Summary
The duplication of eukaryotic genomes involves the replication of DNA from up to several hundred thousand origins of replication. In S phase, two sister replisomes are assembled at each active origin, and they replicate DNA in opposite directions. Little is known about the functional relationship between sister replisomes in S phase. Some data imply that they travel away from one another and thus function independently. However, other studies suggest that sister replisomes form a stationary, functional unit that draws parental DNA towards itself. If this ‘double replisome’ model is correct, an extended DNA molecule whose ends are fixed in space should not undergo replication. To test this prediction, lambda DNA (50 kb) was stretched and immobilized at both ends within a microfluidic flow cell. Upon exposure to replication-competent Xenopus egg extracts, this DNA underwent extensive replication by a single pair of diverging replisomes. The data show that there is no obligatory coupling between sister replisomes and, together with other studies, imply that genome duplication is carried out by individual, autonomously functioning replisomes.
INTRODUCTION
The spatial and functional relationship between the two sister replisomes (Figure 1A-i) that emanate from each origin of DNA replication is not understood ([Bochman and Schwacha, 2009] and [Takahashi et al., 2005]). In one scenario, sister replisomes move apart after initiation and function independently (Figure 1A-ii). In other models, sister replisomes must remain physically coupled after origin firing to allow unwinding by the replicative helicase ([Falaschi, 2000], [Sclafani et al., 2004], [Weisshart et al., 1999] and [Wessel et al., 1992]) (Figure 1A-iii). In bacteria, high-resolution imaging demonstrated that there is no physical interaction between sister replisomes (Reyes-Lamothe et al., 2008). In archaea and eukaryotes, the picture is less clear. Live-cell imaging in Saccharomyces cerevisiae (Kitamura et al., 2006) and pulse-chase experiments in HeLa cells (Ligasová et al., 2009) indicate that sister replisomes reside close to one another within the nucleus, consistent with a physical association. The most prominent example of physical coupling involves the Simian Virus 40 (SV40) in which the replicative DNA helicase large T-antigen (T-ag) is proposed to form obligatory double hexamers ([Alexandrov et al., 2002], [Weisshart et al., 1999] and [Wessel et al., 1992]). In archaea, the MCM helicase forms a complex of two hexamers in solution (Fletcher et al., 2003) and in Saccharomyces cerevisiae, the MCM2-7 helicase complex is loaded onto origins as stable, double hexamers ([Evrin et al., 2009] and [Remus et al., 2009]). However, analysis of endogenous ([Gambus et al., 2006] and [Moyer et al., 2006]) and recombinant (Ilves et al., 2010) MCM2-7 complexes suggests that the active MCM2-7 complex might be a single hexamer. In summary, it is not known whether eukaryotic sister replisomes function independently or as dimeric complexes. This distinction is crucial to elucidate the architecture and mechanism of eukaryotic replisomes, and to understand the spatio-temporal coordination of replication involving a large number of origins.
Figure 1. Single-molecule visualization of eukaryotic replication.
(A) Possible configurations of sister replisomes. The sister replisomes assembled at each origin (i) travel away from each other (ii) or remain physically coupled (iii). Doubly-tethered DNA is replicated efficiently by independently functioning replisomes (iv) but not physically coupled replisomes, which stall after available slack in the DNA is consumed (v). (B) Protocol to induce replication of λ DNA immobilized at one end in a flow cell. (C) Visualization of replicated DNA by TIRF microscopy. λ DNA was incubated with extracts lacking (i, ii) or containing (iii, iv) Geminin and stained with anti-digoxigenin or SYTOX, as indicated.
The physical coupling model envisions that DNA is pumped towards the associated sister replisomes, and that newly replicated DNA is extruded laterally from the replisomereplisome interface (Figure 1A-iii). If this model is correct, a constrained DNA template whose ends are fixed should not undergo efficient DNA replication due to tension that accumulates on the unreplicated portions of the molecule (Figure 1A-v). In contrast, independently functioning replisomes should travel apart and copy a constrained DNA template (Figure 1A-iv). Thus, to differentiate between these two models, we used Xenopus egg extracts to replicate DNA that was constrained at one or both ends, and replication was visualized at the single molecule level. The data show that in this vertebrate model system, efficient replication is independent of physical coupling between sister replisomes.
