Summary
The intracellular protozoan parasite Toxoplasma gondii divides by a unique process of internal budding that involves the assembly of two daughter cells within the mother. The cytoskeleton of Toxoplasma, which is composed of microtubules associated with an inner membrane complex (IMC), has an important role in this process. The IMC, which is directly under the plasma membrane, contains a set of flattened membranous sacs lined on the cytoplasmic side by a network of filamentous proteins. This network contains a family of intermediate filament-like proteins or IMC proteins. In order to elucidate the division process, we have characterized a 14-member sub-family of Toxoplasma IMC proteins that share a repeat motif found in proteins associated with the cortical alveoli in all alveolates. By creating fluorescent protein fusion reporters for the family members we determined the spatio-temporal patterns of all 14 IMC proteins through tachyzoite development. This revealed several distinct distribution patterns and some provide the basis for novel structural models such as the assembly of certain family members into the basal complex. Furthermore we identified IMC15 as an early marker of budding and, lastly, the dynamic patterns observed throughout cytokinesis provide a timeline for daughter parasite development and division.
Keywords: Toxoplasma, alveolates, IMC, intermediate filament-like, endodyogeny
Introduction
Toxoplasma gondii is a member of the Apicomplexa, a phylum comprised predominantly of obligate intracellular protozoan parasites. Toxoplasma is an opportunistic pathogen of humans and primarily causes disease in immunocompromised individuals and the fetus where fast cycles of parasite replication and the ensuing immune response cause tissue lesions. The structural maintenance of the cell shape by the cortical cytoskeleton is critical for pathogenesis (Morrissette et al., 2002). The cortical cytoskeleton is composed of two main components: the microtubules and the inner membrane complex (IMC). The microtubules underlie the IMC, which is composed of membrane sacs called alveoli lined with a meshwork of 8–10 nm intermediate filament-like proteins or IMC proteins (Mann et al., 2001).
The first four intermediate filament-like IMC proteins, IMC1, 2, 3, and 4, were identified in different labs and through different screening techniques (Gubbels et al., 2004, Hu et al., 2006, Mann et al., 2001), leaving open the possibility of the existence of more IMC proteins. Gould et al analyzed genome sequence data for all the Alveolata (ciliates, dinoflagellates and apicomplexans (Leander et al., 2003)) and defined a family of proteins related to IMC1, 3 and 4, which all carry a series of conserved repeats called alveolin repeats (Gould et al., 2008). The alveolin motif contains conserved valine- and proline-rich domains composed of “EKIVEVP” and “EVVR” or “VPV” subrepeats (Gould et al., 2008). IMC2 does not have an alveolin motif and is unrelated to this group.
In apicomplexans the IMC serves as support for the actin-myosin motor apparatus that is required for host cell invasion (Gaskins et al., 2004, Mann et al., 2001). Furthermore in apicomplexans the IMC plays a critical role in cell division, which is driven by assembly of the cortical cytoskeleton and serves as a scaffold for organellogenesis and organelle partitioning (Nishi et al., 2008). Direct demonstration of the critical role of IMC1a and IMC1b in Plasmodium berghei cell division and structural support was provided by genetic knock-outs (Khater et al., 2004, Tremp et al., 2008). We have reported previously that Toxoplasma IMC3 is enriched in the assembling daughter complexes (Gubbels et al., 2004), which appears not to be the case for IMC1 and IMC4 (Hu et al., 2006, Hu et al., 2002). The importance of the IMC to the parasite and the possibility that the individual IMC proteins may behave differently led us to hypothesize that these proteins could fulfill different functions within various stages of the parasite’s life cycle including cytoskeletal assembly, maturation, maintenance, and disassembly.
Here we identified 14 alveolin domain containing genes in the T. gondii sequenced genome database. Subsequently we studied the subcellular localizations of each using YFP-tagged constructs and specific antisera. We identified a number of unique spatio-temporal patterns in addition to those previously described, such as a group of IMC proteins exclusively localizing to the basal complex, an IMC protein co-localizing with the centrosomes, and a group of IMC proteins exclusively localizing to the mature cytoskeleton. Taken together, a surprising level of diversity in the localization and potential function of the alveolin-repeat IMC proteins outlines the developmental stages of the Toxoplasma tachyzoite cell division at an unprecedented spatio-temporal resolution.
