Abstract
The Poisson-Boltzmann equation (PBE) is an established model for the electrostatic analysis of biomolecules. The development of advanced computational techniques for the solution of the PBE has been an important topic in the past two decades. This paper presents a matched interface and boundary (MIB) based PBE software package, the MIBPB solver, for electrostatic analysis. The MIBPB has a unique feature that it is the first interface technique based PBE solver that rigorously enforces the solution and flux continuity conditions at the dielectric interface between the biomolecule and the solvent. For protein molecular surfaces which may possess troublesome geometrical singularities, the MIB scheme makes the MIBPB by far the only existing PBE solver that is able to deliver the second order convergence, i.e., the accuracy increases four times when the mesh size is halved. The MIBPB method is also equipped with a Dirichlet-to-Neumann mapping (DNM) technique, that builds a Green's function approach to analytically resolve the singular charge distribution in biomolecules in order to obtain reliable solutions at meshes as coarse as 1Å — while it usually takes other traditional PB solvers 0.25Å to reach similar level of reliability. The present work further accelerates the rate of convergence of linear equation systems resulting from the MIBPB by utilizing the Krylov subspace (KS) techniques. Condition numbers of the MIBPB matrices are significantly reduced by using appropriate Krylov subspace solver and preconditioner combinations. Both linear and nonlinear PBE solvers in the MIBPB package are tested by protein-solvent solvation energy calculations and analysis of salt effects on protein-protein binding energies, respectively.
1 Introduction
Under physiological conditions, almost all important biological processes, for example, signal transduction, DNA specification, transcription, post transcription modification, translation, protein folding and protein ligand binding, occur in water which comprises 65-90% of cellular mass. An elementary prerequisite for the quantitative description and analysis of the above-mentioned processes is the understanding of solvation, which involves energetics of interactions between solute molecules and solvent molecules or ions in aqueous environment. Solute-solvent interactions are typically classified as the polar type and the non-polar type. Although widely used, this classification is arbitrary and has caveats associated with the non-unique descriptions, as well as the intrinsic coupling between these two types of interactions. The polar type of solute-solvent interactions is the main interest of the present work. It originates from electrostatic effects, which play important roles in biophysics, biochemistry, structural biology, electrochemistry and electrophoresis. The solvent has a substantial volume and a significant contribution to electrostatics via numerous mobile ions. However, it is the solvated solute molecule that is the focus of the interest in most research. As such, the solute is described in atomic or electronic detail, while atomic details of the solvent and mobile ions are approximated by a mean-force description and probability distribution, respectively. This multiscale treatment, denoted as implicit solvent method, can greatly reduce the computational cost of the traditional explicit solvent methods, in which a microscopic description of the solvent is retained. Various implicit solvent models are available to describe polar solvation [58, 62, 22, 15, 3]. The most widely-used methods are currently the generalized Born method [23, 28, 86, 49, 15], polarizable continuum [19, 69, 35] and Poisson-Boltzmann equation (PBE) [1, 39, 62, 22] models. The use of polarizable continuum models is mostly restricted to small molecular systems. Generalized Born methods are very fast but are only heuristic models for estimating polar solvation energies of biomolecular structures. These methods are often used in high-throughput applications such as molecular dynamics simulations [70, 63, 27]. PBE models can be formally derived from Maxwell's equations [7] and offer a somewhat slower, but more accurate way for evaluating polar solvation properties [21, 52, 6]. Additionally, PBE techniques are often used to parameterize and assess the accuracy/performance of generalized Born models [52, 52, 71]. Finally, unlike most generalized Born methods, PB models provide a global solution for the electrostatic potential and field within and around a biomolecule, therefore make them uniquely suited to visualization and other analysis [44, 9] that require global information about electrostatic properties. One of the primary quantitative applications of implicit solvent models in computational biology and chemistry is the calculation of thermodynamic properties via a pre-equilibration [58]. An example of such pre-equilibration approach is the MM/PBSA model [66, 48] which combines implicit solvent models with molecular mechanical approaches to evaluate binding free energies from an ensemble of biomolecular structures. Other important applications of implicit solvent models include the assignment of protein titration states, the calculation of binding energies, and the estimation of solvation energies [50, 68, 50]. One more application area for implicit solvent methods is the evaluation of biomolecular kinetics where implicit solvent models are generally used to provide solvation forces for molecular Langevin dynamics [56, 46], Brownian dynamics [47, 29, 61], or continuum diffusion simulations [17, 64, 65]. A major qualitative use of implicit solvent methods in experimental work is the visualization and qualitative analysis of electrostatic potentials on and around biomolecular surfaces [55, 4, 1], which is now a standard procedure for the analysis of biomolecular structures.
Although the PBE can be analytically solved for a few simple cases [37], it relies on numerical approaches to obtain useful solutions for realistic biological systems. A vast variety of computational approaches, such as finite difference methods (FDMs) [33, 46, 73, 34], finite element methods (FEMs) [5], and boundary integral methods (BIMs) [10, 79], have been developed in the past few decades. Each of these methods is subject to certain inherent advantages and limitations due to its associated underlying formulations. In general, FEMs have advantages in applications that require the rapid adaptation of grid points to account for the structural variation of biomolecules. However, the generation of unstructured grids with good quality for complex biomolecular interfaces is very time-consuming, especially for large biomolecules. BIMs have several intrinsic advantages such as fewer unknowns, exact far-field treatment, and good representation of surface geometry and charge singularity. In light of these advantages, BIMs, especially when accelerated with fast methods such as treecode and fast multiple methods, can also provide an efficient computational approach. However, BIMs are not very efficient in dealing with the nonlinear term in the PB model. FDMs have been the main workhorse for solving the PBE in computational structural biology for their convenience of using 3D Cartesian coordinates to save the cost on mesh generation and electrostatic potential mapping, as well as its adaptability of existing linear algebraic solvers. FDMs based PB solvers, particularly in conjunction with advanced linear algebraic solvers, can offer the best combination of speed, accuracy, and efficiency, therefore make them the most popular approaches in structural biology [2].
Many computational technologies for the PBE were incorporated into popular molecular simulation software packages, such as DelPhi [38], ZAP [56], UHBD [47], MEAD [25], APBS [31], AMBER [45, 46] and CHARMM [36, 12]. These software packages deliver PBE solvers to users who are interested in the study of electrostatics in solution, making the PBE model a widely accepted approach in structural biology. The aim of this paper is to introduce an interface technique based PBE solver, the matched interface and boundary method based PB solver (MIBPB), with online software sharing information. Compared with other existing PB solvers, the MIBPB provides rigorous mathematical treatment of interface jump conditions, geometric singularities and charge singularities as described in the following paragraphs.
The implicit solvent models require an interface definition to indicate the separation of solute atoms from the surrounding solvent. All of the physical properties of interests, such as electrostatic free energies, biomolecular surface areas, molecular cavitation volumes, solvation free energies, and pKa values are very sensitive to the interface definition [24, 51, 67]. The van der Waals surface, the solvent accessible surface [40] and the molecular surface (MS) [57, 20] are often used for this purpose. Different dielectric constants of the solvent and molecular domain lead to discontinuous coefficients in the PBE, resulting in non-smoothness of the solution. Additionally, the MS admits geometric singularities such as cusps and self-intersecting surfaces[20, 60]. Explicit interface treatment of geometric singularities has not been considered in the popular PB solvers. It is desirable to have an interface-based solver that is able to deliver highly accurate solutions to the PBE in the presence of such solution and geometric singularities. The third type of singularity comes from the singular charge terms, i.e., the delta functions at the right hand side of the PBE, often reduce the accuracy of the numerical solution. The treatment of charge singularities is also an important issue in solving the PBE. Finally, due to the complex matrix structure of the linear systems resulting from PBE solvers, the choice of appropriate linear system solvers and the selection of matrix acceleration algorithms are very important issues as well.
The development of the MIBPB solver focuses on resolving the above-mentioned difficulties or singularities. The adaption of the molecular surface, the treatment of discontinuity of coefficients and flux jumps require the application of interface methods [54, 26, 11, 42, 13, 41]. Commonly seen interface techniques were not used in the biomolecular context due to complexity of the biomolecular boundaries. The MIB method [81, 85, 84, 76, 75] has been developed for solving elliptic equations with discontinuous interfaces. It is of arbitrarily high-order accuracy in principle and up to sixth-order accurate MIB schemes have been constructed [75, 85]. The MIB has been successfully applied to the analysis of mechanical structures [78, 82], waveguides [82], biomedical imaging [14], and electromagnetic waves [80]. Three generations of MIB based PB solvers, the MIBPB-I [83], the MIBPB-II [74] and the MIBPB-III [30] have been developed (http://www.math.msu.edu/~wei/MIBPB/). The MIBPB-I is the first PB solver that directly enforces the flux continuity conditions at the dielectric interface in the biomolecular context. However, it cannot maintain its designed order of accuracy in the presence of MS singularities, such as cusps and self-intersecting surfaces. This problem was addressed in the MIBPB-II by utilizing an advanced MIB technique developed by Yu et al. [75] who offer special treatments for geometric singularities. However, the MIBPB-II loses its accuracy when the mesh size is as large as half of the smallest van der Waals radius, because of the interference of the interface and singular charges. To split the singular charge part of the solution, a Dirichlet to Neumann mapping approach [18] was designed in the MIBPB-III, which is by far the most accurate and reliable PB solver. This new solver remains accurate at the smallest van der Waals radius, i.e., about 1.0 Å grid resolution for proteins. Comparing to traditional PB solvers, the MIPPB-III is a few orders of magnitude more accurate at a given mesh size and about three times faster at a given accuracy [74, 30]. The MIBPB is the first and still the only known second-order convergent PB solver for the singular molecular surfaces of biomolecules, where the second order convergence means that the accuracy of the solution improves four times when the mesh size is halved.
Apart from the accuracy, the efficiency of linear system solvers is another important issue crucial to many applications. Previous MIBPB solvers are typically slow in comparing with other FDMs that do not invoke an interface treatment. In this paper, we have paid special efforts on the strategies for the selection of most suitable linear system solvers for the resulting MIBPB matrices. Two linear solver libraries, the SLATEC (http://people.sc.fsu.edu/~burkardt/f_src/slatec/slatec.html) and the PETSc (http://www.mcs.anl.gov/petsc/petsc-as/) are considered in the exploration of linear solvers. Another remaining issue in previous MIBPB solvers is the treatment of the nonlinearity in the PBE. Although this issue was tackled in a dissertation [77], no reliable MIBPB nonlinear solver was produced. The present work develops a reliable nonlinear solver for the PBE with a salt solvent.
The rest of this paper is organized as follows. Section 2 is devoted to the theoretical formulation and computational algorithm of the MIBPB solver. Krylov subspace technique accelerated MIBPB solvers are constructed in Section 3. Extensive experimental validation is given to the different combinations of solvers and pre-conditioners. Section 4 illustrates the usage of the MIBPB software package with some example applications, such as calculating the free energy of solvation of biomolecular systems and salt effect on protein-protein binding. Concluding remarks are provided in Section 5. Finally, a brief introduction to the linear algebraic systems generated from the MIBPB discretization matrices and theoretical underpinnings of the Krylov subspace methods are presented in Appendices.