RESULTS
Replication of Immobilized DNA in Xenopus Egg Extracts
Biotinylated λ phage DNA (48.5 kb) was coupled at one or both 3’ ends to the streptavidin-coated, bottom surface of a microfluidic flow cell (Figure S1A). To replicate these DNA molecules, we used a soluble cell-free system derived from Xenopus egg extracts (Walter et al., 1998). DNA is first exposed to a high-speed supernatant (HSS) of egg cytoplasm that supports ORC-dependent but sequence-independent recruitment of MCM2-7 complexes to DNA. A second, nucleoplasmic extract (NPE) is then added, which supports Cdk2-dependent activation of the MCM2-7 helicase, origin unwinding, replisome assembly, and replication of the DNA.
We first examined replication kinetics of λ phage molecules immobilized at only one end, leaving the DNA template unconstrained. After coupling λ phage DNA to the surface, HSS was drawn into the flow cell and allowed to incubate for 10 minutes (Figure 1B-i; for details, see Materials and Methods). Subsequently, HSS was exchanged with NPE containing digoxigenin labeled dUTP (dig-dUTP). After a further 15 minutes, proteins were removed by washing the flow cell with SDS-containing buffer, fluorescein-conjugated anti-digoxigenin antibody (anti-dig) was added, and the DNA was stretched by buffer flow (Figure 1B-ii). Using total internal reflection fluorescence (TIRF) microscopy, we observed co-linear tracts of anti-dig, indicating that replication of the immobilized λ phage DNA had occurred (Figure 1C-i). We also imaged the DNA using SYTOX Orange (“SYTOX”), a fluorescent DNA intercalating dye that labels duplex DNA in a sequence-independent fashion. SYTOX staining revealed that the immobilized λ phage DNA contained alternating tracts of weak and strong fluorescence intensity (Figure 1C-ii). The strong tracts were twice as intense as the weak tracts (Figure S1B-i, orange trace), and they co-localized with anti-dig staining (Figure 1C, compare i and ii; Figure S1B-i) suggesting the strong tracts were due to the presence of two daughter duplexes (schematically depicted in Figure 1B). Both the anti-dig tracts and intense SYTOX tracts disappeared in the presence of Geminin, an inhibitor of MCM2-7 loading (Figure 1C-iii and iv and Figure S1B-ii), indicating that these two signals reflect chromosomal DNA replication. Our results demonstrate that Xenopus egg extracts can efficiently replicate DNA templates immobilized within a microfluidic flow cell, and that this process is readily detected by two independent means (see Figure S2A for additional examples).
Replication kinetics of singly-tethered DNA molecules
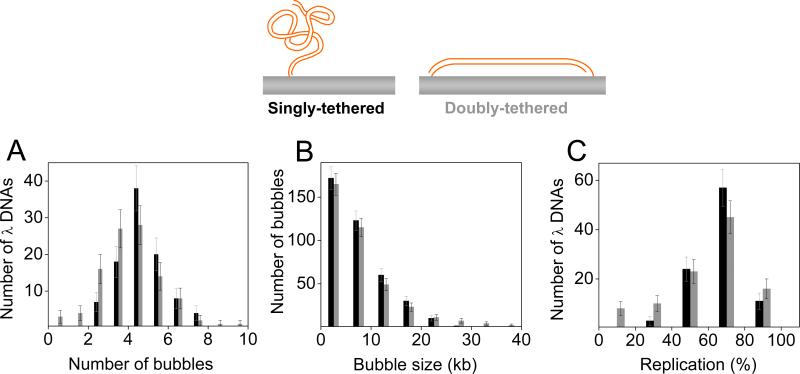
To characterize replication of singly-tethered molecules, we quantified several properties of the replication products (Figure 2A-C, black bars). As shown in Figure 2A, the average number of bubbles per λ DNA was 4.54 ± 1.82, suggesting an average inter-origin distance of 10.7 kb, which agrees well with previous measurements in Xenopus egg extracts (Blow et al., 2001). The lengths of the replication bubbles showed an exponential distribution (Figure 2B), implying that initiation events occurred randomly in time (Herrick et al., 2000). Most λ DNA molecules were more than 60% replicated after 15 minutes in NPE (Figure 2C). These observations suggest that DNA replication of singly-tethered DNA molecules is similar to what was previously observed in Xenopus egg extracts (see also below).
