Abstract
Background
Zymomonas mobilis ZM4 is a Gram-negative bacterium that can efficiently produce ethanol from various carbon substrates, including glucose, fructose, and sucrose, via the Entner-Doudoroff pathway. However, systems metabolic engineering is required to further enhance its metabolic performance for industrial application. As an important step towards this goal, the genome-scale metabolic model of Z. mobilis is required to systematically analyze in silico the metabolic characteristics of this bacterium under a wide range of genotypic and environmental conditions.
Results
The genome-scale metabolic model of Z. mobilis ZM4, ZmoMBEL601, was reconstructed based on its annotated genes, literature, physiological and biochemical databases. The metabolic model comprises 579 metabolites and 601 metabolic reactions (571 biochemical conversion and 30 transport reactions), built upon extensive search of existing knowledge. Physiological features of Z. mobilis were then examined using constraints-based flux analysis in detail as follows. First, the physiological changes of Z. mobilis as it shifts from anaerobic to aerobic environments (i.e. aerobic shift) were investigated. Then the intensities of flux-sum, which is the cluster of either all ingoing or outgoing fluxes through a metabolite, and the maximum in silico yields of ethanol for Z. mobilis and Escherichia coli were compared and analyzed. Furthermore, the substrate utilization range of Z. mobilis was expanded to include pentose sugar metabolism by introducing metabolic pathways to allow Z. mobilis to utilize pentose sugars. Finally, double gene knock-out simulations were performed to design a strategy for efficiently producing succinic acid as another example of application of the genome-scale metabolic model of Z. mobilis.
Conclusion
The genome-scale metabolic model reconstructed in this study was able to successfully represent the metabolic characteristics of Z. mobilis under various conditions as validated by experiments and literature information. This reconstructed metabolic model will allow better understanding of Z. mobilis metabolism and consequently designing metabolic engineering strategies for various biotechnological applications.
Background
The impact of biotechnology on industry and society is dramatically gaining momentum, particularly in the field of agriculture-food, medicine and chemical production. For the chemical industry, which aims to producing value-added chemicals and fuels in a sustainable way, efforts have been put into strain improvement of microorganisms, utilizing many newly emerging state-of-art techniques, for the overproduction of chemicals of interest [1-6]. Yet, the most common problem encountered in strain improvement is that the microorganisms are not naturally optimized for the overproduction of the target compounds desired for human use. Instead, these compounds are produced in small amounts which are sufficient for the microorganism's purpose. Therefore, it is necessary to engineer the microorganism so that the metabolic fluxes are redirected towards overproducing the target products without significantly hampering the overall cellular behavior.
In silico genome-scale metabolic modeling and simulation have proven to be useful in the field of systems metabolic engineering. This approach has successfully contributed to the design of strategies for engineering microorganisms for the production of amino acids, including L-valine [7] and L-threonine [8], lycopene [9], succinic acid [10], ethanol [11], and polylactic acid [12]. Genome-scale modeling and constraints-based flux analysis enables the calculation of intracellular fluxes based on the complex stoichiometric relationship of metabolites constituting the metabolic network. The strength of genome-scale modeling is that it not only predicts the effects of genetic and environmental perturbations on cellular metabolism from a holistic point of view, but can also be used in combination with other high-throughput techniques, for instance gene expression data [7,13].
Zymomonas mobilis, a Gram-negative bacterium, metabolizes glucose, fructose and sucrose via the Entner-Doudoroff (ED) pathway, and is capable of producing up to 12% (w/v) ethanol at a faster rate than yeast [14,15]. In addition to its high ethanol producing ability, its fast sugar consumption and processing rate, and high ethanol tolerance of up to 16% (vol/vol) have attracted attention to Z. mobilis, as a host for industrial biotechnology [15,16]. Recently, Seo et al. [15] first reported the complete sequence and the annotation of the Z. mobilis ZM4 genome, and Yang et al. [17] updated the data, enabling subsequent systematic studies of this organism and hence applications. Spurred with this complete genome sequence and annotation of Z. mobilis ZM4 genome, its genome-scale modeling and simulation could be employed for systematic analyses to understand the characteristics of its metabolism and to design efficient metabolic strategies.
A small scale model of engineered Z. mobilis has already been constructed by Tsantili et al. [18]. This small scale model consists of the central metabolism supported by a few key metabolic reactions that are a lumped representation of cellular functions. However, with the genome-scale metabolic model, we aim to describe the overall metabolic characteristics of Z. mobilis with greater accuracy and scope of its metabolic functions. Pinto et al. [19] reported a study about data integration process for the metabolic network reconstruction of the Z. mobilis. Pinto et al took genome annotation data about Z. mobilis from the NCBI, and obtained the reaction list with stoichiometry data from KEGG [20], BioCyc [21] and BRENDA [22]. However, Pinto et al only focused on the first steps for the collection and processing of the information related to the reconstruction of genome-scale metabolic network. For the reconstruction of promising genome-scale metabolic model, several steps such as proofreading process, determination of biomass composition, and in silico simulations and their validations of the model based on experiments should be encompassed, as depicted on Figure 1. Here, we present a genome-scale metabolic model of the Z. mobilis ZM4, ZmoMBEL601, composed of 601 reactions and 579 metabolites, for systematic characterization of this organism (Table 1; Additional file 1 and 2).
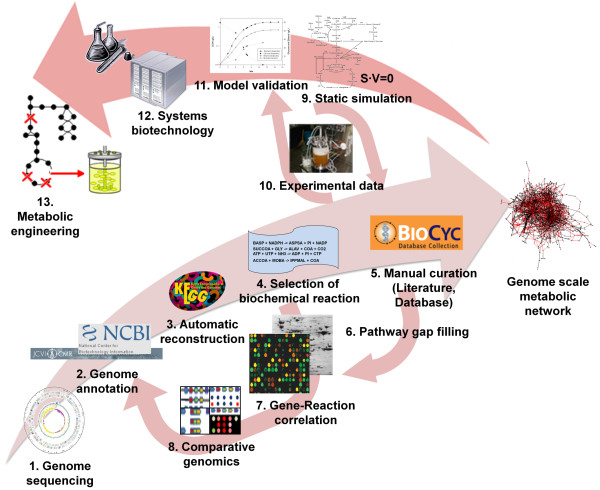
Figure 1.
Procedure for the reconstruction of genome-scale metabolic network in Z. mobilis and its application to metabolic engineering. Automatic reconstruction of metabolic network based on genome sequence and annotation data (1-2-3-4). Manual curation of the metabolic network using literatures, databases, gene-reaction correlation, and comparative genomics to correct errors and fill gaps in the pathways (4-7-8-2 or 5-6-8-2). Determination of biomass composition and validation of metabolic model in comparison with experimental data (9-10-11). Systems metabolic engineering for strain improvement by combining experimental and in silico approaches (12-13). The detailed information about fermentation profile (11), utilized to refine reconstructed metabolic model is in additional file 7.
Table 1.
