Abstract
Background
A low level of response to alcohol during an individual’s early experience with alcohol is associated with an increase risk for alcoholism. A family-based genome-wide linkage analysis using sibling pairs that underwent an alcohol challenge where the level of response to alcohol was measured with the Subjective High Assessment Scale (SHAS) implicated the 10q terminal region. CYP2E1, a gene known for its involvement with ethanol metabolism, maps to this region.
Methods
Variance component multipoint linkage analysis was performed on a combined map of single nucleotide polymorphism (SNP) and microsatellite data. To account for the heterogeneity evident in the dataset, a calculation assuming locus heterogeneity was made using the HLOD (heterogeneity LOD) score. Association between SNP marker allele counts and copy number and SHAS scores were evaluated using a logistic regression model.
Results
Linkage analysis detected significant linkage to CYP2E1 which was diminished due to apparent locus heterogeneity traced to a single family with extreme phenotypes. In retrospect, circumstances recorded during testing for this family suggest that their phenotype data are likely to be unreliable. Significant allelic associations were detected for several CYP2E1 polymorphisms and the SHAS score. DNA sequencing from families that contributed the greatest evidence for linkage did not detect any changes directly affecting the primary amino acid sequence. With the removal of a single family, combined evidence from microsatellites and SNPs offer significant linkage between the level of response to alcohol and the region on the end of chromosome 10.
Conclusion
Combined linkage and association indicate that sequence changes in or near CYP2E1 affect the level of response to alcohol providing a predictor of risk for alcoholism. The absence of coding sequence changes indicates that regulatory sequences are responsible. Implicating CYP2E1 in the level of response to alcohol allows inferences to be made about how the brain perceives alcohol.
Keywords: level of response to alcohol, linkage analysis, association analysis, combined linkage and association, CYP2E1, locus heterogeneity
Introduction
While a number of phenotypic factors can affect the risk for alcoholism, one of the most studied endophenotypes is an individual’s level of response to alcohol during their early experience with alcohol (Heath et al., 1999). The level of response to alcohol can be reliably measured with the Subjective High Assessment Scale (SHAS) during an alcohol challenge or by the Self-Rating of the Effects of Alcohol (SRE) which uses recall to establish the number of drinks required to reach an effect. Children of alcoholics have a greater risk for alcoholism when they have a lower level of response (Pollock, 1992; Schuckit et al., 2000; Schuckit and Smith, 1996). A low level of response established early in an individual’s drinking career can lead to higher future drinking levels (Ehlers et al., 1999; Heath et al., 1999; Volavka et al., 1996). Populations at historically higher risk for alcoholism, such as Native American or Korean, need to consume larger amounts of alcohol to become intoxicated (Garcia-Andrade et al., 1997; Luczak et al., 2002; Wall et al., 1999) compared to those with lower risk (Monteiro et al., 1991) who exhibit a more intense level of response to alcohol. Several studies have implicated genes showing association with the level of response to alcohol (GABA, 5-HT, and KCNMA1) (Barr et al., 2003; Dick et al., 2006; Schuckit et al., 2005). The evidence proving association for these genes is weak by current standards that have been developed as a consequence from the technological advances enabling genome-wide association studies. Even though these genes may affect the level of response to alcohol to some degree, it is possible that these reported associations reflect the typical reporting bias seen in candidate gene studies.
Initially, data were collected from 139 sibling pairs (Wilhelmsen et al., 2003). Variance component analysis found a significant LOD (Log of Odds) score peak of 3.2 for the SHAS score at the 10q terminal region. Of the genes located at 10qter, CYP2E1 has a known involvement with ethanol metabolism. The CYP2E1 enzyme metabolizes ethanol and acetaminophen, as well as many toxicologic and carcinogenic compounds and can be induced by ethanol and nicotine (Tanaka et al., 2000). In the second stage of the study, when 99 newly collected sibling pairs were added (Schuckit et al., 2005), the peak at 10qter was significantly diminished. As will be described in this paper, it was initially assumed that the diminishment was due to locus heterogeneity, but ultimately the reduced evidence for linkage was explained by a single family with extreme and unreliable phenotypes.
Most of the ethanol that is consumed is oxidized by the liver using alcohol dehydrogenase (ADH). At the high concentrations associated with chronic alcohol consumption, metabolism of ethanol to acetaldehyde increases while the subsequent conversion into acetate is decreased, leading to even higher levels of acetaldehyde. It was shown in rats that chronic consumption reduced the oxidation of acetaldehyde in the liver, thus providing an explanation for the high blood acetaldehyde levels measured after chronic use in human subjects (Lieber, 1999). Acetaldehyde is toxic and highly reactive (Zakhari, 2006), binding to nearby proteins thus creating an antibody response, decreased DNA repair, and glutathione depletion ultimately reducing the ability of the liver to clear free radicals (Lieber, 1997). As a result from the oxidation, NAD+ is reduced with the addition of an electron to form NADH (Zakhari, 2006) used by mitochondria for ATP synthesis. At high concentrations, ethanol is oxidized by ADH at a higher rate leading to an increase in the NADH/NAD+ ratio (Zakhari, 2006).
CYP2E1 is part of the Microsomal Ethanol Oxidizing System (MEOS) accounting for up to 10% of ethanol oxidation in the liver (Tanaka et al., 2000). Once the ADH pathway becomes saturated due to high ethanol concentrations, the MEOS pathway activity increases (Lieber, 1997). By the MEOS pathway, CYP2E1 metabolizes ethanol and other substrates into toxic metabolites creating free radicals in the form of reactive oxygen (O2) intermediates creating oxidative stress leading to liver damage. CYP2E1 uses O2 to oxidize ethanol to aldehyde and NADPH to NADP+. While generally used biosynthetically, NADPH can be regenerated from NADP+ with the conversion of NADH to NAD+. In the absence of NADPH, oxidation of ethanol to aldehyde by CYP2E1 results in superoxides (Zakhari, 2006). An excess of reduced NADH in addition to the increased activity of hydrogen shuttles in mitochondria, results in an increased intake of electrons leading to an increase of superoxide anions (Seitz and Stickel, 2007). The increased creation of Reactive Oxygen Species, or ROS, as a result from the shift in cellular redox state, coupled with the reduced ability to clear these free radicals, due to the increase in acetaldehyde, is thought to be a major driving force in the development of alcohol related liver disease.