Figure 2. Replication kinetics of immobilized λ DNA.
Quantification of replication on singly-tethered (black) and doubly-tethered (grey) DNA after 15 minutes incubation in NPE. (A) Number of replication bubbles (anti-dig tracts) per immobilized λ DNA. (B) Length distributions of replication bubbles. (C) Percent replication of individual λ DNA molecules. Error bars indicate standard deviations.
Stretched λ DNA is efficiently replicated in extracts from multiple origins
To test whether physical coupling between sister replisomes is required for their function, we repeated the experiment on DNA that was stretched and doubly-tethered. To achieve this condition, DNA molecules biotinylated at both 3’ ends were introduced into the flow cell at high flow rates. Under these conditions, DNA molecules attached to the surface of the flow cell at one end, whereupon they were instantaneously stretched by buffer flow before binding to the surface at the other end. Using this procedure, we achieved end-to-end distances corresponding to ~90% of the expected contour length of B-form λ-DNA (Figure S2C, see Materials and Methods for details). Importantly, such stretched (doubly-tethered) DNA molecules replicated efficiently from multiple origins (Figure 2A-C, grey bars; Figure S2B). When doubly-tethered DNA molecules were incubated in extracts supplemented with Geminin, there was no DNA replication, demonstrating that replication of constrained DNA molecules is also MCM2-7 dependent (data not shown). Importantly, there was no significant difference in the replication of singly-tethered and doubly-tethered molecules (Figure 2A-C, compare black and grey bars), suggesting that no physical contact between sister replisomes is required for replication in our system.
A single pair of diverging forks replicates stretched λ DNA
Given that there was on average about 10% slack present in the doubly-tethered λ DNA, the replication observed above could have involved many short, neighboring replicons synthesized by physically coupled replisomes. To address this caveat, we examined whether a single pair of diverging sister forks can replicate stretched λ DNA to an extent larger than the slack originally present. To ensure that only a single pair of replisomes was activated on each DNA molecule, we used p27Kip, a Cdk2 inhibitor that blocks new initiations but does not affect elongation (Walter and Newport, 2000). Thus, 2-5 minutes after replication was initiated with NPE, we flowed in fresh NPE containing p27Kip (Figure 3A). Under these conditions, the majority of DNA molecules exhibited one or no replication bubbles (Figure 3C). To verify that the observed bubbles were produced by two diverging replication forks, we supplied dig-dUTP 15 minutes after the initial NPE addition and allowed replication to proceed for a further 25 minutes (Figure 3A).
Figure 3. Replication of stretched DNA by a single pair of diverging forks.
(A) Scheme used to limit replication initiation to a single event on each λ DNA molecule and to visualize bidirectional replication. (B) SYTOX (top), anti-dig (middle), and merged (bottom) images of three mechanically stretched λ DNA molecules containing a single replication bubble. Extent of slack and replication are indicated. (C) Number of replication bubbles per monomeric λ DNAs (n=39). (D) Extent of replication versus the amount of slack on individual λ DNA molecules that underwent single initiations. The amount of slack was calculated by comparing the end-to-end distance of doubly-tethered DNA molecule to the B-form contour length of λ DNA (16.5 μm). The solid line depicts the extent of replication expected if replication stops when the slack initially present in the λ DNA is used up, as expected for physically coupled forks.