2 Theory and algorithm
2.1 Implicit solvent model: The Poisson-Boltzmann Equation
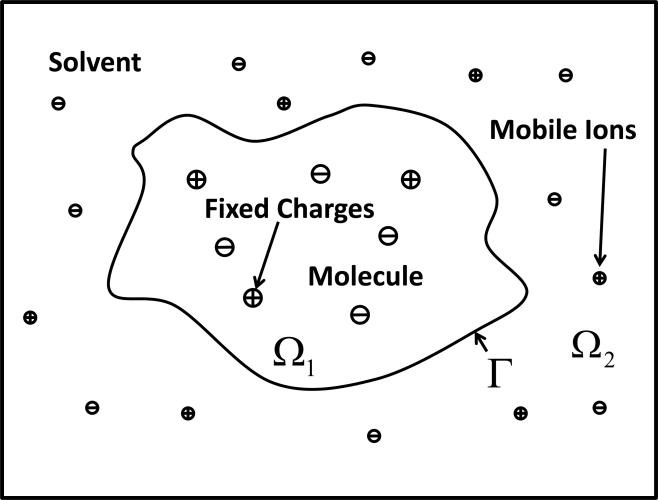
In the implicit solvent model, the solvent is treated as a continuous medium while the description for solute is kept at the atomic level. The electrostatic potential ϕ of a solvent-solute system can be determined by the Poisson-Boltzmann equation (PBE) in a regular domain Ω whose dimension usually has the order from 10Å3 to 500Å3 for biomolecular applications. Figure 1 gives the sketch of the protein-solute system and the computational domain.
Figure 1.
The implicit protein-solvent system
The protein region and the solvent region are denoted as Ω1 and Ω2, respectively. Naturally the whole computational domain is , and the molecular surface is labeled as Γ. For simplicity, the ion-exclusive layer is ignored in the present model. Although mobile ions in the solvent are explicitly indicated in the figure, the whole solvent region is actually modeled by an implicit continuum. Under these assumptions, the PBE reads
| (1) |
| (2) |
where u = ecϕ/kBT is the dimensionless electrostatic potential for computational simplicity, ec the electron charge, kB is the Boltzmann constant and T is the temperature, ε and are dielectric constant and modified Debye-Hückel screening function describing the ion strength, respectively. Here qi is the charge fraction of the fixed charge in the protein and xi denotes the position of the fixed charge, and Nm is the total number of fractional charges. The constant is resulting from dimensionless procedure and εs is for the dielectric constant of the solvent.
In principle, the electrostatics u(x) satisfies the boundary condition at infinity, i,e,:
| (3) |
However, the practical computation has to be restricted onto a bounded domain Ω. Usually, it is taken as a cuboid that contains the target protein and ∂Ω represents the boundary. In this approximation, proper boundary conditions need to be imposed and various treatments are employed upon different numerical schemes. Eq. (2) describes the Dirichlet boundary condition which is widely used in finite difference method. The biological meaning of the rationale is the (screened) Coulomb potential originating from all the fixed charges and mapping on the walls of the cuboid Ω.
The hyperbolic term sinh(u(x)) takes into account the salt effect with the Boltzmann distribution theory at the equilibrium state. Therefore, Eq. (1) is a nonlinear partial differential equation (PDE) of elliptic type. Such a nonlinear term can be linearized under the weak potential approximation, i.e, when , sinh(u(x)) ~ u(x). Thus the linear approximation of Eq. (1) is
| (4) |
Typically, for biomolecular systems of given ranges of temperature and ionic strength, the PBE is solved with the following coefficient bounds [32]
The molecular surface of the solute (protein) is considered as an interface. We partition the whole domain into the solute region and the solvent region, on which the ε(x) takes different values. Similar situation happens to the parameter . Since it represents the ionic strength of the solvent, is nonzero in the solvent region but zero in the solute region. In other words, the spatial-dependent coefficients ε(x) and are discontinuous across the molecular surface. It is a challenge to solve such an elliptic equation with high accuracy because the regularity of its solution is reduced due to the interface and geometric singularity. For this class of problems, numerical accuracy and low convergence rate are typically low without special interface treatments. Another challenge is the singular source term which contains many Delta functions, which are infinity at their spatial locations. Accurate approximation to the point-supported singular functions is an important topic in computational mathematics. The above two difficulties hinder the numerical solution to the PB equation. To maintain a given accuracy, the grid spacing of the discretization has to be sufficiently small because of the low regularity of the solution. On the other hand, a small grid spacing implies millions of variables even for a middle-size protein. For example, the cube embedding a 2800-atom protein may have a dimension of 50 × 50 × 50(Å3), which leads to 1 × 106 variables if the resolution is 0.5Å. This gives rise to a major obstacle for PB applications, especially for the calculation of thermodynamic properties via either the molecular dynamics or pre-equilibrium approaches. In the following sections a robust and efficient mathematical algorithm, the MIB method, is introduced and applied to the solution of the PBE.
2.2 MIB method for the Poisson-Boltzmann equation
Mathematically, the PBE admits at least three types of singularities that hinder one's attempt to obtain highly accurate numerical solutions. The first one is the non-smooth solution singularities. Based on the statement in the previous section, the potential u(x) is naturally restricted to the two subregions as u1 and u2, respectively. The solution of the PBE is subject to the interface continuity condition and flux continuity condition along the interface Γ
| (5) |
| (6) |
where un denotes the normal derivative of the function u(x) and εm is the dielectric constant of the solute molecule. Interface conditions (5) and (6) indicate that the continuity of the potential function and the potential flux across the molecular surface. However, different dielectric values of εs and εm in (6) imply that the normal derivatives of the potential function differ from each other across the surface — in other words, the solution is non-smooth. The second singularity comes from the solvent-solute interface Γ. The interface generated based on the current molecular surface model inevitably introduces geometric singularities such as cusps and self-intersecting surfaces, especially for large proteins. This singularity has been a challenging issue to many traditional interface techniques designed for solving elliptic interface problems. The third singularity is the source term of the PBE, which includes the summation of Delta functions. Second order numerical implementation of Dirac delta functions on the Cartesian grid points is feasible with appropriate approximation. However, the overlap of grid points carrying redistributed charges and those involved in the treatment of geometric interface singularities leads to the accuracy reduction, especially for a coarse mesh. These singularities pose challenges in the numerical implementation of the PBE and make it difficult to balance numerical accuracy and efficiency.
The MIB scheme and the Dirichlet-to-Neumann mapping (DNM) method are employed to deal with the above mentioned three singularities and to achieve high accuracy and efficiency. In the MIB method, the molecular surface is considered as the interface. Interface jump conditions (5) and (6) are enforced at each intersecting point of the interface and mesh lines. As such, the ingredients of the MIB scheme also include local coordinates of the interface in order to overcome geometric singularity constrains. The necessary information of each local coordinate contains the location (x0, y0, z0) and the normal direction n of the intersecting point where the interface meets a mesh line of the grid. Here, n is parameterized as
| (7) |
where θ and ψ are the azimuth and zenith angles with respect to the normal vector, notation T represents the transpose for a vector. This information allows us to set up a local coordinate system at every intersecting point and to define its relation to the global Cartesian grid.
The basic idea of the MIB scheme is to define sets of regular and irregular grid points near the interface according to the desired convergence order. At each regular point, the standard central difference scheme of a given order is applied, while at irregular points, special treatments are to be taken. First, the restricted potential function u1 (and u2) is smoothly extended from one subregion to another on all of the irregular points. Then artificially introduced function values, named as fictitious values, are defined on the irregular points. As smooth extensions of the function, fictitious values are employed in FD schemes at irregular points to guarantee necessary smoothness conditions.
Interface condition (5) is further decomposed into two parts in order to give flexibility to the calculation of fictitious values
| (8) |
where τ1 and τ2 are the two tangential vectors derived from (7)
| (9) |
Through jump conditions (5)-(8), fictitious values are determined as the combinations of the to-be-determined numerical solutions on grid points. While a pair of fictitious values are determined along a mesh at a time, an iterative procedure can be used to determine all the required fictitious values for higher-order schemes by repeatedly using the lowest order jump conditions. The essential strategy of the MIB method is to locally reduce a 2D or a 3D interface problem into 1D-like ones [81, 85, 84, 83]. The essence of the MIB scheme is the use of fictitious values, which entirely assemble the interface conditions, the local geometry of the interface and all the necessary interface information. Therefore, the MIB scheme is very robust for different protein surface and successfully overcomes the first two types of singularities [74, 75, 76]. Rigorous second order MIB schemes have been developed to solve the PBE with geometric singularities of molecular surfaces.
The source term singularity is removed by the DNM. In Ref. [30], the solution u(x) of the PBE is decomposed into singular part and regular part. The singular part of the solution comes from singular delta functions, and is obtained analytically as the Green's function. As a consequence, this separation generates an extra Neumann jump condition at the interface for regular part. Therefore, after the separation, one only needs to solve the remaining homogeneous Poisson-Boltzmann equation subject to corresponding Neumann jump conditions at the interface. This procedure is also called Dirichlet to Neumann mapping. Consequently, truly second-order accurate solution to the PBE with molecular surfaces and singular charges can be obtained with a relatively large grid spacing [30].
3 Pre-conditioner accelerated MIBPB solvers
In practice, when one pursues the numerical solution of the PBE, the discretization of the PBE results in a linear equation system
| (10) |
where h is the discretization resolution, Lh and fh represent the matrix and right hand side generated via the MIB and DNM schemes, Uh is the solution vector. It is worth pointing out that in standard FDMs, the matrix Lh only depends on the grid resolution and the dielectric constants. However, in the MIBPB scheme, the structure of Lh also depends on the molecular surface of a specific protein. Due to this reason, we also call Lh the matrix of a protein for simplicity. The MIB and DNM successfully overcome the equation singularities and promise a high accuracy convergence order by taking into account all the local interface information. However, as a trade-off, the structure of matrix Lh is much more complicated than that from standard FDMs. Specifically, the matrix loses symmetry and may not be positive-definite any more. The lose of these properties will lead to more computational time and memory. Therefore, the selection of appropriate linear solvers becomes subtle when computational efficiency is sought as well.
The review of several basic linear solvers are summarized in Appendix A. However, the matrices from the MIB and the DNM scheme can barely take any advantage from the described methods due to their notoriously complicated structures. Therefore, we put our emphasis on choosing other methods and accelerating techniques. In Appendix B, we include a brief description of Krylov subspace (KS) techniques. Based on the KS theory, proper linear solvers and acceleration techniques (preconditioners) are chosen and compared in this section for the numerical efficiency of MIBPB linear systems. Two KS solvers, the stabilized biconjugate gradient method (BiCG) and the generalized minimal residual method (GMRES), are potentially effective iterative solvers for matrices with general structures. Several preconditioning strategies, the Jacobi preconditioner (JAC), the blocked Jacobi preconditioner (BJAC) and the incomplete LU factorization preconditioner (ILU) are available to incorporate with the two solvers to accelerate the solution of the linear system.
Matrices generated from a set of proteins are employed to test the performance of various KS solver-preconditioner combinations. For each matrix, the condition number, linear system iteration number and iteration time are used to characterize numerical efficiency. All these measurements of matrices are analyzed numerically by the PETSc (http://www.mcs.anl.gov/petsc/petsc-as/). The grid resolution is taken as 1.0Å in the following tests unless otherwise specified. The stopping criterion of all KS solvers are taken as 1 × 10−6 in order to get more accurate solutions while in practical biological applications the criterion can be relaxed to 1 × 10−3 to save CPU time but satisfactory results are also achieved.
