Abstract
Background
Members of the Sox gene family isolated from both vertebrates and invertebrates have been proved to participate in a wide variety of developmental processes, including sex determination and differentiation. Among these members, Sox30 had been considered to exist only in mammals since its discovery, and its exact function remains unclear.
Results
Sox30 cDNA was cloned from the Nile tilapia by RT-PCR and RACE. Screening of available genome and EST databases and phylogenetic analysis showed that Sox30 also exists in non-mammalian vertebrates and invertebrates, which was further supported by synteny analyses. Tissue expression in human, mouse and tilapia suggested that Sox30 was probably a gonad-specific gene, which was also supported by the fact that Sox30 EST sequences were obtained from gonads of the animal species. In addition, four alternatively spliced isoforms were isolated from tilapia gonad. Their temporal and spatial expression patterns during normal and sex reversed gonadal development were investigated by RT-PCR and in situ hybridization. Our data suggest that expressions of Sox30 isoforms are related to stage and phenotypic-sex, observed in the germ cells of male gonad and in somatic cells of the female gonad.
Conclusions
Sox30 is not a gene only existed in mammals, but exists widely throughout the animal kingdom as supported by our bioinformatic, phylogenetic and syntenic analyses. It is very likely that Sox30 is expressed exclusively in gonads. Expression analyses revealed that Sox30 may be involved in female and male gonadal development at different stages by alternative splicing.
Background
Since the discovery of Sry [1-3], numerous genes encoding proteins containing HMG (High Mobility Group) domain have been identified throughout the animal kingdom [4-6]. Genes with their HMG domains similar to that of Sry (testis determining gene in mammals) by at least 50% were named as Sox (SRY-related HMG-box) genes [5]. At present, Sox genes encoding transcription factors, count about 30 members in vertebrates [6,7]. Based on differing levels of structural, organizational similarities and other features, Sox genes can be subdivided into 10 groups, named from A to J [8]. Members of the same group are similar to each other within and outside the HMG-box. However, members from different groups share lower degree of identity in the HMG-box, and no significant identity outside this domain. Many of the Sox proteins, like Sry, Sox2, Sox3, Sox11 and Sox22, are encoded by a single exon [9-12]. In contrast, others, such as Sox6, Sox9, Sox17 and Sox30, consist of multiple exons [13-16]. In addition, the Sox genes are scattered on different chromosomes and are not clustered [5]. Apart from the Sry and Sox3 located on the Y- and X-chromosome respectively, other Sox genes are autosomal [17].
Sox30 (SRY-box containing gene 30), the only member of group H, has been characterized merely in a few species so far. It was firstly isolated from mouse (Mus musculus) and human (Homo sapiens). In these species, two transcripts of Sox30 mRNA were found, one of which encodes a truncated protein lacking partial C-terminal region due to a frame shift [16]. Sox30 was considered to be involved in mammalian spermatogonial differentiation and spermatogenesis [16,18]. As no sequences of Sox30 were available from any other groups of animals, including the sequenced fish genomes at that time, some scholars even speculated that Sox30 was specific to mammals [19]. In zebrafish (Danio rerio), a Sox30 gene has been reported [20], however, it was proved to be Sox21 by GenBank blast and phylogenetic analysis. Interestingly, in our present study, Sox30 was isolated from the Nile tilapia (Oreochromis niloticus) accidentally when cloning Sox9b. Subsequently, by blast search and data mining, Sox30 was also found to exist in genome sequences of the chicken (Gallus gallus), green anole lizard (Anolis carolinensis), western clawed frog (Xenopus tropicalis), sea squirts (Ciona intestinalis), lancelets (Branchiotoma belcheri), acorn worm (Saccoglossus kowalevskii), sea anemone (Nematostella vectensis), and ESTs (Expressed Sequence Tags) of the channel catfish (Ictalurus punctatus), guppy (Poecilia reticulata), fathead minnow (Pimephales promelas), little skate (Leucoraja erinacea), dogfish (Squalus acanthias), the California mussel (Mytilus californianus) and snail (Lottia gigantean). Thus we report here that Sox30 exists widely throughout the animal kingdom rather than specific to mammals as reported previously. More importantly, we isolated four alternatively spliced isoforms of Sox30, and studied their temporal and spatial expression patterns during normal and sex reversed gonadal development in tilapia. Our data suggest that Sox30 may play key roles in gonadal development, and the roles may be different at different stages and sexes.
Methods
Animals
The Nile tilapias for this study were reared in large tanks with a circulating aerated freshwater system. Fish were maintained at ambient temperature (26°C) under natural photoperiod. All genetic females (XX) and males (XY) were obtained by artificial fertilization of eggs from normal females (XX) with sperms from either sex reversed males (XX) or super males (YY), respectively [21]. Then, artificially fertilized eggs were cultured in recirculating water at 26°C to obtain fry. All animal experiments conformed to the Guide for Care and Use of Laboratory Animals and were approved by the Institutional Committee of Laboratory Animal Experimentation at Southwest University, China.
Drug treatment
XX males and XY females were generated by treating fry with estrogen receptor antagonist tamoxifen (TAM) (Novartis Company, AG, Switzerland) and 17β-estradiol (E2) (Sigma, USA), respectively. Fish feed were sprayed with 95% ethanol containing TAM, 25 μg/g; E2, 50 μg/g. Control fish were fed with 95% ethanol sprayed feed. Drug treatment was applied to the fry from 5 to 30 days after hatching (dah), the critical period of the Nile tilapia sex differentiation [22]. E2 (XY) and TAM (XX) treatment resulted in complete sex reversal in tilapia, respectively [23].
Cloning of tilapia Sox30 cDNA and genomic DNA
The SMART RACE cDNA of the gonads, which was used to perform both 5' and 3'-rapid implication of cDNA ends (RACE) to obtain full-length cDNA of tilapia Sox30, were synthesized according to the manufacturer's instruction (Clontech). The Sox30 5' cDNA end was amplified accidentally with primers for Sox9b (partial sequence 225 bp, accession number: DQ632575), and confirmed by blastx http://blast.ncbi.nlm.nih.gov/. Subsequently, 3' RACE primers were designed to amplify 3' cDNA end for tilapia Sox30. Finally, to verify the full-length sequences of Sox30 cDNA, end-to-end PCR was performed and four mRNA isoforms were identified by using 5'- and 3'-UTR primers (T30UTR-F, T30UTR-R). In order to determine the gene structure of the tilapia Sox30, the genomic DNA sequence of this gene was amplified. Genomic DNA was prepared from mature testis and ovary. A 3790 bp Sox30 gene fragment with complete cds (coding sequences) and four introns, 102, 1518, 106 and 980 bp in length, was obtained by genomic PCR using end primers T30UTR-F and T30UTR-R. All the primers and sequences used in the present study were listed in Table 1. The PCR products were resolved on a 1.2% agarose gel and the target DNA fragments were purified using QIAquick Gel Extraction Kit (Qiagen, Germany).