The catalase pathway can oxidize ethanol in conjunction with hydrogen peroxide generating systems, such as NADPH oxidase (Zakhari, 2006). The catalase pathways plays a larger roles in the oxidation of ethanol in the brain, where little ADH oxidation occurs (Zakhari, 2006). In a study by Vasiliou et al. (2006), it was found that animals with a knockout of either catalase or CYP2E1 were more sensitive to the sedative effects of ethanol than control, wild-type animals. The study found that CYP2E1 did not contribute significantly to ethanol clearance in the brain, but was instead involved with ethanol processing in the brain affecting sensitivity.
High ethanol concentrations can interfere with the ability of CYP2E1 to metabolize other substrates due to competition from the shared oxidation pathway leading to reduced drug clearance and elevated drug concentrations (Tanaka et al., 2000). The interaction of certain drugs with alcohol will lead to a long-lasting, enhanced drug effect, often leading to overdose. A similar relationship is thought to exist with nicotine. It has been shown that smokers have a more rapid ethanol clearance than non-smokers, suggesting a biological basis for the correlation of tobacco and alcohol consumption seen in alcoholics (Schoedel and Tyndale, 2003).
A number of polymorphisms in CYP2E1 have been tested in relation to alcoholism and a number of related disorders, including many types of cancer, with varying, often conflicting, results. Carriers of the c2 allele of CYP2E1*5B have increased risk for alcoholic liver disease and are more likely to consume excessive amounts of alcohol possibly due to the higher transcriptional levels of CYP2E1 seen with this allele (Grove et al., 1998; Pirmohamed et al., 1995; Watanabe et al., 1994). Variants in the gene have been implicated in the increased risk of different types of cancer relating to the respiratory and digestive systems (Bouchardy et al., 2000; Liu et al., 2001; Yu et al., 1995).
Due to the previously described relationship between CYP2E1 and the metabolism of ethanol and positive linkage results concerning the level of response to alcohol in relation to alcoholism, a number of single nucleotide polymorphisms (SNP) were genotyped in the CYP2E1 to further elucidate the gene’s role in alcohol response. Both genotype and copy number were tested for association with the level of response to alcohol as measured by the SHAS questionnaire. Combined linkage and association analysis was performed to determine whether a single marker or haplotype could account for the linkage signal seen at 10qter.
Methods
Alcohol Challenge
The data collection protocol was approved by the Human Subjects Protection Committee at the University of California in San Diego and used written, informed consent. The design for the alcohol challenge is fully described in the initial report by Wilhelmsen et al. (2003). Male and female subjects ranging in age from 18 to 29 years old were recruited from a population of college students. Chosen sibling pairs reported having an alcohol dependent parent, but were not alcohol dependent themselves. The siblings included 43.7% males and 56.3% females. They had an average age of 22.4 years and 14.2 years of education. 72.2% were Caucasian, 20.0% were Hispanic, and 7.8% were African-American. For 85.0% of the subjects, the alcohol-dependent parent was the father, whereas for 4.4% it was the mother, in 4.0% it was both parents, and in 6.6% the more intensive interview revealed that neither parent met full criteria for dependence.
To measure each participant’s response to alcohol, the Subjective High Assessment Scale (SHAS) questionnaire was administered. For the challenge, each subject was given 8 minutes to consume a 20% by volume solution of 95% ethanol, at 0.75 ml/kg for women and 0.9 ml/kg for men. Baseline levels for each score (SHAS, body sway, and breath alcohol level) were measured prior to the challenge and then were measured at multiple set time intervals throughout the 3 hour challenge. Ultimately the changes in SHAS score and body sway at 1 hour after the challenge were used as phenotypes for the genome-wide genetic analysis. Genotyping was performed on 811 microsatellite markers across the genome.
Taqman Genotyping
Genomic DNA was extracted from whole peripheral blood samples. Genotyping was performed on 10 SNPs with Taqman genotyping assays with locus specific PCR primers and fluorescent allele specific probes designed by Applied Biosystems. Standard Taqman protocol was followed and endpoint amplification intensity was measured by the 7900 ABI Sequence Detector. The position of the genotyped markers in relation to CYP2E1 can be found in Figure 1. The HapMap Consortium reported three major haplotypes in the Caucasian population, as seen in Figure 1, which could be distinguished by the initial two SNPs that were genotyped. Table 4 lists the names and positions of the genotyped SNPs.
Figure 1. Location of genotyped SNPs in relation to CYP2E1 on chromosome 10.

Figure 1 The top of the figure shows the position on chromosome 10 with each SNP location indicated by triangles. The middle part of the figure shows the position of CYP2E1 with exons represented as yellow rectangles and introns as the lines between. At the bottom, phased haplotypes derived from the HapMap Caucasian (CEU) population are shown. Each vertical block represents a SNP genotyped in HapMap. Not all of these markers were genotyped in the study, so vertical black lines through the haplotype figure indicate actual genotyped SNPs.
Table 4. Translation of identification values for genotyped SNPs and position.
A listing of the various identification names for the SNPs genotyped in the study based on the Applied Biosystems ID. Included under “other names” are names commonly used for specific markers.
| rs ID | ABI Assay ID | Build 129 position (bp) |
Other names |
|---|---|---|---|
| rs10776687 | hCV2431881 | 135184332 | |
| rs9418990 | hCV2431878 | 135187956 | |
| rs2070673 | hCV2431871 | 135190557 | CYP2E1*7_-333T>A |
| -- | hCV30633979 | 135192024 | CYP2E1*2,g.1132G>A |
| rs943975 | hCV7468406 | 135192250 | |
| rs6413421 | hCV25594214 | 135195801 | |
| rs915909 | hCV7468401 | 135197387 | CYP2E1_6498C>T(I321I) |
| rs2515641 | hCV16026002 | 135201352 | CYP2E1_10463T>C(F421F) |
| rs2480258 | hCV2431850 | 135202090 | |
| rs2249695 | hCV2431848 | 135202158 |
Copy number analysis
The copy number of CYP2E1 was determined for each sample in quadruplicate through the amplification of both a probe specific to CYP2E1 and a standard probe by real-time PCR using the standard Gene Dosage protocol provided by Applied Biosystems (Livak and Schmittgen, 2001). Preliminary amplification showed the two probes used for analysis had different efficiencies of amplification, which was corrected by a standard dilution curve added to each plate. The fold increase after n number of cycles was calculated by (efficiency)n and the ratio of this increase between the target and reference genes provided copy number. Standard copy number quantification assumes equal amplification efficiencies, but this is not always a valid assumption. Correcting for even small differences in amplification efficiencies leads to less variability among the quadruplicate samples and lowered standard error in overall copy number determination.