In the replicated molecules, two tracts of dig-dUTP were visible whose outer edges coincided with the boundaries of the intense SYTOX tract, as expected for bi-directional replication from a single origin (Figure 3B and S3A). Figure 3D shows that the extent of replication on every λ molecule examined was much greater than the slack present in the substrate. For a single pair of physically coupled replisomes to produce such large replication bubbles, the λ molecule would have to be stretched well beyond the contour length of B-form DNA. Because the force required to overstretch DNA to such an extent (van Mameren et al., 2009) is larger than any reported for individual DNA motors (Smith et al., 2001), and because such overstretching would almost entirely denature the DNA substrate (van Mameren et al., 2009), it is very unlikely that sister replisomes remained connected during replication in this system. We conclude that sister replisomes can function independently on immobilized λ DNA molecules.
Uncoupling of sister replisomes does not affect fork rates
To test whether the uncoupling of sister replisomes affects their replication dynamics, we compared fork rates on stretched, doubly-tethered and relaxed, singly-tethered DNAs that had undergone a single initiation event (Figure S3). Dividing the lengths of the anti-dig tracts by the duration of the dig-dUTP pulse yielded a mean fork rate of 267 ± 160 bp/min for stretched DNA (n = 91; Figure 4A, grey bars) and 268 ± 161 bp/min for relaxed DNA (n = 98; Figure 4A, black bars). The presence of dig-dUTP did not affect the rate of DNA synthesis (Figure S3C). The measured rates were close to the lower estimates of fork rates in conventional, nuclear assembly Xenopus egg extracts ([Lu et al., 1998] and [Mahbubani et al., 1992]). Thus, replisomes on constrained and unconstrained molecules move at the same rates, demonstrating that replisome uncoupling does not adversely affect replication fork progression.
Figure 4. Analysis of fork-rates.
(A) Length of anti-dig tracts under single-initiation conditions on singly-tethered (black) and doubly-tethered (grey) λ DNA molecules. Error bars indicate standard deviations. (B) Lengths of sister anti-dig tracts of the rightward fork versus the leftward fork on singly (black) and doubly-tethered (grey) DNA molecules. The dashed line represents perfectly correlated sister forks.
Correlation between rates of sister forks on singly-tethered but not doubly-tethered DNA
Previous studies in different experimental systems showed varying but significant degrees of correlation between the rates at which sister replication forks progress ([Conti et al., 2007], [Dubey and Raman, 1987] and [Tapper and Depamphilis, 1980]). We looked for a correlation between progression of sister forks in our system. Figure 4B plots the length of the left versus right anti-dig tracts within single replication bubbles on singly and doubly-tethered DNA. On singly-tethered DNA, there was a weak, positive correlation between the rates at which the two sister replisomes moved (Figure 4B, black squares, n=48; R = 0.26, p = 0.07). Since uncoupling of sister replisomes did not affect fork progression in our system, the correlation between sister forks in relaxed DNA is unlikely to be related to a functional interaction between sisters. Consistent with this, termination of one replication fork by a double-strand DNA break in yeast does not affect progression of the sister fork (Doksani et al., 2009). Therefore, correlations that we and others observed likely represent chromatin microenvironments that result in similar activity of nearby replisomes (Conti et al., 2007), perhaps due to similar concentrations of key replication factors. Consistent with this idea, sister replisomes moving on stretched DNA, which are separated in space, showed no correlation (Figure 4B, grey squares, n=45; R = -0.1, p = 0.47).
Discussion
It has been proposed that sister replisomes function as an obligatory dimeric complex ([Falaschi, 2000], [Kitamura et al., 2006], [Ligasová et al., 2009] and [Sclafani et al., 2004]). However, our data demonstrates that no physical association is required between sister replisomes on λ DNA replicating in Xenopus egg extracts suggesting that replisomes can function independently during vertebrate DNA replication. Together with previous results which failed to find evidence of MCM2-7 double hexamers in S phase using co-IP approaches (Gambus et al., 2006), and recent experiments using purified MCM2-7 holocomplexes (Ilves et al., 2010, Moyer et al., 2006), our data suggest that sister MCM2-7 helicases (and replisomes) normally uncouple upon activation, as seen in bacteria (Reyes-Lamothe et al., 2008), even though they may co-localize within replication foci (Kitamura et al., 2006, Ligasová et al., 2009).