Table 1.
Sequence of primers used in the present study
| Primer | Sequence | Purpose |
|---|---|---|
| R1 | GTAGTCCGGGTGATCCTTCTTGTGC | RACE |
| R2 | AGACGCTCAGCCTCCTCCACGAA | RACE |
| R3 | AGCCTGAGCCCACACCATGAACG | RACE |
| F1 | TCAGCGTCAGCCCAGGTCAGGAGA | RACE |
| F2 | TGTTTCAGGGCTGTGGAGTTGGTGG | RACE |
| F3 | GCAAACGCAAACTGGCAGAGACGC | RACE |
| UTR-F | CGGTCTCACAATGGGAACCAACTC | Full-length cDNA amplification and tissue distribution |
| UTR-R | GCTGAACATCTCCTAGAGCAGTCCA | Full-length cDNA amplification and tissue distribution |
| β-actinF | GGCATCACACCTTCTACAACGA | Internal control |
| β-actinR | ACGCTCTGTCAGGATCTTCA | Internal control |
All the fragments obtained were cloned into pGEM-T Vector (Promega, USA) and bi-directional sequencing was performed by the dideoxy chain termination method using an ABI PRISM 377 DNA genetic analyzer (Sangon, Shanghai, China).
Data mining and sequence analyses
The following genome databases were searched to explore the presence of Sox30-like sequences in other vertebrate and invertebrate species, including the UCSC Genome Bioinformatics at: http://genome.ucsc.edu/, the NCBI databases at http://blast.ncbi.nlm.nih.gov/, the Silk DB databases at http://silkworm.genomics.org.cn/, the JGI databases at http://genome.jgi-psf.org/Nemve1/Nemve1.home.html, the Ensembl databases http://www.ensembl.org/biomart/martview/ and the Gene DB databases at http://www.genedb.org/genedb/smansoni/. Sox30-like sequences were identified from seven species by blast search against genome sequences and further confirmed in EST databases, i.e., chicken (XP_414564, CN227447, CN229457, CO771932 and BU460467), anole lizard (scaffold_69: 1510867-1526492), frog (scaffold_ 177: 129323-130299, CX934092 and CX934091), sea squirts (NM_001078355), lancelets (chrUn: 451615932-451618210, BW716506, BW714230, BW695368 and BW697885), acorn worm (scaffold_16907: 1180000-1220000) and sea anemones (scaffold_134:263216-265501), but failed to identify it from other 80 animal species with genome sequence information. Partial or complete Sox30-like sequences were also identified by blast search against the NCBI EST databases from channel catfish (FD337967), guppy (ES386169), fathead minnow (DT310961, DT345945, DT310040, DT100872 and DT266812), little skate (GH161005 and CV222391), dogfish (CX196372), the California mussel (ES397482) and snail (FC701316, FC674522, FC687599 and FC698472).
The orientation and chromosomal position of Sox30 and its adjacent genes Thg1l (tRNA-histidine guanylyltransferase 1-like) and Adam19 (ADAM metallopeptidase domain 19) were determined manually from the gene orientations listed in BIOMART from the Ensembl database. Synteny analyses of Sox30, Thg1l and Adam19 in human, mouse, chicken, anole lizard and frog were performed by comparing the cDNA sequences with genome sequence using BLAST search from the UCSC Genome Bioinformatics or by search in BIOMART from the Ensembl database.
The multiple alignment software DNAstar and ClustalX were employed to analyze the nucleotide sequences and their deduced amino acid (aa) sequences. Based on the alignment results, a phylogenetic tree of Sox30 proteins was constructed using the fruitfly (Drosophila melanogaster) SoxF as an outgroup. The Sox30 sequences isolated from chicken, anole lizard, frog, channel catfish, guppy, fathead minnow, little skate, dogfish, sea squirts, lancelets, acorn worm, the California mussel, snail and sea anemones were also included in the phylogenetic analysis. The tree was constructed using the neighbor-joining method. The scores represent bootstrap values of 1000 trials, indicating the credibility of each branch. All the aa sequences used in the phylogenetic analysis were obtained from GenBank, UCSC and JGI Genome Bioinformatics, except that from tilapia. The GenBank accession numbers and scaffold numbers of these sequences are listed in Table 2.
Table 2.
Sequences used by this study
| Name | Species | Group | Accession No. | Pubmed No. |
|---|---|---|---|---|
| SoxF (Sox15) |
fruitfly, Drosophila melanogaster |
F | NM_079015 | 10731132 |
| Sox30 | sea squirts,Ciona intestinalis | H | NM_001078355 | 15269171 |
| Sox30* | lancelet, Branchiostoma floridae | H | chrUn:45161593 2-451618210 |
unpublished |
| Sox30* | western clawed frog, Xenopus tropicalis | H | scaffold_177:129323-130 299, CX934091 and CX934092 |
unpublished |
| Sox30 | mouse, Mus musculus | H | NP_775560 | 16564520 |
| Sox30 | chicken, Gallus gallus | H | XP_414564 | unpublished |
| Sox30 | the Nile tilapia, Oreochromis niloticus | H | GQ463453 | unpublished |
| Sox30 | human, Homo sapiens | H | NP_848511 | 10359848 |
| Sox30 | cattle, Bos taurus | H | NM_001046429 | 19393038 |
| Sox30* | mussel, Mytilus californianus | H | ES397482 | unpublished |
| Sox30* | catfish, Ictalurus punctatus | H | FD337967 | unpublished |
| Sox30* | guppy, Poecilia reticulata | H | ES386169 | unpublished |
| Sox30 | fathead minnow, Pimephales promelas | H |
DT310961, DT345945 DT310040, DT100872 and DT266812 |
unpublished |
| Sox30* | snail, Lottia gigantean | H |
FC701316, FC674522 FC687599 and FC698472 |
unpublished |
| Sox30 | sea anemone, Nematostella vectensis | H | scaffold_134:2632 16-265501 |
unpublished |
| Sox30 | acorn worm, Saccoglossus kowalevskii | H | scaffold_134:1180000 -1220000 |
unpublished |
| Sox30* | little skate, Leucoraja erinacea | H | GH161005 and CV222391 | unpublished |
| Sox30* | dogfish, Squalus acanthias | H | CX196372 | unpublished |
| Sox30 | anole lizard, Anolis carolinensis | H | scaffold_69: 1510867-1526492 | unpublished |
Note. * Partial sequences identified from databases and first named in this report.
