Abstract
The central role of metabolic perturbation to the pathology of malaria, the promise of antimetabolites as antimalarial drugs and a basic scientific interest in understanding this fascinating example of highly divergent microbial metabolism has spurred a major and concerted research effort towards elucidating the metabolic network of the Plasmodium parasites. Central carbon metabolism, broadly comprising the flow of carbon from nutrients into biomass, has been a particular focus due to clear and early indications that it plays an essential role in this network. Decades of painstaking efforts have significantly clarified our understanding of these pathways of carbon flux, and this foundational knowledge, coupled with the advent of advanced analytical technologies, have set the stage for the development of a holistic, network-level model of plasmodial carbon metabolism. In this review we summarize the current state of knowledge regarding central carbon metabolism and suggest future avenues of research. We focus primarily on the blood stages of Plasmodium falciparum, the most lethal of the human malaria parasites, but also integrate results from simian, avian and rodent models of malaria that were a major focus of early investigations into plasmodial metabolism.
Keywords: Plasmodium, malaria, central carbon metabolism, metabolic network, metabolomics
Introduction
While the process of evolving into a parasitic niche seems to have resulted in a `paring down' of many Plasmodium metabolic pathways, including the wholesale loss of de novo amino acid and purine biosynthesis, pioneering early work suggested that most of the core conserved components of carbon metabolism – glycolysis [1, 2], the pentose phosphate pathway [3], lipid biogenesis [4, 5], glycosylation [6, 7] and at least some components of citric acid metabolism [8, 9] – were present in some form. However, efforts to precisely map the flow of carbon through the metabolic network of the malaria parasite were often stymied by difficulties pertaining to the isolation and culturing of parasites, complications arising from the interconnected nature of parasite and host cell metabolism, technical limitations of metabolic tracing using classical methodologies and in some cases marked divergence between pathways in Plasmodium spp. and the model organisms in which they were first elucidated (reviewed in [10–12]). Dedicated efforts by a host of researchers, however, have resolved most of these technical problems and filled in many of the blank spots in the metabolic map.
The picture that arises from these studies is of a network that is both more streamlined and more modular than that found in free-living protozoa. This seems to be a consequence of dispensing with the flexibility that a free-living microbe must maintain in order to dynamically regulate its metabolic network to consume any of a wide variety of possible combinations of nutrients. Within the nutrient-rich and homeostatic environment of the blood stages, the parasite appears to consume a defined set of nutrients (glucose, glutamine, free fatty acids, etc.) via several discrete pathways (glycolysis/pentose phosphate pathway, carboxylic acid metabolism, fatty acid elongation and modification) with a low degree of interconnectivity. A fine-grained understanding of plasmodial metabolism will hopefully aid in exploiting this metabolic rigidity when selecting drug targets and designing antimalarials.
We expect the broad outline of central carbon metabolism to be conserved between the Plasmodium spp. due to the markedly similar life cycles, comparable drug sensitivities and low levels of divergence in metabolic enzymes at the genome level. However, we caution that the metabolism of the rodent and avian malaria parasites in particular may well be more complicated given their proclivity to invade immature, nucleated erythrocytes (reticulocytes) that are more metabolically complex than the mature erythrocytes favored by simian parasites.
Glycolysis
Based on numerous classical experiments, carbon metabolism of the malaria parasites has been considered largely synonymous with carbohydrate metabolism, principally the Embden-Meyerhof-Parnas (EMP) pathway of glycolysis. It has long been clear from in vitro studies of blood-stage Plasmodium parasites that they are voracious consumers of glucose (blood sugar). The host cell, at least in the human parasites, is the mature erythrocyte (red blood cell, or RBC), whose metabolism has been exhaustively studied and is comparatively simple enough to be simulated by comprehensive kinetic models [13, 14]. The RBC lacks a mitochondrion and therefore is entirely dependent on glucose fermentation. As a nonproliferative cell with modest energetic needs, RBC glucose consumption is relatively low, on the order of 5 μmol glucose / 24 hours / 109 RBCs [15]. Upon invasion by a Plasmodium parasite, however, the glucose consumption rate is estimated to increase up to 100-fold at the most metabolically active trophozoite and schizont stages [12].
In vitro culture experiments using several other sugars have established that, besides glucose, only fructose can support continuous growth (albeit at a reduced rate) [16, 17]. Most of the glucose consumed (60–70%) by P. falciparum is incompletely oxidized to lactic acid and excreted [15], although the exact percentage varies between different Plasmodium species and the atmospheric culture conditions used. This glucose consumption contrasts with the >90% glucose-to-lactate conversion observed in uninfected RBCs and reflects the increased flux of glucose carbon into biomass (nucleic acids, lipids, glycosylated proteins) required for proliferating parasites. Only a very small fraction of the total glucose is completely oxidized to carbon dioxide, at least in mammalian malaria parasites [1, 18, 19], which has generally been taken to indicate the absence of a functional citric acid cycle contributing to energy generation. This is in keeping with the observation that in vitro cultures of P. falciparum exhibit only minimal oxygen consumption [20] (and in fact prefer microaerophilic conditions of ~5% oxygen, being growth-inhibited by normal atmospheric oxygen concentrations [21].) These results strongly suggest that blood-stage Plasmodium rely primarily upon glucose fermentation for their energetic needs. Accordingly, inhibitors of mitochondrial respiration have only a small effect on parasite ATP pools [22].
The parasite accommodates this vastly increased need for glucose by expressing at least one essential hexose transporter to the surface of the infected cell [23], increasing the hexose permeability of the erythrocyte membrane [24]. Such modifications of the host cell raise the question of precisely how the host glycolytic machinery interacts with that of the parasite. All the enzymes required for a complete EMP pathway are 1) encoded in the parasite genome [25], 2) expressed during the blood stages [26], and 3) detected in infected cells, in some cases substantially increasing the activity normally observed in RBCs (reviewed in [27]). Free glucose should be quickly phosphorylated to glucose-6-phosphate by the host cell hexokinase upon entry into the RBC cytosol, rendering it membrane-impermeant and effectively trapping it within the host compartment. Its import into the parasite may depend on dephosphorylation by an acid phosphatase (PFI0880c) that is trafficked to the erythrocyte and cleaves phosphate from a diversity of small molecules [28]. This nonspecific cleavage of high-energy phosphate bonds might have the effect of draining energy from the host cytosol, despite the fact that the parasite seems to require the host cell to remain “viable” (maintaining its redox state, etc.) until lysis occurs in order to successfully complete its developmental cycle. Malaria parasites may circumvent this difficulty by actively supplying the host with metabolically useful molecules such as ATP [29] and glutathione [30]. This secreted phosphatase may also help explain the decline observed in infected cells in the levels of 2,3-diphosphoglycerate [31–33], an allosteric regulator of hemoglobin oxygen affinity produced by an enzyme (diphosphoglycerate mutase) present in the erythrocyte but not the parasite [25, 34].
Other carbon sources
Many microbes that prefer sugars as a carbon source maintain the ability to metabolize other compounds such as acetate, pyruvate, ethanol or amino acids depending on their availability. Generating the 5- and 6-carbon sugars necessary for growth alternatively depends on gluconeogenesis, in which the catabolic reactions of glycolysis are essentially reversed through the use of different enzymes at the regulated thermodynamic control steps (reviewed in [35]). However, the complete sequencing of the P. falciparum genome revealed no homologs of fructose bisphosphatase, an enzyme normally required for gluconeogenesis [25]. Strangely, the parasite does possess phosphoenolpyruvate carboxykinase (PEPCK, PF13_0234) [36], an enzyme converting oxaloacetate and ATP to PEP and carbon dioxide that usually functions in supplying citric acid cycle or amino acid-derived carbon to gluconeogenesis. The parasites also encode a PEP carboxylase (PEPC, PF14_0246), which has almost the opposite gene expression profile (peak expression at 18 and 47 hours post invasion for PEPCK and PEPC, respectively) [26]. PEPC has been purified from P. berghei [37], and essentially runs the reverse reaction: PEP and carbon dioxide converted to oxaloacetate and inorganic phosphate. The role of this enzyme remains unclear, but would suggest a regulated carbon fixation step.
It is well-established through 14CO2 incorporation experiments that all studied Plasmodium spp. possess the ability to fix carbon, although the nature of the end products differs between species [11]. Our own metabolomic investigation of carbon flux in P. falciparum showed clear evidence of PEPC activity, as parasites fed uniformly 13C-labeled glucose generated oxaloacetate/malate labeled at three of the four carbons (indicative of the carboxylation of PEP) [38] (Fig. 1). The purpose of this side pathway is unclear since we have found that the majority of the malate produced in this fashion is excreted from the cell. This results in an energetic cost of one ATP molecule for every molecule of PEP that does not complete glycolysis. Perhaps PEPC acts in vivo to supply oxaloacetate as a carbon skeleton for the generation of aspartate by aspartate transaminase (PFB0200c) when aspartate concentrations are limiting. However, in cell culture, aspartate levels are relatively high and this flux may be redirected to malate, which is then excreted due to overflow metabolism. In our studies, aspartate was clearly interconverting with cytosolic oxaloacetate/malate pools, although we were unable to detect any gluconeogenic PEPCK activity in the asexual stages, since 13C-labeled aspartate failed to label PEP, pyruvate or lactate [38]. Interestingly, the up-regulation of PEPCK in P. falciparum gametocytes and zygotes has prompted the hypothesis that there is a switch to gluconeogenic metabolism at these stages [36]. How this can be achieved in the absence of fructose bisphosphatase remains unclear.