CYP2E1 resequencing
Index cases from the 96 families with the greatest evidence for linkage to the 10q terminal region were selected for resequencing. Each coding sequence exon was resequenced using primers from Applied Biosystems using the standard provided procedure.
Linkage allowing for heterogeneity
A map of the positions of the genotyped SNPs relative to the microsatellite markers was created with Fastlink. Variance component methods were used to recalculate LOD scores using SOLAR v4.0.7 with the identity by descent provided through pedigree information and estimating multipoint identity by descent sharing probabilities (Almasy and Blangero, 1998). Variance component linkage analysis uses correlation in the phenotype to partition out variance between relative pairs into the effects of the genes in the region of interest, additive genetic effects of other genes, and non-shared environmental variance. To account for the heterogeneity evident in the dataset, a calculation assuming locus heterogeneity was made using the HLOD (heterogeneity LOD) score to identify the cause of the lowered peak at the end of chromosome 10.
Association analysis
Association between SNP marker allele counts and copy number and SHAS scores were evaluated using a logistic regression model through the SAS statistical package testing for statistical inferences using a generalization of the standard linear model. Family ID and marker genotype were used as classification variables and the effects of copy number and genotype were modeled against the SHAS score. Genotypes were classified based in the count of the minor frequency allele.
Combined Linkage and Association
Combined linkage and association analysis using SOLAR v4.0.7 was performed to include identity by state information similar to the approach used by Almasy et al. (1999) where the variance in the SHAS score that cannot be accounted for by the covariate parameter based on the number of minor alleles was decomposed into the standard variance components. To see whether the linkage signal could be explained by the allele effects of a single SNP, each SNP marker was individually tested by including the number of minor alleles as a covariate. Polygenic covariate screening was used to calculate the significance level after the inclusion of any particular SNP and multipoint analysis was used to calculate the multipoint LOD score for the SHAS score.
By combining linkage and association approaches, a disease loci position can be confined to a region finer than linkage analysis alone and avoid false positive association results due to admixture. Assuming the linkage is not over-estimated, if a measured variant is the actual functional variant affecting the phenotype and no other variants nearby confer any additional risk, linkage analysis conditional on the genotype of such a variant should provide no evidence for linkage. However if the suspected variant is in some degree of linkage disequilibrium with the actual causal variant, the evidence for linkage will be reduced proportional to the degree of LD.
Results
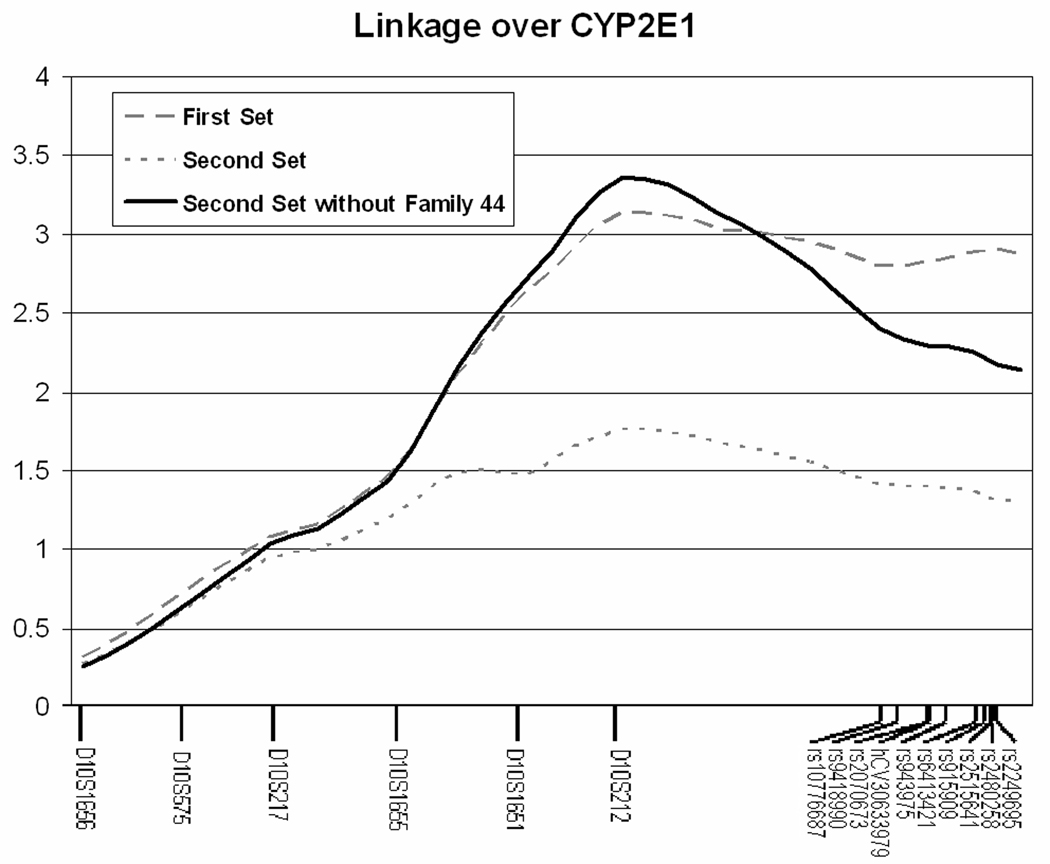
With a combined map of microsatellite and SNP markers the dataset was reanalyzed, as described in the original linkage study (Schuckit et al., 2005; Wilhelmsen et al., 2003), using SOLAR. When divided into the two stages of sample sets from the previous study, significant linkage was found in stage 1 samples with a peak LOD score of 3.14; however, when combining the initial 139 sibling pairs with the additional 99 sibling pairs in stage 2, the linkage signal lowered to a peak LOD score of 1.61. The LOD score plot can be seen in Figure 2.
Figure 2. LOD score plot showing linkage in the region surrounding CYP2E1.
Figure 2 Shows the LOD score plot highlighting the linkage near CYP2E1. The line labeled as First Set represents the initial 139 sibling pairs. The line labeled as Second Set represents the complete set of 238 sibling pairs. Once the family with questionable phenotypes was removed, the strength of the linkage signal was restored to the level provided by the First Set samples. Locations of microsatellite markers and SNPs are shown on the X-axis.