Analysis of Sox30 expression by RT-PCR and semiquantitative RT-PCR
Total RNAs (2.0 μg) were isolated from various tissues of adults (240dah) (XX and XY fish, for tissue distribution analysis) and the gonads of monosex (XX and XY fish at 5, 10, 15, 35, 90, 150, 240dah, for ontogeny analysis). Thereafter, total RNAs were treated with DNase I to eliminate the genomic DNA contamination. Then first strand cDNAs were synthesized and RT-PCR was carried out to check the expression levels of tilapia Sox30 according to methods described previously [24]. The templates for positive and negative controls were set with Sox30 plasmid DNA and deionized water, respectively. A 342-bp fragmentofβ-actin was amplified as internal control to test the quality of the cDNAs used in the PCR. The PCR products were subjected to agarose gel (1.2%) electrophoresis.
To examine the influence of drug treatment on Sox30 gene expression, gonads of 5 individuals were collected from each control group (XX female, XY male) and drug treated group (TAM-induced XX male and E2-induced XY female) when the fish were adult at 240dah. Semi-quantitative RT-PCR [25] was performed to measure the mRNA levels using the primers (T30UTR-F, T30UTR-R) described in the previous section. A series of PCRs with different cycle numbers (from 22 to 36, with an interval of 2) were performed to determine the linear phase of the amplification using 1 to 5 diluted cDNA template. Based on these pilot experiments, 28 cycles for β-actin and 34 cycles for the target gene were chosen and applied to the subsequent semi-quantitative RT-PCR analyses. Band intensities resulting from the PCR amplification were analyzed using the image analysis software Quantity One (Bio-Rad). Sox30 alternatively spliced mRNA levels were expressed relative to that of β-actin in each sample. In our study, β-actin expression was found to be unaffected by the drug treatment [23]. Results obtained were expressed as mean values ± S.E.M. from five individual fish. Data analyses were performed using one-way ANOVA and the least significant difference on the GraphPad Prism 4 software (GraphPad Software, San Diego, CA, USA).
In situ hybridization (ISH)
Tilapia gonads from 10, 120 and 210dah were dissected and fixed in 4% paraformaldehyde (PFA) in 0.85× PBS (pH 7.4) at 4°C overnight. After fixation, gonads were embedded in paraffin. Cross-sections of 5 μm were cut with a sliding microtome. Probes of both sense and antisense digoxigenin (DIG)-labeled RNA strands were transcribed in vitro from a linearized tilapia Sox30 plasmid, using the RNA labeling kit (Roche Diagnostics GmbH, Mannheim, Germany). In situ hybridization was carried out as follows: sections were deparaffinized, hydrated and treated with proteinase K (10 μg/ml, Amersco, USA) and then hybridized with the sense or antisense DIG-labeled RNA probe at 60°C for 16-22 h. The hybridization signals were then detected using alkaline phosphatase-conjugated anti-DIG antibody (Roche, Germany) and NBT as the chromogen [26].
Results
Molecular cloning of tilapia Sox30
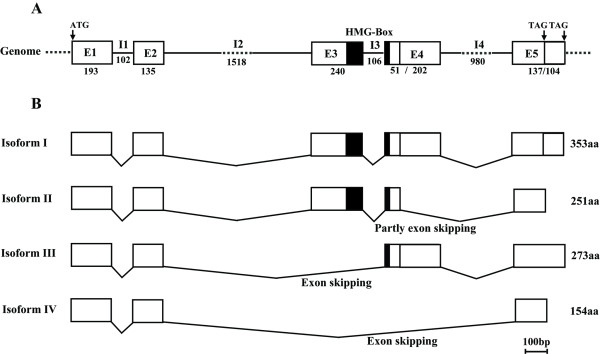
The Sox30 genomic DNA and four alternatively spliced cDNAs were cloned from the Nile tilapia. The genomic DNA is 3790 bp long (GenBank accession no. GQ463457), containing 5 exons, 193, 135, 240, 253 and 241 bp, respectively, and four introns (two in the N-terminal region, one in the C-terminal region and one within the HMG-box). All introns share the typical intron characteristics (gt----ag). The isolated full-length Sox30 cDNA (isoform-I) is 1474 bp (GenBank accession no. GQ463453), containing an open reading frame (ORF) of 1062 bp encoding a putative protein of 353 aa, a 116 bp 5' untranslated region (UTR), and a 296 bp 3' UTR. The other three alternatively spliced isoforms (isoform-II, -III and -IV, GenBank accession nos. GQ463454, GQ463455 and GQ463456) amplified by the same pair of primers were 306, 240 and 597 bp shorter than the ORF of isoform-I due to exon skipping. The four isoforms share the same 5'-UTR, while only isoform-I and -III share the same 3'-UTR. Isoform-II and -IV have 3'-UTRs 104 bp longer than those of isoform-I and -III due to the reading frame shift and introduction of a stop codon which is 104 bp upstream of the stop codon found in isoform-I and -III transcripts. Further analysis suggested that these isoforms (I-IV) contain 5, 5, 4 and 3 exons, respectively. Compared with the longest isoform (isoform-I, which contains 5 exons), part of exon 4, exon 3, exon 3 and exon 4 was spliced off in Isoform-II (with an ORF of 756 bp encoding 251aa), -III (822 bp encoding 273aa), and -IV (465 bp encoding 154aa), respectively (Figure 1).
Figure 1.
Tilapia Sox30 gene structure (A) and its four alternatively spliced isoforms (B). Rectangle, solid line and dotted line represent exon, intron and omitted sequences, respectively. Numbers represent the lengths of nucleotide and protein sequences. E and I stand for exon and intron, respectively. ATG and TAG are start and stop codon, respectively. The HMG-box is shaded.