Features of the in silico metabolic model of Z. mobilis ZM4
| Features | Number |
|---|---|
| Genome featurea | |
| Genome size (base pair, bp) | 2,056,363 |
| Total genes | 1,808 |
| Open reading frames (ORFs) | 1,728 |
| In silico metabolic model | |
| Total reactions | 601 |
| Biochemical reactions | 571 |
| Transport reactions | 30 |
| Metabolites | 579 |
| ORFs assigned in metabolic network | 348 |
| ORF coverageb | 20.14% |
Results and discussion
Genome-scale reconstruction of Z. mobilis ZM4 metabolic model
To reconstruct the Z. mobilis metabolic model, the NCBI [23], CMR [24], and ExPASy [25] databases are utilized first to obtain information regarding Z. mobilis's genome sequence and its annotation. Then, the data on the metabolic reactions and metabolites were obtained from several databases and literatures. KEGG [20], which contains diverse information about biological pathways, was predominantly utilized in the construction of the draft metabolic model (Figure 1). The TCDB [26] and TransportDB [27] were employed in the collection of information regarding transport systems, which KEGG is deficient in, present in Z. mobilis to implement the nutrient uptake and product secretion systems between the intracellular and external environments. Meanwhile, the BioCyc [21] database was used to define each reaction's reversibility. Additionally, the BioSilico database [28] was applied to compare and integrate previously obtained information. Reactions, which are not assigned to a gene in Z. mobilis or for which no evidence is available for its presence in Z. mobilis, can also be added to the metabolic model. In the former case, literature evidence would allow for the addition of the metabolic reaction to the metabolic model. In the latter case, there are instances where the metabolic reaction where no literature evidence is present but is necessary to achieve the feasible flux distribution in the reconstructed metabolic model. In this case the metabolic reaction is added and noted as a point of interest in further studies into Z. mobilis.
The resulting ZmoMBEL601 metabolic model comprises 579 metabolites and 601 metabolic reactions, comprising of 571 biochemical conversions and 30 transport reactions. A total of 347 open reading frames (ORFs) were included in the metabolic model, which represents approximately 20.1% of the ORFs with assigned function in the Z. mobilis ZM4 genome (Table 1; Additional file 1 and 2). This ORF coverage in the ZmoMBEL601 metabolic model is similar to other reported genome-scale metabolic models (Additional file 3) [29-38] due to its small genome size. Once reconstructed, ZmoMBEL601 was dissected in detail to further characterize the metabolic network.
General features of the Z. mobilis ZM4 metabolic model
The central carbon metabolism of Z. mobilis is different compared to other known gram-negative microorganisms, such as E. coli. Z. mobilis is known to metabolize only glucose, fructose and sucrose through the ED pathway, producing ethanol and CO2, and is unable to utilize the glycolytic pathway due to the absence of 6-phosphofructokinase, which converts fructose-6-phosphate into fructose-1,6-bisphosphate (Figure 2A) [16,39]. Z. mobilis ZM4 also does not have two enzymes in the tricarboxylic acid (TCA) cycle: 2-oxoglutarate dehydrogenase and malate dehydrogenase (Figure 2A). Despite the absence of these enzymes, Z. mobilis is still able to produce important building blocks including oxaloacetate, malic acid, and fumaric acid through alternative metabolic pathways; phosphoenolpyruvate carboxylase (phophoenolpyruvate + CO2 → oxaloacetate + orthophosphate) and citrate lyase (citrate → acetate + oxaloacetate) for oxaloacetate production, malic enzyme (malate ↔ pyruvate + CO2) for malic acid production, and fumarate dehydratase (malate ↔ fumarate) for fumaric acid production. These characteristic features of central metabolism in Z. mobilis ZM4 were reflected in ZmoMBEL601.
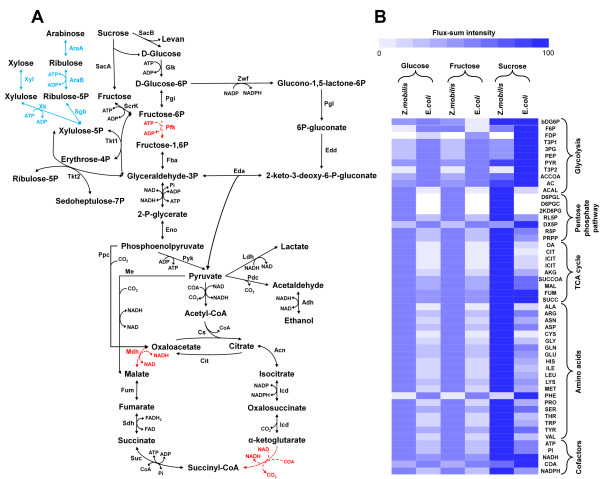
Figure 2.
Central metabolism and analysis of flux-sum intensity in Z. mobilis. (A) Central metabolism, including ED Pathway and TCA cycle in Z. mobilis based on annotated genes. Reactions for utilizing pentose sugars in Z. mobilis are introduced and represented to blue lines. The red lines indicate the missing reactions in Z. mobilis. (B) Flux-sum intensity of selected metabolites, including cofactors, amino acids and others, in central metabolism for Z. mobilis and E. coli, respectively, for three carbon sources (i.e. glucose, fructose, and sucrose). Uptake rate of each carbon source is fixed to 10 mmol/gDCW/h, and reaction for oxygen uptaking was deleted to describe anaerobic condition in both organisms. NGAME (i.e. non-growth associated maintenance energy) value for both organisms was eliminated. Upper constraints (i.e. limit the flux on fumaric acid, acetic acid, malic acid, acetoin, and acetaldehyde production) in Z. mobilis metabolic model for regulating the flux more realistically are relieved. Results are normalized by maximum value of each metabolite. Darker color indicates stronger flux-sum intensity.
The equation for biomass formation, which requires precursors from a set of biosynthetic reactions, was constructed to describe the cell growth (Additional file 1 and 4). The lipid composition of Z. mobilis was found to have a unique feature compared to other organisms, which allows for tolerance to higher alcohol levels. Because short-chain alcohols (e.g. methanol, ethanol) directly interacts with the lipid bilayer and increases its fluidity, the cell tries to maintain its membrane fluidity by changing the composition of the lipid layer; increasing the amount of long-chain fatty acids to create a rigid cell wall and synthesizing hopanoids, a pentacyclic lipid compounds, which can adjust the cell membrane permeability. This abundance of hopanoids and vaccenic acid (C18:1) in Z. mobilis was proposed to explain the evolutionary adaptation of Z. mobilis to survive in the presence of high ethanol level [40]. It has been reported that the lipids of Z. mobilis are composed of three groups of compounds: phospholipids, hopanoids, and nonpolar lipids [41]. Hopanoids are further categorized into five types: tetrahydroxybacteriohopanetetrol (THBH), tetrahydroxybacteriohopane-glucosamine (THBH-GA), tetrahydroxybacteriohopane-ether (THBH-ET), diplopterol, and dopene [41-43]. However, genes involved in the biosynthesis of the five types of hopanoids have not been annotated. Therefore, hopanoids biosynthetic pathways were constructed and incorporated into the metabolic model based on the results previously reported [42,44-46] (Additional file 1 and 4). Reactions biosynthesizing 5 different types of phospholipids [40,41,47] and 9 different types of nonpolar lipids [40] in the membrane of Z. mobilis were included in the same manner based on existing reports.
Physiological characteristics of Z. mobilis related to aerobic metabolism
Z. mobilis ZM4 is a facultative anaerobic microorganism, and has been reported to have a reduced growth rate and ethanol production rate under aerobic conditions compared to anaerobic condition [15]. This phenomenon can be explained by the presence of two key enzymes of the Z. mobilis metabolism; a type 2 NADH oxidoreductase and an NADH oxidase.