Multipoint linkage analysis allowing for locus heterogeneity was performed for the SHAS phenotype using microsatellite and SNP data from 10qter. The family specific heterogeneity score (α) and LOD score was inspected for each family. Most of the families had α scores of 0.99, one family stood out with an α of 0.37 indicating poor support for linkage to the region. The individual pedigree LOD score for this family alone was −0.97 accounting for most of the reduction in LOD score seen between stage 1 and stage 2. Based on the level of alleles shared between siblings in this family, it was expected that these siblings should have more similar SHAS scores. The sibling pair had a large difference in phenotype despite having inherited the same chromosome Identical By Descent from both of their parents (IBD of 2). While one sibling reported a SHAS score of 26.75 (z = 2.147), the other reported a SHAS score of 4 (z = −0.7161).
Comments from the observers of the study propose phenotyping error as the cause of the extreme phenotype difference. The more sensitive sibling felt nauseated during the challenge. This is a common response reported by many subjects, and the SHAS score for this individual is around the same value reported by other individuals with a similar response. The other sibling, with SHAS scores indicating they were insensitive to alcohol, had a blood alcohol level close to the predicted value, indicating that they were appropriately dosed. This sibling fainted briefly during blood draw. The researchers involved in testing these subjects take special care to avoid fainting and report that it only happens 2% of the time. Although the subject woke up quickly and admittedly felt fine, this fainting spell could have contributed to the low response indicated by a low SHAS score. Therefore it is likely that the reduction in LOD score by the inclusion of the discordant family is due to a phenotyping error. While the inclusion of this family has dramatic affect on the linkage analysis it has a negligible affect on the association analysis. Due to irregularity from the reported testing, the whole family was removed from analysis. When the single family was removed, the original LOD score peak was reestablished with the maximum LOD score for 10qter at 3.40.
SNP genotypes could not be called automatically using the manufacture supplied program due to copy number differences which affected the allele signal intensities causing samples to fall between the heterozygous and homozygous genotypes. Genotype calls were made manually, assuming that the intermediate samples were either AAB or ABB depending on the location relative to the heterozygote cluster. The best estimate of copy number and genotype was made by integrating the real-time PCR measured copy number, Taqman derived genotype and pedigree structure. While the majority of subjects (85%) have 2 copies, 11% had 3 copies, and 4% had 1 copy. A small number (<2%) were considered to have greater than 3 copies based on real time PCR, but these measurements were not considered plausible based on Taqman derived genotype and pedigree information.
Logistic regression analysis using a mixed model to control for the relatedness of subjects within families was used to investigate the association between copy number genotype and level of response to alcohol. Copy number had little effect on the level of response to alcohol. The presence of at least one copy of a relatively rare allele for several SNPs is associated with a more intense response to alcohol. The best evidence for association was found for the first three markers, which lie near the beginning of the gene near the promoter region, when considering genotype alone. The SNP rs10776687 showed the greatest evidence for association with a p value of 0.007 and was able to explain 4.6% of the variance of the SHAS score phenotype (r2=0.046). In this case, copy number was ignored and all genotypes were assigned as biallelic. When considering copy number and genotype together, none of the markers were significant. Copy number alone (1, 2, or 3) was not significantly associated. Interestingly, when modeling the effect of genotype and copy number on the number of cigarettes smoked per day, copy number of CYP2E1 was associated with this smoking phenotype with an average p value of 0.014. Association p values for logistic regression analysis are shown in Table 1.
Table 1. Association p values for logistic regression analysis between the SHAS score and CYP2E1 genotype alone or genotype considering copy number.
Logistic regression was used to test for association between the genotyped SNPs and the SHAS quantity representing the level of response to alcohol. P-values < 0.05 are in bold. The three markers near the 3’ end and two from the 5’ end were most associated with the level of response to alcohol. The best evidence for association came from the initial SNP, rs10776687. None of the SNPs were associated when the genotype call was made considering copy number.
| Genotype | Genotype considering copy number | Minor allele frequency |
|
|---|---|---|---|
| rs10776687 | 0.007 | 0.103 | 0.056 |
| rs9418990 | 0.024 | 0.125 | 0.244 |
| rs2070673 | 0.015 | 0.077 | 0.238 |
| hCV30633979 | 0.215 | 0.253 | 0.004 |
| rs943975 | 0.182 | 0.274 | 0.131 |
| rs6413421 | 0.133 | 0.123 | 0.057 |
| rs915909 | 0.058 | 0.081 | 0.007 |
| rs2515641 | 0.45 | 0.261 | 0.176 |
| rs2480258 | 0.04 | 0.187 | 0.252 |
| rs2249695 | 0.024 | 0.139 | 0.268 |
Intuitively we would expect that inclusion of a causal SNP as a covariate would reduce the residual linkage due to IBD to zero (Almasy et al., 1999), but this is an area of active research. After each SNP marker was separately tested in the variance component model, it was found that inclusion of the number of minor alleles of any single SNP was not able to explain all variation in the SHAS phenotype. When considering the dataset after removing family 44, the peak LOD score was 3.36. The marker that lowered this LOD score the most (by 1.21 LOD units) when included as a covariate was rs10776687, the marker located closest to the linkage peak. Other SNPs lowered the LOD score by lesser amounts, as seen in the Table 2 below. Combined linkage and association analysis indicates that no single locus tested is likely to be the only causal allele. When testing haplotypes, none of the three haplotypes were able to completely account for the linkage signal.
Table 2. Results of variance component linkage analysis for combined linkage and association.