Sequence, phylogeney and synteny analyses
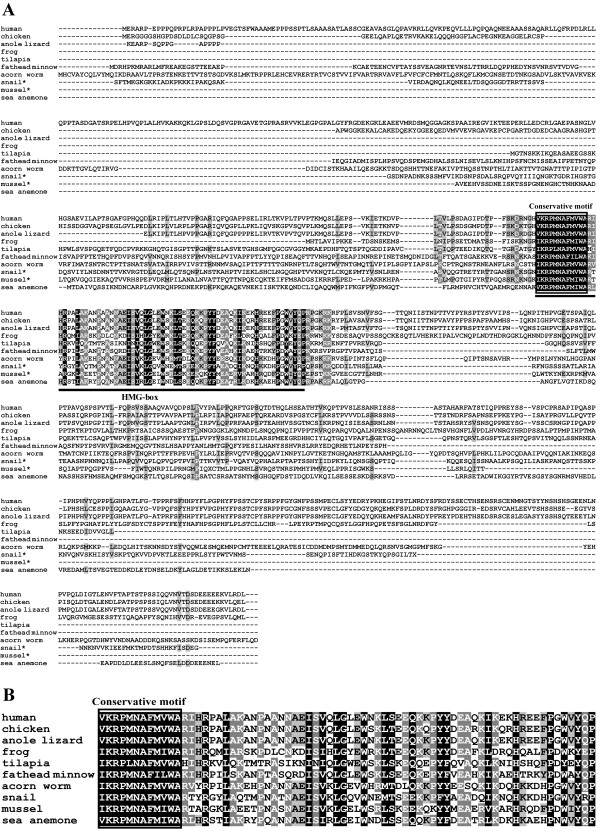
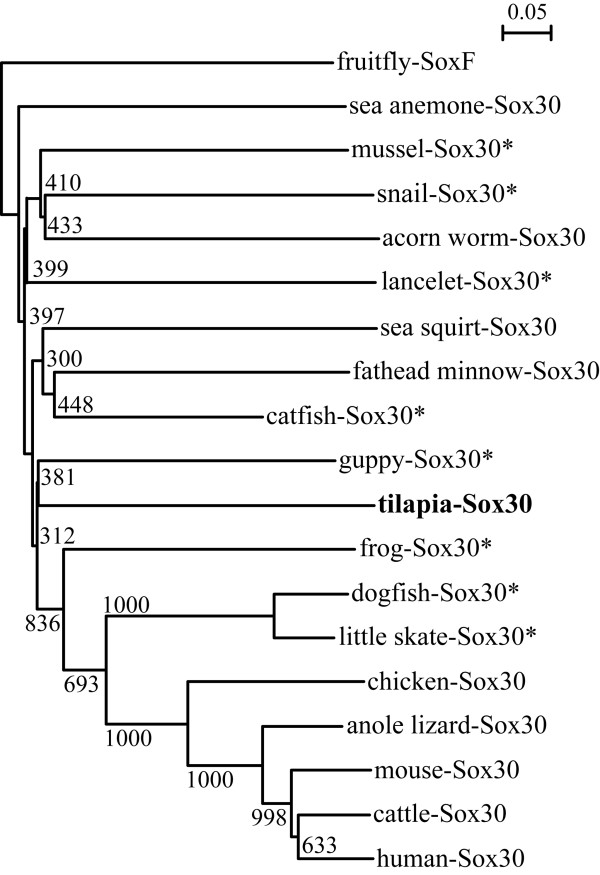
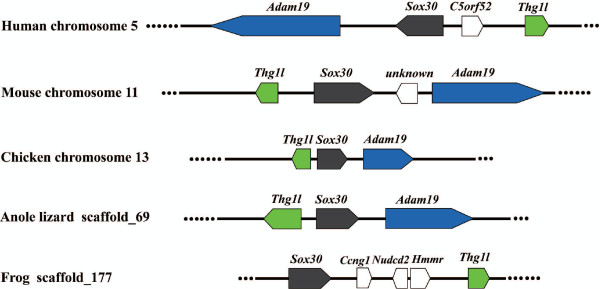
Multiple alignment of the aa sequences of Sox30 s from various species revealed that Sox30 was poorly conserved except in the HMG-box, the characteristic DNA binding domain (Figure 2). The HMG-boxes of tilapia and guppy Sox30 shared the highest similarity (about 52.9%), whereas the whole peptides of both shared rather low similarity (about 28%). Compared with Sox30 from mammals, tilapia Sox30 is shorter in both N- and C-terminal regions (Figure 2A). As was shown in Figure 3, Sox30 s isolated from tilapia and other species were clustered into the same clade with the mammalian Sox30 s, indicating that they are genuine Sox30 s. Synteny analyses showed that Sox30 and its adjacent gene Thg1l and Adam19 were located on the same chromosome/scaffold in the same gene order in human, mouse, chicken, anole lizard and frog (Figure 4).
Figure 2.
Alignment of the amino acid sequences of Sox30 s (A) and their HMG-boxes (B). The HMG-box is underlined. The conservative motif in HMG-box is boxed. * indicates partial sequences.
Figure 3.
Phylogenetic analysis of Sox30 s based on alignment of their full-length amino acid sequences. This tree was generated by the default settings of Clustal × alignment program and constructed using fruitfly SoxF as an outgroup. The values in the tree represent bootstrap scores out of 1000 trials, indicating the credibility of each branch. Branch lengths are proportional to the number of amino acid changes on the branch. * indicates partial sequences. The little skate and dogfish Sox30 s were clustered in a wrong position because the isolated two sequences are very short, including only partial N- terminal and HMG-box regions.
Figure 4.
Synteny analyses of Sox30 and its adjacent gene Thg1l and Adam19 in human, mouse, chicken, anole lizard and frog. Rectangles represent genes in chromosome/scaffold, dotted lines represent omitted genes of the chromosome/scaffold, and the arrow represents gene-coding direction. Sox30, Thg1l and Adam19 are shown in grey, green and blue, respectively. The other genes are shown in white.
Tissue expression of Sox30 gene
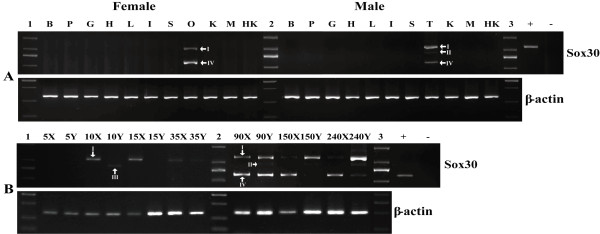
Tissue distribution analysis using RT-PCR revealed that only three of the four isoforms of tilapia Sox30 (isoform-I, -II and -IV) were expressed in adult. They were expressed exclusively in gonads, with isoform-I higher in the testis; isoform-IV higher in the ovary; while isoform-II only in the testis, not in the ovary (Figure 5A).
Figure 5.
Expression patterns of Sox30 in the Nile tilapia. (A) RT-PCR analysis of Sox30 from various tissues of adult fish. B, brain; P, pituitary; G, gill; H, heart; L, liver; I, intestine; S, spleen; O, ovary; K, kidney; M, muscle; HK, Head kidney; T, testis; 1, 2, and 3, marker; +, positive control; -, negative control; β-actin was used as the internal control; (B) Ontogeny (5-240 dah) of Sox30 expression in tilapia gonads by RT-PCR. 1, 2, marker; X, female; and Y, male; +, positive control; -, negative control; I, II, III, IV indicate isoform-I, -II, -III and -IV, respectively.