Results
Genome mining for genes encoding IMC proteins
Since the identification of IMC1, 3, and 4 was not as part of a comprehensive search for IMC proteins we identified additional IMC genes through querying the fully sequenced T. gondii genome in ToxoDB using IMC1, 3, and 4 as seeds. This approach identified a total of 14 related IMC genes containing an alveolin domain (Fig. 1A and Table S1). We validated the splicing of IMC genes 7, 9, and 12–15 by RACE PCR and DNA sequencing. Only IMC14 and IMC15 varied from the predicted gene models: IMC14 has several splice variants in the Prugniaud strain but only one in the RH strain (the RH sequence was used in this study) and IMC15 has different start and stop codons (Fig. S1). The remaining IMC genes have consistent gene prediction models and we were able to amplify their open reading frames from tachyzoite cDNA to confirm these annotations.
Fig. 1.
The IMC proteins and their expression during tachyzoite development. (A) Full length and validated open reading frames of each IMC protein in order by IMC number and their corresponding ToxoDB gene name. The alveolin repeat regions are represented in yellow and the N- and C-termini in green. Cysteines are indicated in red and predicted palmytoylation sites marked with blue asterisks. (B) Affymetrix array expression pattern of the IMC mRNAs through two cycles of tachyzoite development. RH strain parasites expressing the herpes simplex thymidine kinase (TK) were synchronized by a thymidine block. Cell cycle stages and timing of budding are indicated at the top. Expression levels are normalized to internal controls on the Affymetrix array. (C) Maximum and minimum expression levels of the IMC genes in the second cycle represented in (B) (hours 6–12). Expression level is shown as the raw fluorescence hybridization data.
Coordinated expression of the IMC proteins in tachyzoite development
Coordinated gene expression patterns among proteins with related functions have been observed during the erythrocytic development of the apicomplexan Plasmodium falciparum (Bozdech et al., 2003, Le Roch et al., 2003). To see if this applies to the IMC proteins, the expression patterns were assembled for each of the identified IMC genes from genome-wide microarray expression data (data kindly provided by Drs. Michael White and Michael Benhke, Montana State University). The majority of the IMC genes (IMC1, 3–6, 8–11, 13, and 15) display a highly coordinated expression profile reaching a maximum coinciding with budding, whereas IMC7 and 12 exhibit a profile opposing the majority and IMC14 trails the majority pattern by 1 hour (Fig. 1B). When comparing the absolute mRNA expression levels, only IMC9 and 15 stand out with maximum expression levels less than the minimum levels of almost all the other IMC genes (Fig. 1C). Furthermore, many mRNAs displayed an up to a 10-fold dynamic range between minimum and maximum suggesting that although timing of expression is coordinated for most IMC genes, their expression levels are quite variable.
Comparisons of cortical IMC proteins in mature and budding parasites
Although IMC proteins 1, 3, and 4 had been characterized previously, it was never formally established how these three proteins localize relative to one another. When IMC1 and IMC3 are compared it is clear that more IMC3 is associated with the budding daughter cytoskeletons than with the cytoskeleton of the mother cell as opposed to the more equal distribution that IMC1 exhibits (Fig. 2A,B,E,F). The high intensity of IMC3 is maintained in the recently emerged but then drops to lower basal levels in fully mature daughters (Fig. 2A and Fig. S2). Comparable results were obtained with parasites expressing YFP-tagged versions of IMC1 and IMC3 indicating that the observed differences were not due to masking of the antibody binding site (Fig. S3A,B). When IMC4 is compared to IMC1 and IMC3 using a parasite line expressing YFP-IMC4 it is found to localize like IMC1 with equal distribution in the mother and daughter buds (Fig. S4A,B). This pattern is corroborated with a C-terminal fusion, IMC4-YFP, as well (Fig. S5A).
Fig. 2.
Relative localization of the cortical IMC proteins throughout tachyzoite development. (A–B) Mature (A) and budding (B) parasites co-stained with antibodies against IMC1 (green) and IMC3 (red). (C) Budding parasites expressing ptub driven YFP-IMC6. (D) Budding parasites expressing ptub driven YFP-IMC10. (E) Intensity profile across the budding daughters indicated in (B) panel 3 marked by arrow “E”. Arrowheads indicate specific localization of IMC1 in the mature mother parasites not detected for IMC3. (F) Mature mother intensity profile along the arrow indicated in (C) panel 3 marked with “F”. (G–H) Intensity profiles as indicated by arrows “G” and “H” in (C) panel 3. Relative distance is shown along the length and direction of the arrows in (B,C) on the x-axis and relative intensity is shown on the y-axis.