The double hexamer model for MCM2-7 was largely inspired by the analysis of SV40 T-ag. Electron microscopy showed that T-ag loads onto the SV40 origin as two hexamers, which associate through their N-termini (Valle et al., 2000). In addition, during T-ag-mediated DNA unwinding, a fraction of DNAs adopt a “rabbit ear” conformation, in which two loops of single-stranded DNA emanate from the T-ag complex, implying an association of two hexamers (Wessel et al., 1992). Finally, mutations in T-ag that compromise double-hexamer formation inhibit DNA unwinding, and double-hexamers of T-ag possess higher unwinding activity than single hexamers ([Alexandrov et al., 2002] and [Weisshart et al., 1999]). Taken together, these studies strongly suggest that double-hexamer formation is crucial for SV40 replication. However, it is not clear whether the interactions between two hexamers are essential only for replication initiation or also during fork elongation.
The independent action of sister replisomes has significant advantages for eukaryotic cells. First, the multi-replicon model, in which sister replisomes complete DNA synthesis at different times would be difficult to reconcile with obligatory physical coupling between sisters. Second, single replisomes, having smaller dimensions than double replisomes, may be more adept at bypassing certain roadblocks and navigating a highly populated nuclear environment (Takahashi et al., 2004). The single-molecule techniques described here should be suitable to investigate the requirement for physical association between sister replisomes in other systems.
EXPERIMENTAL PROCEDURES
DNA immobilization in the microfluidic flow cell
Sample flow cells were prepared as described previously (Lee et al., 2006). Briefly, coverslips were functionalized with partially biotinylated high-molecular weight poly(ethylene glycol) and incubated with 1 mg/ml streptavidin. Flow cells were assembled using these coverslips, double-adhesive tape and glass slides with pre-drilled holes (Figure S1A). Outlet polyethylene tubing (0.03 inch inner diameter) was attached to an automated syringe pump (Harvard Instruments) to provide constant flow. Inlet tubing with 5 cm length and 0.015 inch inner diameter reduced dead volume. To prevent nonspecific DNA sticking to the surface, the flow cell was incubated with blocking buffer (20 mM Tris pH 7.5, 50 mM NaCl, 2 mM EDTA, 0.2 mg/ml BSA) for at least 15 minutes.
To attach λ DNA (New England Biolabs) to the streptavidin coated surface at one end, the single-stranded 5’ tails of λ DNA were annealed and ligated to complementary oligonucleotides 5’-AGGTCGCCGCCC-Biotin-3’ and 5’-GGGCGGCGACCT-3’ (Integrated DNA Technologies). For double tethering, both oligos contained biotin at the 3’ end. Biotinylated λ DNA (15-50 pM) in blocking buffer was injected into the flow cell at a constant rate of 20-100 μl/min. At a flow rate of 50-100 μl/min, λ DNA biotinylated at both ends was stretched to 70-80% of its contour length (16.5 μm) (Figure S2C i-iii). To stretch DNA further, we used chloroquine, which intercalates into and extends the pitch of dsDNA (Cohen and Yielding, 1965). When injected at 100 μl/min in the presence of 100 μM chloroquine in blocking buffer, end-to-end distance of doubly-tethered λ DNAs was 85-95% of its contour length (Figure S2C-iv). After DNA injection and before addition of extract, chloroquine was removed by extensive washing of the flow cell with blocking buffer (5 minutes at 100 μl/min).
To limit our analysis to DNA molecules that remained doubly-tethered during the entire replication reaction, we used a reduced flow rate (25 μl/min) for all buffer exchanges following replication. In this way, even if a DNA molecule that detached from one end in extracts became doubly-tethered during subsequent washes, it would be stretched to a much lesser extent than molecules that remained stretched throughout the experiment. Thus, λ DNA stretched to 85-95% of its contour length at the end of the experiment must have stayed doubly-tethered during replication. Therefore, we analyzed only those molecules that were stretched to 85% or more as doubly-tethered.