Ontogeny of Sox30 expression in tilapia gonads
Sox30 expression was detected in gonads of tilapia during sex determination, differentiation and later development by RT-PCR. Isoform-I mRNA was detected consistently in the gonads at 10, 15, 35, 90, 150 and 240 dah with a very interesting expression profile: ovary only at 10 and 15 dah; both ovary and testis from 35 dah onwards, higher in testis than in ovary from 90 dah to adult (Figure 5B). Isoform-II showed a male-specific expression from 90 dah to adult. Isoform-III was detected exclusively in male gonad at 10 dah only, and no expression was detected during any other developmental stages. Isoform-IV started to express in both testis and ovary from 90 dah onwards, with higher level in the ovary from 150 dah to adult.
Expression of Sox30 in sex-reversed gonads
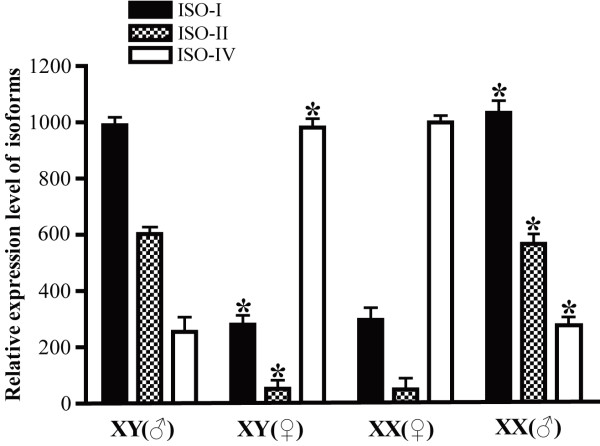
Consistent with tissue distribution results, three isoforms (I, II and IV but not III) were detected in adult gonads by semi-quantitative RT-PCR. Isoform-I and -IV mRNA were detected in gonads of all four groups, i.e. XX (♀) and XY (♂) control fish and sex reversed XX (♂, induced by TAM treatment) fish and sex reversed XY (♀, induced by E2 treatment) fish. Isoform-I was significantly down-regulated in sex reversed XY (♀) gonads, but up-regulated in sex reversed XX (♂) gonads, being consistent with the result of tissue distribution and ontogeny analyses that isoform-I was expressed higher in the testis; Isoform-IV was up-regulated in sex reversed XY (♀) gonads, but down-regulated in sex reversed XX (♂) gonads, which was consistent with that isoform-IV was expressed higher in the ovary. Isoform-II mRNA was only detected in normal XY (♂) and sex reversed XX (♂) gonads but not in normal XX (♀) and sex reversed XY (♀) gonads, consistent with that isoform-II showed a male gonad-specific expression in tissue distribution and ontogeny analyses. These data indicated that the expressions of Sox30 are phenotypically related rather than genotypically related (Figure 6).
Figure 6.
Relative expression levels of tilapia Sox30 isoforms in gonad of control and sex reversed adult tilapia by semi-quantitative RT-PCR. Results were expressed as mean values ± S.E.M. from five individual fish (*P < 0.01 significantly different as compared with the respective control by one-way ANOVA). Iso is the abbreviation of isoform. XX (♂), XX male; XY (♀), XY female; XY (♀) fish, male-to-female sex reversal induced by 17β-estradiol (E2); XX (♂) fish, female-to-male sex reversal induced by tamoxifen (TAM). The doses of the drugs used are described in the Materials and Methods section.
Expression of Sox30 by in situ hybridization in tilapia gonads
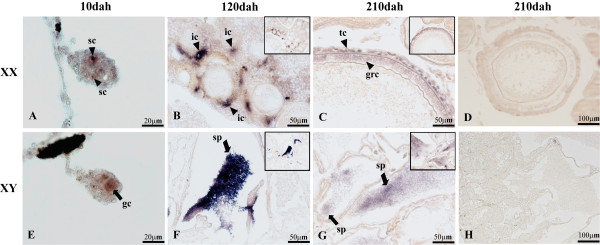
To ascertain which population of cells in the developing gonads expresses Sox30, ISH was performed using gonads at 5, 10, 120 and 210 dah. Specific signals were observed in the male (XY) germ cells at 10 dah (Figure 7E) and the sperms of the testis at 120 and 210 dah (Figure 7F, G). In contrast, specific signals were detected in the female (XX) somatic cells at 10 dah (Figure 7A), interstitial cells of the ovary at 120 dah (Figure 7B), and theca and granulosa cells of the ovary at 210 dah (Figure 7C), but no signal was detected in both sexes at 5 dah (data not shown). Taking the ontogeny results into consideration, we conclude that isoform-III is restricted to the germ cells in male gonads and isoform-I is expressed in the somatic cells of the female gonads at 10 dah.
Figure 7.
Expression of tilapia Sox30 in gonads examined by in situ hybridization with antisense (A-C, E-G) and sense (D, H) probes. At 10 dah, Sox30 expression appears in germ cells (gc) of the male gonad (E), and in a few round somatic cells (sc) of the female gonad (A); at 120 dah, Sox30 was expressed in sperms (sp) of the testis (F) and interstitial cells (ic) of the ovary (B); at 210 dah, Sox30 was expressed in sperms of the testis (G), theca (tc) and granulosa cells (grc) of the ovary (C). The arrow and arrowhead indicate signals of Sox30 in the XY and XX gonads, respectively.
However, due to the overlapping probes (The probes used in this study recognize all the four Sox30 isoforms. It is impossible to design isoform-specific probe for all of them), we failed to distinguish the signals detected at 120 and 210 dah gonads. At least three and two isoforms could produce the signals detected in the testis and the ovary, respectively at these stages.
Discussion
Bioinformatics and phylogeney analyses of Sox30
In the present study, a novel Sry-related gene was isolated from the Nile tilapia accidentally when cloning Sox9b. It was characterized as Sox30 by subsequent blast against GenBank for its relatively high identity with its mammalian orthologs, which was further confirmed by phylogenetic and syntenic analyses. To date, Sox30 has been reported only in mammals [16,18,19], but not in any non-mammalian vertebrates (including fish) and invertebrates. Our data, for the first time, provided the solid proofs for the existence of Sox30 in a teleost fish, the Nile tilapia.