First of all, the matrix condition numbers are examined. The condition number can predict the level of difficulty in solving the linear system before it is really solved. The magnitude of a matrix condition number depends on the size and structure of a biomolecule. More specifically, under the same grid resolution, a molecule which has a larger number of atoms needs a larger computational domain and a larger matrix size. Meanwhile, a molecule which has a more complex surface geometry leads to more interface conditions and a larger matrix size. Both cases contribute to higher condition numbers. Therefore, the size and complexity of a biomolecule usually affect the numerical efficiency of the MIBPB solver.
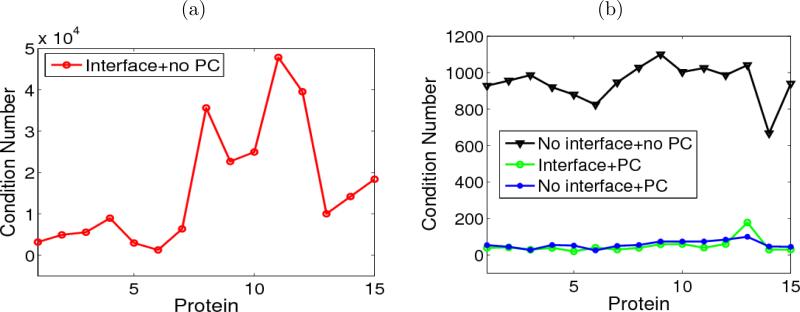
Figure 2(a) presents condition numbers of matrices corresponding to 15 protein structures and indicates the numerical difficulties of solution without proper acceleration techniques. The horizontal axis lists proteins. Protein data bank (PDB) identification numbers (IDs) are listed in the figure. The numbers of atoms of these proteins range from 500 to 2000. Discretizing the PBE with the MIB scheme, without any preconditioner (PC) applied, the condition numbers are usually in the order of 104, about one order larger than those of the matrices generated from the standard FD discretization, i.e., without the interface treatment. This is expected because the use of the molecular surface as the interface and all included local information around the interface, the MIBPB matrices do not maintain the symmetry and are not positive definite. The MIB matrices generally have larger condition numbers and require more CPU time [83, 74, 30].
Figure 2.
Condition numbers over 15 proteins (PDB IDs from protein 1 to protein 15: 1ajj, 1vii, 1cbn, 1bbl, 1fca, 1sh1, 1vjw, 1fxd, 1bpi, 1a2s, 1frd, 1svr, 1a63, 2erl, 2pde). (a) Condition numbers for unpreconditioned (unPCed) MIBPB matrices; (b) Comparisons of condition numbers under three settings.
By using of preconditioners (PCs), the magnitudes of condition numbers of MIBPB matrices are significantly reduced to less than one hundred, as shown in the circle plot of Fig. 2(b). The triangle plot in Fig. 2(b) gives the condition number magnitudes of the matrices from standard FDMs without PC, revealing the huge differences among different treatments. The circle and dot plots are condition number magnitudes for matrices with PC, from both the MIB scheme and the FDM, respectively. Interestingly, it can be concluded that the condition number magnitudes of two schemes are reduced to almost the same level by using the PCs. We can safely say that the difficulty of solving the linear system generated from the interface based MIBPB scheme is actually comparable to that from standard FD discretization. Under almost the same numerical efficiency, MIB scheme and DNM are able to obtain higher accuracy because all the local geometry information of the molecular surface has been taken into account.
Quantitatively, for a specific KS solver such as GMRES, the iteration numbers and computing time of linear systems for 7 proteins are listed in Table 1. It is well-known that condition numbers can only be mathematically estimated for large matrices, then the listed condition numbers calculated by PETSc solvers may not be exact. Despite this fact, we can still have a sense from the numbers how the PC significantly reduces the difficulties of solving the linear systems.
Table 1.
Iteration numbers and CPU time for the discretization matrices of proteins
| Proteins | Preconditioned iteration |
Un-Preconditioned iteration |
|||||
|---|---|---|---|---|---|---|---|
| ID | atoms | number | time | condition number | number | time | condition number |
| 1mbg | 903 | 19 | 0.3 | 40 | 5404 | 54 | 118900 |
| 1r69 | 997 | 18 | 0.3 | 40 | 5400 | 58 | 250400 |
| 1bor | 832 | 20 | 0.3 | 30 | 2152 | 23 | 138850 |
| 1vii | 596 | 17 | 0.2 | 42 | 3963 | 28 | 4963 |
| 1fxd | 824 | 19 | 0.3 | 39 | 7084 | 80 | 35637 |
| 2erl | 573 | 17 | 0.2 | 29 | 4858 | 36 | 14223 |
| 1a2s | 1272 | 23 | 0.6 | 61 | 10000 | 156 | 24981 |
As stated earlier, two KS iterative methods, the stabilized BiCG and the GMRES, are associated with three types of preconditioners, JAC, BJAC and ILU. Table 2 compares the effect of combinations of these KS solvers and preconditioners. For different preconditioning strategies, since the ways of counting iteration numbers are different, only the iteration time for each combination is listed in the table. Sample proteins of various sizes are presented in this table, from small size (less than 1000 atoms), middle size (1000-3000 atoms) to large size (around 8000 atoms). It can be concluded that the GMRES performs slightly better than the stabilized BiCG does for small-sized proteins but stabilized BiCG take the lead in middle- and big-sized proteins. Among the three kinds of preconditioners, the BJAC and the ILU almost have the same effects and are slightly better than the JAC. Therefore, the combination of stabilized BiCG and BJAC is recommended and set as the default option in the MIBPB package.
Table 2.
Iteration time for different combinations of KS solvers and preconditioners
| Protein |
BiCG |
GMRES |
|||||
|---|---|---|---|---|---|---|---|
| ID | atom | BJAC | ILU | JAC | BJAC | ILU | JAC |
| 1ajj | 519 | 0.24 | 0.24 | 0.26 | 0.23 | 0.23 | 0.38 |
| 1vjw | 828 | 0.29 | 0.29 | 0.35 | 0.26 | 0.26 | 0.44 |
| 1a2s | 1272 | 0.56 | 0.56 | 0.51 | 0.57 | 0.56 | 0.84 |
| 1a7m | 2809 | 1.69 | 1.67 | 1.77 | 1.93 | 1.91 | 2.85 |
| 1f6w | 8243 | 3.90 | 3.88 | 4.48 | 4.70 | 4.65 | 7.19 |
As indicated at the beginning, all the mathematical algorithms and techniques are enforced to maintain the high order convergence of the MIBPB solver. Table 3 lists the numerical evidence of the second order convergence through a set of given protein surfaces, atomic coordinates, radii and charges, where protein surfaces are generated by MSMS, and the standard CHARMM force field parameters are used. A special analytical solution was designed and given in [30] for the convergence order check of all proteins. In this table, the numerical error is defined as , where is the numerical solution of the PBE at grid resolution h while uexact is the designed exact solution. The numerical experiments are implemented under resolutions h = 1.0Å, 0.5Å and 0.25Å. The numerical error is supposed to be reduced by four times as the grid size is halved and this is clearly demonstrated in the table.
Table 3.
Convergence test of MIBPB solver with a set of proteins
| Proteins ID | Error h=1.0Å | h=0.5Å | Order | Error h=0.25Å | Order |
|---|---|---|---|---|---|
| 1ajj | 6.52E-2 | 1.13E-2 | 2.52 | 1.76E-3 | 2.68 |
| 1a23 | 1.026E1 | 1.72E-1 | 2.57 | 2.74E-2 | 2.65 |
| 1b4l | 1.19E-1 | 1.25E-2 | 3.25 | 2.07E-3 | 2.59 |
| 1bbl | 1.32E-1 | 1.86E-2 | 2.82 | 1.81E-3 | 3.36 |
| 1bor | 9.44E-2 | 1.31E-2 | 2.84 | 1.97E-3 | 2.73 |
| 1fca | 1.20E-1 | 1.20E-2 | 3.31 | 1.78E-3 | 2.76 |
| 1frd | 7.93E-2 | 1.24E-2 | 2.67 | 2.02E-3 | 2.61 |
| 1fxd | 7.66E-2 | 1.19E-2 | 2.68 | 2.00E-3 | 2.57 |
| 1hpt | 8.05E-2 | 1.37E-2 | 2.50 | 1.77E-3 | 2.90 |
| 1mbg | 1.35E-1 | 1.08E-2 | 3.64 | 1.69E-3 | 2.67 |
| 1neq | 8.52E-2 | 1.27E-2 | 2.74 | 1.83E-3 | 2.79 |
| 1r69 | 7.95E-2 | 1.15E-2 | 2.39 | 1.92E-3 | 2.96 |
| 1svr | 7.94E-2 | 1.17E-2 | 2.21 | 1.94E-3 | 3.13 |
| 1uxc | 7.55E-2 | 1.27E-2 | 2.57 | 2.02E-3 | 2.65 |
| 1vjw | 7.22E-2 | 1.20E-2 | 2.58 | 2.23E-3 | 2.43 |
| 2pde | 1.12E-1 | 1.64E-2 | 2.77 | 5.46E-3 | 1.58 |
The above mentioned tests are carried out in conjunction with the PETSc software package, whose installation may not be so straightforward. An alternative is to use the SLATEC, which is easier to implement and also includes tens of linear system solvers with different preconditioners. To compare the performance of the PETSc and the SLATEC, we show the computation time of ten methods in the SLATEC for five proteins, whose atom number varies from five hundreds to eight thousands in Table 4. All methods are listed in the form: preconditioner/solver. Here GS, DS, BiGS, and OM represent the Gauss-Seidel, the diagonal scaling, the biconjugate gradient squared method, and the orthomin sparse iterative method, respectively. The combination of the ILU/BiCG is used in the PETSc. From the table, it can be seen that the iteration time of the PETSc is slightly shorter than that of most solvers in the SLATEC for small-sized proteins. The last column of the table lists the averaged CPU time for the PETSc and solvers in SLATEC. The averaged time, which in some sense could reflect the abilities of solvers for proteins in various sizes, is the sum of the CPU time for each corresponding protein and weighted by the atom number. By checking the averaged CPU time one can generally conclude that the ILU/BiCG of the PETSC takes less iteration time than the SLATEC schemes do. Moreover, according to our experience, the PETSc is more stable than the SLATEC for large proteins. However, the SLATEC can be easily incorporated in the MIBPB package. Whereas, the PETSc needs to be pre-installed by the user as discussed below.
Table 4.