To assess the subcellular localizations of the novel IMC proteins we generated N-terminal YFP fusions for each protein and expressed them in T. gondii. The expressed fusion proteins were verified through Western blot (Fig. S6). Live parasites were imaged to identify the subcellular distribution patterns of the fusion proteins and, in addition, their localization dynamics were determined in all cases relative to IMC3 with an IMC3 antibody. Among the novel IMC proteins we identified two, IMC6 and IMC10, which show a localization pattern like IMC3 (Fig. 2C,D,G,H and Fig. S4C-F). This pattern is the same with C-terminal YFP fusions (Fig. S5B,D). Taken together IMC3, 6, and 10 comprise a related group of cortical IMC proteins distinct from IMC1 and 4 based on their behavior during parasite development.
IMC7, IMC12, and IMC14 localize exclusively to the mature cytoskeleton
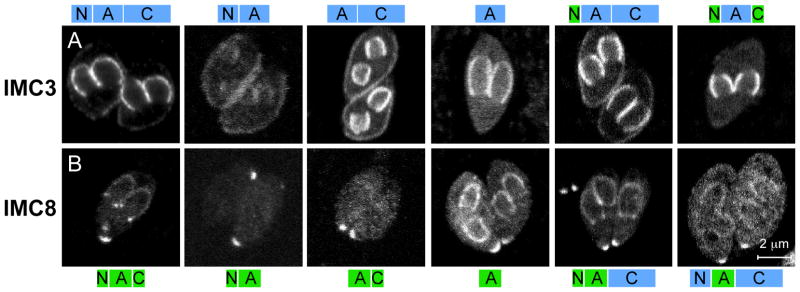
In contrast to the IMC proteins described above, IMC7, 12, and 14 are only found in the mature cortical IMC. The cortical localization of IMC7, 12, and 14 does not correspond with the emergence of mature daughters (Fig. 3A,D and Fig. S7C,G) but instead occurs sometime before the initiation of budding (Fig. 3A,B,C; Fig. S7A,B,E,F; and Movies S1 and S2). C-terminal fusions of IMC7 and 12 confirm these observed patterns (Fig. S5C,E) and this behavior is not unexpected based on the delays in the expression profiles of IMC7, 12, and 14 (Fig. 1B). To verify that expression of YFP-IMC7, 12, and 14 do not interfere with maturation we used a glideosome component, GAP45, which is incorporated into the IMC upon daughter maturation (Agop-Nersesian et al., 2009). As shown in Figure 3F, GAP45 did associate with the cortex of the emerging daughters expressing cytoplasmically localized YFP-IMC7, while some IMC7 is still in the cortex of the disassembling mother. The results are the same for YFP-IMC12 and 14 (Fig. S7D,H). It is important to note that IMC7 and 14 are expressed under their native promoters instead of the α-tubulin (ptub) promoter as ptub closely matches the expression patterns of the majority of the IMC proteins but not these (Fig. 1B; Drs. Michael Behnke, John Wootton and Michael White, paper in preparation). The IMC12 expression pattern is the same under the ptub promoter and its native promoter but under the native promoter it fades rapidly during imaging; therefore, ptub driven YFP-IMC12 is presented here (localization data for pimc12 driven constructs is provided in Fig. S8).
Fig. 3.
IMC7, 12, and 14 associate exclusively with mature cytoskeletons. (A) Selected frames from time-lapse images of YFP-IMC12 (Movie S2). (B–E) Three stages of tachyzoite development are shown for YFP-IMC7 (green) co-stained with IMC3 antibody (red): (B) mature, (C) mid budding, and (D) emerging daughters. (E) Intensity profile as described in Figure 2 across the arrow shown in panel 3 of (C). (F) YFP-IMC7 (green) expressing parasites co-stained with GAP45 antibody (red). (G,H) YFP-IMC7, 12, and 14 associate with the cortical cytoskeleton in the G1-phase of the cell cycle. YFP-IMC7, 12, and 14 expressing parasites were co-stained with centrin antibody (red) and DAPI (blue). YFP-IMC7 is provided as an example in (G). The following three stages of development were defined: cytoplasmic YFP with 1 centrosome (cyto, 1 cent), cortical YFP with 1 centrosome (cort, 1 cent), and cortical YFP with duplicated centrosomes (cort, 2 cent). All stages were blindly counted and G1 distributions plotted in (H) as percentage of parasites in G1. Results from three independent experiments counting 60–160 parasites per experiment are shown; error bars denote standard deviation.