Subsequently, genome databases of fishes, including zebrafish, medaka (Oryzias latipes), stickleback (Gasterosteus aculeatus), takifugu (Takifugu rubripes) and tetraodon (Tetraodon nigroviridis) were searched for possible Sox30-like genes. However, it seems that this gene does not exist in any database of these teleosts. EST data bases were also searched afterward, and complete or partial Sox30 sequences were successfully isolated from the channel catfish, guppy, fathead minnow, little skate and dogfish. Further screening of the genome sequences of other animals showed that Sox30 was also present in other chordates, including chicken, anole lizard, frog, sea squirts, lancelets and acorn worm (also called as SoxH in the last three species), and non-chordate invertebrates, including the California mussel, snail and sea anemone (also called as SoxH), while not found in the nematode (Caenorhabditis elegans), fruitfly and sea lamprey. A recent report stated the existence of Sox30-like sequence in sea urchin (Strongylocentrotus purpuratus) [27], but we failed to find in the genome and EST databases by blast search. These findings suggested that Sox30 is not specific to mammals, but also exists in other vertebrates and invertebrates. Then, why we failed to find Sox30-like sequences in all sequenced teleost genomes? There are two possible explanations. One is that the genome sequences from these species have not covered all the genome yet; and the other is that Sox30 was secondarily lost in these species during evolution. However, the chance for the latter possibility is quite small because of the following two reasons, 1) cyprinids and ictalurids, such as fathead minnow and channel catfish, are relatively primitive teleosts while cichlids and poeciliids, such as tilapia and guppy, are relatively advanced teleosts. It is unlikely that Sox30 was lost in all the other teleosts except the primitive and advanced species; 2) both fathead minnow and zebrafish belong to the same family, Cyprinidae. It is difficult to believe that Sox30 was secondarily lost in the zebrafish but not in another closely related species, fathead minnow. Therefore, we tend to accept the former explanation. To confirm this, further cloning Sox30 from other teleosts is required.
Sox30 was found not only in chordates such as human, mouse, chicken, frog, tilapia, channel catfish, guppy, fathead minnow, little skate, dogfish, sea squirts, lancelets and acorn worm, but also in non-chordate invertebrate species, such as the California mussel, snail and sea anemone, indicating that it had already appeared before or with the emergence of coelenterates. Then, it is of interests to know from which ancestor gene Sox30 was derived. Group H, which only consists of Sox30, shared the highest similarity to group F in 10 Sox groups (Additional file 1, Fig.S1), suggesting that they may share a common ancestor. However, the exact timing for the divergence of the two groups is still unclear. Its presence in the relatively primitive invertebrate species, sea anemones (no Sox30 or other Sox genes have been found in protozoa and bacteria at present) and its position (the outermost clade) in the phylogenetic tree (Additional file 1, Fig. S1) suggested that Sox30 seemed to be one of the oldest members in the Sox family, which is consistent with the previous conclusion [28].
Sequence and gene structure analyses of Sox30
Compared with its counterparts from mammals, tilapia Sox30 genomic sequence was smaller in size due to compression of intron sequences. Similar phenomenon has been found for Sox30 in sea squirts, lancelets and acorn worm. Alignment of aa sequences indicated that Sox30 s was poorly conserved, even in the highly conserved HMG-box among both distant and closely related species, suggesting high diversity and rapid evolution of these proteins. This also explains why Sox30 has not been successfully isolated from non-mammalian animals until the accident cloning of tilapia Sox30 in this study. Gene structure analysis showed that tilapia Sox30, like some other Sox group members, such as Sox8, Sox9 and Sox10 [29], was also characterized by the splitting of the HMG-box by an intron. The same is true to Sox30 of acorn worm, lancelets, sea squirts, frog, chicken, mouse and human, and the location of the intron is conserved among these species. These findings indicated that this intron had already been fixed before or with the emergence of chordates.
Alternative splicing (AS) is emerging as one of the most important mechanisms to control vertebrate gene expression. Existing data indicate that as much as 76% of genes generate alternatively spliced products [30]. AS has been associated with a regulatory system in tissue- or stage-specific splicing mechanisms by which expression may be regulated. This regulation may be achieved by the introduction of premature stop codons that function as an on-off gene expression switch [31]. Previous study showed that two different mRNA isoforms of Sox17, Sox30 have been isolated in mouse and two different mRNA isoforms of Sox9 in frog because of AS [15,16,32]. In mice, the Sox17 isoform is expressed at a low level in the testis throughout postnatal development, while the t-Sox17 isoform is expressed abundantly in the testis, predominantly in postmeiotic germ cells [15]. Based on these results, they suggested that a switch from Sox17 to the t-Sox17 isoform may alter the function of Sox17 at the meiotic and postmeiotic phases during spermatogenesis in mice [15]. Our data demonstrated that tilapia Sox30 has four alternatively spliced isoforms. Early termination of the stop codon was found in the isoform-II and -IV transcripts of tilapia Sox30, suggesting that the mechanism mentioned above may be also responsible for regulation of Sox30 gene expression in tilapia. Besides, similar to mouse Sox17, four isoforms of Sox30 were also expressed in a non-parallel manner during gonadal development of tilapia. Thus, there may be some switches from one to the other isoform(s) to alter the function of Sox30 during gonadal development. It is worthwhile to note that the HMG-box region was deleted partially in isoform-III and completely in isoform-IV, respectively, which resulted in truncated proteins lacking most parts or all of the HMG-box domain and the nuclear localization signal.
It is interesting to know whether these protein isoforms are ever made, but now there is no antibody available because Sox30 is poorly conserved among species of different classes of animals, and therefore, the available antibody against mammalian Sox30 is not suitable for usage in fish. There are reports showing that alternatively spliced isoforms of Sox17 and Sox30, without functional DNA binding domain or C-terminus, very similar to tilapia Sox30 isoforms, were translated into proteins in mammals [15,16]. Based on those findings, we speculated that all isoforms of Sox30 in tilapia may also be translated into the protein products. As Sox30 gene, like Sry and some other Sox genes, uses the single HMG-box for DNA-binding, it would be very interesting to know what these truncated isoforms do and how they function. Whether this would result in different capacity of transcription activation, e.g. complete non-functional factors or dominant negative mutants of the wild type Sox30, is still an open question.
The expression pattern and functional relevance of Sox30 in tilapia
Previous reports in mammals showed that human and mouse Sox30 are exclusively expressed in normal adult testis and specifically in germ cells [16]. This expression pattern suggests that Sox30 may be involved in mammalian spermatogonial differentiation and spermatogenesis [18] However, it is still unclear in which cell type Sox30 is expressed in the mammalian ovary even though there was report indicating its possible expression in mouse oocytes [33]. In the present study, Sox30 started to express in gonads from 10 dah, earlier than the morphological gonadal differentiation period (about 25 dah) in tilapia and showed a gonad specific expression pattern at least in adults. Of course, expression of Sox30 in extra-gonadal tissues in other stages of development cannot be completely excluded. Meanwhile, expression of each alternatively spliced isoform showed a clear sexual dimorphism in gonads. However, none of the four isoforms of Sox30 were detected at 5 dah, the critical period of tilapia molecular sex determination, which excluded its role as the sex determining gene in tilapia. In sex reversed adult gonads, three types of Sox30 also exhibited a phenotypic sex-related expression pattern. Sox30 expression was restricted to the germ cells at 10 dah and later to sperms in male gonads, indicating its possible involvement in spermatogonial differentiation and spermatogenesis in male fish, like in human and mouse. In female, it was expressed in the somatic cells in female gonads at 10, 120 and 210 dah, respectively.