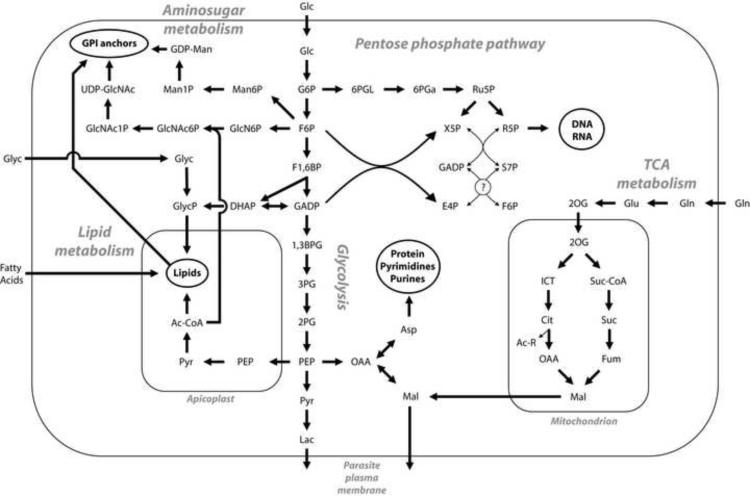
Fig 1.
An integrated map of carbon flow through the metabolic network of Plasmodium falciparum. Arrows show the proposed direction of flux through the corresponding enzymatic reaction as suggested by experimental evidence; note that this is only intended to indicate net flux, and that the reaction in question might be reversible. Cofactors (ATP, NADH, etc.) are not shown for the sake of clarity. Text in circles represent major biomass components; the circled question mark indicates uncertainty about the existence of the enzyme transaldolase. Abbreviations: Glc, glucose; G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; F1,6BP, fructose-1,6-bisphosphate; DHAP, dihydroxyacetone phosphate; GADP, glyceraldehyde-3-phosphate; 1,3BPG, 1,3-bisphosphoglycerate; 3PG, 3-phosphoglycerate; 2PG, 2-phosphoglycerate; PEP, phosphoenolpyruvate; Pyr, pyruvate; Lac, lactate; Ac-CoA, acetyl-CoA; Ac-R, either acetate or acetyl-CoA; GlycP, glycerol-3-phosphate; Glyc, glycerol; Man6P, mannose-6-phosphate; Man1P, mannose-1-phosphate; GDP-Man, GDP-mannose; GlcN6P, glucosamine-6-phosphate; GlcNAc6P, N-acetyl-glucosamine-6-phosphate; GlcNAc1P, N-acetylglucosamine-1-phosphate; UDP-GlcNAc, UDP-N-acetyl-glucosamine; 6PGL, 6-phosphoglucono-δ-lactone; 6PGa, 6-phosphogluconate; Ru5P, ribulose-5-phosphate; R5P, ribose-5-phosphate; X5P, xylulose-5-phosphate; S7P, sedoheptulose-7-phosphate; E4P, erythrose-4-phosphate; Asp, aspartate; Gln, glutamine; Glu, glutamate; 2OG, 2-oxoglutarate; ICT, isocitrate; Cit, citrate; OAA, oxaloacetate; Mal, malate; Suc-CoA, succinyl-CoA; Suc, succinate; Fum, fumarate; GPI, glycophosphatidylinositol.
Several early reports suggested that various Plasmodium spp. possess at least a limited ability to metabolize a number of other substrates, such as glycolytic end-products or tricarboxylic acid cycle intermediates [39–42]. However, most of these experiments rely on the stimulation of oxygen uptake as a measure of nutrient consumption, which can be problematic given the unclear relationship between this metric and parasite growth. Possible contaminating sources for this activity have been extensively discussed elsewhere [10–12]. Other experiments using simian and avian malaria parasites directly demonstrated the conversion of radioactively labeled lactate and pyruvate to glycolytic intermediates, organic acids and volatiles such as formate and acetate [1, 9, 39]. However, we caution that since these experiments used non-physiological glucose-free conditions and/or involved erythrocyte-free parasite preparations, special care should be taken in weighing their relationship to in vivo parasite metabolism, where 1) the parasite resides within a selectively permeable host erythrocyte, 2) serum glucose levels are robustly maintained by the host, and 3) lactate is rapidly excreted.
Plasmodium spp. also lack the ability to generate carbohydrate stores in the form of polysaccharides such as glycogen [18, 19, 43]. Thus the blood-stage parasites are obligately dependent on the fermentation of a constant and abundant supply of glucose. This adaptation is sensible given the parasite's adaptation to a peculiar evolutionary niche: glucose is the most abundant nutrient in human serum and its homeostasis is maintained by a powerful regulatory system. The availability of carbohydrates or other potential carbon sources in the other stages of the life cycle is difficult to study given the general intractability of culturing these stages, but it has been shown that the hemolymph of Anopheles stephensi is rich in glucose and the storage carbohydrate trehalose [44] and the essential P. berghei hexose transporter (PB000562.01.0) is expressed throughout development in the mosquito [23].
Pentose phosphate pathway
The pentose phosphate pathway (PPP, also known as the hexose monophosphate shunt), a critical conserved pathway in virtually all cells capable of metabolizing carbohydrates as a carbon source, is composed of two interconnected branches. The oxidative arm, in which glucose-6-phosphate is ultimately oxidized to ribose-5-phosphate, generates both the riboses needed for nucleic acid synthesis, as well as NADPH, which is used for redox control and as a cofactor for biosynthetic reactions. The non-oxidative arm, comprising a series of reversible reactions interconverting 3, 4, 5, 6 and 7-carbon sugar phosphates can either recycle ribose-5-phosphate generated by the oxidative arm back into glycolytic intermediates (when NADPH is required but nucleotide synthesis is not) or else converts glycolytic intermediates into ribose-5-phosphate without concomitant NADPH production (Fig. 1). It is one of the major metabolic pathways in human erythrocytes, consuming 3–11% of the glucose metabolized under normal conditions [45], as it is the only source of the NADPH required to reduce glutathione in response to oxidative stress. Though counterintuitive given its indispensable nature in proliferating cells, the existence of a complete PPP in the malaria parasites was for decades a point of controversy due to difficulties in detecting the necessary enzymes in parasite extracts [11] and early reports of very slight increases in pathway activity in RBCs infected with simian, avian and rodent malaria parasites [3, 9, 19]. However, the presence of the first and rate-limiting enzyme in the pathway, glucose-6-phosphate dehydrogenase (G6PDH, PF14_0511), was eventually established [46, 47]. The remaining enzymes are also encoded in the genome [25] and expressed [26], with the singular exception of transaldolase, for which no homolog has yet been identified [25].
The infected RBC commits roughly the same fraction of glucose to the oxidative PPP as does a normal RBC [48]. Since glucose consumption is massively increased during infection [12], the absolute flux through the PPP is likewise increased. The most recent investigation of this pathway in P. falciparum determined that the activity of the oxidative branch increased 78-fold by the trophozoite stages [48]. 82% of this activity could be recapitulated in free parasites, indicating that the parasite is responsible for the majority of the flux but also that the erythrocyte PPP is up-regulated 24-fold, to levels roughly similar to those observed when subjecting uninfected RBCs to oxidative insult. Two reports, one using P. falciparum [48] and the other in the simian parasite P. knowlesi [3], have found that significant amounts of radioactivity are incorporated into parasite nucleic acids from both 1- and 6- 14C-glucose. Since the carbon at position 6 is lost to decarboxylation in the oxidative PPP and so cannot be incorporated into ribose, this implies that the non-oxidative PPP (in which glycolytic intermediates can be converted to riboses) contributes significantly to the total pool of ribose-5-phosphate.
Despite the observation that the parasite PPP predominates in terms of flux, clear evidence for the importance of the host pathway is found in the fact that G6PDH deficiency, the most common human enzymopathy, is associated with resistance to clinical malaria (reviewed in [49]). The mechanistic basis for this protection remains somewhat controversial; in some cases slowed growth in G6PDH-deficient erythrocytes has been directly observed in vitro [50], while another group disputes this claim and suggests that the protection is mediated instead by an increase in oxidative damage to the host cell membrane, marking it for early phagocytosis by macrophages [51]. Nevertheless, the parasite requires a certain degree of viability from its host cell during the progression through blood-stage development, which includes maintenance of an adequate glutathione pool, with a sufficiently high ratio of reduced (GSH) to oxidized glutathione (GSSG, glutathione disulfide), within the host cell cytosol [52]. The regeneration of GSH from GSSG is fueled by NADPH from the RBC's pentose phosphate metabolism, whose flux is limited by G6PDH. Intriguingly, it has been reported that the parasite also actively supplies GSH to the host compartment while inducing the excretion of GSSG [30], suggesting that a combined effort encompassing both recycling by the host and biosynthesis by the parasite may be required to sustain the infected cell.