Comparison of CPU time for the PETSc and the SLATEC schemes
| Protein ID | 1ajj | 1vjw | 1a2s | 1a7m | 2ade | Averaged |
|---|---|---|---|---|---|---|
| Atoms | 519 | 828 | 1272 | 2809 | 8344 | CPU time |
| PETSc | 0.235 | 0.272 | 0.529 | 1.729 | 3.777 | 2.72 |
| | ||||||
| GS/GS | 0.866 | 1.222 | 2.225 | 9.512 | 55.016 | 35.58 |
| ILU/ILU | 0.523 | 0.883 | 1.344 | 5.854 | 32.479 | 21.07 |
| DS/BiCG | 0.331 | 0.467 | 1.041 | 3.140 | 14.015 | 9.27 |
| ILU/BiCG | 0.262 | 0.401 | 0.701 | 2.038 | 7.846 | 5.27 |
| DS/BiGS | 0.243 | 0.313 | 0.602 | 2.900 | 8.879 | 6.05 |
| ILU/BiGS | 0.187 | 0.393 | 0.410 | 1.433 | 6.575 | 4.34 |
| DS/OM | 0.206 | 0.420 | 0.496 | 3.338 | 21.993 | 14.08 |
| ILU/OM | 0.179 | 0.291 | 0.389 | 1.25 | 5.993 | 3.95 |
| DS/GMRES | 0.417 | 0.559 | 0.999 | 3.856 | 26.262 | 16.84 |
| ILU/GMRES | 0.198 | 0.279 | 0.439 | 1.615 | 7.685 | 5.05 |
4 Usage illustration and application
4.1 Work flow of the MIBPB package
The MIBPB solver package incorporates with two packages to accomplish the electrostatic potential calculation. First, molecular structures are prepared via Python software package PDB2PQR (http://pdb2pqr.sourceforge.net/): it accomplishes many common tasks of preparing structures for continuum electrostatic calculations, such as adding a limited number of missing heavy atoms to biomolecular structures, determining side-chain pKas, placing missing hydrogens, etc. Users can either submit the protein PDB index to the online server (http://pdb2pqr.sourceforge.net/) or download the executable file to prepare the molecular structure.
Once the molecular structure is prepared, the computational domain Ω will be automatically generated based on the coordinates of the protein atoms: first a smallest cuboid that contains the protein will be calculated and then each length of the cuboid is symmetrically extend at two ends by 5 to 10Å, depending on the protein size. This strategy usually employed in many FDMs is verified to be reasonable in practices and also the extension of the cuboid can be customized easily. The larger size of Ω is of course closer to real biological situation. However, the solution of the PBE is not sensitive to this change while the computational cost will be increased.
Additionally, the geometry of the molecular surface used in the MIB scheme is generated by the MSMS (http://www.scripps.edu/sanner/html/msms_home.html). Given the information of the coordinates and radius of each atom in the molecule, surfaces are generated at given water probe radius in a triangulation form. The intersection of each triangle with the meshing lines and the normal direction extracted from the surface information are key ingredients of the MIBPB scheme. For the MSMS parameter, the water molecule probe radius is recommended as 1.4 and the vertex density is 10. These parameters are enough to generate the molecular surface with good quality, various 3D Cartesian grid resolutions in current use can obtain necessary surface information under this setting.
There are two options for choosing KS solvers and preconditioners in solving MIBPB matrices. One is to use the SLATEC, which has been incorporated in our MIBPB package. The other way is to use the PETSc. According to our tests, the PETSc is generally more stable and reliable than the SLATEC, particularly for large proteins. It needs to be pre-installed by the user if one chooses the PETSc matrix acceleration option.
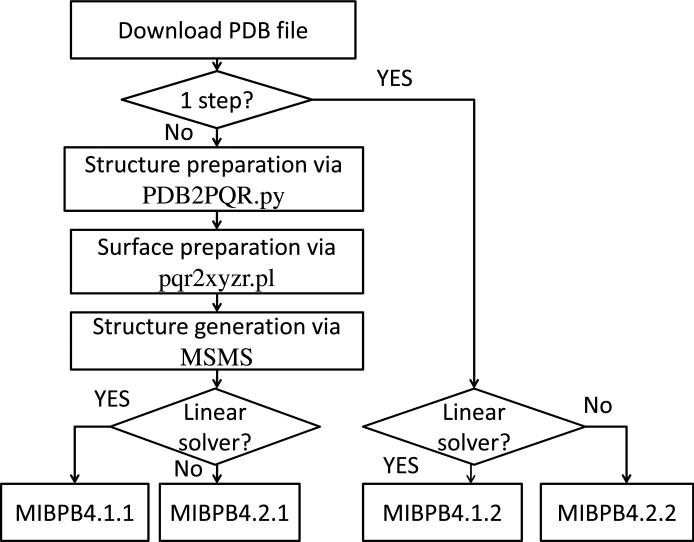
The current MIBPB package offers half stand-alone solvers in which users have to prepare the molecular structures and generate the surfaces on their own with desired parameters. The package also has one-step solvers which have integrated all the steps with default parameter settings. Either the half stand-alone or one-step solver is further classified into linear solver and nonlinear solver. Therefore, there are in total four executable MIBPB files in the package. Additionally, two other auxiliary small Perl scripts, the pqr2xyzr.pl and dat2dx.pl are included in the MIBPB package to accomplish the molecular surface preparation and ultimate data visualization. Figure (3) is the work flow of the MIBPB package usage. Users are referred to http://www.math.msu.edu/~wei/MIBPB/ for detailed instructions.
Figure 3.
Work flow of the MIBPB package
For a clearer demonstration, we use one specific protein example to illustrate the procedure. Protein with ID 1ajj is assumed to have been downloaded from the Protein Data Bank and saved as file 1ajj.pdb.
- Prepare the protein structure
- Input file: 1ajj.pdb
- Command line: python pqd2pqr.py –ff=CHARMM 1ajj.pdb 1ajj_apbs.pqr
- Output file: 1ajj_apbs.pqr.
- Remark: For full usage of pqb2pqr.py, users are referred to the corresponding link.
- Molecular surface preparation
- Input file: 1ajj_apbs.pqr
- Command line: pqr2xyzr 1ajj
- Output files: 1ajj.xyzr, 1ajj.pqr
- Remark: 1ajj.xyzr file stores the coordinates and radii of the atoms in the protein, 1ajj.pqr stores the coordinates and partial charges. They are necessary files for the MSMS to generate molecular surfaces.
- Molecular surface generation
- Input files: 1ajj.xyzr, 1ajj.pqr
- Command line: msms -if 1ajj.xyzr -prob 1.4 -de 10 -of 1ajj
- Output files: 1ajj.vert, 1ajj.face. Now the molecular surface is generated in the triangulation form. The vertices and normal direction of each triangle are saved in these files.
- Remark: water probe radius and triangulation density are set as default values 1.4 and 10, respectively. They are adjustable parameters.
- MIBPB implementation
- Linear solver: mibpb4.1.1 1ajj eps1=1 eps2=80 h=0.5
- Nonlinear solver: mibpb4.2.1 1ajj eps1=1 eps2=80 kappa=1.0 h=0.5
- Output file: 1ajj_pbe.dat
- Remark: Above command lines give the standard format. Parameters are adjustable.
4.2 Work flow for the display of the surface electrostatic potential
After the electrostatic potential file is obtained by running the MIBPB solver, we can display it on the molecular surface by using the VMD (http://www.ks.uiuc.edu/Research/vmd/), a molecular visualization program. We are able to visualize the potential distribution on the surface by implementing a file transformation via the Perl script dat2dx.pl. Moreover, by taking the difference of surface electrostatic potentials under different grid resolutions h, we are also able to check the convergence of the solutions and therefore suggest a proper grid resolution for balancing high numerical accuracy and efficiency. The procedure is shown as the following.
- Visualization file preparation.
- MIBPB package generates output file [pdbname]_pbe.dat, in which the electrostatic potentials on grid points of the protein-solvent system are stored. Before displaying the electrostatic potential on the molecular surface, one needs to use dat2dx.pl script to transform the data file to the [pdbname].dx file.
- For example, for protein 1ajj, one gets 1ajj_pbe.dat file from the MIBPB package. Then use the command: dat2dx.pl 1ajj [dcel] [xleft] [xright] [yleft] [yright] [zleft] [zright] where [dcel] is the mesh size (we assume a uniform mesh). Here [xleft],[xright], [yleft], [yright], [zleft] and [zright] prescribe the span of computational domain in x, y, z direction, respectively. Here xleft, xright, yleft, yright, zleft, and zright should be the same as those used in calculating the potential.
- Molecular surface drawing
- Load the PDB data file into the VMD
- Set drawing parameters in the Graphical Representation window: choose the “Volume” option for coloring method and the “Surf” option for drawing method.
- Surface electrostatic potential drawing
- Load the [pdbname].dx format potential file into the VMD. In the Molecular File Browser window, load [pdbname].dx file for the same protein as that in molecular surface (instead of for new molecular).
- Set drawing parameters in the same Graphical Representation window as that in the second step. Choose the “Volume” option for coloring method and the “Surf” option for drawing method. Adjust the Color Scale Data Range to see different color effects.
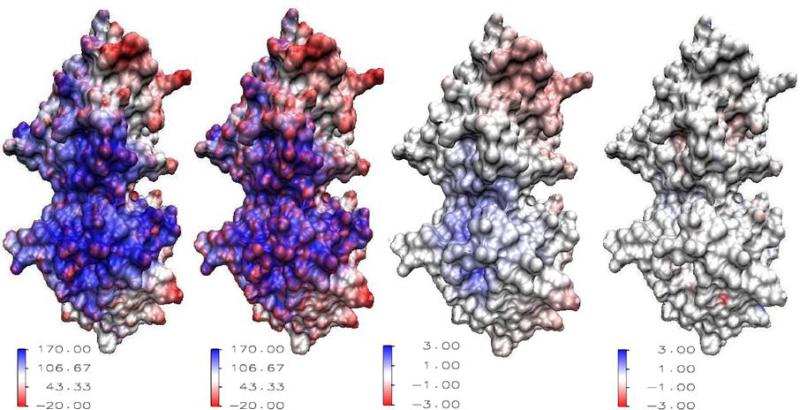
Figure 4 illustrates the visualization of electrostatic potential calculated from the MIBPB package, using protein 1beb as an example. The potentials calculated via both the linear MIBPB solver and the nonlinear MIBPB solver are plotted on the molecular surface via the VMD through the above procedure. Figure 4(a) displays the potential distribution on the surface of protein 1beb when the solvent is water. In this case the linear MIBPB solver is implemented because is set as zero. While Fig. 4(b) presents the potential distribution when , in which case the nonlinear MIBPB solver is employed. These two calculations are carried out when grid resolution h is taken as 1.0Å. Figure 4(c) gives the difference of electrostatics in (a) and (b), from which the salt effect on electrostatic distribution may be observed. Figure 4(d) reveals the potential difference in solvent when the calculations are under resolutions h = 1.0Å and 0.5Å, i.e. the error |ϕh – ϕh/2|. It can be found that the error is almost zero around the molecular surface, this fact indicates that at h = 1.0Å, the result is accurate enough so that reducing grid resolution to 0.5Å does not give too much improvement. Mathematically speaking, the result is almost convergent between mesh size 1.0Å and 0.5Å, which is the recommended grid resolution range in the MIBPB package.
Figure 4.
Visualizations of surface electrostatic potentials of protein 1beb. From left to right: (a) Surface electrostatic potential (I = 0, h = 1.0Å); (b): Surface electrostatic potential with the ionic strength I = 1 (h = 1.0Å); (c): The difference of surface electrostatic potentials between in an ionic solvent (I = 1) and in a water solvent (I = 0); (d): The difference of surface electrostatic potentials in water (I = 0) between grid meshes h = 1.0Å and h = 0.5Å.