Sox30 is a strong regulator of the Sf-1 [33], the most important steroidogenic factor found in all vertebrates, including fish [34,35]. Our in situ hybridization data demonstrated clearly that Sox30 is expressed in the somatic cells, especially steroidogenic cells of the ovary, colocalized with Sf-1 in tilapia [34]. Taken together, we speculate that Sox30 may be an important regulator for somatic differentiation and steroidogenesis in female fish as well. Both isoform I and IV are expressed in the ovary after 35 dah. Although isoform IV is expressed much higher than isoform I it lacks the DNA binding HMG-box, and therefore, it is unlikely that this isoform regulates Sf-1 expression by direct binding to the promoter. However, the possibility that isoform IV might function as a dominant negative mutant of the wild type molecule (isoform I) can not be excluded because it still has the other functional domains, such as transactivation domain. More works need to be done to unravel this.
In addition to the gonad specific expression of Sox30 in human, mouse and tilapia, all Sox30 EST sequences were also derived from gonads (testis and ovary in chicken and channel catfish, testis in frog, guppy and fathead minnow, male gonads in snail) demonstrating that Sox30 may also be a gonad specific gene in these species. These data further supported that Sox30 plays a key role in gonadal differentiation and development, which might be conserved among species in the animal kingdom. Moreover, four alternatively spliced Sox30 isoforms exhibited different temporal and spatial expression patterns in tilapia gonads. Alternative splicing of Sox30 mRNAs and different temporal expression pattern of the spliced isoforms had also been reported in human and mouse [16]. Therefore, we speculate that Sox30 may be involved in gonadal differentiation and development in different sexes, at different stages and in different cell types of gonads in the animal kingdom by AS. Further study on Sox30 in other species is required to confirm this.
The low similarity between tilapia Sox30 and its mammalian counterpart raises a question of whether tilapia Sox30 is a genuine Sox30 or just a new Sox member of the group H. To answer this, we analyzed the synteny of Sox30 and its adjacent genes in human, mouse, chicken, anole lizard and frog (tilapia was not included because its genome sequences has not finished and open yet). Sox30 and its adjacent gene Thg1l and Adam19 were found to be located on the chromosome 5, 11, 13 and scaffold_69 and _177 in human, mouse, chicken, anole lizard and frog, respectively. The conservation in synteny between non-mammalian species and mammals indirectly support that our isolated Sox30 s are genuine orthologs of mammalian Sox30.
Conclusion
For the first time, Sox30 was found to exist in non-mammalian vertebrates and invertebrates by cloning, genome and EST database analyses. The probable gonad specific expression in all species analyzed and the temporal and spatial expression of the alternatively spliced isoforms indicated that Sox30 may play a key role in gonadal differentiation and development, and the function of Sox30 may be regulated by AS and expression in different gender, different stages and different cell types in gonads in a wide variety of animals.
Authors' contributions
FH carried out most of experiments, data mining and drafted the manuscript. ZJW co-carried out most of experiments and co-draft the manuscript. FRW participated in ISH. ZHL participated in some of expression study. BFH participated in gene cloning. DSW designed and supervised the experiment, analyzed data and critically edited the manuscript. All authors read and approved the final manuscript.
Contributor Information
Fei Han, Email: han16897723@yahoo.com.cn.
Zhijian Wang, Email: wangzj@swu.edu.cn.
Fengrui Wu, Email: fengruiwu@126.com.
Zhihao Liu, Email: minenut@163.com.
Baofeng Huang, Email: hbf410@163.com.
Deshou Wang, Email: wdeshou@swu.edu.cn.
Acknowledgements
This work was supported by grants from the National High Technology Research and Development Program (863 program) of China (No. 2007AA10Z165), the National Natural Science Foundation of China (Nos. 30770272, 30871905, 30831160508 and 31030063), the National Basic Research Program of China (2009CB941200), the Program for Changjiang Scholars and Innovative Research Team in University (IRT0859), the Natural Science Foundation Project of CQ CSTC (Nos. 2008BB1006 and 2008AC1016) and the Specialized Research Fund for the Doctoral Program of Higher Education of China (No. 20090182110008).