Replication of immobilized DNA
In a separate line of investigation, we recently discovered that DNA replication in Xenopus egg extracts requires a minimum threshold concentration of DNA (~1 ng/μl) in HSS and NPE (Lebofsky et al., in press). Since the effective concentration of λ DNA immobilized in the flow cell was extremely low, we supplemented HSS (Walter et al., 1998) and NPE (Walter et al., 1998) with “carrier” plasmid to raise the overall DNA concentration to levels that are compatible with DNA replication. Thus, after immobilizing λ DNA on the functionalized surface, HSS containing carrier plasmid (5-10 ng/μl of pBluescript II KS (-)) was injected at 10 μl/min for 2 minutes and further incubated for 8 minutes without flow. Next, a 2:1 mixture of NPE and HSS supplemented with 5-10 ng/μl of pBS (“replication extract”) was flowed in at 10 μl/min for 80 seconds, followed by incubation for different lengths of time without flow, as indicated. 7 μM dig-dUTP (Roche Inc.) was also included in the replication extract for labeling of replicated regions. All reactions were carried out at room temperature (22 °C).
To observe bidirectional replication involving single initiation events, immobilized λ DNA was incubated with HSS/Carrier plasmid and subsequently replaced with replication extract (lacking dig-dUTP). After the time specified, a second replication extract containing 66 μg/ml p27Kip was injected and incubated further. Finally, a replication extract containing 66 μg/ml p27Kip and 7 μM dig-dUTP was introduced. In each case, replication extract was injected at 10 μl/min for 80 seconds.
To stop the replication reaction, the flow cell was washed with SDS buffer (20 mM Tris pH 7.5, 50 mM NaCl, 12 mM EDTA, 0.1% SDS) for 10 minutes at 25 μl/min. To label dig-dUTP, anti-digoxigenin-fluorescein antibody (Roche Inc.) diluted to 0.4 μg/ml with buffer (10 mM Hepes pH 7.7, 2.5 mM MgCl2, 50 mM KCl, 0.2 mg/ml BSA) was drawn into the flow cell for 20 minutes at 25 μl/min. Excess antibody was removed by washing the flow cell with blocking buffer. Finally, blocking buffer containing 15 nM SYTOX Orange (Invitrogen) was introduced to fluorescently label dsDNA.
Use of oligonucleotides as carrier DNA
As described above, DNA replication of tethered λ DNA in the flow cell was carried out using egg extracts (HSS and NPE) that were supplemented with high concentrations of a carrier plasmid. Like the immobilized λ DNA, the free carrier plasmid is also expected to undergo replication in the flow cell. To ensure that the activity of replisomes on the immobilized λ DNA was not dependent on interactions with replisomes on the carrier plasmid, we replaced the carrier plasmid with a short, double-stranded oligonucleotide (29 bp; Lebofsky et al., in press) that does not support loading of MCM2-7 helicase due its small size and therefore cannot undergo replication (Edwards et al., 2002). We have shown that such a nucleotide is able to replace carrier plasmid to promote licensing in HSS (Lebofsky et al., in press).
Doubly-tethered λ DNA was licensed with HSS containing 10 ng/μl of the oligo duplex. Subsequently, standard replication extract (NPE/HSS/Carrier plasmid) was injected. Importantly, the carrier plasmid supplied with the replication extracts did not undergo DNA replication because it was added to a mixture of HSS and NPE, and NPE contains very high concentrations of Geminin, which block DNA replication (Figure S4A) (Walter et al., 1998). Thus, in this sequence, none of the added carrier DNA (oligo or plasmid) underwent DNA replication, yet single initiations on many stretched λ DNA molecules still produced bubbles larger than the slack present on stretched λ DNA (Figure S4B). These observations confirm that replication of carrier plasmid does not mediate replication of immobilized λ DNA molecules.