4.3 Application to solvation energy calculations
One of the most important applications of the PBE model is solvation energy calculations for the protein-solvent systems. In this section, solvation energies of 28 proteins are calculated and compared with popular PBE solvers to examine the feasibility, usefulness and robustness of the linear solver in the MIBPB package. These proteins have a wide range of numbers of atoms, from around 500 up to 10,000. The corresponding spatial dimensions extend from about 30Å × 30Å × 30Å to almost 100Å × 100Å × 100Å. Among these calculations, the dielectric constant is set to 1 for proteins and 80 for the solvent. The ion strength is set to zero because no ion is considered for the moment.
The calculation of electrostatic solvation energy ΔGelec is to sum all the fixed charges {qi} of the solute in the solvent, weighted by the reaction field potential ϕrf(x):
| (11) |
where xi is the position of each charge. Based on the continuum electrostatics, the reaction field potential is the difference between the electrostatic potential in the solvent environment ϕs(x) and the reference electrostatic potential ϕref(x), i.e, ϕrf(x) = ϕs(x) − ϕref(x). Here ϕrf(x) can be computed by solving the PBE twice with corresponding settings. Specifically, ϕref(x) is calculated by setting a uniform dielectric constant in the whole computational domain, while ϕs(x) is calculated by setting the dielectric constants for solute and solvent differently. Therefore, ϕref(x) can be obtained by the standard linear PB equation with the Dirichlet boundary condition via the standard finite difference or FFT methods but ϕs(x) is solved by using the MIBPB algorithm.
The performance of the MIBPB method for calculating solvation energies has already been examined in our previous work [30]. It has been shown that the MIBPB solver has high accuracy and good convergence order because of the use of interface treatments but has relatively low numerical efficiency due to the absence of appropriate matrix acceleration techniques. The MIBPB matrix requires longer CPU time to solve. The Krylov theory and associated preconditioners discussed in the present work make the MIBPB solver more efficient. Here the new results are presented for various proteins.
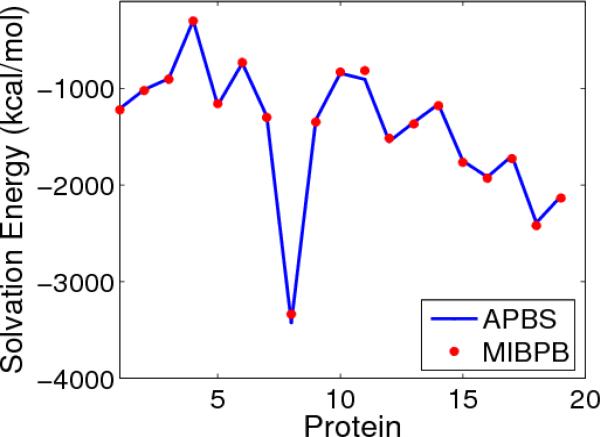
Figure 5 gives the comparison of the calculated solvation energies from the MIBPB and the APBS packages. It is seen that the solvation energies calculated from the MIBPB agree very well with those from the APBS. The mesh sizes of h = 1Å is used in the MIBPB and h = 0.25Å is used in the APBS methods, respectively. The reader is referred to Ref. [30] for a more detailed comparison among the MIBPB, the APBS and the PBEQ methods.
Figure 5.
Comparison of solvation energies of proteins (From protein 1 to 19: 1ajj,1bbl,1vii,1cbn,2pde,1sh1,1fca,1fxd,1vjw,1bor,1hpt,1bpi,1mbg,1r69,1neq,1a2s,1svr,1a63,1a7m) calculated by using the MIBPB and the APBS methods.
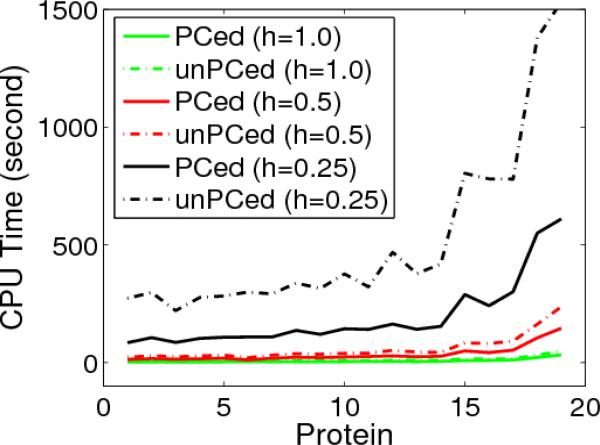
Once the preconditioning techniques are applied, the required CPU time is significantly reduced. Figure 6 illustrates the differences of the CPU time needed to calculate solvation energies for nineteen moderately large proteins at three different grid resolutions. The solid lines are the CPU time for preconditioned (PCed) systems and dashed lines are for unpreconditioned (unPCed) systems. It can be concluded that at each grid resolution, preconditioners can save more than half of the overall CPU time.
Figure 6.
Comparison of CPU time of preconditioned (PCed) and un-preconditioned (unPCed) MIBPB methods for 19 proteins (from protein 1 to 19: 1ajj,1bbl,1vii,1cbn,2pde,1sh1,1fca,1fxd,1vjw,1bor,1hpt,1bpi,1mbg,1r69,1neq,1a2s,1svr,1a63,1a7m).
Table 5 lists the results for the tested proteins at different grid resolutions and compares the values with the original MIBPB-III scheme in terms of solvation energies and CPU time. For each protein case from different grid resolutions, the CPU time increases in nonuniform pattern from less than 10 seconds for h =1.0Å, several tens of seconds for h =0.5Å, to a few hundreds of seconds for h =0.25Å. Note that there is an eight times increase in the number of unknowns when the mesh size is halved. The increase in the CPU time is roughly linear with the increase in the number of unknowns.
Table 5.
Solvation energies and CPU time for proteins
| Solvation energy (kcal/mol) |
CPU time (second) |
|||||||
|---|---|---|---|---|---|---|---|---|
| MIBPB+KS | MIBPB | MIBPB+KS | MIBPB | |||||
| h(Å) | 1.0 | 0.5 | 0.25 | 0.25 | 1.0 | 0.5 | 0.25 | 0.25 |
| 1ajj | -1141.9 | -1136.3 | -1136.6 | -1137.2 | 2.3 | 15 | 85 | 273 |
| 2pde | -826.2 | -819.9 | -817.2 | -820.9 | 3.4 | 17 | 108 | 283 |
| 1vii | -914.6 | -901.5 | -902.8 | -901.2 | 2.8 | 15 | 86 | 221 |
| 1cbn | -311.1 | -303.6 | -303.7 | -303.8 | 2.9 | 16 | 102 | 277 |
| 1bor | -858.8 | -854.3 | -857.9 | -853.7 | 4.5 | 24 | 143 | 377 |
| 1bbl | -998.3 | -986.9 | -988.7 | -986.8 | 2.8 | 16 | 106 | 298 |
| 1fca | -1215.5 | -1199.9 | -1200.0 | -1200.1 | 3.5 | 19 | 109 | 292 |
| 1uxc | -1157.6 | -1138.1 | -1139.1 | -1138.7 | 4.2 | 21 | 127 | 347 |
| 1sh1 | -755.7 | -728.0 | -751.4 | -753.3 | 3.3 | 12 | 109 | 300 |
| 1mbg | -1368.4 | -1349.8 | -1352.4 | -1346.1 | 4.8 | 25 | 142 | 378 |
| 1ptq | -893.5 | -871.8 | -872.2 | -873.1 | 3.9 | 22 | 133 | 376 |
| 1vjw | -1250.9 | -1236.9 | -1236.9 | -1237.9 | 4.1 | 26 | 120 | 315 |
| 1fxd | -3309.7 | -3299.7 | -3301.6 | -3300.0 | 4.2 | 31 | 138 | 338 |
| 1r69 | -1111.0 | -1086.5 | -1087.9 | -1089.5 | 5.5 | 32 | 154 | 419 |
| 1hpt | -827.3 | -810.9 | -812.7 | -814.3 | 4.9 | 26 | 141 | 322 |
| 1bpi | -1320.8 | -1298.9 | -1301.3 | -1301.9 | 5.4 | 50 | 164 | 469 |
| 1a2s | -1928.8 | -1913.1 | -1913.6 | -1913.5 | 9.6 | 47 | 242 | 780 |
| 1frd | 2879.8 | -2851.4 | -2856.3 | -2851.9 | 10.8 | 51 | 284 | 707 |
| 1svr | -1741.6 | -1709.8 | -1710.7 | -1711.2 | 11.1 | 57 | 301 | 779 |
| 1neq | -1765.6 | -1729.1 | -1732.7 | -1730.1 | 9.1 | 50 | 289 | 804 |
| 1a63 | -2420.8 | -2371.2 | -2370.2 | -2373.5 | 22 | 113 | 550 | 1376 |
| 2erl | -964.2 | -948.2 | -949.3 | -948.8 | 2.3 | 15 | 101 | 276 |
It is found that, at resolution of 0.25Å, the results from the MIBPB+KS and from the original MIBPB-III have less than 0.1% disagreement. This is due to the use of different convergence norms in the KS solvers and the regular solver. The solvation energy calculations show a correct convergence tendency. The values from resolutions of 0.25Å and 0.5Å are pretty close, while calculations at h = 0.25Å cost much more CPU time. Therefore, we can conclude that grid resolution between 0.5Å and 1.0Å is sufficient for the calculation and can guarantee the accuracy.
Table 6 shows the robustness and efficiency of the MIBPB package for calculating solvation energies of large proteins which exceed 7000 atoms. For time efficiency, all the calculations are carried out at the grid resolution of h = 1.0Å. Note that the reliability of these solvation free energies has been cross-checked with other popular PB solvers. The reported CPU time can be used as a reference.
Table 6.
Solvation energies (kcal/mol) and CPU time (second) for large proteins
| Protein |
MIBPB (h = 1.0Å) |
||
|---|---|---|---|
| ID |
atoms |
Solvation energy |
CPU time (s) |
| 1cbg | 7838 | -5659.9 | 181 |
| 1c4k | 11439 | -9901.9 | 398 |
| 1e24 | 7776 | -9506.4 | 231 |
| 1f6w | 8243 | -5611.2 | 225 |
| 1po7 | 7796 | -5471.2 | 206 |
4.4 Application to salt effects on protein-protein binding
In this section, the ability of the MIBPB package to solve the nonlinear PBE is tested by checking solvent salt effect on protein-protein binding. The nonlinear PBE has had considerable success in describing biomelocular electrostatics with salt effects on the binding of ligands, peptides and proteins to nucleic acids, membranes and proteins. The binding free energies reflect the non specific salt dependence of the formation of macromolecular complex and the measurement is the binding affinity. Some experimental data are available and the binding affinity is calculated as the ratio between salt dependent binding energy ΔΔGel(I) at a specific salt strength I and the natural logarithm of I. In the present work, we have implemented the nonlinear version of the PBE solver in the MIBPB package. Simulation results are obtained by varying the ionic strengths.
The binding energy (ΔGel) has several components while the one related to electrostatics is the difference of the electrostatic free energies of the complex and each of its free molecules [8]
| (12) |
where , and represent the electrostatic free energies of the complex AB, component A and component B, respectively, at a given ionic strength I.