Z. mobilis possesses a type 2 NADH oxidoreductase, which does not pump protons during electron transport in aerobic respiration, unlike the more common type 1 NADH oxidoreductase [48]. Thus, type 2 NADH oxidoreductase does not contribute to the proton gradient of the cellular membrane, which is the driving force in generating ATP. While this type 2 NADH oxidoreductase does not pump protons while passing electrons through the electron transport chain in Z. mobilis, other membrane proteins, such as cytochrome bc1 complex, electron transfer flavoprotein, and ubiquinone protein, are present to generate the proton gradient and thereby drive ATP generation under aerobic condition. Therefore, Z. mobilis can grow under aerobic conditions but at a lower growth rate due to the decreased supply of ATP. The Z. mobilis NADH oxidase catalyzes the oxidization of NADH (NADH + 0.5 O2 → NAD) under aerobic condition. Because of this enzyme, the pool of NADH, which is used for ethanol production, is decreased, resulting in a decrease in capacity for producing ethanol under aerobic condition [48].
These characteristics of aerobic shift, which is the environmental change from anaerobic to aerobic condition, are reflected in ZmoMBEL601, and were validated by performing simulations using constraints-based flux analysis and comparing the results to published data. The growth rate and ethanol production rate in Z. mobilis were modeled at several different values of the constraints (0, 1, 2, and 2.8 mmol/gDCW/h) applied to the NADH oxidase activity during the simulation under aerobic condition (Figure 3A). The predicted growth and ethanol production rate were found to decrease gradually as the flux rate of NADH oxidase increased (Figure 3A). Additionally, because the NADH oxidase utilizes O2 to oxidize NADH, the activity level of NADH oxidase is directly correlated to the O2 uptake rate. Therefore, it can say that the production rate of ethanol and growth rate decreased as the O2 uptake rates increased [16,49]. These consistent simulation outcomes validate ZmoMBEL601.
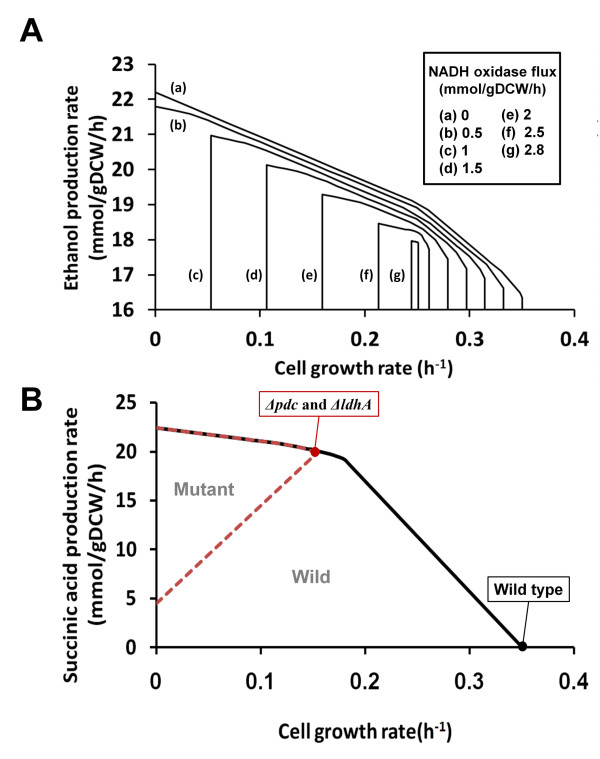
Figure 3.
Characteristics of aerobic shift and strategies for succinic acid production in Z. mobilis. (A) Trade-off curves of Z. mobilis for the flux rate of NADH oxidase reaction, ethanol production rate, and growth rate under aerobic condition. The relationship between growth rate and ethanol production rate was investigated for several flux levels of NADH oxidase reaction constrained by 0, 1, 2, and 2.8 mmol/gDCW/h under aerobic condition. The vertical lines of each graph that meet x-axis drop to zero. (B) Trade-off curves of wild type and mutant (Δpdc and ΔldhA) strains of Z. mobilis for succinic acid production under anaerobic condition. The black solid line and the red dotted line indicate the flux solution space of the wild type and mutant strain of Z. mobilis, respectively. Each circle denotes the state of the wild type and mutant strain of Z. mobilis on each maximal growth rate, respectively.
Analysis of flux-sum intensity between Z. mobilis and E. coli
The analysis of flux-sum intensity between Z. mobilis and E. coli was carried out with the three carbon sources, glucose, fructose, and sucrose, under anaerobic condition with defined minimal medium (Figure 2B) [50,51]. The flux-sum is defined as half of the summation of all consumption and generation fluxes around a particular metabolite under pseudo-steady state [50,51]. As the flux-sum is closely related to the turnover rate of metabolites, the metabolic state of the system can be elucidated through flux-sum analysis that examines the interconversion pattern of specific metabolites comprising of the network [51].
Metabolites, including some cofactors, amino acids, and others involved in central carbon metabolic pathways, were selected and categorized into a few groups to display the characteristic of each pathway. Flux-sum intensity of all the aforementioned metabolites, except for F6P in Z. mobilis, and for bDG6P, F6P and FDP in E. coli shows equal levels whether glucose or fructose was fed as a carbon source. The reason for the different flux-sum intensity of bDG6P, F6P, and FDP under two different carbon sources (i.e. glucose and fructose) is that glucose is metabolized to glucose-6-phosphate via glucokinase, and fructose is metabolized to glucose-6-phosphate via fructokinase and phosphoglucose isomerase. The strongest intensity value for the flux-sum was obtained from growth on sucrose because of its disaccharide chemical structure of glucose and fructose, resulting in having the effects of two carbon sources at the same time.
The different flux-sum intensity pattern of the metabolites in Z. mobilis and E. coli can be explained through the differences in the structure of their metabolic network. The metabolic network of Z. mobilis is small and even if some of the branch points are to be deleted, such as Pfk (phosphofructokinase) in glycolytic pathway, SucA (2-oxoglutarate dehydrogenase), and Mdh (malate dehydrogenase) in TCA cycle, there is little chance for the carbon flux to be distributed through other metabolic pathways (Figure 2A). Z. mobilis essentially has a linear like central metabolic pathway, including ED pathway and incomplete TCA cycle, and this allows for the metabolites in Z. mobilis to display flux-sum intensities of similar level. On the other hand, E. coli has a relatively large and robust metabolic network, which allows the uptake carbon flux to be redistributed through alternate pathways other than the central metabolic pathway. Thus, the variation of flux-sum intensity about metabolites in E. coli is larger than that of Z. mobilis. Flux-sum intensity of metabolites in glycolytic pathway in E. coli was found to be stronger than Z. mobilis under the three carbon sources. This was expected as Z. mobilis has an incomplete glycolytic pathway and the carbon flux of Z. mobilis flows through the ED pathway. For the same reason, Z. mobilis shows a stronger flux-sum intensity of metabolites in the ED pathway compared to that of E. coli. As E. coli possesses the pathway from acetyl-CoA to acetic acid through acetyl phosphate, which is absent in Z. mobilis, the acetyl-CoA pool of E. coli is converted to the acetic acid under anaerobic condition to generate ATP via substrate-level phosphorylation. Therefore, the flux-sum intensity of TCA cycle metabolites in E. coli is lower than that of Z. mobilis, which distributes its acetyl-CoA pool to the TCA cycle. Additionally, precursors of many amino acids that are generated through the TCA cycle subsequently display a relatively more intensive flux-sum value in Z. mobilis compared to E. coli.