TIRF Microscopy
Immobilized λ DNA molecules were imaged on an objective-type total internal reflection fluorescence configuration using an inverted microscope (IX-71; Olympus) equipped with a 60x oil objective (PlanApo, N.A.=1.45; Olympus) and a 1.6x magnification unit. A multi-wavelentgh Ar-Kr ion laser (Innova 70C-Spectrum, Coherent Inc.) was used for illumination. SYTOX Orange and fluorescein were excited with 568 and 488-nm laser light, respectively, using varying intensities and 100 ms exposures per frame. Images were acquired using an Andor iXon back-illuminated electron-multiplying CCD camera (Andor Technology) at 2 Hz. Singly-tethered molecules were imaged at a flow rate of 100-125 μl/min (for stretching), while doubly-tethered DNAs were imaged in the absence of flow since they were already stretched.
Image processing
To improve the signal-to-noise ratio on fluorescence images of replicated λ DNA, multiple (5-30) consecutive images of SYTOX and fluorescein were averaged separately using ImageJ and merged using Adobe Photoshop. The end-to-end distance of each DNA molecule was measured via the SYTOX image and the size of a replication bubble was determined using the SYTOX or the fluorescein signal.
Supplementary Material
Acknowledgements
We thank Ronald Lebofsky for communicating unpublished results and Charles C. Richardson, Ronald Lebofsky, and Jerard Hurwitz for critical reading of the manuscript. This work was supported by NIH grant GM62267 and a Leukemia and Lymphoma Scholar Award to J.C.W. A.M.v.O. acknowledges support from American Cancer Society grant RSG-08-234-01 and Searle Scholarship 05-L-104. A. B. L. was supported by NIH/NIGMS Molecular Biophysics Training Grant T32 GM008313.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- First cell-free system for single molecule analysis of eukaryotic DNA replication.
- Nanomanipulation of DNA molecules used as substrates for eukaryotic replication.
- Sister replisomes can uncouple during replication and function independently.
- Uncoupling of sister replisomes does not affect fork progression.
REFERENCES
- Alexandrov AI, Botchan MR, Cozzarelli NR. Characterization of Simian Virus 40 T-antigen Double Hexamers Bound to a Replication Fork. J. Biol. Chem. 2002;277:44886–44897. doi: 10.1074/jbc.M207022200. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Gillespie PJ, Francis D, Jackson DA. Replication Origins in Xenopus Egg Extract Are 5-15 Kilobases Apart and Are Activated in Clusters That Fire at Different Times. J. Cell Biol. 2001;152:15–26. doi: 10.1083/jcb.152.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A. The Mcm Complex: Unwinding the Mechanism of a Replicative Helicase. Microbiol. Mol. Biol. Rev. 2009;73:652–683. doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SN, Yielding KL. Spectrophotometric Studies of the Interaction of Chloroquine with Deoxyribonucleic Acid. J. Biol. Chem. 1965;240:3123–3131. [PubMed] [Google Scholar]
- Conti C, Sacca B, Herrick J, Lalou C, Pommier Y, Bensimon A. Replication Fork Velocities at Adjacent Replication Origins Are Coordinately Modified during DNA Replication in Human Cells. Mol. Biol. Cell. 2007;18:3059–3067. doi: 10.1091/mbc.E06-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y, Bermejo R, Fiorani S, Haber JE, Foiani M. Replicon Dynamics, Dormant Origin Firing, and Terminal Fork Integrity after Double-Strand Break Formation. Cell. 2009;137:247–258. doi: 10.1016/j.cell.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey DD, Raman R. Do sister forks of bidirectionally growing replicons proceed at unequal rates? Exp. Cell Res. 1987;168:555–560. doi: 10.1016/0014-4827(87)90028-0. [DOI] [PubMed] [Google Scholar]
- Edwards MC, Tutter AV, Cvetic C, Gilbert CH, Prokhorova TA, Walter JC. MCM2-7 Complexes Bind Chromatin in a Distributed Pattern Surrounding the Origin Recognition Complex in Xenopus Egg Extracts. J. Biol. Chem. 2002;277:33049–33057. doi: 10.1074/jbc.M204438200. [DOI] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. U. S. A. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falaschi A. Eukaryotic DNA replication: a model for a fixed double replisome. Trends Genet. 2000;16:88–92. doi: 10.1016/s0168-9525(99)01917-4. [DOI] [PubMed] [Google Scholar]
- Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat. Struct. Biol. 2003;10:160–167. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Herrick J, Stanislawski P, Hyrien O, Bensimon A. Replication fork density increases during DNA synthesis in X. laevis egg extracts. J. Mol. Biol. 2000;300:1133–1142. doi: 10.1006/jmbi.2000.3930. [DOI] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 Helicase by Association with Cdc45 and GINS Proteins. Mol. Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E, Blow JJ, Tanaka TU. Live-Cell Imaging Reveals Replication of Individual Replicons in Eukaryotic Replication Factories. Cell. 2006;125:1297–1308. doi: 10.1016/j.cell.2006.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebofsky R, van Oijen AM, Walter JC. DNA is a co-factor for its own replication in Xenopus egg extracts. Nucleic Acids Res. doi: 10.1093/nar/gkq739. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-B, Hite RK, Hamdan SM, Sunney Xie X, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- Ligasová A, Raska I, Koberna K. Organization of human replicon: Singles or zipping couples? J. Struct. Biol. 2009;165:204–213. doi: 10.1016/j.jsb.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZH, Sittman DB, Romanowski P, Leno GH. Histone H1 Reduces the Frequency of Initiation in Xenopus Egg Extract by Limiting the Assembly of Prereplication Complexes on Sperm Chromatin. Mol. Biol. Cell. 1998;9:1163–1176. doi: 10.1091/mbc.9.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahbubani HM, Paull T, EIder JK, Blow JJ. DNA replication initiates at multiple sites on plasmid DNA in Xenopus egg extracts. Nucl. Acids Res. 1992;20:1457–1462. doi: 10.1093/nar/20.7.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proceedings of the National Academy of Sciences. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JFX. Concerted Loading of Mcm27 Double Hexamers around DNA during DNA Replication Origin Licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. Independent Positioning and Action of Escherichia coli Replisomes in Live Cells. Cell. 2008;133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA, Fletcher RJ, Chen XS. Two heads are better than one: regulation of DNA replication by hexameric helicases. Genes Dev. 2004;18:2039–2045. doi: 10.1101/gad.1240604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- Takahashi TS, Wigley DB, Walter JC. Pumps, paradoxes and ploughshares: mechanism of the MCM2-7 DNA helicase. Trends Biochem. Sci. 2005;30:437–444. doi: 10.1016/j.tibs.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat. Cell Biol. 2004;6:991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- Tapper DP, Depamphilis ML. Preferred DNA sites are involved in the arrest and initiation of DNA synthesis during replication of SV40 DNA. Cell. 1980;22:97–108. doi: 10.1016/0092-8674(80)90158-0. [DOI] [PubMed] [Google Scholar]
- Valle M, Gruss C, Halmer L, Carazo JM, Donate LE. Large T-Antigen Double Hexamers Imaged at the Simian Virus 40 Origin of Replication. Mol. Cell. Biol. 2000;20:34–41. doi: 10.1128/mcb.20.1.34-41.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mameren J, Gross P, Farge G, Hooijman P, Modesti M, Falkenberg M, Wuite GJL, Peterman EJG. Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18231–18236. doi: 10.1073/pnas.0904322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Newport J. Initiation of Eukaryotic DNA Replication: Origin Unwinding and Sequential Chromatin Association of Cdc45, RPA, and DNA Polymerase alpha. Mol. Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Walter J, Sun L, Newport J. Regulated Chromosomal DNA Replication in the Absence of a Nucleus. Mol. Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- Weisshart K, Taneja P, Jenne A, Herbig U, Simmons DT, Fanning E. Two Regions of Simian Virus 40 T Antigen Determine Cooperativity of Double-Hexamer Assembly on the Viral Origin of DNA Replication and Promote Hexamer Interactions during Bidirectional Origin DNA Unwinding. J. Virol. 1999;73:2201–2211. doi: 10.1128/jvi.73.3.2201-2211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel R, Schweizer J, Stahl H. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J. Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.