Glycosylation and aminosugars
The malaria parasite expresses a variety of glycoconjugated proteins (circumsporozoite protein, the merozoite surface proteins), several of which are essential for invasion and virulence [53]. However, the range of glycoconjugates produced, and the corresponding biosynthetic enzymes, appears to be severely restricted, limited almost entirely to the production of glycophosphatidylinositol (GPI) anchors required for protein-membrane association (reviewed in [54]). These moieties are assembled stepwise in the lumen of the endoplasmic reticulum on a phosphatidylinositol core through the sequential addition of glucosamine (as its activated form, UDP-N-acetyl-glucosamine, subsequently deacetylated) and four mannose molecules. The inositol is acylated, typically by myristic acid, at the 2-O position prior to mannosylation; the third mannose in the chain serves as the site for addition of ethanolamine phosphate, through which the GPI anchor is then attached to a protein.
This pathway thus draws carbon compounds from a variety of sources: hexose sugars (mannose, the glucose moiety of glucosamine), inositol biosynthesis, lower glycolysis (glycerol phosphate from the reduction of dihydroxy-acetone-phosphate, acetyl-CoA from the oxidation of pyruvate), ethanolamine from the host, and lipids from scavenging and modification (Fig. 1). We have recently shown that the acetyl moieties utilized for the synthesis of UDP-N-acetyl-glucosamine ultimately derive from glucose, presumably through the action of the pyruvate dehydrogenase (PDH) complex on glycolytic pyruvate [38]. Thus it is surprising that the PDH E1-alpha and E3 subunit genes of P. yoelii can be disrupted with no obvious growth phenotype until the liver stages [55], given that GPI biosynthesis is essential for blood-stage survival [56]. It is possible that in the event of PDH inactivation a separate acetyl-CoA-generating mechanism (discussed below) might compensate, indicating a degree of metabolic flexibility within this pathway. Nonetheless, the importance of GPI-anchored proteins to parasite development and the unique nature of certain of the enzymatic steps has suggested this pathway as a potential drug target [54].
Lipid biogenesis
The highly proliferative growth of the blood stage parasite depends on manufacturing significant quantities of new membrane. However, early experiments tracking the incorporation of radio-labeled glucose revealed that the amount of carbon from this nutrient incorporated into the lipid fraction of P. knowlesi biomass was low and occurred mainly in the glycerol backbone instead of the acyl chains [5]. This suggests that de novo fatty acid biosynthesis, or at least that using carbohydrates as a carbon source, is not a significant component of parasite metabolism. Interest in fatty acid metabolism has recently resurged with the discovery that the parasite genome encodes the complete suite of enzymes necessary for Type II (bacterial) fatty acid synthesis (FAS). These enzymes are all targeted to the apicoplast, a nonphotosynthetic plastid-like organelle derived from a secondary endosymbiotic event that is suspected to play a role in a number of plant-derived metabolic pathways [25, 57]. Type II FAS is an attractive drug target and considerable excitement has been generated by the discovery that triclosan, an antibiotic targeting bacterial enoyl-acyl carrier protein reductase (ENR), efficiently inhibits both the incorporation of 14C-acetate into parasite fatty acids and P. falciparum growth in culture [58]. However, the transcripts for various enzymes in the Type II FAS pathway show very low to undetectable blood-stage expression levels in microarray analyses [26], and subsequent investigations determined that several of these, including ENR, could be genetically disrupted in both P. berghei and P. falciparum without any discernible affect on blood-stage growth [59, 60]. However, the FAS pathway is essential during the liver stages, as these mutants exhibit a block in liver cyst development. Therefore, these studies conclusively demonstrate that ENR is not the blood-stage target of triclosan. That triclosan remains lethal to these ENR knockout parasites, and in fact still inhibits acetate incorporation [58], imply that it targets another aspect of parasite metabolism, perhaps acyl chain extension.
A more complete understanding of the structure of parasite fatty acid metabolism derives from a series of experiments involving metabolic tracers and different lipid precursor supplementations [61]. P. falciparum is typically cultured in serum-free medium supplemented with lipid-rich albumin, which is required for growth due to the complex mixture of fatty acids it provides. By systematically testing different combinations of free fatty acids for their ability to support parasite growth, Mi-ichi et al. were able to determine six combinations of C14, C16, C18 fatty acids and several desaturation products thereof that enable continuous culture of P. falciparum. No single fatty acid was required in every combination because the parasite possesses a limited ability to modify exogenously supplied fatty acids as needed. Specifically, metabolic labeling experiments using radioactive fatty acid species determined that the parasite can elongate *****C14 to C16 and C16 to C18, as well as desaturate acyl chains (primarily at the ω–9 position).
A model that arises from the data above is of a lipid metabolism in which preformed fatty acids are 1) scavenged from the host cell and serum, 2) subjected to a limited set of modifications by parasitic elongases and desaturases, and 3) incorporated into membrane glycerides with a glycerol backbone derived ultimately from glucose through the EMP pathway (Fig. 1). The two-carbon units (in the form of acetyl-CoA) necessary for elongation might be produced from glycolytic pyruvate in the apicoplast by the PDH complex localized to that compartment [62]. However, this enzyme is not essential during the blood stages [55]. It is possible that a glutamine-driven pathway (discussed below) could supply acetyl-CoA to lipid elongation, either under normal circumstances or else solely in the PDH mutant. This remains to be demonstrated however, since the few metabolic labeling experiments to robustly label acyl chains in parasite cultures have used very high levels of free 14C-acetate (P. knowlesi [5], P. falciparum [58]). In these experiments, only a small fraction (~15%) of the total radioactivity incorporated into lipids from 14C-glucose in P. knowlesi was detected in the acyl chain [5]. In addition, we have recently used gas chromatography-mass spectrometry profiling of lipids in parasites fed either 13C-glucose or 13C-glutamine, but were unable to measure any 13C incorporation into fatty acids [38]. Fatty acids acquired through scavenging from the extracellular environment evidently cannot serve as carbon sources themselves due to the absence from the genome of any of the β-oxidation enzymes necessary for the breakdown of acyl chains. Whole, preformed lipids might in theory be scavenged directly from the host cell membrane or host serum, but there is no evidence of lipid (as opposed to fatty acid) scavenging from the extracellular environment. Moreover, there is a net 6-fold increase in the phospholipid content of the infected erythrocyte [63], indicating significant amounts of phospholipid biosynthesis on the part of the parasite. The `bewildering' series of plasmodial pathways responsible for assembling complex phospholipids, and the open questions pertaining to their study, have been very well reviewed elsewhere [64].
Tricarboxylic acid metabolism
Mitochondrial tricarboxylic acid (TCA) metabolism has long been considered a “black box” within the rest of the malaria parasite's metabolism, unclear both in terms of its function as well as its basic architecture. In most free-living microbes this pathway acts as a versatile central hub of carbon metabolism in which carbon derived from glycolysis or other nutrients (ethanol, acetate, amino acids, etc.) is fully oxidized to carbon dioxide while generating energy (by oxidative phosphorylation), reducing equivalents and biosynthetic precursors (such as for amino acid synthesis). However, from the earliest reports in the malaria literature onwards a more confusing and contradictory picture emerges. Electron micrographic observations of the human malaria parasites' morphology revealed the single mitochondrion to be only minimally cristate [65], calling into question its role as a site of active metabolism (although cristae are observed in the mitochondria of avian parasites [65, 66]). The low levels of oxygen consumption observed in culture and the centrality of anaerobic glycolysis to energy metabolism discussed above further implied that the primary mitochondrial function of oxidative phosphorylation was absent or of minimal importance. In fact, the absence of obvious homologs to the F0 a and b subunits of the F1F0 ATP synthase in the parasite genome led to speculation that this enzyme complex may be incapable of generating ATP [25], although candidate homologs have recently been identified bioinformatically [67]. Malaria parasites have also dispensed with de novo amino acid biosynthesis, instead acquiring them through scavenging from the host serum or hemoglobin catabolism [68], which generates such an excess of amino acids that most are excreted from the infected cell as waste [69]. Biochemical and informatic studies suggest that the parasite malate dehydrogenase is actually cytosolic [70, 71], and so cannot contribute to a mitochondrial TCA cycle. Furthermore, as mentioned above the absence of the PDH enzyme complex from the mitochondrion [62], where it generally serves as the fundamental link between carbohydrate and mitochondrial carboxylic acid metabolism, converting glycolytic pyruvate to mitochondrial acetyl-CoA for citrate synthesis, raised the question of how, if at all, carbon enters the TCA cycle. This coincides with early reports that only very minimal amounts of 14CO2 and keto acids are produced from 14C –glucose during intraerythrocytic growth of both simian and avian parasites [1, 2, 9].