Analysis of maximum in silico yields of ethanol between Z. mobilis and E. coli
Z. mobilis has attracted attention for its high ethanol producing ability, and therefore many studies have been conducted regarding this topic [14,52,53]. Through in-depth researches, pyruvate decarboxylase in Z. mobilis, which converts pyruvate to acetaldehyde, has been shown to be one of the important reasons for the high ethanol producing capability in Z. mobilis [54]. Additionally, there are reports of genetically engineered E. coli possessing the pyruvate decarboxylase gene from Z. mobilis having increased ethanol production capability compared to the wild type strain [55-58]. Therefore, the role of pyruvate decarboxylase enzyme in ethanol production was examined using the Z. mobilis metabolic model ZmoMBEL601 by analysis of the maximum in silico yields of ethanol, and compared the results with analysis from the E. coli metabolic models EcoMBEL979 and iAF1260 [30,59].
Maximum in silico yields of ethanol in the Z. mobilis and E. coli metabolic models were 2 and 1.85 mol ethanol/mol glucose, respectively where EcoMBEL979 and iAF1260 showed the same yield. The result was in agreement with reports showing Z. mobilis is more capable of producing ethanol [56,60]. To evaluate the role of pyruvate decarboxylase about ethanol production in Z. mobilis, the simulation, where the pyruvate decarboxylase in Z. mobilis was removed from the Z. mobilis metabolic model, was performed by knocking out the respective metabolic reaction in the Z. mobilis metabolic model. The results of the simulation showed that the capacity for ethanol production dropped when pyruvate decarboxylase was removed from the Z. mobilis metabolic model (i.e. 0.03 mol ethanol/mol glucose from 2 mol ethanol/mol glucose). This outcome was similar to the reported experimental result of Seo et al. [61] that the pyruvate decarboxylase knockout mutant did not produce ethanol by fermentation process. Thus, it can be concluded that the pyruvate decarboxylase reaction is the essential part of producing ethanol in Z. mobilis. Next, the pyruvate decarboxylase reaction was introduced into the E. coli metabolic model to verify its role for ethanol production in E. coli. With the addition of the pyruvate decarboxylase reaction, the maximum in silico yield in the E. coli metabolic model improved to 2 mol from 1.85 mol ethanol/mol glucose. To further investigate the exact impact of pyruvate decarboxylase, other reactions of the E. coli metabolism, which can be utilized in the production of ethanol, but not found in Z. mobilis, were removed to determine its role in the production of ethanol. If the removal of the reactions resulted in a decrease in ethanol production, then the pyruvate decarboxylase reaction was concluded to not be the sole factor for the improved yield in E. coli. The significant differences of reactions between Z. mobilis and E. coli are that E. coli has only the phosphofructokinase reaction in glycolytic pathway, the 2-oxoglutarate dehydrogenase reaction and the malate dehydrogenase reaction in TCA cycle. Therefore, these reactions, which do not exist in Z. mobilis, were removed from the E. coli metabolic model. Despite the removal of all these reactions, the maximal in silico ethanol yields remained at 2 mol ethanol/mol glucose through alternative available metabolic pathways. Furthermore, the elimination of other reactions, such as glutamate dehydrogenase, ethanolamine ammonia-lyase, and acetaldehyde dehydrogenase, did not affect the maximum in silico yield of ethanol in E. coli metabolic model. Through these simulation results, it can be concluded that pyruvate decarboxylase is the main factor that allows Z. mobilis to have a greater capacity for ethanol production than E. coli. The presence of the other reactions were not seen to have direct correlation with the ethanol production capacity in E. coli.
The reaction above not found in Z. mobilis, were examined in their role in producing ethanol in E. coli using the wild type E. coli metabolic model, which lacks pyruvate decarboxylase. The reaction corresponding to the enzyme of transaldolase was removed, and resulted in the declined ethanol production yield, 1.78 mol ethanol/mol glucose. Because pentose phosphate pathway generates NADPH, the deletion of transaldolase reaction resulted in the decreased supply of reducing power. Therefore, it implies that the pentose phosphate pathway has an important role for producing ethanol in the E. coli wild type strain. It should be noted that EcoMBEL979 and iAF1260 produced the same results for this study.
Expanding carbon source utilization of Z. mobilis
Z. mobilis cannot utilize pentose sugars due to incomplete metabolic pathways of pentose [16,39,62,63]. Because the feedstock holds a significant portion of cost in bioprocess, developing pentose sugar fermenting strains can reduce the cost by enabling strains to use cheap, renewable, lignocellulosic biomass. Several genes involved in pentose sugar metabolism, including those encoding xylose isomerase, xylulokinase, and transaldolase for utilizing xylose and arabinose isomerase, ribulokinase, ribulose-5-phosphate-4-epimerase, and transaldolase for utilizing arabinose, respectively, are absent in Z. mobilis. However, Zhang et al. [64] successfully introduced four genes (xylose isomerase, xylulokinase, transaldolase, and transketolase) into Z. mobilis for utilizing xylose as a carbon source. De Graaf et al. [63] also reported that Z. mobilis CP4 produces ethanol with xylose as a carbon source by introducing the same enzymes. Deanda et al. [65] reported that five genes (arabinose isomerase, ribulokinase, ribulose-5-phosphate-4-epimerase, transaldolase, transketolase) were necessary for Z. mobilis to utilize arabinose as a carbon source. According to these results, new pathways (three pathways for xylose and four pathways for arabinose; the transketolase gene was annotated and originally included in the metabolic model) were introduced in the metabolic model ZmoMBEL601 (Figure 2A; Additional file 1). Then, xylose and arabinose in the metabolic model were converted to xylulose 5-phophate via xylulose and ribulose 5-phosphate, respectively, and the fluxes through the pentose phosphate pathway were increased through producing more ribose-5-phosphate from ribulose 5-phosphate [63]. By utilizing the metabolic model, in silico simulations can be performed to predict the capability of Z. mobilis to metabolize carbon sources on anaerobic growth, and thereby allow for the expansion in the number of available carbon source [16,39,63-65].
For further application of engineered Z. mobilis metabolic model, the simulations were performed to investigate the maximum in silico yield of ethanol for three carbon sources (i.e. glucose, xylose, arabinose) in ZmoMBEL601 (Additional file 5A). Because of the difference in the substrates' carbon number, glucose showed a higher maximum in silico yield of ethanol and growth rate than those of xylose and arabinose. Additionally, to examine the relationship between biomass and ethanol production, single reaction knockout simulation was performed and the trade-off curves for each were plotted (additional file 5B). As the reactions in central metabolic pathway (i.e. glycolytic pathway, pentose phosphate pathway, and TCA cycle) have significant importance in the metabolism, the range of simulation (i.e. the place of knockout reaction) was limited to the central metabolic pathway. The results displayed 15 different cases in trade-off curves for glucose and 13 for arabinose and xylose. Out of these cases we focused on four of them: reactions which are essential to growth but not to ethanol production (i.e. type 2), reactions which are essential to ethanol production but not to growth (i.e. type 3), reactions which are essential to both growth and ethanol production (i.e. type 4), and reactions which are not essential to both growth and ethanol production but result the rapid decrease in ethanol production when deleted (i.e. type 7). In the case of type 2 reactions, the lists of reactions are the same for both xylose and arabinose, but glucose has an additional reaction in the list: ribose 5-phosphate isomerase. Alcohol dehydrogenase was the only reaction in the type 3 case and is essential for the ethanol production regardless of the carbon source. In the case of type 4 reactions, the reaction lists of each carbon were directly related to its initial utilization pathway: reactions in ED pathway for every carbon sources and additional reactions in pentose phosphate pathway for xylose and arabinose. Thus, both growth and ethanol production were impossible when these reactions are deleted. Pyruvate decarboxylase is a type 7 reaction and its deletion resulted in a drop but not complete elimination of the ethanol production rate.