Combined linkage and association analysis showed that a single marker was unable to account for all of the variation in the signal. This was accomplished by adding each SNP individually into the model as a covariate. With no covariates, the LOD score was 3.36. The SNP that lowered the score the most when added as a covariate was rs10776687and was able to explain 5.3% of the variance in the SHAS score.
| Covariate | LOD score |
link+assoc | covar p | Variance |
|---|---|---|---|---|
| None | 3.36 | |||
| rs10776687 | 2.15 | 4.85 | 4.25E-04 | 0.0531 |
| rs9418990 | 2.46 | 4.13 | 5.65E-03 | 0.021 |
| rs2070673 | 2.54 | 4.25 | 5.04E-03 | 0.0311 |
| rs943975 | 2.23 | 2.87 | 8.74E-02 | 0.0105 |
| rs2515641 | 2.88 | 3.69 | 5.30E-02 | 0.0076 |
| rs2480258 | 2.42 | 3.77 | 1.26E-02 | 0.0268 |
| rs2249695 | 2.54 | 3.99 | 9.58E-03 | 0.02 |
In addition to testing SNP markers to determine whether a single SNP could directly influence the SHAS phenotype, a number of smoking and drinking phenotypes were analyzed by including them as covariates when modeling their relationship with SHAS. Three of these phenotypes (average number of cigarettes per day, the maximum amount drank in one day, and the average number of drinks consumed on days the subject consumed alcohol) improved the overall peak LOD score compared to the model without covariates. The other tested phenotypes (number of days the subject drank in the last week, amount consumed 24 hours prior to challenge, number of days of smoking in a month, and number of cigarettes smoked on days where the subject smoked), lowered the peak LOD score presumably because these phenotypes are correlated with the SHAS score. All LOD scores are shown in the Table 3 below.
Table 3. Results of variance component linkage analysis with the inclusion of several drinking and smoking covariates.
A number of smoking and drinking phenotypes were analyzed by including them as covariates when modeling their relationship with SHAS. Three of these phenotypes (average number of cigarettes per day, the maximum amount drank in one day, and the average number of drinks consumed on days the subject drank) improved the overall peak LOD score compared to the model without covariates.
| covariate LOD |
link+assoc LOD |
variance | |
|---|---|---|---|
| number of days in the last week where subject drank | 2.19 | 5.07 | 0.078 |
| amount consumed in the last 24 hours | 2.82 | 3.68 | 0.028 |
| days smoking per month in previous 6 months | 3.03 | 4.81 | 0.046 |
| cigarettes per day, on smoking days in previous 6 months | 3.27 | 4.95 | 0.036 |
| Average number of cigarettes per day | 3.37 | 4.49 | 0.026 |
| maximum amount drank in one day | 3.47 | 6.92 | 0.084 |
| Average number of drink on days they drank | 3.77 | 6.24 | 0.066 |
To examine whether previously unidentified missense sequence changes could be responsible for the association detected, coding sequence exons were resequenced for 96 index cases from the families with the strongest evidence of linkage. No missense changes were found and no novel polymorphisms were identified.
Discussion
Alcoholism is a complex disease with potentially many genetic influences. Investigators have tried to minimize heterogeneity by choosing a narrowly defined phenotype such as the level of response to alcohol that was measured with the SHAS score in this study. Significant evidence for linkage to 10qter was observed for the SHAS score in subjects from an alcohol challenge only after the removal of one family that retrospectively should not have been included in the analysis. After linkage was found at the end of chromosome 10, several SNPs genotyped in CYP2E1 were associated through logistic regression with the level of response to alcohol as reported by the SHAS score. Copy number did not appear to affect the SHAS score even though copy number differences were found between individuals across the CYP2E1 gene.
When considered separately, linkage was found over the region containing CYP2E1 and SNPs from the gene were found to be associated with the SHAS score. If inclusion of the causal variant as a covariate in variance component analysis always reduces the residual LOD score to zero, we were unable to implicate a causal variant directly influencing the level of response to alcohol in the families studied. It is possible that an unidentified polymorphism nearby could play a causal role in the level of response to alcohol as the degree of signal reduction is largest in the marker closest to the linkage peak and then declines for markers farther away. Another possibility is that a single marker cannot account for the entire linkage signal because many markers in the region play a role in the response. Instead of a single polymorphism causing variation, combinations of polymorphisms across the region may work together to contribute to the variation seen in our dataset. Support from the heterogeneity LOD score calculation showing that all families showed evidence for linkage combined with the independently derived association analysis imply that the LOD score peak was not over-estimated. It still can be concluded the regulatory sequences near CYP2E1 appear to play a role in the level of response to alcohol.
Variance component linkage analysis for the level of response to alcohol was significantly affected by including covariates for recent drinking and smoking behavior. Since CYP2E1 expression is inducible by alcohol and nicotine (Joshi and Tyndale, 2006), this further supports the role of CYP2E1 in level of response to alcohol. CYP2E1 represents a metabolic intersection between these substances of abuse (Schoedel and Tyndale, 2003). It was initially surprising that while an association was not found between the level of response to alcohol and copy number of CYP2E1, an association was found between nicotine use and copy number. Studies have shown that neither ethanol nor nicotine increase the level of CYP2E1 mRNA in rat hepatic tissue (Howard et al., 2001). Ethanol likely changes the activity of CYP2E1 by interacting with the active site leading to increased protein stabilization and reduced clearance by degradation. Given the induction of CYP2E1 by nicotine requires multiple doses and does not interact with the catalytic function of CYP2E1, it is thought that the mechanism behind nicotine induction is not through protein stabilization. (Micu et al., 2003; Schoedel and Tyndale, 2003) Since the molar concentrations are vastly different it is unlikely that both nicotine and alcohol could stabilize CYP2E1 by the same mechanism. Since ethanol and nicotine likely induce CYP2E1 through different mechanisms, these two drugs may have an additive effect on CYP2E1 induction and function (Schoedel and Tyndale, 2003). The ability to induce CYP2E1 activity by nicotine, but not alcohol, could be dependent on basal transcription rates that could be affected by gene copy number.
The four polymorphisms commonly tested in CYP2E1, CYP2E1*5B (c2), CYP2E1*6 (C), CYP2E1*1B (A1), and CYP2E1*1D (1C), have been found to be associated with alcoholism and related disorders in a number of studies. Several of these variants are rare in the Caucasian population (see below). Carriers of the c2 allele of *5B have often been found to have increased risk for alcoholic liver disease (Grove et al., 1998) possibly due to the increased tendency to consume excessive amounts of alcohol (Pirmohamed et al., 1995). The C allele of *6 was shown to be associated with the predisposition for alcoholism in Japanese men (Iwahashi et al., 1998). The A1 allele of *1B was found to have a significantly higher allele frequency in alcoholics than in nonalcoholic individuals from a Mexican Indians population (Montano Loza et al., 2006). The 1D variant allele was shown to be associated with elevated CYP2E1 activity after alcohol consumption (McCarver et al., 1998). For every association found with CYP2E1 variants, a number of studies found no association between the variants and alcohol consumption or risk of alcoholism which could be due to differing phenotype categorization or population allele frequencies (Carr et al., 1996; Itoga et al., 2001; Pastorelli et al., 2001; Plee-Gautier et al., 2001; Vidal et al., 2004).