However, other lines of evidence indicate that some form of mitochondrial carboxylate metabolism occurs during the blood stage. The simian malaria parasite P. knowlesi and avian parasite P. lophurae fix 14CO2 into metabolites including citrate, malate and succinate [8, 72]. With the advent of the genome sequence it was realized that the parasite encodes homologs to all of the enzymes necessary for a complete TCA cycle [25], which are all are roughly coexpressed during the intraerythrocytic stage [26] and possess putative mitochondrial signal sequences save for malate dehydrogenase. In place of malate dehydrogenase, a second enzyme capable of essentially running the same reaction (malate:quinone oxidoreductase, PFF0815w) is expressed and appears to be mitochondrially targeted. In addition, the P. falciparum citrate synthase homolog (PF10_0218) [73], aconitase (PF13_0229) [74] and isocitrate dehydrogenase (IDH, PF13_0242) [38] have been localized entirely or partially to the mitochondrion. Intriguingly, while most eukaryotes possess at least three isoforms of IDH (mitochondrial NADP-dependent, mitochondrial NAD-dependent and cytosolic NADP-dependent), the parasite encodes only the mitochondrial NADP-dependent enzyme. Enzymatic activity for the TCA cycle enzymes succinate dehydrogenase (PFL0630w and PF10_0334) [75], aconitase [74] and IDH (in P. knowlesi) [76] have been detected in parasite extracts, and IDH has been cloned and characterized [77, 78]. Furthermore, the Plasmodium spp. possess an essential de novo heme biosynthetic pathway which presumably requires succinyl-CoA to function [79]. As the only known source of this precursor in the parasite's metabolic network, at least a subset of the TCA cycle enzymes (2-oxoglutarate dehydrogenase or succinyl-CoA synthetase) must be active. Finally, a metabolomic survey of the P. falciparum blood-stage developmental cycle discovered that the levels of several TCA cycle intermediates (ketoglutarate, iso/citrate, aconitate) oscillate periodically over the intraerythrocytic developmental cycle roughly in phase with the TCA cycle enzymes, suggesting that they are actively synthesized [80]. In order to explain these disparate strands of data, several models have been put forward. One possibility is that the full TCA pathway is active during different parasite life cycle stages, with the parasite's branched-chain alpha-keto-acid dehydrogenase complex possibly supplying mitochondrial acetyl-CoA through amino acid catabolism [81], or perhaps by acting as a surrogate pyruvate dehydrogenase [82].
Efforts to address this enigma in our laboratory have taken advantage of recent advances in mass spectrometry-based high throughput chemimetric technologies [83–85]. These metabolomic platforms have the advantage of permitting rapid and simultaneous quantification of a large panel of cellular metabolites, and, using mass spectrometry for detection, allows for relatively simple metabolic tracing experiments using stable isotope-labeled nutrients in which the number and position of labeled atoms can be determined. By feeding P. falciparum cultures 13C-glucose and analyzing isotopic distributions we have confirmed that carbohydrate metabolism does not contribute significantly to the TCA cycle, though there is clear evidence of cytosolic CO2 fixation through PEP carboxylase to oxaloacetate and malate [38]. 13C-glutamine, by contrast robustly labels all TCA cycle intermediates measured. It seems that glutamine is efficiently taken up by the infected cell [86] and converted to glutamate (likely through glutamate synthase, PF14_0334), which is itself converted to 2-oxoglutarate by glutamate dehydrogenase (GDH). P. falciparum encodes 3 distinct GDH genes: an NADP-dependent enzyme presumably targeted to the apicoplast (PF14_0286) [87], which may supply NADPH for biosynthetic reactions in that compartment; another NADP-dependent GDH lacking a cleavable signal sequence, and so presumably cytosolic (PF14_0164) [88]; and a large, fungal-type NAD-dependent GDH (PF08_0132) [87] with no predicted signal sequences. While we can only speculate at this point which of these predominates in terms of flux, one hypothesis that has been put forward has it that the cytosolic NADP-GDH might be a major source of NADPH for redox control [12], which would concomitantly generate cytosolic 2-oxoglutarate. Studies in our lab have determined that a large amount of 2-oxoglutarate is effluxed from infected cells as an apparent waste product, but some fraction appears to enter the mitochondrion, possibly through a malate:oxoglutarate antiporter homolog (PF08_0031), and serves as the carbon source driving TCA metabolism [38].
Surprisingly, the observed isotope labeling patterns when cells are fed 13C-glutamine indicated that this metabolism comprises two largely independent pathways: an oxidative branch running successively from 2-oxoglutarate to succinyl-CoA, succinate, fumarate and malate (essentially half of a canonical TCA cycle), and a reductive branch running from 2-oxoglutarate to isocitrate, citrate, oxaloacetate and malate (Fig. 1). We find that the resulting malate is excreted along with malate produced during the cytosolic carbon fixation process. The oxidative branch is unlikely to carry a very high flux but should generate reducing power (NADH) for the electron transport chain, GTP (via substrate-level phosphorylation) and, critically, the succinyl-CoA required for heme biosynthesis. The reductive branch, by contrast, produces two-carbon units during the citrate cleavage step rather than consuming two-carbon units as acetyl-CoA.
These results help to clarify a long-standing mystery in the field of Plasmodium metabolism by revealing a significantly diverged architecture for this fundamental pathway. Several other puzzling observations become explicable in the context of this model. For example, losing the canonical TCA cycle's NAD-dependent IDH while retaining the NADP-dependent isoform may be explained by noting that the reaction catalyzed by the NAD-dependent isoform is typically irreversible in the oxidative direction [89] while the NADP-dependent reaction is freely reversible in a wide variety of organisms [89–92]. Thus this permits the pathway to run in the reductive direction. Also, the presence of the putative mitochondrial malate:oxoglutarate antiporter has been difficult to interpret given the absence of any identifiable glutamate:aspartate antiporter, as the pair normally function together to complete the metabolic circuit known as the malate-aspartate shuttle [81]. However, if the mitochondrion is consuming 2-oxoglutarate and excreting malate then this transporter's role would simply be to exchange substrate for product. Other questions remain open: our data suggest that the parasite has evolved (at least) two independent pathways for the production of acetyl-CoA, and that these two pathways therefore might play different metabolic roles. However, it is not completely clear at this point whether this reductive pathway localizes entirely to the mitochondrion, or how essential it is for parasite growth. Further study is required to elucidate these and other puzzles.
Systems level studies
A variety of recent research efforts have aimed to elevate our understanding of plasmodial biology and metabolism to a systems level through the use of high-throughput “-omics” approaches, both in in vitro and in vivo settings. Though generally more difficult to interpret than classical approaches directed at one to a few nodes in a biological pathway, such datasets offer the potential to capture behaviors of the entire network. A study of the transcriptional profiles of P. falciparum samples harvested directly from clinical malaria patients for example, found that the parasites may exist in three broad clusters that seem to reflect a difference in metabolism [93]. Based on the gene expression of glycolytic and TCA cycle genes, the authors concluded that one cluster resembled normal anaerobic fermentative metabolism commonly seen in culture, while another seemed to show evidence of a starvation response and a switch to oxidative energy generation. Our biochemical evidence described above renders it doubtful that oxidative phosphorylation plays a significant energetic role, although it is possible that these data indicate an up-regulation of mitochondrial metabolism to generate two-carbon units in under conditions of reduced glucose availability in starved parasites (although other investigators have challenged these results as artifacts of analysis [94]). Several groups have reported that TCA cycle enzymes are present during all life cycle stages of the parasite analyzed, and may be particularly up-regulated in gametocytes, ookinetes and sporozoites of P. falciparum and P. yoelii [95–99]. The functional implications of this remain mysterious, given both the unlikely nature that this pathway plays an energetic role and the unclear metabolic requirements at the stages of development when the parasite is not actively proliferating.
Of course, metabolomic methodologies will be critical in the push towards dissecting the interlocking carbon metabolic pathways of the Plasmodium parasites and their host cells (reviewed in [64, 100]). Such techniques have been used to probe metabolic perturbations induced by malarial infection in in vitroP. falciparum cultures [80] and in vivo rodent malaria models [101, 102]. More directly, the ability of analytic technologies such as mass spectrometry and nuclear magnetic resonance (NMR) spectroscopy to trace the flow of heavy isotope-labeled nutrients through pathways is proving to be exceptionally valuable in this regard. Our own pathway analysis of carbon flow in P. falciparum, discussed in detail above, resulted in a new model architecture for mitochondrial carboxylic acid metabolism [38]. An investigation of labeling from 13C-glucose by NMR has suggested the intriguing possibility that, in addition to severely increasing the glucose consumption of the infected cell, P. falciparum is capable of somehow inhibiting glucose utilization by uninfected erythrocytes (seemingly through the inhibition of the host phosphofructokinase and pyruvate kinase by an excreted soluble factor) [33, 103]. If this finding can be confirmed and the inhibitory molecule identified it would be a fascinating insight into the parasitic modulation of host metabolism. Another group has reported, again using NMR-based measurements of 13C-labeled glucose metabolism, that in vitro cultures of P. falciparum produce glycerol as a major metabolite of glucose [104]. This finding is surprising given the rarity of glycerol production among eukaryotes and the absence of any enzyme in the current P. falciparum genome annotation capable of generating glycerol, such as a glycerol-3-phosphatase or a reversible glycerol kinase, although this activity is found in other parasitic protozoa such as the Trypanosomes [105–107]. These authors hypothesize that a functional glycerol-3-phosphate shuttle is therefore present in Plasmodium spp. and that a reappraisal of parasite redox metabolism may be necessary. However, since no quenching or deproteination step was used during metabolite extraction, it is possible that the observed glycerol may be the product of a nonspecific phosphatase acting on glycerol-3-phosphate produced by the parasite (for example, the excreted acid phosphatase discussed above was found to cleave phosphate from a wide variety of small-molecule substrates, including glycerol-2-phosphate [28]). Nevertheless, these recent observations suggest that the parasite may have some further metabolic tricks up its sleeve and that our current views of redox control and carbon metabolism should be interpreted with an open mind.