Strategies for succinic acid production in Z. mobilis
Z. mobilis has the potential for the overproduction of a chemical that is of industrial value by redirecting metabolic pathways upon gene knockout. One of the examples would be succinic acid, a four carbon dicarboxylic acid and intermediate of TCA cycle. Additionally, it is an important industrial product useful for pharmaceutical and chemical intermediates, additives in the food industry, fertilizers, solvents, and polymers [66,67]. The biotechnological overproduction of succinic acid in Z. mobilis was investigated through gene knockout simulation using constraints-based flux analysis implemented by constraining the flux value of the respective knockout reaction to zero. For single gene knockout simulation, satisfactory targets that produce succinic acid were not obtained, compared with previous knowledge on the production level of succinic acid. The best result of the single-gene knockout simulation for succinic acid production gave low yields of succinic acid only 0.15% of theoretical maximum yield for succinic acid production (2 mol succinic acid/mol glucose). Simulations of double gene knockout, however, resulted in the combination of pyruvate decarboxylase and D-lactate dehydrogenase as the best targets for succinic acid overproduction (Figure 3B), and the pairs of gene targets are presented in the additional file 6. Functional and physical relationship among reactions in metabolic network, utilized to discover potential combinatorial engineering targets, can be revealed by applying grouping reaction constraint to constraints-based flux analysis [68]. Hence, additional gene knockout simulations that contain grouping reaction constraints were performed, and obtained the same results. These analyses revealed that the inactivation of pyruvate decarboxylase essential for ethanol production redirected the metabolic fluxes towards lactic acid production, and the inactivation of D-lactate dehydrogenase redirected the metabolic fluxes toward succinic acid production in TCA cycle. These results agree well with the work of Seo et al. [61], which obtained a result of a 95% theoretical yield of succinic acid by performing the gene knockout for pyruvate decarboxylase and D-lactate hydrogenase. On the other hand, E. coli metabolic model did not show significant increment in the succinic acid production when the same target D-lactate dehydrogenase was knocked out; E. coli does not have pyruvate decarboxylase. This is because of the many other alternative pathways in E. coli which can complement and relieve the constraints of gene deletion. Therefore, E. coli requires its own specific gene target combinations, which are distinct from Z. mobilis. As the reconstructed ZmoMBEL601 was able to design gene knockout strategy for succinic acid production successfully, it could suggest the strategies for other biochemicals as well, such as lactic acid, fumaric acid, and malic acid.
Conclusions
We have presented the genome-scale reconstruction and analysis of metabolic network in Z. mobilis ZM4. The metabolic model was constructed systematically via four steps; automatic building, manual curation, rational determination of the biomass composition, and completion of the metabolic model. The metabolic model reflects the physiological characteristics of Z. mobilis, including the ED pathway, incomplete pentose phosphate pathway, oxidative phosphorylation mechanisms, and high ethanol producing ability. The metabolic model was utilized to investigate the characteristics of ethanol production and was further characterized through the comparison with the E. coli metabolic model. The metabolic model allowed for the development of strategies for strain improvement, including the addition of several pathways to the metabolic model to allow for the metabolism of pentose sugar in Z. mobilis ZM4, strategies for ethanol and succinic acid production in Z. mobilis ZM4, and constraints-based flux analysis to give an accurate representation of phenotypes that match with reported data. As a consequence of analysis, validation, and application mentioned above, the genome-scale reconstructed metabolic model of Z. mobilis ZM4, ZmoMBEL601, is expected to be useful for systematical designing of strain development strategy for biotechnology applications.
Materials and methods
Reconstruction of metabolic model
Metabolic model of the Z. mobilis ZM4 was constructed by using the combined information from many different sources, including public databases, literatures, and experiments. The construction of the metabolic model was carried out with four distinct steps: First, automatic building of a draft metabolic model with data retrieved from databases; from genomes to metabolic pathways. Second, the draft metabolic model is manually curated through literature information. Third, rational determination of the biomass composition and maintenance requirements are done by experimental and literature data. Finally, validation of the metabolic model with the information obtained from previous steps and correctly modify with literatures and experimental results (Figure 1).
For efficient construction of the Z. mobilis ZM4 metabolic model, we used different softwares and databases, which include NCBI [23], CMR [24], KEGG [20], TCDB [26], TransportDB [27], ExPASy [25], BioCyc [21], BioSilico [28], MFAML [69], and MetaFluxNet [70].
Constraints-based flux analysis
Constraints-based flux analysis is a method for studying metabolic networks with the assumption of a pseudo-steady state and constraints are imposed by mass balance of the metabolites [4,71]. This pseudo-steady state approximation is generally valid because the metabolites concentrations tend to reach to equilibrium much faster compared to genetic regulation [72]. This results in a stoichiometric model Sij • vj = 0, in which Sij is a stoichiometric coefficient of a metabolite i in the jth reaction and vj is the flux of the jth reaction given in mmol/gDCW/h. Additional constraints can also be introduced to represent measured or imposed values for metabolites and are implemented as inequalities. Thus, fermentations were conducted to observe cellular behaviors and to measure key metabolites concentrations, which were used in constraints-based flux analysis (Figure 1; Additional file 7). With the uptake rate and the by-product secretion rate, we imposed upper and lower constraints in the Z. mobilis metabolic model (i.e. limit the flux on fumaric acid, acetic acid, malic acid, acetoin, and acetaldehyde production) for regulating the flux more realistically. The calculation of the maximum in silico yields of ethanol was done using the metabolic model of Z. mobilis (ZmoMBEL601) and E. coli (EcoMBEL979 and iAF1260). As the pyruvate dehydrogenase reaction in E. coli has low activity under anaerobic conditions [73], the pyruvate dehydrogenase reaction was inactivated in the E. coli metabolic models. Uptake rate of glucose is fixed to 10 mmol/gDCW/h, and reaction for oxygen uptaking was deleted to describe anaerobic condition in both genome-scale metabolic models. The NGAME (i.e. non-growth associated maintenance energy) values for both genome-scale metabolic models were eliminated. Ethanol production rate are normalized to the glucose uptake rate.
Analysis of flux-sum intensity
The flux-sum is defined as the half of the summation of all consumption and generation rate around a particular metabolite under pseudo steady-states [50,51]. To understand the state of a cellular metabolism, the intensity of flux-sum for each metabolite was analyzed, as a good property for investigating the interconversion of metabolites. The flux-sum of metabolite i is formulated as , where Sij is the stoichiometric coefficient of a metabolite i in the jth reaction and vj is the flux of the jth reaction. The Sij • vj means the absolute values of consumption or generation rate of metabolite i in the jth reaction. The flux-sum of metabolite i is calculated by dividing the Sij • vj by one half. The flux-sum intensities of metabolites from central metabolism in Z. mobilis were analyzed to investigate the turnover rate of each metabolite using the ZmoMBEL601 model and compared to the flux-sum intensity of each metabolite in E. coli using the EcoMBEL974 model. The analysis of flux-sum intensity between both organisms was carried out with the three kinds of carbon sources including glucose, fructose, and sucrose.