Of the markers measured in the current study, the most associated SNP with level of response to alcohol, rs10776687, is in complete linkage disequilibrium (LD) with the c1 allele of CYP2E1*5B, rs2031920, implying that this marker is associated with the level of response to alcohol as well. A homozygous genotype of the minor allele c2 of CYP2E1*5B is associated with an increase in gene transcription (Watanabe et al., 1994). Another marker, rs2515641, is in complete LD with rs2070676, also known as CYP2E1*1B.
As CYP2E1 is involved with the metabolism of many carcinogenic compounds, it is not surprising that variants in the gene have been implicated in a number of different types of cancer. The generation of ROS as a result of CYP2E1 oxidation will lead to the creation of lipid peroxidation products such as 4-hydroxynonenal which reacts with DNA to form DNA adducts leading to highly mutagenic cells resistant to apoptosis (Carr et al., 1996). The metabolism of procarcinogens by CYP2E1 commonly found in alcohol, tobacco, and industrial chemicals can be enhanced through chronic ethanol (Bailey and Cunningham, 2002).
While a number of CYP2E1 variants have been analyzed in relation to cancer development, CYP2E1*5B is most often considered. Many of these associations are enhanced by alcohol or nicotine intake which further supports the role of CYP2E1 in the metabolism of these substances. The c1/c1 genotype of the CYP2E1*5B variant increased risk of hepatocellular carcinoma in smokers from a Taiwanese population (Yu et al., 1995) and oral cavity cancer in heavy smokers from Caucasians and African Americans populations (Liu et al., 2001). Conversely other studies have found evidence for the minor c2 allele leading to an increased risk of hepatocellular carcinoma in ethanol users with chronic liver disease and oral cavity cancer in combination with heavy drinking (Bouchardy et al., 2000). Others have found no association between the CYP2E1*5B variant and the same types of cancer including a number of studies for hepatocellular carcinoma (Kato et al., 1995; Lee et al., 1997; Wong et al., 2000). Many CYP2E1 association studies did not detect an association because the c2 risk allele is rare in Caucasians (2–3%) (Kato et al., 1992; Pastorelli et al., 2001) and African Americans (0.3–7%) (Kato et al., 1992; London et al., 1996; Wu et al., 1997), but much more common for Asian (24–30%) (Lee et al., 1997; Tan et al., 2000; Watanabe et al., 1990) and Mexican American populations (15%) (Wu et al., 1997).
In aggregate it appears that alleles that increase CYP2E1 expression increase level of response to alcohol and risk for cancer, presumably by allowing the activation of procarcinogens or the production of ROS. Previous evidence for the involvement of CYP2E1 with alcohol metabolism and the incidence of several alcohol related cancers, strongly supports the conclusion that CYP2E1 alleles are associated with the level of response to alcohol and ultimately the development of alcohol use disorders. With multiple lines of evidence linking CYP2E1 to alcohol intake and subsequent outcomes, this gene can be an important predictor of risk for alcoholism and provide us with a better understanding of how the brain perceives alcohol. Drugs that affect the expression of this gene and, subsequently, the perception of alcohol, could reduce intoxication or limit consumption and thus moderate the development of alcoholism.
Acknowledgements
We wish express our thanks to the participants of the alcohol challenge for their willingness to partake in this investigation. This work was supported by funds provided by the State of California for medical research on alcohol and substance abuse through the University of California, San Francisco; the Veterans Affairs Research Service; National Institute on Alcohol Abuse and Alcoholism [grant numbers 05526, 08403], a grant from the CompassPoint Addiction Foundation, and the Bowles Center for Alcohol Studies at UNC [grant number 5P600AA011605]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Williams JT, Dyer TD, Blangero J. Quantitative trait locus detection using combined linkage/disequilibrium analysis. Genet Epidemiol. 1999;17:S31–S36. doi: 10.1002/gepi.1370170706. [DOI] [PubMed] [Google Scholar]
- Bailey SM, Cunningham CC. Contribution of mitochondria to oxidative stress associated with alcoholic liver disease. Free Radic Biol Med. 2002;32:11–16. doi: 10.1016/s0891-5849(01)00769-9. [DOI] [PubMed] [Google Scholar]
- Barr CS, Newman TK, Becker ML, Champoux M, Lesch KP, Suomi SJ, Goldman D, Higley JD. Serotonin transporter gene variation is associated with alcohol sensitivity in rhesus macaques exposed to early-life stress. Alcohol Clin Exp Res. 2003;27:812–817. doi: 10.1097/01.ALC.0000067976.62827.ED. [DOI] [PubMed] [Google Scholar]
- Bouchardy C, Hirvonen A, Coutelle C, Ward PJ, Dayer P, Benhamou S. Role of alcohol dehydrogenase 3 and cytochrome P-4502E1 genotypes in susceptibility to cancers of the upper aerodigestive tract. Int J Cancer. 2000;87:734–740. [PubMed] [Google Scholar]
- Carr LG, Yi IS, Li TK, Yin SJ. Cytochrome P4502E1 genotypes, alcoholism, and alcoholic cirrhosis in Han Chinese and Atayal Natives of Taiwan. Alcohol Clin Exp Res. 1996;20:43–46. doi: 10.1111/j.1530-0277.1996.tb01041.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Plunkett J, Wetherill LF, Xuei X, Goate A, Hesselbrock V, Schuckit M, Crowe R, Edenberg HJ, Foroud T. Association between GABRA1 and drinking behaviors in the collaborative study on the genetics of alcoholism sample. Alcohol Clin Exp Res. 2006;30:1101–1110. doi: 10.1111/j.1530-0277.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Garcia-Andrade C, Wall TL, Cloutier D, Phillips E. Electroencephalographic responses to alcohol challenge in Native American Mission Indians. Biol Psychiatry. 1999;45:776–787. doi: 10.1016/s0006-3223(98)00113-9. [DOI] [PubMed] [Google Scholar]
