3016(03)00188-3. [DOI] [PubMed] [Google Scholar]
- 42.Hoeller U, Borgmann K, Bonacker M, et al. Individual radiosensitivity measured with lymphocytes may be used to predict the risk of fibrosis after radiotherapy for breast cancer. Radiother Oncol. 2003;69:137–144. doi: 10.1016/j.radonc.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 43.Deckbar D, Birraux J, Krempler A, et al. Chromosome breakage after G2 checkpoint release. J Cell Biol. 2007;176:749–755. doi: 10.1083/jcb.200612047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang H, Fletcher L, Beeharry N, et al. Abnormal cytokinesis after X-irradiation in tumor cells that override the G2 DNA damage check-point. Cancer Res. 2008;68:3724–3732. doi: 10.1158/0008-5472.CAN-08-0479. [DOI] [PubMed] [Google Scholar]