Media and cultivation
Frozen Z. mobilis ZM4 strain stock (with 70% glycerol) at -70°C was inoculated into a sealed tube containing 10 mL of ZM medium. ZM medium contains per liter: 50 g glucose, 2 g KH2PO4, 1 g (NH4)2SO4, 1 g MgSO4·7H2O, 10 g yeast extract. After incubating at 30°C for 12 hrs, the culture was transferred to a 500 mL sealed flask containing 100 mL ZM medium for subculture. Fermentation was carried out at 30°C in a 6.6 L bioreactor (BioFlo 3000, New Brunswick Scientific, Edison, NJ, USA) containing 2 L of ZM medium. The pH was controlled at 5.5 with 5 M NaOH. Anaerobic condition was achieved by 50 rpm agitation speed and flushing the bioreactor with oxygen-free CO2 gas (Kosock gas, Daejeon, Korea). In case of aerobic culture, the dissolved oxygen concentration was maintained above 40% of air saturation by supplying air at 1 vvm (air volume/working volume/minute) and by automatically controlling the agitation speed up to 1,000 rpm.
Analytical procedures
The cell growth was monitored by measuring the absorbance at 600 nm using an Ultrospec 3000 spectrophotometer (Pharmacia Biotech., Uppsala, Sweden). Metabolites were analyzed by high-performance liquid chromatography (Varian ProStar 210, Palo Alto, CA, USA), equipped with an Aminex HPX-87H column (300 mm × 7.8 mm, Bio-Rad Laboratories, Herculules, CA, USA), UV/VIS (Varian ProStar 320, Palo Alto, CA, USA) and a refractive index (Shodex RI-71, Minato-Ku, Tokyo, Japan) detectors. The column was eluted isocratically at 50°C at a flow rate of 0.5 mL/min with 0.01 N H2SO4.
Biomass composition
Biosynthetic pathways and composition of each molecules/metabolites of proteins, DNA, RNA, lipids, small molecules pool, and cell wall were formulated with literature data or reasonably assumed as described in additional file 4. Many types of data were collected not only from Z. mobilis ZM4 and various Z. mobilis strains (e.g., Z. mobilis CP4) but also from other species for this purpose, because it was not possible to describe all the formulation mechanism with only the data from Z. mobilis ZM4. The data was properly manipulated and the biomass composition and reaction of Z. mobilis ZM4 strain was determined (Additional file 4).
Lists of abbreviations
The abbreviations are represented as follows: Acn: aconitate hydratase; Adh: alcohol dehydrogenase; AraA: L-arabinose isomerase; AraB: L-ribulokinase; Cit: citrate lyase; Cs: citrate synthase; Eda: 2-dehydro-3-deoxyphosphogluconate aldolase; Edd: phosphogluconate dehydratase; Eno: enolase; Fba: fructose-bisphosphate aldolase; Fum: fumarate hydratase; Glk: glucokinase; Icd: isocitrate dehydrogenase; Ldh: L-lactate dehydrogenase; Mdh: malate dehydrogenase; Me: Malic enzyme; Pdc: pyruvate decarboxylase; Pfk: 6-phosphofructokinase; Pgi: glucose-6-phosphate isomerase; Pgl: 6-phosphogluconolactonase; Ppc: phosphoenolpyruvate carboxylase; SacA: beta-fructofuranosidase; SacB: levansucrase; ScrK: fructokinase; Sdh: succinate dehydrogenase; Sgb: L-ribulose-5-phosphate 4-epimerase; Suc: succinyl-CoA synthetase; SucA: 2-oxoglutarate dehydrogenase; Tkt: transketolase; Xk: xylulokinase; Xyl: xylose isomerase; Zwf: glucose-6-phosphate 1-dehydrogenase; DCW: dry cell weight;.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
KYL, JMP, TYK, HSY, and SYL generated ideas; KYL, JMP, TYK, and SYL designed research; KYL, JMP, and SYL performed research; KYL and JMP performed analytical experiments; KYL, JMP, and SYL analyzed data; KYL, JMP, TYK, HSY, and SYL wrote the paper. All authors read and approved the final version of the manuscript.
Supplementary Material
List of metabolic reactions in the genome-scale metabolic model of Zymomonas mobilis ZM4
List of metabolites in the genome-scale metabolic model of Zymomonas mobilis ZM4
ORF (Open Reading Frame) coverage of reconstructed metabolic models
Biomass composition of Zymomonas mobilis ZM4
Trade-off curves of (A) engineered strain of Zymomonas mobilis for the utilization of xylose and arabinose and (B) single gene knockout mutants based on the engineered strain for glucose, xylose, and arabinose as a carbon source.
Double gene knockout targets for succinic acid production identified by constraints-based flux analysis
Batch culture profile of Zymomonas mobilis ZM4
Contributor Information
Kyung Yun Lee, Email: kyungyunlee@kaist.ac.kr.
Jong Myoung Park, Email: kaiHman@kaist.ac.kr.
Tae Yong Kim, Email: kimty@kaist.ac.kr.
Hongseok Yun, Email: hsyun@kaist.ac.kr.
Sang Yup Lee, Email: leesy@kaist.ac.kr.
Acknowledgements
This work was supported by the Korean Systems Biology Research Project (20100002164) of the Ministry of Education, Science and Technology (MEST). Further support by the World Class University Program (R32-2009-000-10142-0) through the National Research Foundation of Korea funded by the MEST is appreciated.
References
- Lee SY, Lee DY, Kim TY. Systems biotechnology for strain improvement. Trends Biotechnol. 2005;23:349–358. doi: 10.1016/j.tibtech.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Park JH, Lee SY. Towards systems metabolic engineering of microorganisms for amino acid production. Curr Opin Biotechnol. 2008;19:454–460. doi: 10.1016/j.copbio.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Cameron DC, Tong IT. Cellular and metabolic engineering. An overview. Appl Biochem Biotechnol. 1993;38:105–140. doi: 10.1007/BF02916416. [DOI] [PubMed] [Google Scholar]
- Lee SY, Papoutsakis ET. Metabolic engineering. New York: Marcel Dekker; 1999. [Google Scholar]
- Parekh S, Vinci VA, Strobel RJ. Improvement of microbial strains and fermentation processes. Appl Microbiol Biotechnol. 2000;54:287–301. doi: 10.1007/s002530000403. [DOI] [PubMed] [Google Scholar]
- Park JH, Lee SY, Kim TY, Kim HU. Application of systems biology for bioprocess development. Trends Biotechnol. 2008;26:404–412. doi: 10.1016/j.tibtech.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Park JH, Lee KH, Kim TY, Lee SY. Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation. Proc Natl Acad Sci USA. 2007;104:7797–7802. doi: 10.1073/pnas.0702609104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Park JH, Kim TY, Kim HU, Lee SY. Systems metabolic engineering of Escherichia coli for L-threonine production. Mol Syst Biol. 2007;3:149. doi: 10.1038/msb4100196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper H, Jin YS, Moxley JF, Stephanopoulos G. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab Eng. 2005;7:155–164. doi: 10.1016/j.ymben.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Song H, Lee SY. Genome-based metabolic engineering of Mannheimia succiniciproducens for succinic acid production. Appl Environ Microbiol. 2006;72:1939–1948. doi: 10.1128/AEM.72.3.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bro C, Regenberg B, Forster J, Nielsen J. In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab Eng. 2006;8:102–111. doi: 10.1016/j.ymben.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Jung YK, Kim TY, Park SJ, Lee SY. Metabolic engineering of Escherichia coli for the production of polylactic acid and its copolymers. Biotechnol Bioeng. 2010;105:161–171. doi: 10.1002/bit.22548. [DOI] [PubMed] [Google Scholar]