- Garcia-Andrade C, Wall TL, Ehlers CL. The firewater myth and response to alcohol in Mission Indians. Am J Psychiatry. 1997;154:983–988. doi: 10.1176/ajp.154.7.983. [DOI] [PubMed] [Google Scholar]
- Grove J, Brown AS, Daly AK, Bassendine MF, James OF, Day CP. The RsaI polymorphism of CYP2E1 and susceptibility to alcoholic liver disease in Caucasians: effect on age of presentation and dependence on alcohol dehydrogenase genotype. Pharmacogenetics. 1998;8:335–342. doi: 10.1097/00008571-199808000-00007. [DOI] [PubMed] [Google Scholar]
- Heath AC, Madden PA, Bucholz KK, Dinwiddie SH, Slutske WS, Bierut LJ, Rohrbaugh JW, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychol Med. 1999;29:1069–1081. doi: 10.1017/s0033291799008909. [DOI] [PubMed] [Google Scholar]
- Howard LA, Micu AL, Sellers EM, Tyndale RF. Low doses of nicotine and ethanol induce CYP2E1 and chlorzoxazone metabolism in rat liver. J Pharmacol Exp Ther. 2001;299:542–550. [PubMed] [Google Scholar]
- Itoga S, Harada S, Nomura F. Polymorphism of the 5'-flanking region of the CYP2E1 gene: an association study with alcoholism. Alcohol Clin Exp Res. 2001;25:11S–15S. doi: 10.1097/00000374-200106001-00004. [DOI] [PubMed] [Google Scholar]
- Iwahashi K, Ameno S, Ameno K, Okada N, Kinoshita H, Sakae Y, Nakamura K, Watanabe M, Ijiri I, Harada S. Relationship between alcoholism and CYP2E1 C/D polymorphism. Neuropsychobiology. 1998;38:218–221. doi: 10.1159/000026544. [DOI] [PubMed] [Google Scholar]
- Joshi M, Tyndale RF. Induction and recovery time course of rat brain CYP2E1 after nicotine treatment. Drug Metab Dispos. 2006;34:647–652. doi: 10.1124/dmd.105.008029. [DOI] [PubMed] [Google Scholar]
- Kato S, Onda M, Matsukura N, Tokunaga A, Tajiri T, Kim DY, Tsuruta H, Matsuda N, Yamashita K, Shields PG. Cytochrome P4502E1 (CYP2E1) genetic polymorphism in a case-control study of gastric cancer and liver disease. Pharmacogenetics. 1995;5(Spec No):S141–S144. doi: 10.1097/00008571-199512001-00016. [DOI] [PubMed] [Google Scholar]
- Kato S, Shields PG, Caporaso NE, Hoover RN, Trump BF, Sugimura H, Weston A, Harris CC. Cytochrome P450IIE1 genetic polymorphisms, racial variation, and lung cancer risk. Cancer Res. 1992;52:6712–6715. [PubMed] [Google Scholar]
- Lee HS, Yoon JH, Kamimura S, Iwata K, Watanabe H, Kim CY. Lack of association of cytochrome P450 2E1 genetic polymorphisms with the risk of human hepatocellular carcinoma. Int J Cancer. 1997;71:737–740. doi: 10.1002/(sici)1097-0215(19970529)71:5<737::aid-ijc8>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Lieber CS. Cytochrome P-4502E1: its physiological and pathological role. Physiol Rev. 1997;77:517–544. doi: 10.1152/physrev.1997.77.2.517. [DOI] [PubMed] [Google Scholar]
- Lieber CS. Microsomal Ethanol-Oxidazing System (MEOS): The First 30 Years (1968–1998)—A Review. Alcohol Clin Exp Res. 1999;23:991–1007. [PubMed] [Google Scholar]
- Liu S, Park JY, Schantz SP, Stern JC, Lazarus P. Elucidation of CYP2E1 5' regulatory RsaI/Pstl allelic variants and their role in risk for oral cancer. Oral Oncol. 2001;37:437–445. doi: 10.1016/s1368-8375(00)00099-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- London SJ, Daly AK, Cooper J, Carpenter CL, Navidi WC, Ding L, Idle JR. Lung cancer risk in relation to the CYP2E1 Rsa I genetic polymorphism among African-Americans and Caucasians in Los Angeles County. Pharmacogenetics. 1996;6:151–158. doi: 10.1097/00008571-199604000-00002. [DOI] [PubMed] [Google Scholar]
- Luczak SE, Elvine-Kreis B, Shea SH, Carr LG, Wall TL. Genetic risk for alcoholism relates to level of response to alcohol in Asian-American men and women. J Stud Alcohol. 2002;63:74–82. [PubMed] [Google Scholar]
- McCarver DG, Byun R, Hines RN, Hichme M, Wegenek W. A genetic polymorphism in the regulatory sequences of human CYP2E1: association with increased chlorzoxazone hydroxylation in the presence of obesity and ethanol intake. Toxicol Appl Pharmacol. 1998;152:276–281. doi: 10.1006/taap.1998.8532. [DOI] [PubMed] [Google Scholar]
- Micu AL, Miksys S, Sellers EM, Koop DR, Tyndale RF. Rat hepatic CYP2E1 is induced by very low nicotine doses: an investigation of induction, time course, dose response, and mechanism. J Pharmacol Exp Ther. 2003;306:941–947. doi: 10.1124/jpet.103.052183. [DOI] [PubMed] [Google Scholar]
- Montano Loza AJ, Ramirez Iglesias MT, Perez Diaz I, Cruz Castellanos S, Garcia Andrade C, Medina Mora ME, Robles Diaz G, Kershenobich D, Gutierrez Reyes G. Association of alcohol-metabolizing genes with alcoholism in a Mexican Indian (Otomi) population. Alcohol. 2006;39:73–79. doi: 10.1016/j.alcohol.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Monteiro MG, Klein JL, Schuckit MA. High levels of sensitivity to alcohol in young adult Jewish men: a pilot study. J Stud Alcohol. 1991;52:464–469. doi: 10.15288/jsa.1991.52.464. [DOI] [PubMed] [Google Scholar]
- Pastorelli R, Bardazzi G, Saieva C, Cerri A, Gestri D, Allamani A, Airoldi L, Palli D. Genetic determinants of alcohol addiction and metabolism: a survey in Italy. Alcohol Clin Exp Res. 2001;25:221–227. [PubMed] [Google Scholar]