References
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;364:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Münsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- Wilson MJ, Dearden PK. Evolution of the insect Sox genes. BMC Evol Biol. 2008;8:120. doi: 10.1186/1471-2148-8-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny LH, Lovell-Badge R. Sox genes find their feet. Curr Opin Genet Dev. 1997;7:338–344. doi: 10.1016/S0959-437X(97)80147-5. [DOI] [PubMed] [Google Scholar]
- Schepers GE, Teasdale RD, Koopman P. Twenty pairs of Sox: extent, homology, and nomenclature of the mouse and human Sox transcription factor gene families. Dev Cell. 2002;3:167–170. doi: 10.1016/S1534-5807(02)00223-X. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Dumitriu B, Penzo-Méndez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related highmobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- Hacker A, Capel B, Goodfellow P, Lovell-Badge R. Expression of Sry, the mouse sex determining gene. Development. 1995;121:1603–1614. doi: 10.1242/dev.121.6.1603. [DOI] [PubMed] [Google Scholar]
- Collignon J, Sockanathan S, Hacker A, Cohen-Tannoudji M, Norris D, Rastan S, Stevanovic M, Goodfellow PN, Lovell-Badge R. A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development. 1996;122:509–520. doi: 10.1242/dev.122.2.509. [DOI] [PubMed] [Google Scholar]
- Jay P, Goze C, Marsollier C, Taviaux S, Hardelin JP, Koopman P, Berta P. The human SOX11 gene: cloning, chromosomal assignment and tissue expression. Genomics. 1995;29:541–545. doi: 10.1006/geno.1995.9970. [DOI] [PubMed] [Google Scholar]
- Jay P, Sahly I, Goze C, Taviaux S, Poulat F, Couly G, Abitbol M, Berta P. SOX22 is a new member of the SOX gene family, mainly expressed in human nervous tissue. Hum Mol Genet. 1997;6:1069–1077. doi: 10.1093/hmg/6.7.1069. [DOI] [PubMed] [Google Scholar]
- Wright E, Hargrave MR, Christiansen J, Cooper L, Kun J, Evans T, Gangadharan U, Greenfield A, Koopman P. The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat Genet. 1995;9:15–20. doi: 10.1038/ng0195-15. [DOI] [PubMed] [Google Scholar]
- Cohen-Barak O, Hagiwara N, Arlt MF, Horton JP, Brilliant MH. Cloning, characterization and chromosome mapping of the human SOX6 gene. Gene. 2001;265:157–164. doi: 10.1016/S0378-1119(01)00346-8. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Kanai-Azuma M, Noce T, Saido TC, Hayashi Y, Yazaki K. Identification of two Sox17 messenger RNA isoforms, with and without the high mobility group box region, and their differential expression in mouse spermatogenesis. J Cell Biol. 1996;133:667–681. doi: 10.1083/jcb.133.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaki E, Inazawa J, Copeland NG, Gilbert DJ, Jenkins NA, Ohsugi M, Tezuka T, Yoshida M, Semba K. Identification of a novel Sry-related gene and its germ cell-specific expression. Nucleic Acids Res. 1999;27:2503–2510. doi: 10.1093/nar/27.12.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanovic M, Lovell-Badge R, Collignon J, Goodfellow PN. SOX3 is an X-linked gene related to SRY. Hum Mol Genet. 1993;2:2013–2018. doi: 10.1093/hmg/2.12.2013. [DOI] [PubMed] [Google Scholar]
- Ballow D, Meistrich ML, Matzuk M, Rajkovic A. Sohlh1 is essential for spermatogonial differentiation. Dev Biol. 2006;294:161–167. doi: 10.1016/j.ydbio.2006.02.027. [DOI] [PubMed] [Google Scholar]
- Koopman P, Schepers G, Brenner S, Venkatesh B. Origin and diversity of the SOX transcription factor gene family: genome-wide analysis in Fugu rubripes. Gene. 2004;17:177–186. doi: 10.1016/j.gene.2003.12.008. [DOI] [PubMed] [Google Scholar]
- De Martino SP, Errington F, Ashworth A, Jowett T, Austin CA. Sox30: a novel zebrafish sox gene expressed in a restricted manner at the midbrain-hindbrain boundary during neurogenesis. Dev Genes Evol. 1999;209:357–362. doi: 10.1007/s004270050264. [DOI] [PubMed] [Google Scholar]
- Wang DS, Jiao B, Hu C, Huang X, Liu Z, Cheng CH. Discovery of a gonad-specific IGF subtype in teleost. Biochem Biophys Res Commun. 2008;367:336–341. doi: 10.1016/j.bbrc.2007.12.136. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Kajiura-Kobayashi H, Nagahama Y. Induction of XY sex reversal by estrogen involves altered gene expression in a teleost, tilapia. Cytogenet Genome Res. 2003;101:289–294. doi: 10.1159/000074351. [DOI] [PubMed] [Google Scholar]
- Zhang WL, Zhou LY, Senthilkumaran B, Sudhakumari CC, Kobayashi T, Nagahama Y, Wang DS. Molecular cloning of two isoforms of 11β-hydroxylase and their expressionsin the Nile tilapia, Oreochromis niloticus. Gen Comp Endocrinol. 2010;165:34–41. doi: 10.1016/j.ygcen.2009.05.018. [DOI] [PubMed] [Google Scholar]
- Zhou L, Wang D, Kobayashi T, Yano A, Paul-Prasanth B, Suzuki A, Sakai F, Nagahama Y. A novel type of P450c17, lacking the lyase activity is responsible for C21-steroid biosynthesis in the fish ovary and head kidney. Endocrinology. 2007;148:4282–4291. doi: 10.1210/en.2007-0487. [DOI] [PubMed] [Google Scholar]
- Jiao BW, Huang XG, Chan CB, Zhang L, Wang DS, Cheng CHK. The co-existence of two growth hormone receptors in teleost fish and their differential signal transduction, tissue distribution and hormonal regulation of expression in seabream. J Mol Endocrinol. 2006;36:23–40. doi: 10.1677/jme.1.01945. [DOI] [PubMed] [Google Scholar]
- Wang D, Kobayashi T, Zhou L, Nagahama Y. Molecular cloning and gene expression of Foxl2 in the Nile tilapia, Oreochromis niloticus. Biochem Biophys Res Commun. 2004;320:83–89. doi: 10.1016/j.bbrc.2004.05.133. [DOI] [PubMed] [Google Scholar]
- Phochanukul N, Russell S. No backbone but lots of Sox: Invertebrate Sox genes. Int J Biochem Cell Biol. 2010;42:453–464. doi: 10.1016/j.biocel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Nagai K. Molecular evolution of Sry and Sox gene. Gene. 2001;270:161–169. doi: 10.1016/S0378-1119(01)00479-6. [DOI] [PubMed] [Google Scholar]
- Hett AK, Ludwig A. SRY-related (Sox) genes in the genome of European Atlantic sturgeon (Acipenser sturio) Genome. 2005;48:181–186. doi: 10.1139/g04-112. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Smith CW, Valcárcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/S0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- Takase M, Noguchi S, Nakamura M. Two Sox9 messenger RNA isoforms: isolation of cDNAs and their expression during gonadal development in the frog, Rana rugosa. FEBS Lett. 2000;466:249–254. doi: 10.1016/S0014-5793(00)01078-4. [DOI] [PubMed] [Google Scholar]
- Sakai N, Terami H, Suzuki S, Haga M, Nomoto K, Tsuchida N, Morohashi K, Saito N, Asada M, Hashimoto M, Harada D, Asahara H, Ishikawa T, Shimada F, Sakurada K. Identification of NR5A1 (SF-1/AD4BP) gene expression modulators by large-scale gain and loss of function studies. J Endocrinol. 2008;198:489–497. doi: 10.1677/JOE-08-0027. [DOI] [PubMed] [Google Scholar]
- Wang DS, Kobayashi T, Zhou LY, Paul-Prasanth B, Ijiri S, Sakai F, Okubo K, Morohashi K, Nagahama Y. Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with ad4 binding protein/steroidogenic factor 1. Molecular Endocrinology. 2007;21:712–725. doi: 10.1210/me.2006-0248. [DOI] [PubMed] [Google Scholar]
- Wang DS, Zhou LY, Kobayashi T, Matsuda M, Shibata Y, Sakai F, Nagahama Y. Dmrt1 repression of aromatase transcription, a possible mechanism favoring the male pathway in tilapia. Endocrinology. 2010;151:1331–1340. doi: 10.1210/en.2009-0999. [DOI] [PubMed] [Google Scholar]