- Shlomi T, Cabili MN, Herrgard MJ, Palsson BO, Ruppin E. Network-based prediction of human tissue-specific metabolism. Nat Biotechnol. 2008;26:1003–1010. doi: 10.1038/nbt.1487. [DOI] [PubMed] [Google Scholar]
- Rogers PL, Lee KJ, Skotnicki ML, Tribe DF. Ethanol production by Zymomonas mobilis. Adv Biochem Eng. 1982;23:37–84. [Google Scholar]
- Seo JS, Chong H, Park HS, Yoon KO, Jung C, Kim JJ, Hong JH, Kim H, Kim JH, Kil JI. et al. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat Biotechnol. 2005;23:63–68. doi: 10.1038/nbt1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swings J, De Ley J. The biology of Zymomonas. Bacteriol Rev. 1977;41:1–46. doi: 10.1128/br.41.1.1-46.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Pappas KM, Hauser LJ, Land ML, Chen GL, Hurst GB, Pan C, Kouvelis VN, Typas MA, Pelletier DA. et al. Improved genome annotation for Zymomonas mobilis. Nat Biotechnol. 2009;27:893–894. doi: 10.1038/nbt1009-893. [DOI] [PubMed] [Google Scholar]
- Tsantili IC, Karim MN, Klapa MI. Quantifying the metabolic capabilities of engineered Zymomonas mobilis using linear programming analysis. Microb Cell Fact. 2007;6:8. doi: 10.1186/1475-2859-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto JP, Dias O, Lourenço A, Carneiro S, Ferreira EC, Rocha I, Rocha M. In: Advances in Soft Computing (ASC) Corchado JM, De Paz JF, Rocha MP, Riverola FF, editor. Vol. 49. Berlin Heidelberg: Springer-Verlag; 2009. 2nd International Workshop on Practical Applications of Computational Biology and Bioinformatics (IWPACBB 2008): Data Integration Issues in the Reconstruction of the Genome-Scale Metabolic Model of Zymomonas Mobillis; pp. 92–101. full_text. [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M. et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg I, Chang A, Hofmann O, Ebeling C, Ehrentreich F, Schomburg D. BRENDA: a resource for enzyme data and metabolic information. Trends Biochem Sci. 2002;27:54–56. doi: 10.1016/S0968-0004(01)02027-8. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information (NCBI) http://www.ncbi.nlm.nih.gov
- Davidsen T, Beck E, Ganapathy A, Montgomery R, Zafar N, Yang Q, Madupu R, Goetz P, Galinsky K, White O, Sutton G. The comprehensive microbial resource. Nucleic Acids Res. 2010;38:D340–345. doi: 10.1093/nar/gkp912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q, Chen K, Paulsen IT. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 2007;35:D274–279. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou BK, Kim JS, Jun JH, Lee DY, Kim YW, Chae S, Roh M, In YH, Lee SY. BioSilico: an integrated metabolic database system. Bioinformatics. 2004;20:3270–3272. doi: 10.1093/bioinformatics/bth363. [DOI] [PubMed] [Google Scholar]
- David H, Ozcelik IS, Hofmann G, Nielsen J. Analysis of Aspergillus nidulans metabolism at the genome-scale. BMC Genomics. 2008;9:163. doi: 10.1186/1471-2164-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BO. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol. 2007;3:121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Dal'Molin CG, Quek LE, Palfreyman RW, Brumbley SM, Nielsen LK. AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol. 2010;152:579–589. doi: 10.1104/pp.109.148817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo ML, Palsson BO, Herrgard MJ. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst Biol. 2009;3:37. doi: 10.1186/1752-0509-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvarasu S, Karimi IA, Ghim GH, Lee DY. Genome-scale modeling and in silico analysis of mouse cell metabolic network. Mol Biosyst. 2010;6:152–161. doi: 10.1039/b912865d. [DOI] [PubMed] [Google Scholar]
- Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BO. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Herrgard MJ, Palsson BO. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res. 2004;14:1298–1309. doi: 10.1101/gr.2250904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David H, Akesson M, Nielsen J. Reconstruction of the central carbon metabolism of Aspergillus niger. Eur J Biochem. 2003;270:4243–4253. doi: 10.1046/j.1432-1033.2003.03798.x. [DOI] [PubMed] [Google Scholar]
- Kim HU, Kim TY, Lee SY. Genome-scale metabolic network analysis and drug targeting of multi-drug resistant pathogen Acinetobacter baumannii AYE. Mol Biosyst. 2010;6:339–348. doi: 10.1039/b916446d. [DOI] [PubMed] [Google Scholar]
- Schilling CH, Covert MW, Famili I, Church GM, Edwards JS, Palsson BO. Genome-scale metabolic model of Helicobacter pylori 26695. J Bacteriol. 2002;184:4582–4593. doi: 10.1128/JB.184.16.4582-4593.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekaran P, Chandra Raj K. Ethanol fermentation technology - Zymomonas mobilis. Current Sci. 1999;77:56–68. [Google Scholar]
- Carey VC, Ingram LO. Lipid composition of Zymomonas mobilis: effects of ethanol and glucose. J Bacteriol. 1983;154:1291–1300. doi: 10.1128/jb.154.3.1291-1300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau RA, Powell MJ, Fett WF, Whitaker BD. The effect of ethanol and oxygen on the growth of Zymomonas mobilis and the levels of hopanoids and other membrane lipids. Curr Microbiol. 1997;35:124–128. doi: 10.1007/s002849900224. [DOI] [PubMed] [Google Scholar]
- Hermans MA, Neuss B, Sahm H. Content and composition of hopanoids in Zymomonas mobilis under various growth conditions. J Bacteriol. 1991;173:5592–5595. doi: 10.1128/jb.173.17.5592-5595.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau RA, Powell MJ, Osman SF, Whitaker BD, Fett WF, Roth L, O'Brien DJ. Analysis of intact hopanoids and other lipids from the bacterium Zymomonas mobilis by high-performance liquid chromatography. Anal Biochem. 1995;224:293–301. doi: 10.1006/abio.1995.1043. [DOI] [PubMed] [Google Scholar]
- Vincent SP, Sinay P, Rohmer M. Composite hopanoid biosynthesis in Zymomonas mobilis: N-acetyl-D-glucosamine as precursor for the cyclopentane ring linked to bacteriohopanetetrol. Chem Commun. 2003. pp. 782–783. [DOI] [PubMed]
- Chiara JL, Storch de Gracia I, Bastida A. Synthesis of the aminocyclopentitol moieties of the hopanoids of Zymomonas mobilis and 'Anacystis montana'. Chem Commun. 2003. pp. 1874–1875. [DOI] [PubMed]
- Perzl M, Reipen IG, Schmitz S, Poralla K, Sahm H, Sprenger GA, Kannenberg EL. Cloning of conserved genes from Zymomonas mobilis and Bradyrhizobium japonicum that function in the biosynthesis of hopanoid lipids. Biochim Biophys Acta. 1998;1393:108–118. doi: 10.1016/s0005-2760(98)00064-2. [DOI] [PubMed] [Google Scholar]