Suggested research avenues and concluding remarks
The preceding decades of malaria research have vastly clarified our understanding of the biology of the parasite and filled in many of the blank spaces of its metabolic map. The challenge now facing the malaria community is to build on the foundation of these results a comprehensive understanding of Plasmodium physiology, metabolism and host-parasite interactions both at the cellular and organismal level. One major stumbling block on the way toward this goal is our poor understanding of the metabolic interconnection between the RBC and its invader. The infected cell is highly compartmentalized and in one sense the essence of intracellular parasitism is the exchange of nutrients and wastes between parasite, host cell and environment. This is due largely to the significant difficulty inherent to compartmental analysis of metabolite concentrations and fluxes and a paucity of information about the localization and even identity of plasmodial proteins (most of which are unannotated). One clear starting point to unraveling this web of interactions is a concerted effort to identify and localize the many putative transporters of unknown function expressed during the blood stage [108]. A more complete understanding of the transport capabilities of the infected cell will significantly aid in efforts to understand how Plasmodium spp. mediate their exchange with the environment through the host cytoplasm.
Another fruitful area of inquiry is the genetic control of metabolism. Many regulatory interactions are difficult to study in malarial systems due to the lack of genetic tools. However, the progeny of phenotypically distinct strains have been used in conjunction with quantitative trait loci (QTL) analysis to uncover the loci governing traits such as drug resistance [109] and mRNA expression level [110]. This tool has also been used with metabolomics to probe metabolic regulation in Arabidopsis thaliana [111, 112]. Such an approach has enormous potential in malaria research to reveal interactions not accessible by other methods.
A more fine-scaled understanding of not just the architecture of the metabolic pathways of the infected cell but their dynamics is achievable through the use of fluxomics to map out the fluxes through the metabolic network. Such studies entail using a variation on classical pulse-chase techniques, rather using stable isotope-labeled nutrients and analytical platforms (MS or NMR-based) capable of measuring changes in the isotopic labeling pattern of downstream intermediates. Such studies measure both steady-state fluxes and the transient change in fluxes induced by a perturbation, both of which provide significant insight into the dynamic regulation and enzyme kinetics at play in the system [113–115]. Again, several technical challenges must be resolved before such studies can be carried out with current in vitro malaria culture techniques, which are complicated by the presence of host-cell compartments, enzymes, the presence of uninfected cells and other, more subtle peculiarities of the system. For example, our initial investigations into the TCA cycle were complicated by the observation that citrate concentrations in our RBC extracts were far in excess of the negligible erythrocyte citrate content reported in the literature [116]. We determined that this was due to citrate entering the RBCs during storage in the high citrate (>10 mM final concentration) anticoagulant solutions routinely used during blood collection. This issue was easily resolved using blood collected in sodium heparin solutions [38] but points to the special care that must be taken to minimize the effect of metabolic perturbations resulting from the nutrient environment during in vitro tissue culturing.
The avenues above will ultimately prove their worth by contributing to a systems level model for Plasmodium metabolism. Flux-balanced (stoichiometric) in silico metabolic networks have been developed for well-studied model organisms such as yeast [117] and E. coli [118] as well as several pathogens [119–121], and recent efforts by several groups have focused on constructing such an informatic tool for P. falciparum [122–124]. At present these are limited by our lack of understanding about the structure, dynamics and compartmentalization of the infected cell's metabolism, and such models must be refined by experimental evidence such as described above. A sufficiently accurate model will permit immediate in silico experiments to determine vulnerable drug targets (enzymes and transporters) and multi-drug dosing strategies, potentially significantly speeding the process of rational drug design. A fundamental understanding of central carbon metabolism in Plasmodium spp., and the unique adaptations differentiating the parasite and its host provide exciting new avenues for future therapeutic intervention strategies.
Abbreviations
- EMP
Embden-Meyerhof-Parnas
- RBC
red blood cell
- PEP
phosphoenolpyruvate
- PEPCK
PEP carboxykinase
- PEPC
PEP carboxylase
- PPP
pentose phosphate pathway
- G6PDH
glucose-6-phosphate dehydrogenase
- GPI
glycophosphatidylinositol
- ENR
enoyl-acyl carrier protein reductase
- PDH
pyruvate dehydrogenase
- TCA
tricarboxylic acid
- IDH
isocitrate dehydrogenase
- NMR
nuclear magnetic resonance
- GSH
reduced glutathione
- GSSG
oxidized glutathione (glutathione disulfide)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Scheibel LW, Pflaum WK. Carbohydrate metabolism in Plasmodium knowlesi. Comparative biochemistry and physiology. 1970;37:543. [Google Scholar]
- [2].Sherman IW, Ruble JA, Ting IP. Plasmodium lophurae: (U-14C)-glucose catabolism by free Plasmodia and duckling host erythrocytes. Exp Parasitol. 1969;25:181–92. doi: 10.1016/0014-4894(69)90064-2. [DOI] [PubMed] [Google Scholar]
- [3].Shakespeare PG, Trigg PI, Kyd SI, Tappenden L. Glucose metabolism in the simian malaria parasite Plasmodium knowlesi: activities of the glycolytic and pentose phosphate pathways during the intraerythrocytic cycle. Ann Trop Med Parasitol. 1979;73:407–15. doi: 10.1080/00034983.1979.11687280. [DOI] [PubMed] [Google Scholar]
- [4].Rock RC. Incorporation of 14 C-labelled fatty acids into lipids of rhesus erythrocytes and Plasmodium knowlesi in vitro. Comp Biochem Physiol B. 1971;40:893–906. doi: 10.1016/0305-0491(71)90035-6. [DOI] [PubMed] [Google Scholar]
- [5].Rock RC. Incorporation of 14 C-labelled non-lipid precursors into lipid of Plasmodium knowlesi in vitro. Comp Biochem Physiol B. 1971;40:657–69. doi: 10.1016/0305-0491(71)90141-6. [DOI] [PubMed] [Google Scholar]
- [6].Udeinya IJ, Van Dyke K. Labelling of membrane glycoproteins of cultivated Plasmodium falciparum. Bull World Health Organ. 1980;58:445–8. [PMC free article] [PubMed] [Google Scholar]
- [7].Schmidt-Ullrich R, Wallach DF, Lightholder J. Metabolic labelling of P. knowlesi-specific glycoproteins in membranes of parasitized rhesus monkey erythrocytes. Cell Biol Int Rep. 1980;4:555–61. doi: 10.1016/0309-1651(80)90021-1. [DOI] [PubMed] [Google Scholar]
- [8].Sherman IW, Ting IP. Carbon dioxide fixation in malaria (Plasmodium lophurae) Nature. 1966;212:1387–8. doi: 10.1038/2121387a0. [DOI] [PubMed] [Google Scholar]
- [9].Sherman IW, Ting IP, Tanigoshi L. Plasmodium lophurae: glucose-1-14C and glucose-6-14C catabolism by free plasmodia and duckling host erythrocytes. Comp Biochem Physiol. 1970;34:625–39. doi: 10.1016/0010-406x(70)90289-6. [DOI] [PubMed] [Google Scholar]
- [10].Homewood CA, Neame KD. Biochemistry of malarial parasites. In: Kreier J, editor. Malaria. Academic Press; New York: 1980. pp. 346–8. [Google Scholar]
- [11].Scheibel LW. Plasmodial metabolism and related organellar function during various stages of the life cycle: carbohydrates. In: Wernsdorfer W, McGregor I, editors. Malaria, Principles and Practices of Malariology. Churchill Livingstone; Edingurgh, United Kingdom: 1988. pp. 172–6. [Google Scholar]
- [12].Roth E., Jr. Plasmodium falciparum carbohydrate metabolism: a connection between host cell and parasite. Blood Cells. 1990;16:453–60. discussion 61–6. [PubMed] [Google Scholar]
- [13].Jamshidi N, Edwards JS, Fahland T, Church GM, Palsson BO. Dynamic simulation of the human red blood cell metabolic network. Bioinformatics. 2001;17:286–7. doi: 10.1093/bioinformatics/17.3.286. [DOI] [PubMed] [Google Scholar]
- [14].Joshi A, Palsson BO. Metabolic dynamics in the human red cell. Part I--A comprehensive kinetic model. J Theor Biol. 1989;141:515–28. doi: 10.1016/s0022-5193(89)80233-4. [DOI] [PubMed] [Google Scholar]