The electrostatic free energy can be further split into three components
| (13) |
where Gcoul is the Coulomb energy calculated in a homogeneous medium, Grxn is the corrected reaction field energy and Gsalt(I) is the electrostatic energy contributed by mobile ions. Among the three terms in Eq. (13), only Gsalt(I) is salt dependent. Thus, the salt dependence of the binding free energy ΔΔGel(I) is electrostatic component of the binding energy in Eq. (12) calculated at some salt strength I minus the one calculated at the zero salt concentration [8]
| (14) |
where various energy terms are calculated at different ionic strengths by using the MIBPB package.
To verify our nonlinear solver, one hetero-dimeric and one homo-dimeric complexes are studied in this work. The experiments on these two protein complexes can be found in [59, 72] and biological features(1emv and 1beb) are listed in Table 7. The first four columns describe the properties of proteins and the last two columns are the slopes (binding affinity) of the lines in Fig. 7. It can be seen in a quantitative view that the slopes obtained from experiments and simulations are very close to each other. The calculations were performed assuming that all Arg, Asp, Glu and Lys residues are ionized in both free and bound states. The results are obtained with dielectric constants of 2 and 80 for the solute and the continuum solvent, respectively. The parameter is determined by:
| (15) |
where ec, kBT are the same as those defined in Eq. (1), Na is the Avogadro's number. After a simple derivation, is given by
| (16) |
for T = 298K. Here the ion strength I is in the unit of mole.
Table 7.
Binding affinity
| Complex | PDB ID | Complex charge | Surface area (Å2) | Charge of the free monomers | Experimental data | MIBPB (h = 1.0Å) |
|---|---|---|---|---|---|---|
| E9Dnase-Im9 (10)(B-A) | 1emv | -3 | 1465 | B=+5; A=-8 | 2.17 | 2.42 |
| Lactoglobulin dimer (57)(A-B) | 1beb | +26 | 1167 | A=B=+13 | -1.62 | -1.90 |
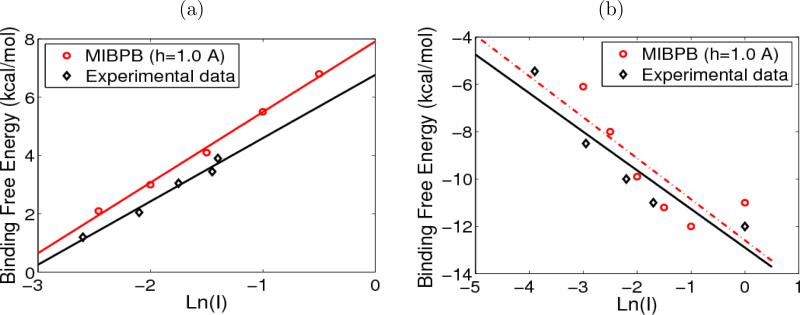
Figure 7.
(a) Binding affinity of 1emv; (b) Binding affinity of 1beb.
Figure 7 depicts the experimental and calculated salt dependence of the binding free energies ΔΔGel(I) for the two complexes. The ΔΔGel(I) are plotted against the logarithm of the salt strength I. The salt dependence is assumed as in a linear pattern therefore the least square fitting line is applied to calculate the binding affinity, which is the slope of the line. It should be explained that experimental data dots for ΔΔGel(I) are read from the graphs in Ref. [8], while the fitting line slope is explicitly given based on the experimental data with error bars. The diamond points and solid line are experimental data and the corresponding fitting line, respectively. The circle points and dashed line are numerical stimulations.
In the homo-dimeric complex, the experimentally observed binding free energies decrease with the increase of ionic strength, while for the hetero-diemric complex, the experimental measurement had detected an increase. Our results obtained from the MIBPB package reproduced these observations. The calculated magnitudes of the slopes of the salt dependence are in quite good agreement with experimental results within the range of errors, as the fitting lines are almost parallel. The discrepancies between the experimental data and simulation results are also expected: the PBE, no matter in linear or full nonlinear form, only gives the ideal situation of the solute-solvent system but many details, such as the “protein conformation change, pKa shifts upon complexation or possible ionization states”, are lacking [8]. Despite these approximation, the application of PBE for static protein structures is suggested in general by these good agreements with experimental data.
5 Conclusion
This paper introduces the matched interface and boundary based Poisson-Boltzmann solver (MIBPB) software package and its work flow for practical applications in biochemistry, biophysics and structural biology. Two applications, the solvation free energy calculation and the salt dependence binding affinity calculation are carried out to justify the robustness, accuracy and efficiency of solving the linear and nonlinear PBEs. The results of solvation free energy calculations are compared with those from a traditional PBE solver and the results of binding affinity are compared with experimental data. The MIBPB solver is verified to be highly accurate – by far the only existing second-order accurate method for solving the Poisson-Boltzmann (PB) equation with discontinuous dielectric constants, singular charge sources and geometric singularities from the molecular surfaces of biomolecules. More specifically, the MIBPB has built in advanced interface techniques that are able to deal with discontinuous solvent-solute interfaces and geometric singularities of molecular surfaces. The Dirichlet to Neumann mapping, or Green's function approach, has been developed in the MIBPB solver to analytically resolve the singular point charges. Consequently, the MIBPB solver is able to deliver high accuracy at relatively coarse meshes. The MIBPB solver provides reliable electrostatic potentials at the mesh of about 1Å, whereas traditional methods have to resort to about 0.25Å mesh resolution to achieve a similar level of reliability.
In the present work, we further equip the MIBPB solver with advanced Krylov subspace techniques to accelerate the speed of the convergence of solving linear equation systems originated from the MIBPB discretization. The performances of various solver-preconditioner combinations have been carefully examined through mathematical analysis and numerical experiments. Dramatic reductions in condition numbers are found when appropriate preconditioners are utilized. Upon the use of appropriate combinations of preconditioners and solvers, significant reductions in the CPU time are found in solving the PB equation for large proteins. Both the PETSc and the SLATEC are employed in the present MIBPB package to speed up the convergence rate of the iterations of the linear systems. The PETSc package is found to be more reliable and efficient. In the present work, the structure preparation of proteins is conducted via the PDB2PQR software package, while the MSMS software package is utilized for the molecular surface generation.
Additionally, the nonlinear MIBPB solver has been developed in the present work. This is achieved via the standard inexact Newton method, assisted by the Krylov subspace acceleration techniques. The present nonlinear MIBPB solver has been tested and applied to the salt dependence analysis of protein-protein binding interactions. Our results of binding a nities are compared with experimental data.
Acknowledgments
This work was supported in part by NSF grants DMS-0616704 and CCF-0936830, and NIH grants GM-090208 and CA-127189.
Appendices
A Linear equation systems and MIBPB matrix
A system of linear algebraic equations is formed after discretizing the PB model
| (17) |
where Lh is a real non-singular n by n matrix under grid spacing h, uh is the numerical solution vector and fh is the source term vector. The matrix Lh is viewed as a linear operator mapping into , the space being a linear space equipped with an inner-product (·, ·) inducing a norm defined as follows
where ui represents the i-th component of the vector u.
Generally, systems of linear algebraic equations are commonly solved by using direct methods and iterative methods. Direct methods, such as Gaussian elimination, and LU decomposition work for general matrices with arbitrary structure but require large computer memory. Therefore, they are not computationally efficient and hence unsuitable for solving the 3D PB model of biomolecules, even for small proteins.
Some of the iterative methods such as Richardson, Jacobi, Gauss-Seidel and SOR iterations, also work well for general structured matrices but they are barely employed due to the reduced robustness for large protein system. The classic linear iteration methods for solving Eq. (17) can be viewed as the following form
| (18) |
where B is matrix approximating in some sense. Different construction of matrix B results in a different iterative method. The necessary and sufficient condition for the convergence of algorithm (18) is that the spectrum ρ of the error propagation operator must be smaller than 1, i.e., E = Ih − BLh and ρ(E) < 1 [53], where Ih is the identity operator associated with the grid resolution h. The smaller value of ρ(E) indicates the better convergence of the method. The spectra of this family of iteration methods can be expressed as ρ(E) = 1 − O(h2), which implies that as grid spacing gets smaller, these methods converge more and more slowly. This property severely restricts the wide applications of these methods for large linear systems.
The conjugate gradient (CG) method is a very efficient iterative method if the matrix is symmetric and positive definite. Actually it is the main workhorse of most popular PBE solvers since the matrices from the standard FDMs or FEMs satisfy these good properties. The multigrid (MG) method is an accelerating technique and can be applied in combination with any of commonly used solvers. Using a hierarchy of discretizations, MG shifts the computation between coarser and finer grids by extrapolation and restriction, and thus accelerates the convergence. It is almost the fastest accelerating technique known so far and applied in many popular PB solvers, such as the APBS.
Unfortunately, the matrix Lh from the MIB can barely take advantages from the described methods due to its notoriously complicated structure. For the discretization of the Laplace operator in the PBE by standard FDMs, every grid point except the boundary ones takes the following form:
| (19) |
where i, j, k represent the discretization indices along the x, y, z directions, respectively. The coefficients cm,m = 0, 1, ...6 only depend ε and grid spacing h. The symmetric structure of Eq. (19) and the facts and c1 = c2 = c3 = c4 = c5 = c6 make the whole matrix symmetric and positive definite.
However, since the MIB scheme takes into account the interface treatment and at all the irregular grid points near the interface, discetizations are modified. For the simplest case, assume that only one fictitious point is needed and without the loss of generality, the modification is in the form:
| (20) |
Note that the fictitious value f* is used in Eq. (20) for the smooth extension of the function. The fictitious value f* can further be expanded as the linear combination of the unknown function values.
| (21) |
where is the nearby function values around are the corresponding coefficients. Usually M = 10 in second order MIB scheme for a smooth interface but could be bigger for interface with singularities. The choice of and calculation of totally depend on the local information of the interface. The introduction of the fictitious values gives high accuracy for the interface problems but also ruins the good properties such as symmetry and positive-definiteness of the overall matrix.
To solve the matrices generated from the MIB scheme, the direct methods and regular iterative methods will be ruled out from the beginning due to the poor convergence for huge systems. The CG method also does not work because the unpredictably general matrix structures. Meanwhile, the direct application of the multigrid method, which is an important accelerating technique, also has a potential problem due to the shift of irregular point locations during grid refinement cycles. Ref [16] showed the poor behavior of the algebraic multigrid method (AMD) and proposed a new multigrid scheme based on the local interface problem but the interpolation operator at the interface will cost much extra work.
Therefore, we put more emphasis on looking for suitable solvers and accelerating techniques in the Krylov subspace theory. Stabilized biconjugate gradient method (BiCG) and generalized minimal residual method (GMRES) are two examples in Krylov subspace methods, which deal with the general nonsingular matrix that does not have to be symmetric and positive definite. In the following section, the Krylov subspace (KS) methods and their analysis are briefly introduced. Different types of preconditioners are associated to the KS solvers. These combinations are tested in Section 3 to achieve the optimal convergence rate for solving the linearized or nonlinear MIBPB matrices.