- Pirmohamed M, Kitteringham NR, Quest LJ, Allott RL, Green VJ, Gilmore IT, Park BK. Genetic polymorphism of cytochrome P4502E1 and risk of alcoholic liver disease in Caucasians. Pharmacogenetics. 1995;5:351–357. doi: 10.1097/00008571-199512000-00003. [DOI] [PubMed] [Google Scholar]
- Plee-Gautier E, Foresto F, Ferrara R, Bodenez P, Simon B, Manno M, Berthou F, Lucas D. Genetic repeat polymorphism in the regulating region of CYP2E1: frequency and relationship with enzymatic activity in alcoholics. Alcohol Clin Exp Res. 2001;25:800–804. [PubMed] [Google Scholar]
- Pollock VE. Meta-analysis of subjective sensitivity to alcohol in sons of alcoholics. Am J Psychiatry. 1992;149:1534–1538. doi: 10.1176/ajp.149.11.1534. [DOI] [PubMed] [Google Scholar]
- Schoedel KA, Tyndale RF. Induction of nicotine-metabolizing CYP2B1 by ethanol and ethanol-metabolizing CYP2E1 by nicotine: summary and implications. Biochim Biophys Acta. 2003;1619:283–290. doi: 10.1016/s0304-4165(02)00487-7. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL. An 8-year follow-up of 450 sons of alcoholic and control subjects. Arch Gen Psychiatry. 1996;53:202–210. doi: 10.1001/archpsyc.1996.01830030020005. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Kalmijn J, Tsuang J, Hesselbrock V, Bucholz K. Response to alcohol in daughters of alcoholics: a pilot study and a comparison with sons of alcoholics. Alcohol Alcohol. 2000;35:242–248. doi: 10.1093/alcalc/35.3.242. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Wilhelmsen K, Smith TL, Feiler HS, Lind P, Lange LA, Kalmijn J. Autosomal linkage analysis for the level of response to alcohol. Alcohol Clin Exp Res. 2005;29:1976–1982. doi: 10.1097/01.alc.0000187598.82921.27. [DOI] [PubMed] [Google Scholar]
- Seitz HK, Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat Rev Cancer. 2007;7:599–612. doi: 10.1038/nrc2191. [DOI] [PubMed] [Google Scholar]
- Tan W, Song N, Wang GQ, Liu Q, Tang HJ, Kadlubar FF, Lin DX. Impact of genetic polymorphisms in cytochrome P450 2E1 and glutathione S-transferases M1, T1, and P1 on susceptibility to esophageal cancer among high-risk individuals in China. Cancer Epidemiol Biomarkers Prev. 2000;9:551–556. [PubMed] [Google Scholar]
- Tanaka E, Terada M, Misawa S. Cytochrome P450 2E1: its clinical and toxicological role. J Clin Pharm Ther. 2000;25:165–175. doi: 10.1046/j.1365-2710.2000.00282.x. [DOI] [PubMed] [Google Scholar]
- Vasiliou V, Ziegler TL, Bludeau P, Petersen DR, Gonzalez FJ, Deitrich RA. CYP2E1 and catalase influence ethanol sensitivity in the central nervous system. Pharmacogenet Genomics. 2006;16:51–58. doi: 10.1097/01.fpc.0000182777.95555.56. [DOI] [PubMed] [Google Scholar]
- Vidal F, Lorenzo A, Auguet T, Olona M, Broch M, Gutierrez C, Aguilar C, Estupina P, Santos M, Richart C. Genetic polymorphisms of ADH2, ADH3, CYP4502E1 Dra-I and Pst-I, and ALDH2 in Spanish men: lack of association with alcoholism and alcoholic liver disease. J Hepatol. 2004;41:744–750. doi: 10.1016/j.jhep.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Volavka J, Czobor P, Goodwin DW, Gabrielli WF, Jr, Penick EC, Mednick SA, Jensen P, Knop J. The electroencephalogram after alcohol administration in high-risk men and the development of alcohol use disorders 10 years later. Arch Gen Psychiatry. 1996;53:258–263. doi: 10.1001/archpsyc.1996.01830030080012. [DOI] [PubMed] [Google Scholar]
- Wall TL, Johnson ML, Horn SM, Carr LG, Smith TL, Schuckit MA. Evaluation of the self-rating of the effects of alcohol form in Asian Americans with aldehyde dehydrogenase polymorphisms. J Stud Alcohol. 1999;60:784–789. doi: 10.15288/jsa.1999.60.784. [DOI] [PubMed] [Google Scholar]
- Watanabe J, Hayashi S, Kawajiri K. Different regulation and expression of the human CYP2E1 gene due to the RsaI polymorphism in the 5'-flanking region. J Biochem. 1994;116:321–326. doi: 10.1093/oxfordjournals.jbchem.a124526. [DOI] [PubMed] [Google Scholar]
- Watanabe J, Hayashi S, Nakachi K, Imai K, Suda Y, Sekine T, Kawajiri K. PstI and RsaI RFLPs in complete linkage disequilibrium at the CYP2E gene. Nucleic Acids Res. 1990;18:7194. doi: 10.1093/nar/18.23.7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen KC, Schuckit M, Smith TL, Lee JV, Segall SK, Feiler HS, Kalmijn J. The search for genes related to a low-level response to alcohol determined by alcohol challenges. Alcohol Clin Exp Res. 2003;27:1041–1047. doi: 10.1097/01.ALC.0000075551.02714.63. [DOI] [PubMed] [Google Scholar]
- Wong NA, Rae F, Simpson KJ, Murray GD, Harrison DJ. Genetic polymorphisms of cytochrome p4502E1 and susceptibility to alcoholic liver disease and hepatocellular carcinoma in a white population: a study and literature review, including meta-analysis. Mol Pathol. 2000;53:88–93. doi: 10.1136/mp.53.2.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Shi H, Jiang H, Kemp B, Hong WK, Delclos GL, Spitz MR. Associations between cytochrome P4502E1 genotype, mutagen sensitivity, cigarette smoking and susceptibility to lung cancer. Carcinogenesis. 1997;18:967–973. doi: 10.1093/carcin/18.5.967. [DOI] [PubMed] [Google Scholar]
- Yu MW, Gladek-Yarborough A, Chiamprasert S, Santella RM, Liaw YF, Chen CJ. Cytochrome P450 2E1 and glutathione S-transferase M1 polymorphisms and susceptibility to hepatocellular carcinoma. Gastroenterology. 1995;109:1266–1273. doi: 10.1016/0016-5085(95)90587-1. [DOI] [PubMed] [Google Scholar]
- Zakhari S. Overview: how is alcohol metabolized by the body? Alcohol Res Health. 2006;29:245–254. [PMC free article] [PubMed] [Google Scholar]