- [15].Jensen MD, Conley M, Helstowski LD. Culture of Plasmodium falciparum: the role of pH, glucose, and lactate. J Parasitol. 1983;69:1060–7. [PubMed] [Google Scholar]
- [16].Geary TG, Divo AA, Bonanni LC, Jensen JB. Nutritional requirements of Plasmodium falciparum in culture. III. Further observations on essential nutrients and antimetabolites. J Protozool. 1985;32:608–13. doi: 10.1111/j.1550-7408.1985.tb03087.x. [DOI] [PubMed] [Google Scholar]
- [17].Woodrow CJ, Burchmore RJ, Krishna S. Hexose permeation pathways in Plasmodium falciparum-infected erythrocytes. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:9931–6. doi: 10.1073/pnas.170153097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Scheibel LW, Miller J. Glycolytic and cytochrome oxidase activity in Plasmodia. Mil Med. 1969;134:1074–80. [PubMed] [Google Scholar]
- [19].Bowman IB, Grant PT, Kermack WO, Ogston D. The metabolism of Plasmodium berghei, the malaria parasite of rodents. 2. An effect of mepacrine on the metabolism of glucose by the parasite separated from its host cell. Biochem J. 1961;78:472–8. doi: 10.1042/bj0780472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Krungkrai J, Burat D, Kudan S, Krungkrai S, Prapunwattana P. Mitochondrial oxygen consumption in asexual and sexual blood stages of the human malarial parasite, Plasmodium falciparum. Southeast Asian J Trop Med Public Health. 1999;30:636–42. [PubMed] [Google Scholar]
- [21].Scheibel LW, Ashton SH, Trager W. Plasmodium falciparum: microaerophilic requirements in human red blood cells. Exp Parasitol. 1979;47:410–8. doi: 10.1016/0014-4894(79)90094-8. [DOI] [PubMed] [Google Scholar]
- [22].Fry M, Webb E, Pudney M. Effect of mitochondrial inhibitors on adenosinetriphosphate levels in Plasmodium falciparum. Comp Biochem Physiol B. 1990;96:775–82. doi: 10.1016/0305-0491(90)90230-q. [DOI] [PubMed] [Google Scholar]
- [23].Slavic K, Straschil U, Reininger L, Doerig C, Morin C, Tewari R, et al. Life cycle studies of the hexose transporter of Plasmodium species and genetic validation of their essentiality. Molecular microbiology. 2010;75:1402–13. doi: 10.1111/j.1365-2958.2010.07060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kirk K, Horner HA, Kirk J. Glucose uptake in Plasmodium falciparum-infected erythrocytes is an equilibrative not an active process. Molecular and biochemical parasitology. 1996;82:195–205. doi: 10.1016/0166-6851(96)02734-x. [DOI] [PubMed] [Google Scholar]
- [25].Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS biology. 2003;1:85–100. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sherman IW. Carbohydrate Metabolism of Asexual Stages. In: Sherman IW, editor. Malaria, Parasite Biology, Pathogenesis and Protection. ASM Press; Washington, D.C.: 1998. pp. 135–43. [Google Scholar]
- [28].Muller IB, Knockel J, Eschbach ML, Bergmann B, Walter RD, Wrenger C. Secretion of an acid phosphatase provides a possible mechanism to acquire host nutrients by Plasmodium falciparum. Cell Microbiol. 2010;12:677–91. doi: 10.1111/j.1462-5822.2010.01426.x. [DOI] [PubMed] [Google Scholar]
- [29].Kanaani J, Ginsburg H. Metabolic interconnection between the human malarial parasite Plasmodium falciparum and its host erythrocyte. Regulation of ATP levels by means of an adenylate translocator and adenylate kinase. J Biol Chem. 1989;264:3194–9. [PubMed] [Google Scholar]
- [30].Atamna H, Ginsburg H. The malaria parasite supplies glutathione to its host cell--investigation of glutathione transport and metabolism in human erythrocytes infected with Plasmodium falciparum. Eur J Biochem. 1997;250:670–9. doi: 10.1111/j.1432-1033.1997.00670.x. [DOI] [PubMed] [Google Scholar]
- [31].Ali SN, Fletcher KA, Maegraith BG. 2:3 diphosphoglycerate in P. berghei-infected blood. Trans R Soc Trop Med Hyg. 1971;65:8–9. [PubMed] [Google Scholar]
- [32].Deslauriers R, Ekiel I, Kroft T, Smith IC. NMR studies of malaria. 31P nuclear magnetic resonance of blood from mice infected with Plasmodium berghei. Biochim Biophys Acta. 1982;721:449–57. doi: 10.1016/0167-4889(82)90101-x. [DOI] [PubMed] [Google Scholar]
- [33].Mehta M, Sonawat HM, Sharma S. Malaria parasite-infected erythrocytes inhibit glucose utilization in uninfected red cells. FEBS Lett. 2005;579:6151–8. doi: 10.1016/j.febslet.2005.09.088. [DOI] [PubMed] [Google Scholar]
- [34].Roth EF, Jr., Calvin MC, Max-Audit I, Rosa J, Rosa R. The enzymes of the glycolytic pathway in erythrocytes infected with Plasmodium falciparum malaria parasites. Blood. 1988;72:1922–5. [PubMed] [Google Scholar]
- [35].Exton JH. Gluconeogenesis. Metabolism. 1972;21:945–90. doi: 10.1016/0026-0495(72)90028-5. [DOI] [PubMed] [Google Scholar]
- [36].Hayward RE. Plasmodium falciparum phosphoenolpyruvate carboxykinase is developmentally regulated in gametocytes. Molecular and biochemical parasitology. 2000;107:227–40. doi: 10.1016/s0166-6851(00)00191-2. [DOI] [PubMed] [Google Scholar]
- [37].McDaniel HG, Siu PM. Purification and characterization of phosphoenolpyruvate carboxylase from Plasmodium berghei. J Bacteriol. 1972;109:385–90. doi: 10.1128/jb.109.1.385-390.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Olszewski KL, Mather MW, Morrisey JM, Garcia BA, Vaidya AB, Rabinowitz JD, et al. Branched tricarboxylic acid metabolism in Plasmodium falciparum. Nature. 2010;466:774–8. doi: 10.1038/nature09301. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [39].Ali SN, Fletcher KA. Carbohydrate metabolism of malarial parasites--I. Metabolism of lactate in Plasmodium knowlesi infected monkey erythrocytes. Comp Biochem Physiol B. 1985;80:725–9. doi: 10.1016/0305-0491(85)90452-3. [DOI] [PubMed] [Google Scholar]
- [40].Nagarajan K. Metabolism of Plasmodium berghei. I. Krebs cycle. Exp Parasitol. 1968;22:19–26. doi: 10.1016/0014-4894(68)90074-x. [DOI] [PubMed] [Google Scholar]
- [41].Maier J, Coggeshall LT. Respiration of Malaria Plasmodia. The Journal of Infectious Diseases. 1941;69:87–96. [Google Scholar]
- [42].Wendel WB. Respiratory and carbohydrate metabolism of malaria parasites. J Biol Chem. 1943;148:21–34. [Google Scholar]
- [43].Dasgupta B. Polysaccharides in the different stages of the life-cycles of certain sporozoa. Parasitology. 1960;50:509–14. doi: 10.1017/s0031182000025580. [DOI] [PubMed] [Google Scholar]
- [44].Mack SR, Samuels S, Vanderberg JP. Hemolymph of Anopheles stephensi from noninfected and Plasmodium berghei-infected mosquitoes. 3. Carbohydrates. J Parasitol. 1979;65:217–21. [PubMed] [Google Scholar]
- [45].Yunis JJ, Yasmineh WG. Glucose metaholism in human erythrocytes. In: Yunis JJ, editor. Biochemical Methods in Red Cell Genetics. Academic Press; New York, NY: 1969. pp. 1–49. [Google Scholar]
- [46].Kurdi-Haidar B, Luzzatto L. Expression and characterization of glucose-6-phosphate dehydrogenase of Plasmodium falciparum. Molecular and biochemical parasitology. 1990;41:83–91. doi: 10.1016/0166-6851(90)90099-8. [DOI] [PubMed] [Google Scholar]
- [47].Buckwitz D, Jacobasch G, Kuckelkorn U, Megow D. Glucose-6-phosphate dehydrogenase from Plasmodium berghei: kinetic and electrophoretic characterization. Biomed Biochim Acta. 1990;49:S295–300. [PubMed] [Google Scholar]
- [48].Atamna H, Pascarmona G, Ginsburg H. Hexose-monophosphate shunt activity in intact Plasmodium falciparum-infected erythrocytes and in free parasites. Molecular and biochemical parasitology. 1994;67:79–89. doi: 10.1016/0166-6851(94)90098-1. [DOI] [PubMed] [Google Scholar]
- [49].Cappellini MD, Fiorelli G. Glucose-6-phosphate dehydrogenase deficiency. Lancet. 2008;371:64–74. doi: 10.1016/S0140-6736(08)60073-2. [DOI] [PubMed] [Google Scholar]
- [50].Roth EF, Jr., Raventos-Suarez C, Rinaldi A, Nagel RL. Glucose-6-phosphate dehydrogenase deficiency inhibits in vitro growth of Plasmodium falciparum. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:298–9. doi: 10.1073/pnas.80.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cappadoro M, Giribaldi G, O'Brien E, Turrini F, Mannu F, Ulliers D, et al. Early phagocytosis of glucose-6-phosphate dehydrogenase (G6PD)-deficient erythrocytes parasitized by Plasmodium falciparum may explain malaria protection in G6PD deficiency. Blood. 1998;92:2527–34. [PubMed] [Google Scholar]
- [52].Ginsburg H, Atamna H. The redox status of malaria-infected erythrocytes: an overview with an emphasis on unresolved problems. Parasite. 1994;1:5–13. doi: 10.1051/parasite/1994011005. [DOI] [PubMed] [Google Scholar]