B Krylov subspace method and preconditioning
Suppose u0 is an initial guess for the solution u in system (17) and defines the initial residual r0 = f − Lu0. For notation simplicity, the subscript h is dropped here. As shown in Ref. [43], the Krylov subspace can be derived from the following projection method. The mth iteration um, m = 1, 2, ... is of the form
| (22) |
where is some m-dimensional space, called the search space. Strictly speaking, Eq. (22) is an abused notation, it means that um can be decomposed as the residual r0 and an element in space . Because of m degrees of freedom, a total of m constraints is required to make um unique. This is achieved by choosing an m-dimensional space , called the constraint space, and by requiring that the mth residual is orthogonal to that space, i.e.,
| (23) |
Here the orthogonality is in the sense of the inner product in the Euclidean space.
If the space is defined as the Krylov subspace , i,e,
| (24) |
then the projection method is the so-called Krylov subspace method. More specifically, if , it is the Galerkin method, which includes the CG method and its generalizations, and if , it yields the GMRES. These are the basic idea of Krylov subspace methods.
For the convergence analysis, note that conditions (22) and (23) imply that the error u − um and the residual rm can be written in the polynomial form
| (25) |
where pm is a polynomial of degree at most m and with value one at the origin. Ref. [43] gives the error bound for Krylov subspace methods
| (26) |
where πm denotes the set of polynomials of degree at most m and with value one at the origin, λk are the eigenvalues of the matrix L. It can be concluded from Eq. (26) that the convergence behavior of the Krylov subspace methods is completely determined by their spectra.
However, as indicated in Ref. [43], it is always difficult to really evaluate the upper bound. Alternatively, it states that the condition number of the matrix is a criteria which although, only partially reveals the practice convergence behavior but is easier to calculate. For matrix L, the condition number is defined as the ratio of the extreme eigenvalues or spectra
| (27) |
Since the rate of the convergence of Krylov projection methods for a particular linear system is strongly dependent on its spectrum, preconditioner is typically used to alter the spectrum and hence accelerate the convergence rate of iterative techniques. Preconditioner can be applied to system (17) by
| (28) |
where ML and MR denote the left and right precondition matrices. Usually if MR = I, the left preconditioned results and the residual is given by
| (29) |
Properly preconditioned matrix M−1L may significantly reduce the condition number of L, hence the rate of convergence is accelerated. The commonly used precondition strategies are Jacobi preconditioner, block preconditioner and incomplete LU factorization. However, preconditioning a large sparse system is an empirical exercise. Different preconditioners work better for different kinds of problems. In Section 3, the combinations of different Krylov subspace solvers and preconditioners are investigated, and the rate of convergence is analyzed via the spectra of preconditioned and un-preconditioned matrices.
The accelerated Krylov methods are also crucial for solving the nonlinear Poisson-Boltzmann equation. The discretization of the nonlinear PB equation results in a linear equation system of the form
| (30) |
where the matrix L is still from the MIBPB scheme, the nonlinear term N(·) is diagonal and . The inexact-Newton method is perhaps one of the most efficient ways to solve nonlinear system (30)
| (31) |
| (32) |
where F′ is the Jacobian matrix [∂Fi(u)/∂uj] and takes the form F′(u) = L + N′(u). Here N′ is the Jacobian matrix of N(u) and is also diagonal .
It is easy to see that the inexact-Newton method is a two-layer iterative algorithm. The correction term vn in outer iteration (32) is considered as a rough solution of inner iteration (31). The scheme converges linearly when ηn, the ratio of the residual rn between the function value F(u), is less than 1, and converges super-linearly as the sequence ηn has the property that limn→∞ ηn = 0. In the MIBPB package, accelerated Krylov subspace methods are applied to inner iteration (31) in order to attain fast convergence.
Literature cited
- 1.Baker NA. Poisson-Boltzmann methods for biomolecular electrostatics. Methods in Enzymology. 2004;383:94–118. doi: 10.1016/S0076-6879(04)83005-2. [DOI] [PubMed] [Google Scholar]
- 2.Baker NA. Improving implicit solvent simulations: a Poisson-centric view. Current Opinion in Structural Biology. 2005;15(2):137–43. doi: 10.1016/j.sbi.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Baker NA, Bashford D, Case DA. Implicit solvent electrostatics in biomolecular simulation. In: Leimkuhler B, Chipot C, Elber R, Laaksonen A, Mark A, Schlick T, Schutte C, Skeel R, editors. New Algorithms for Macromolecular Simulation. Springer; 2006. [Google Scholar]
- 4.Baker NA, McCammon JA. Electrostatic interactions. In: Bourne P, Weissig H, editors. Structural Bioinformatics. John Wiley & Sons, Inc.; New York; 2003. pp. 427–440. [Google Scholar]
- 5.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bashford D, Case DA. Generalized Born models of macromolecular solvation e ects. Annual Review of Physical Chemistry. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 7.Beglov D, Roux B. Solvation of complex molecules in a polar liquid: an integral equation theory. Journal of Chemical Physics. 1996;104(21):8678–8689. [Google Scholar]
- 8.Bertonati C, Honig B, Alexov E. Poisson-boltzmann calculations of nonspecific salt e ects on protein-protein binding free energy. Biophysical Journal. 2007;92:1891–1899. doi: 10.1529/biophysj.106.092122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blomberg N, Gabdoulline RR, Nilges M, Wade RC. Classification of protein sequences by homology modeling and quantitative analysis of electrostatic similarity. Proteins. 1999;37(3):379–387. doi: 10.1002/(sici)1097-0134(19991115)37:3<379::aid-prot6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 10.Boschitsch AH, Fenley MO. Hybrid boundary element and finite di erence method for solving the nonlinear Poisson-Boltzmann equation. Journal of Computational Chemistry. 2004;25(7):935–955. doi: 10.1002/jcc.20000. [DOI] [PubMed] [Google Scholar]
- 11.Bramble J, King J. A finite element method for interface problems in domains with smooth boundaries and interfaces. Adv. Comput. Math. 1996;6:109–138. [Google Scholar]
- 12.Brooks BR, Bruccoleri RE, Olafson BD, States D, Swaminathan S, Karplus M. Charmm: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 13.Cai W, Deng SZ. An upwinding embedded boundary method for maxwell's equations in media with material interfaces: 2d case. J. Comput. Phys. 2003;190:159–183. [Google Scholar]
- 14.Chen D, Wei GW, Cong X, Wang G. Computational methods for optical molecular imaging. Communications in Numerical Methods in Engineering. 2009;25:11371161. doi: 10.1002/cnm.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Brooks CL., III Implicit modeling of nonpolar solvation for simulating protein folding and conformational transitions. Physical Chemistry Chemical Physics. 2008;10:471–81. doi: 10.1039/b714141f. [DOI] [PubMed] [Google Scholar]
- 16.Chen T, Strain J. Piecewise-polynomial discretization and krylov-accelerated multigrid for elliptic interface problems. JOURNAL OF COMPUTATIONAL PHYSICS. 2008;16:7503–7542. [Google Scholar]
- 17.Cheng Y, Suen JK, Radi Z, Bond SD, Holst MJ, McCammon JA. Continuum simulations of acetylcholine di usion with reaction-determined boundaries in neuromuscular junction models. Biophysical Chemistry. 2007;127(3):129–39. doi: 10.1016/j.bpc.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chern I-L, Liu J-G, Weng W-C. Accurate evaluation of electrostatics for macromolecules in solution. Methods and Applications of Analysis. 2003;10(2):309–28. [Google Scholar]
- 19.Chiba M, Fedorov DG, Kitaura K. Polarizable continuum model with the fragment molecular orbital-based time-dependent density functional theory. Journal of Computational Chemistry. 2008;29:2667–2676. doi: 10.1002/jcc.21000. [DOI] [PubMed] [Google Scholar]
- 20.Connolly ML. Analytical molecular surface calculation. Journal of Applied Crystallography. 1983;16(5):548–558. [Google Scholar]
- 21.David L, Luo R, Gilson MK. Comparison of generalized Born and Poisson models: Energetics and dynamics of HIV protease. Journal of Computational Chemistry. 2000;21(4):295–309. [Google Scholar]
- 22.Davis ME, McCammon JA. Electrostatics in biomolecular structure and dynamics. Chemical Reviews. 1990;94:509–21. [Google Scholar]
- 23.Dominy BN, Brooks CL., III Development of a generalized Born model parameterization for proteins and nucleic acids. Journal of Physical Chemistry B. 1999;103(18):3765–3773. [Google Scholar]
- 24.Dong F, Vijaykumar M, Zhou HX. Comparison of calculation and experiment implicates significant electrostatic contributions to the binding stability of barnase and barstar. Biophysical Journal. 2003;85(1):49–60. doi: 10.1016/S0006-3495(03)74453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engels M, Gerwert K, Bashford D. Computational studies on bacteriorhodopsin: Conformation and proton transfer energetics. Biophys. Chem. 1995;56:95. doi: 10.1016/0301-4622(95)00020-x. [DOI] [PubMed] [Google Scholar]
- 26.Fedkiw RP, Aslam T, Merriman B, Osher S. A non-oscillatory Eulerian approach to interfaces in multimaterial flows (the ghost fluid method). J. Comput. Phys. 1999;152:457–492. [Google Scholar]
- 27.Feig M, Brooks I, C. L. Recent advances in the development and application of implicit solvent models in biomolecule simulations. Curr Opin Struct Biol. 2004;14:217–224. doi: 10.1016/j.sbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Feig M, Im W, Brooks I, C. L. Implicit solvation based on generalized Born theory in di erent dielectric environments. Journal of Chemical Physics. 2004;120(2):903–911. doi: 10.1063/1.1631258. [DOI] [PubMed] [Google Scholar]
- 29.Gabdoulline RR, Wade RC. Brownian dynamics simulation of protein-protein di usional encounter. Methods-α Companion to Methods in Enzymology. 1998;14(3):329–341. doi: 10.1006/meth.1998.0588. [DOI] [PubMed] [Google Scholar]
- 30.Geng W, Yu S, Wei GW. Treatment of charge singularities in implicit solvent models. Journal of Physical Chemistry. 2007;127:114106. doi: 10.1063/1.2768064. [DOI] [PubMed] [Google Scholar]
- 31.Holst M, Baker N, Wang F. Adaptive multilevel finite element solution of the Poisson-Boltzmann equation I. algorithms and examples. Journal of Computational Chemistry. 2000;21(15):1319–1342. [Google Scholar]
- 32.Holst MJ. Multilevel Methods for the Poisson-Boltzmann Equation. University of Illinois at Urbana-Champaign, Numerical Computing Group; Urbana-Champaign: 1993. [Google Scholar]
- 33.Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268(5214):1144–9. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 34.Im W, Beglov D, Roux B. Continuum solvation model: electrostatic forces from numerical solutions to the Poisson-Boltzmann equation. Computer Physics Communications. 1998;111(1-3):59–75. [Google Scholar]