- Savvides AL, Kallimanis A, Varsaki A, Koukkou AI, Drainas C, Typas MA, Karagouni AD. Simultaneous ethanol and bacterial ice nuclei production from sugar beet molasses by a Zymomonas mobilis CP4 mutant expressing the inaZ gene of Pseudomonas syringae in continuous culture. J Appl Microbiol. 2000;89:1002–1008. doi: 10.1046/j.1365-2672.2000.01208.x. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Song KB, Rhee SK. A novel aerobic respiratory chain-linked NADH oxidase system in Zymomonas mobilis. J Bacteriol. 1995;177:5176–5178. doi: 10.1128/jb.177.17.5176-5178.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringer S, Finn RK, Sahm H. Effect of oxygen on the metabolism of Zymomonas mobilis. Arch Microbiol. 1984;139:76–381. doi: 10.1007/BF00408383. [DOI] [Google Scholar]
- Kim PJ, Lee DY, Kim TY, Lee KH, Jeong H, Lee SY, Park S. Metabolite essentiality elucidates robustness of Escherichia coli metabolism. Proc Natl Acad Sci USA. 2007;104:13638–13642. doi: 10.1073/pnas.0703262104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BK, Lee DY. Flux-sum analysis: a metabolite-centric approach for understanding the metabolic network. BMC Syst Biol. 2009;3:117. doi: 10.1186/1752-0509-3-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baratti J, Varma R, Bulock JD. High productivity ethanol fermentation on a mineral medium using a flocculent strain of Zymomons mobilis. Biotechnol Lett. 1986;8:175–180. doi: 10.1007/BF01029374. [DOI] [Google Scholar]
- Baratti JC, Bu'lock JD. Zymomonas mobilis: a bacterium for ethanol production. Biotechnol Adv. 1986;4:95–115. doi: 10.1016/0734-9750(86)90006-6. [DOI] [PubMed] [Google Scholar]
- Hoppner TC, Doelle HW. Purification and Kinetic Characteristics of Pyruvate Decarboxylase and Ethanol Dehydrogenase from Zymomonas-Mobilis in Relation to Ethanol-Production. Eur J Appl Microbiol. 1983;17:152–157. doi: 10.1007/BF00505880. [DOI] [Google Scholar]
- Alterthum F, Ingram LO. Efficient ethanol production from glucose, lactose, and xylose by recombinant Escherichia coli. Appl Environ Microbiol. 1989;55:1943–1948. doi: 10.1128/aem.55.8.1943-1948.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram LO, Conway T, Clark DP, Sewell GW, Preston JF. Genetic engineering of ethanol production in Escherichia coli. Appl Environ Microbiol. 1987;53:2420–2425. doi: 10.1128/aem.53.10.2420-2425.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brau B, Sahm H. Cloning and Expression of the Structural Gene for Pyruvate Decarboxylase of Zymomonas mobilis in Escherichia coli. Arch Microbiol. 1986;144:296–301. doi: 10.1007/BF00410966. [DOI] [Google Scholar]
- Ingram LO, Clark DP. Ethanol production using engineered mutant E. coli. United States Patent 5028539. 1991.
- Lee SY, Woo HM, Lee DY, Choi HS, Kim TY, Yun H. Systems-level analysis of genome-scale microbial metabolisms under the integrated software environment. Biotechnol Bioprocess Eng. 2005;10:425–431. doi: 10.1007/BF02989825. [DOI] [Google Scholar]
- Hong SH, Moon SY, Lee SY. Prediction of maximum yields of metabolites and optimal pathways for their production by metabolic flux analysis. J Microbiol Biotechnol. 2003;13:571–577. [Google Scholar]
- Seo JS, Chong H, Kim JY, Kim JH. Method for mass production of primary metabolites, strain for mass production of primary metabolites, and method for preparation thereof. Korean patent number 0725021. 2007.
- Kim IS, Barrow KD, Rogers PL. Kinetic and nuclear magnetic resonance studies of xylose metabolism by recombinant Zymomonas mobilis ZM4(pZB5) Appl Environ Microbiol. 2000;66:186–193. doi: 10.1128/AEM.66.1.186-193.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Graaf AA, Striegel K, Wittig RM, Laufer B, Schmitz G, Wiechert W, Sprenger GA, Sahm H. Metabolic state of Zymomonas mobilis in glucose-, fructose-, and xylose-fed continuous cultures as analysed by 13C- and 31P-NMR spectroscopy. Arch Microbiol. 1999;171:371–385. doi: 10.1007/s002030050724. [DOI] [PubMed] [Google Scholar]
- Zhang M, Eddy C, Deanda K, Finkelstein M, Picataggio S. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science. 1995;267:240–243. doi: 10.1126/science.267.5195.240. [DOI] [PubMed] [Google Scholar]
- Deanda K, Zhang M, Eddy C, Picataggio S. Development of an arabinose-fermenting Zymomonas mobilis strain by metabolic pathway engineering. Appl Environ Microbiol. 1996;62:4465–4470. doi: 10.1128/aem.62.12.4465-4470.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Kim JS, Lee SY, In YH, Choi SS, Rih JK, Kim CH, Jeong H, Hur CG, Kim JJ. The genome sequence of the capnophilic rumen bacterium Mannheimia succiniciproducens. Nat Biotechnol. 2004;22:1275–1281. doi: 10.1038/nbt1010. [DOI] [PubMed] [Google Scholar]
- Song H, Lee SY. Production of succinic acid by bacterial fermentation. Enzyme Microb Technol. 2006;39:352–361. doi: 10.1016/j.enzmictec.2005.11.043. [DOI] [Google Scholar]
- Park JM, Kim TY, Lee SY. Prediction of metabolic fluxes by incorporating genomic context and flux-converging pattern analyses. Proc Natl Acad Sci USA. 2010. [DOI] [PMC free article] [PubMed]
- Yun H, Lee DY, Jeong J, Lee S, Lee SY. MFAML: a standard data structure for representing and exchanging metabolic flux models. Bioinformatics. 2005;21:3329–3330. doi: 10.1093/bioinformatics/bti502. [DOI] [PubMed] [Google Scholar]
- Lee SY, Lee DY, Hong SH, Kim TY, Yun H, Oh YG, Park S. MetaFluxNet, a program package for metabolic pathway construction and analysis, and its use in large-scale metabolic flux analysis of Escherichia coli. Genome Inform. 2003;14:23–33. [PubMed] [Google Scholar]
- Stephanopoulos GN, Aristidou AA, Nielsen J. Metabolic engineering. San Diego: Academic Press; 1998. [Google Scholar]
- Schilling CH, Edwards JS, Palsson BO. Toward metabolic phenomics: analysis of genomic data using flux balances. Biotechnol Progr. 1999;15:288–295. doi: 10.1021/bp9900357. [DOI] [PubMed] [Google Scholar]
- Wang QZ, Ou MS, Kim Y, Ingram LO, Shanmugam KT. Metabolic Flux Control at the Pyruvate Node in an Anaerobic Escherichia coli Strain with an Active Pyruvate Dehydrogenase. Appl Environ Microbiol. 2010;76:2107–2114. doi: 10.1128/AEM.02545-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of metabolic reactions in the genome-scale metabolic model of Zymomonas mobilis ZM4
List of metabolites in the genome-scale metabolic model of Zymomonas mobilis ZM4
ORF (Open Reading Frame) coverage of reconstructed metabolic models
Biomass composition of Zymomonas mobilis ZM4
Trade-off curves of (A) engineered strain of Zymomonas mobilis for the utilization of xylose and arabinose and (B) single gene knockout mutants based on the engineered strain for glucose, xylose, and arabinose as a carbon source.
Double gene knockout targets for succinic acid production identified by constraints-based flux analysis
Batch culture profile of Zymomonas mobilis ZM4