- [53].Sanders PR, Kats LM, Drew DR, O'Donnell RA, O'Neill M, Maier AG, et al. A set of glycosylphosphatidyl inositol-anchored membrane proteins of Plasmodium falciparum is refractory to genetic deletion. Infection and immunity. 2006;74:4330–8. doi: 10.1128/IAI.00054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].von Itzstein M, Plebanski M, Cooke BM, Coppel RL. Hot, sweet and sticky: the glycobiology of Plasmodium falciparum. Trends Parasitol. 2008;24:210–8. doi: 10.1016/j.pt.2008.02.007. [DOI] [PubMed] [Google Scholar]
- [55].Pei Y, Tarun AS, Vaughan AM, Herman RW, Soliman JM, Erickson-Wayman A, et al. Plasmodium pyruvate dehydrogenase activity is only essential for the parasite's progression from liver infection to blood infection. Molecular microbiology. 2010;75:957–71. doi: 10.1111/j.1365-2958.2009.07034.x. [DOI] [PubMed] [Google Scholar]
- [56].Naik RS, Davidson EA, Gowda DC. Developmental stage-specific biosynthesis of glycosylphosphatidylinositol anchors in intraerythrocytic Plasmodium falciparum and its inhibition in a novel manner by mannosamine. J Biol Chem. 2000;275:24506–11. doi: 10.1074/jbc.M002151200. [DOI] [PubMed] [Google Scholar]
- [57].Waller RF, Keeling PJ, Donald RG, Striepen B, Handman E, Lang-Unnasch N, et al. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12352–7. doi: 10.1073/pnas.95.21.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Surolia N, Surolia A. Triclosan offers protection against blood stages of malaria by inhibiting enoyl-ACP reductase of Plasmodium falciparum. Nature medicine. 2001;7:167–73. doi: 10.1038/84612. [DOI] [PubMed] [Google Scholar]
- [59].Vaughan AM, O'Neill MT, Tarun AS, Camargo N, Phuong TM, Aly AS, et al. Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell Microbiol. 2009;11:506–20. doi: 10.1111/j.1462-5822.2008.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yu M, Kumar TR, Nkrumah LJ, Coppi A, Retzlaff S, Li CD, et al. The fatty acid biosynthesis enzyme FabI plays a key role in the development of liver-stage malarial parasites. Cell Host Microbe. 2008;4:567–78. doi: 10.1016/j.chom.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Mi-Ichi F, Kita K, Mitamura T. Intraerythrocytic Plasmodium falciparum utilize a broad range of serum-derived fatty acids with limited modification for their growth. Parasitology. 2006;133:399–410. doi: 10.1017/S0031182006000540. [DOI] [PubMed] [Google Scholar]
- [62].Foth BJ, Stimmler LM, Handman E, Crabb BS, Hodder AN, McFadden GI. The malaria parasite Plasmodium falciparum has only one pyruvate dehydrogenase complex, which is located in the apicoplast. Molecular microbiology. 2005;55:39–53. doi: 10.1111/j.1365-2958.2004.04407.x. [DOI] [PubMed] [Google Scholar]
- [63].Vial HJ, Ancelin ML. Malarial lipids. An overview. Subcell Biochem. 1992;18:259–306. [PubMed] [Google Scholar]
- [64].Besteiro S, Vo Duy S, Perigaud C, Lefebvre-Tournier I, Vial HJ. Exploring metabolomic approaches to analyse phospholipid biosynthetic pathways in Plasmodium. Parasitology. 2010;137:1343–56. doi: 10.1017/S0031182009991934. [DOI] [PubMed] [Google Scholar]
- [65].Rudzinska MA. The fine structure of malaria parasites. Int Rev Cytol. 1969;25:161–99. doi: 10.1016/s0074-7696(08)60203-x. [DOI] [PubMed] [Google Scholar]
- [66].Aikawa M. The fine structure of the erythrocytic stages of three avian malarial parasites, Plasmodium fallax, P. lophurae, and P. cathemerium. Am J Trop Med Hyg. 1966;15:449–71. doi: 10.4269/ajtmh.1966.15.449. [DOI] [PubMed] [Google Scholar]
- [67].Mogi T, Kita K. Identification of mitochondrial Complex II subunits SDH3 and SDH4 and ATP synthase subunits a and b in Plasmodium spp. Mitochondrion. 2009;9:443–53. doi: 10.1016/j.mito.2009.08.004. [DOI] [PubMed] [Google Scholar]
- [68].Liu J, Istvan ES, Gluzman IY, Gross J, Goldberg DE. Plasmodium falciparum ensures its amino acid supply with multiple acquisition pathways and redundant proteolytic enzyme systems. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8840–5. doi: 10.1073/pnas.0601876103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Krugliak M, Zhang J, Ginsburg H. Intraerythrocytic Plasmodium falciparum utilizes only a fraction of the amino acids derived from the digestion of host cell cytosol for the biosynthesis of its proteins. Molecular and biochemical parasitology. 2002;119:249–56. doi: 10.1016/s0166-6851(01)00427-3. [DOI] [PubMed] [Google Scholar]
- [70].Lang-Unnasch N. Purification and properties of Plasmodium falciparum malate dehydrogenase. Molecular and biochemical parasitology. 1992;50:17–25. doi: 10.1016/0166-6851(92)90240-k. [DOI] [PubMed] [Google Scholar]
- [71].Bender A, van Dooren GG, Ralph SA, McFadden GI, Schneider G. Properties and prediction of mitochondrial transit peptides from Plasmodium falciparum. Molecular and biochemical parasitology. 2003;132:59–66. doi: 10.1016/j.molbiopara.2003.07.001. [DOI] [PubMed] [Google Scholar]
- [72].Sherman IW, Ting IP. Carbon dioxide fixation in malaria. II. Plasmodium knowlesi (monkey malaria) Comp Biochem Physiol. 1968;24:639–42. doi: 10.1016/0010-406x(68)91018-9. [DOI] [PubMed] [Google Scholar]
- [73].Tonkin CJ, van Dooren GG, Spurck TP, Struck NS, Good RT, Handman E, et al. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Molecular and biochemical parasitology. 2004;137:13–21. doi: 10.1016/j.molbiopara.2004.05.009. [DOI] [PubMed] [Google Scholar]
- [74].Hodges M, Yikilmaz E, Patterson G, Kasvosve I, Rouault TA, Gordeuk VR, et al. An iron regulatory-like protein expressed in Plasmodium falciparum displays aconitase activity. Molecular and biochemical parasitology. 2005;143:29–38. doi: 10.1016/j.molbiopara.2005.05.004. [DOI] [PubMed] [Google Scholar]
- [75].Suraveratum N, Krungkrai SR, Leangaramgul P, Prapunwattana P, Krungkrai J. Purification and characterization of Plasmodium falciparum succinate dehydrogenase. Molecular and biochemical parasitology. 2000;105:215–22. doi: 10.1016/s0166-6851(99)00180-2. [DOI] [PubMed] [Google Scholar]
- [76].Sahni SK, Saxena N, Puri SK, Dutta GP, Pandey VC. NADP-specific isocitrate dehydrogenase from the simian malaria parasite Plasmodium knowlesi: partial purification and characterization. J Protozool. 1992;39:338–42. doi: 10.1111/j.1550-7408.1992.tb01326.x. [DOI] [PubMed] [Google Scholar]
- [77].Chan M, Sim TS. Recombinant Plasmodium falciparum NADP-dependent isocitrate dehydrogenase is active and harbours a unique 26 amino acid tail. Exp Parasitol. 2003;103:120–6. doi: 10.1016/s0014-4894(03)00090-0. [DOI] [PubMed] [Google Scholar]
- [78].Wrenger C, Muller S. Isocitrate dehydrogenase of Plasmodium falciparum. Eur J Biochem. 2003;270:1775–83. doi: 10.1046/j.1432-1033.2003.03536.x. [DOI] [PubMed] [Google Scholar]
- [79].Rao A, Yeleswarapu SJ, Srinivasan R, Bulusu G. Localization of heme biosynthesis pathway enzymes in Plasmodium falciparum. Indian J Biochem Biophys. 2008;45:365–73. [PubMed] [Google Scholar]
- [80].Olszewski KL, Morrisey JM, Wilinski D, Burns JM, Vaidya AB, Rabinowitz JD, et al. Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell Host Microbe. 2009;5:191–9. doi: 10.1016/j.chom.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].van Dooren GG, Stimmler LM, McFadden GI. Metabolic maps and functions of the Plasmodium mitochondrion. FEMS Microbiol Rev. 2006;30:596–630. doi: 10.1111/j.1574-6976.2006.00027.x. [DOI] [PubMed] [Google Scholar]
- [82].Seeber F, Limenitakis J, Soldati-Favre D. Apicomplexan mitochondrial metabolism: a story of gains, losses and retentions. Trends Parasitol. 2008;24:468–78. doi: 10.1016/j.pt.2008.07.004. [DOI] [PubMed] [Google Scholar]
- [83].Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- [84].Lu W, Kimball E, Rabinowitz JD. A high-performance liquid chromatography-tandem mass spectrometry method for quantitation of nitrogen-containing intracellular metabolites. J Am Soc Mass Spectrom. 2006;17:37–50. doi: 10.1016/j.jasms.2005.09.001. [DOI] [PubMed] [Google Scholar]
- [85].Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem. 2010;82:3212–21. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Elford BC, Haynes JD, Chulay JD, Wilson RJ. Selective stage-specific changes in the permeability to small hydrophilic solutes of human erythrocytes infected with Plasmodium falciparum. Molecular and biochemical parasitology. 1985;16:43–60. doi: 10.1016/0166-6851(85)90048-9. [DOI] [PubMed] [Google Scholar]
- [87].Aparicio IM, Marin-Menendez A, Bell A, Engel PC. Susceptibility of Plasmodium falciparum to glutamate dehydrogenase inhibitors--a possible new antimalarial target. Molecular and biochemical parasitology. 2010;172:152–5. doi: 10.1016/j.molbiopara.2010.04.002. [DOI] [PubMed] [Google Scholar]