- 35.Improta R, Barone V, Scalmani G, Frisch MJ. A state-specific polarizable continuum model time dependent density functional theory method for excited state calculations in solution. Journal of Chemical Physics. 2006;125(054103) doi: 10.1063/1.2222364. [DOI] [PubMed] [Google Scholar]
- 36.Jo S, Vargyas M, Vasko-Szedlar J, Roux B, Im W. Pbeq-solver for online visualization of electrostatic potential of biomolecules. Nucleic Acids Research. 2008;36:W270–W275. doi: 10.1093/nar/gkn314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirkwood JG. Theory of solution of molecules containing widely separated charges with special application to zwitterions. J. Comput. Phys. 1934;7:351–361. [Google Scholar]
- 38.Klapper I, Hagstrom R, Fine R, Sharp K, Honig B. Focussing of electric fields in the active site of cu-zn superoxide dismutase: E ects of ionic strength and amino acid modification. Protein. 1986;1:47–59. doi: 10.1002/prot.340010109. [DOI] [PubMed] [Google Scholar]
- 39.Lamm G. The Poisson-Boltzmann equation. In: Lipkowitz KB, Larter R, Cundari TR, editors. Reviews in Computational Chemistry, pages 147–366. John Wiley and Sons, Inc.; Hoboken, N.J.: 2003. [Google Scholar]
- 40.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 41.LeVeque RJ, Li ZL. The immersed interface method for elliptic equations with discontinuous coe cients and singular sources. SIAM J. Numer. Anal. 1994;31:1019–1044. [Google Scholar]
- 42.Li ZL, Lin T, Wu XH. New Cartesian grid methods for interface problems using the finite element formulation. Numerische Mathematik. 2003;96:61–98. [Google Scholar]
- 43.Liesen J, Tichy P. Convergence analysis of krylov subspace methods. CGAMM-Mitteilungen. 2004;27:153–173. [Google Scholar]
- 44.Livesay DR, Jambeck P, Rojnuckarin A, Subramaniam S. Conservation of electrostatic properties within enzyme families and superfamilies. Biochemistry. 2003;42(12):3464–3473. doi: 10.1021/bi026918f. [DOI] [PubMed] [Google Scholar]
- 45.Lu BZ, Chen WZ, Wang CX, Xu XJ. Protein molecular dynamics with electrostatic force entirely determined by a single poisson-boltzmann calculation. Proteins. 2002;48(3):497–504. doi: 10.1002/prot.10172. [DOI] [PubMed] [Google Scholar]
- 46.Luo R, David L, Gilson MK. Accelerated Poisson-Boltzmann calculations for static and dynamic systems. Journal of Computational Chemistry. 2002;23(13):1244–53. doi: 10.1002/jcc.10120. [DOI] [PubMed] [Google Scholar]
- 47.Madura JD, Briggs JM, Wade RC, Davis ME, Luty BA, Ilin A, Antosiewicz J, Gilson MK, Bagheri B, Scott LR, McCammon JA. Electrostatics and di usion of molecules in solution - simulations with the University of Houston Brownian Dynamics program. Computer Physics Communications. 1995;91(1-3):57–95. [Google Scholar]
- 48.Massova I, Kollman PA. Computational alanine scanning to probe protein-protein interactions: A novel approach to evaluate binding free energies. Journal of the American Chemical Society. 1999;121(36):8133–43. [Google Scholar]
- 49.Mongan J, Simmerling C, McCammon JA, Case DA, Onufriev A. Generalized Born model with a simple, robust molecular volume correction. Journal of Chemical Theory and Computation. 2007;3(1):159–69. doi: 10.1021/ct600085e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nielsen JE, Vriend G. Optimizing the hydrogen-bond network in Poisson-Boltzmann equation-based pK(a) calculations. Proteins. 2001;43(4):403–412. doi: 10.1002/prot.1053. [DOI] [PubMed] [Google Scholar]
- 51.Nina M, Im W, Roux B. Optimized atomic radii for protein continuum electrostatics solvation forces. Biophysical Chemistry. 1999;78(1-2):89–96. doi: 10.1016/s0301-4622(98)00236-1. [DOI] [PubMed] [Google Scholar]
- 52.Onufriev A, Bashford D, Case DA. Modification of the generalized Born model suitable for macromolecules. Journal of Physical Chemistry B. 2000;104(15):3712–3720. [Google Scholar]
- 53.Ortega JM. In Numerical Analysis: A second course. Academic Press; NY: 1999. [Google Scholar]
- 54.Peskin CS. Numerical analysis of blood flow in the heart. Journal of Computational Physics. 1977;25(3):220–52. [Google Scholar]
- 55.Petrey D, Honig B. GRASP2: Visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods in Enzymology. 2003;374:492–509. doi: 10.1016/S0076-6879(03)74021-X. [DOI] [PubMed] [Google Scholar]
- 56.Prabhu NV, Panda M, Yang QY, Sharp KA. Explicit ion, implicit water solvation for molecular dynamics of nucleic acids and highly charged molecules. J. Comput. Chem. 2008;29:1113–1130. doi: 10.1002/jcc.20874. [DOI] [PubMed] [Google Scholar]
- 57.Richards FM. Areas, volumes, packing, and protein structure. Annual Review of Biophysics and Bioengineering. 1977;6(1):151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 58.Roux B, Simonson T. Implicit solvent models. Biophysical Chemistry. 1999;78(1-2):1–20. doi: 10.1016/s0301-4622(98)00226-9. [DOI] [PubMed] [Google Scholar]
- 59.Sakurai K, Oobatake M, Goto Y. Salt-dependent monomerdimer equilibrium of bovine α-lactoglobulin at ph 3. Protein Sci. 2001;10:2325–2335. doi: 10.1110/ps.17001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanner MF, Olson AJ, Spehner JC. Reduced surface: An e cient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 61.Sept D, Elcock AH, McCammon JA. Computer simulations of actin polymerization can explain the barbed-pointed end asymmetry. Journal of Molecular Biology. 1999;294(5):1181–1189. doi: 10.1006/jmbi.1999.3332. [DOI] [PubMed] [Google Scholar]
- 62.Sharp KA, Honig B. Electrostatic interactions in macromolecules - theory and applications. Annual Review of Biophysics and Biophysical Chemistry. 1990;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- 63.Simonson T. Macromolecular electrostatics: continuum models and their growing pains. Current Opinion in Structural Biology. 2001;11(2):243–252. doi: 10.1016/s0959-440x(00)00197-4. [DOI] [PubMed] [Google Scholar]
- 64.Song Y, Zhang Y, Bajaj CL, Baker NA. Continuum di usion reaction rate calculations of wild-type and mutant mouse acetylcholinesterase: Adaptive finite element analysis. Biophysical Journal. 2004;87(3):1558–66. doi: 10.1529/biophysj.104.041517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song Y, Zhang Y, Shen T, Bajaj CL, McCammon JA, Baker NA. Finite element solution of the steady-state Smoluchowksi equation for rate constant calculations. Biophysical Journal. 2004;86(4):2017–2029. doi: 10.1016/S0006-3495(04)74263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Swanson JMJ, Henchman RH, McCammon JA. Revisiting free energy calculations: A theoretical connection to MM/PBSA and direct calculation of the association free energy. Biophysical Journal. 2004;86(1):67–74. doi: 10.1016/S0006-3495(04)74084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Swanson JMJ, Mongan J, McCammon JA. Limitations of atom-centered dielectric functions in implicit solvent models. Journal of Physical Chemistry B. 2005;109(31):14769–72. doi: 10.1021/jp052883s. [DOI] [PubMed] [Google Scholar]
- 68.Tang CL, Alexov E, Pyle AM, Honig B. Calculation of pKas in RNA: On the structural origins and functional roles of protonated nucleotides. Journal of Molecular Biology. 2007;366(5):1475–96. doi: 10.1016/j.jmb.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 69.Tomasi J, Mennucci B, Cammi R. Quantum mechanical continuum solvation models. Chem. Rev. 2005;105:2999–3093. doi: 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- 70.Tsui V, Case DA. Molecular dynamics simulations of nucleic acids with a generalized Born solvation model. Journal of the American Chemical Society. 2000;122(11):2489–2498. [Google Scholar]
- 71.Tsui V, Case DA. Calculations of the absolute free energies of binding between RNA and metal ions using molecular dynamics simulations and continuum electrostatics. Journal of Physical Chemistry B. 2001;105(45):11314–11325. [Google Scholar]
- 72.Wallis R, Moore GR, James R, Kleanthous C. Protein-protein interactions in colicin e9 dnase-immunity protein complexes. 1. di usion-controlled association and femtomolar binding for the cognate complex. Biochemistry. 1995;34:13743–13750. doi: 10.1021/bi00042a004. [DOI] [PubMed] [Google Scholar]
- 73.Warwicker J, Watson HC. Calculation of the electric potential in the active site cleft due to alpha-helix dipoles. Journal of Molecular Biology. 1982;157(4):671–9. doi: 10.1016/0022-2836(82)90505-8. [DOI] [PubMed] [Google Scholar]
- 74.Yu S, Geng W, Wei GW. Treatment of geometric singularities in implicit solvent models. Journal of Chemical Physics. 2007;126:244108. doi: 10.1063/1.2743020. [DOI] [PubMed] [Google Scholar]
- 75.Yu S, Wei GW. Three-dimensional matched interface and boundary (MIB) method for treating geometric singularities. Journal of Computational Physics. 2007;227:602–632. [Google Scholar]
- 76.Yu S, Zhou Y, Wei GW. Matched interface and boundary (MIB) method for elliptic problems with sharp-edged interfaces. Journal of Computational Physics. 2007;224(2):729–756. [Google Scholar]
- 77.Yu SN. Matched interface and boundary (MIB) method for geometric singularities and its application to molecular biology and structural analysis. 2007. Dissertation of Michigan State University.
- 78.Yu SN, Xiang Y, Wei GW. Matched interface and boundary (MIB) method for the vibration analysis of plates. Communications in Numerical Methods in Engineering. in press. [Google Scholar]
- 79.Zauhar RJ, Morgan RS. A new method for computing the macromolecular electric potential. Journal of Molecular Biology. 1985;186(4):815–20. doi: 10.1016/0022-2836(85)90399-7. [DOI] [PubMed] [Google Scholar]
- 80.Zhao S. High order vectorial analysis of waveguides with curved dielectric interfaces. IEEE Microwave and Wireless Components Letters. 2009;19:266–268. [Google Scholar]
- 81.Zhao S, Wei GW. High-order FDTD methods via derivative matching for maxwell's equations with material interfaces. Journal of Computational Physics. 2004;200(1):60–103. [Google Scholar]
- 82.Zhao S, Wei GW. Matched interface and boundary (MIB) for the implementation of boundary conditions in high-order central finite di erences. Int. J. Numer. Meth. Engng. 2009;77:1690–1730. doi: 10.1002/nme.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou YC, Feig M, Wei GW. Highly accurate biomolecular electrostatics in continuum dielectric environments. Journal of Computational Chemistry. 2008;29:87–97. doi: 10.1002/jcc.20769. [DOI] [PubMed] [Google Scholar]
- 84.Zhou YC, Wei GW. On the fictitious-domain and interpolation formulations of the matched interface and boundary (MIB) method. Journal of Computational Physics. 2006;219(1):228–246. [Google Scholar]
- 85.Zhou YC, Zhao S, Feig M, Wei GW. High order matched interface and boundary method for elliptic equations with discontinuous coe cients and singular sources. Journal of Computational Physics. 2006;213(1):1–30. [Google Scholar]
- 86.Zhu J, Alexov E, Honig B. Comparative study of generalized Born models: Born radii and peptide folding. Journal of Physical Chemistry B. 2005;109(7):3008–22. doi: 10.1021/jp046307s. [DOI] [PubMed] [Google Scholar]