- [88].Wagner JT, Ludemann H, Farber PM, Lottspeich F, Krauth-Siegel RL. Glutamate dehydrogenase, the marker protein of Plasmodium falciparum--cloning, expression and characterization of the malarial enzyme. Eur J Biochem. 1998;258:813–9. doi: 10.1046/j.1432-1327.1998.2580813.x. [DOI] [PubMed] [Google Scholar]
- [89].Kornberg A, Pricer WE., Jr. Di- and triphosphopyridine nucleotide isocitric dehydrogenases in yeast. J Biol Chem. 1951;189:123–36. [PubMed] [Google Scholar]
- [90].Grisolia S, Vennesland B. CARBON DIOXIDE FIXATION IN ISOCITRIC ACID. J Biol Chem. 1947;170:461–5. [Google Scholar]
- [91].Ochoa S. BIOSYNTHESIS OF TRICARBOXYLIC ACIDS BY CARBON DIOXIDE FIXATION: III. ENZYMATIC MECHANISMS. J Biol Chem. 1948;174:133–57. [PubMed] [Google Scholar]
- [92].Chung AE, Franzen JS. Oxidized triphosphopyridine nucleotide specific isocitrate dehydrogenase from Azotobacter vinelandii. Isolation and characterization. Biochemistry. 1969;8:3175–84. doi: 10.1021/bi00836a007. [DOI] [PubMed] [Google Scholar]
- [93].Daily JP, Scanfeld D, Pochet N, Le Roch K, Plouffe D, Kamal M, et al. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature. 2007;450:1091–5. doi: 10.1038/nature06311. [DOI] [PubMed] [Google Scholar]
- [94].Lemieux JE, Gomez-Escobar N, Feller A, Carret C, Amambua-Ngwa A, Pinches R, et al. Statistical estimation of cell-cycle progression and lineage commitment in Plasmodium falciparum reveals a homogeneous pattern of transcription in ex vivo culture. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7559–64. doi: 10.1073/pnas.0811829106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–6. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- [96].Tarun AS, Peng X, Dumpit RF, Ogata Y, Silva-Rivera H, Camargo N, et al. A combined transcriptome and proteome survey of malaria parasite liver stages. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:305–10. doi: 10.1073/pnas.0710780104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Hall N, Karras M, Raine JD, Carlton JM, Kooij TW, Berriman M, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–6. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- [98].Young JA, Fivelman QL, Blair PL, de la Vega P, Le Roch KG, Zhou Y, et al. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Molecular and biochemical parasitology. 2005;143:67–79. doi: 10.1016/j.molbiopara.2005.05.007. [DOI] [PubMed] [Google Scholar]
- [99].Lasonder E, Janse CJ, van Gemert GJ, Mair GR, Vermunt AM, Douradinha BG, et al. Proteomic profiling of Plasmodium sporozoite maturation identifies new proteins essential for parasite development and infectivity. PLoS pathogens. 2008;4:e1000195. doi: 10.1371/journal.ppat.1000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kafsack BF, Llinas M. Eating at the table of another: metabolomics of host-parasite interactions. Cell Host Microbe. 7:90–9. doi: 10.1016/j.chom.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Li JV, Wang Y, Saric J, Nicholson JK, Dirnhofer S, Singer BH, et al. Global metabolic responses of NMRI mice to an experimental Plasmodium berghei infection. Journal of proteome research. 2008;7:3948–56. doi: 10.1021/pr800209d. [DOI] [PubMed] [Google Scholar]
- [102].Basant A, Rege M, Sharma S, Sonawat HM. Alterations in urine, serum and brain metabolomic profiles exhibit sexual dimorphism during malaria disease progression. Malar J. 2010;9:110. doi: 10.1186/1475-2875-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Mehta M, Sonawat HM, Sharma S. Glycolysis in Plasmodium falciparum results in modulation of host enzyme activities. J Vector Borne Dis. 2006;43:95–103. [PubMed] [Google Scholar]
- [104].Lian LY, Al-Helal M, Roslaini AM, Fisher N, Bray PG, Ward SA, et al. Glycerol: an unexpected major metabolite of energy metabolism by the human malaria parasite. Malar J. 2009;8:38. doi: 10.1186/1475-2875-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Hammond DJ, Aman RA, Wang CC. The role of compartmentation and glycerol kinase in the synthesis of ATP within the glycosome of Trypanosoma brucei. J Biol Chem. 1985;260:15646–54. [PubMed] [Google Scholar]
- [106].Kralova I, Rigden DJ, Opperdoes FR, Michels PA. Glycerol kinase of Trypanosoma brucei. Cloning, molecular characterization and mutagenesis. Eur J Biochem. 2000;267:2323–33. doi: 10.1046/j.1432-1327.2000.01238.x. [DOI] [PubMed] [Google Scholar]
- [107].Steinborn K, Szallies A, Mecke D, Duszenko M. Cloning, heterologous expression and kinetic analysis of glycerol kinase (TbGLK1) from Trypanosoma brucei. Biological chemistry. 2000;381:1071–7. doi: 10.1515/BC.2000.132. [DOI] [PubMed] [Google Scholar]
- [108].Martin RE, Henry RI, Abbey JL, Clements JD, Kirk K. The `permeome' of the malaria parasite: an overview of the membrane transport proteins of Plasmodium falciparum. Genome Biol. 2005;6:1–22. doi: 10.1186/gb-2005-6-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Wellems TE, Walker-Jonah A, Panton LJ. Genetic mapping of the chloroquine-resistance locus on Plasmodium falciparum chromosome 7. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:3382–6. doi: 10.1073/pnas.88.8.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Gonzales JM, Patel JJ, Ponmee N, Jiang L, Tan A, Maher SP, et al. Regulatory hotspots in the malaria parasite genome dictate transcriptional variation. PLoS biology. 2008;6:e238. doi: 10.1371/journal.pbio.0060238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Keurentjes JJ, Fu J, de Vos CH, Lommen A, Hall RD, Bino RJ, et al. The genetics of plant metabolism. Nat Genet. 2006;38:842–9. doi: 10.1038/ng1815. [DOI] [PubMed] [Google Scholar]
- [112].Rowe HC, Hansen BG, Halkier BA, Kliebenstein DJ. Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell. 2008;20:1199–216. doi: 10.1105/tpc.108.058131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Zamboni N, Fendt SM, Ruhl M, Sauer U. (13)C-based metabolic flux analysis. Nat Protoc. 2009;4:878–92. doi: 10.1038/nprot.2009.58. [DOI] [PubMed] [Google Scholar]
- [114].Yuan J, Fowler WU, Kimball E, Lu W, Rabinowitz JD. Kinetic flux profiling of nitrogen assimilation in Escherichia coli. Nature chemical biology. 2006;2:529–30. doi: 10.1038/nchembio816. [DOI] [PubMed] [Google Scholar]
- [115].Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, et al. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nature biotechnology. 2008;26:1179–86. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Dajani RM, Orten JM. A study of the citric acid cycle in erythrocytes. J Biol Chem. 1958;231:913–24. [PubMed] [Google Scholar]
- [117].Papp B, Pal C, Hurst LD. Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature. 2004;429:661–4. doi: 10.1038/nature02636. [DOI] [PubMed] [Google Scholar]
- [118].Edwards JS, Palsson BO. The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5528–33. doi: 10.1073/pnas.97.10.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Thiele I, Vo TD, Price ND, Palsson BO. Expanded metabolic reconstruction of Helicobacter pylori (iIT341 GSM/GPR): an in silico genome-scale characterization of single- and double-deletion mutants. J Bacteriol. 2005;187:5818–30. doi: 10.1128/JB.187.16.5818-5830.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Heinemann M, Kummel A, Ruinatscha R, Panke S. In silico genome-scale reconstruction and validation of the Staphylococcus aureus metabolic network. Biotechnol Bioeng. 2005;92:850–64. doi: 10.1002/bit.20663. [DOI] [PubMed] [Google Scholar]
- [121].Oberhardt MA, Puchalka J, Fryer KE, Martins dos Santos VA, Papin JA. Genome-scale metabolic network analysis of the opportunistic pathogen Pseudomonas aeruginosa PAO1. J Bacteriol. 2008;190:2790–803. doi: 10.1128/JB.01583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Fatumo S, Plaimas K, Mallm JP, Schramm G, Adebiyi E, Oswald M, et al. Estimating novel potential drug targets of Plasmodium falciparum by analysing the metabolic network of knockout strains in silico. Infect Genet Evol. 2009;9:351–8. doi: 10.1016/j.meegid.2008.01.007. [DOI] [PubMed] [Google Scholar]
- [123].Yeh I, Hanekamp T, Tsoka S, Karp PD, Altman RB. Computational analysis of Plasmodium falciparum metabolism: organizing genomic information to facilitate drug discovery. Genome Res. 2004;14:917–24. doi: 10.1101/gr.2050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Plata G, Hsiao T, Olszewski KL, Llinas M, Vitkup D. Reconstruction and flux-balance analysis of the Plasmodium falciparum metabolic network. Mol Syst Biol. 2010 doi: 10.1038/msb.2010.60. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]