To determine the timing of the shift of IMC7 and 12 to the cortical IMC, time-lapse movies of YFP-IMC7 and YFP-IMC12 parasite lines were collected throughout tachyzoite development (Fig. 3A; Movies S1 and S2). The movies show that the time between the end of cytokinesis and the reappearance of the cortical YFP localization is approximately two hours. Thus the transition of these IMC proteins occurs in G1. Collection of a comparable IMC14 movie failed due to the rapid bleaching of the weak YFP signal. Instead of a movie, we co-stained the parasites with a centrin antibody to mark the centrosome. Centrosome division marks the transition from G1- to S-phase (Hartmann et al., 2006). By counting the number of parasites carrying one centrosome and relating that to the localization of YFP-IMC7, 12 or 14 to either the cytoplasm or cortex we determined the relative timing in G1 for the appearance of the cortical localization (Fig. 3G,H). Counting was performed blind and faint cortical localization surrounding cytoplasmic localization was considered cortical. The relatively large standard deviation across the counts does not allow us to draw definitive conclusions, however the mRNA expression profiles (Fig. 1B) suggest that IMC14 may appear at the cortex before IMC7 or IMC12.
IMC11 co-localizes with PhIL1
Expression of a YFP tagged IMC11 driven by either the ptub or its endogenous promoter, resulted in very weak expression of YFP in an area of the cortex known as the apical cap as well as an even weaker localization to the basal end of the IMC (Fig. S9). The localization to the apical cap was confirmed through co-expression of a CherryRFP tagged version of PhIL1, an IMC-associated protein of unknown function (Gilk et al., 2006) (Fig. S9C). The number of parasites that could be captured with visible basal IMC11 signal was too low to make a firm observation regarding basal co-localization. Using a MORN1 antibody, which highlights the basal complex (Gubbels et al., 2006), the basal localization of YFP-IMC11 is supported (Fig. S9D), but the poor YFP signal again did not permit a firm co-localization assignment. Antiserum was raised against full-length IMC11 because IMC11 is too small to use only the non-alveolin domains. This unfortunately made it cross-reactive with the alveolin motifs in other IMC proteins preventing the confirmation of the YFP fusion.
IMC15 associates with the centrosomes and additional structures in the cytoskeleton
YFP-IMC15, when driven by ptub, localizes to the mature cortical IMC, the budding daughter IMC, the apical cap, and the basal complex in addition to novel localization at the very apical end and the centrosomes (Fig. 4). A ptub driven C-terminal fusion confirms its localization to the centrosomes and early buds, however its expression is faint and cortical localization is only clear in some parasites (Fig. S5G). When under its endogenous promoter IMC15 continues to localize to the centrosomes, budding daughters, and the extreme apical end of the parasite (Fig. S3E). The basal expression is greatly reduced and the cortical expression and cap expression are no longer present, suggesting the latter two might be the result of overexpression. Since the very apical end is relatively intense even under the native promoter we tested if this expression corresponds to the conoid by co-expresssing YFP-IMC15 with myc2-centrin2, a protein present in the apical polar ring (Hu et al., 2006). Centrin2 appears to be more apical than IMC15 and to be surrounded on the bottom by IMC15 (Fig. 4C). This suggests that the bright apical localization of IMC15 is in or around the conoid.
Fig. 4.
IMC15 associates with the duplicated centrosomes and transitions to the budding cytoskeleton. (A) live ptub driven YFP-IMC15 expressing parasites showing a total projection (left) and an optical section (middle) of mature parasites and a parasite with two intensely stained budding daughters (right). Arrows indicate the very basal end of the cytoskeleton, arrowheads the very apical end, and the double arrowheads the cap region. (B) YFP-IMC15 (green) expressing parasites co-stained with IMC3 antibody (red) in mid budding parasites. (C) YFP-IMC15 (green) expressing parasites co-expressing myc2-centrin2 and stained with myc antibody (red) in early budding parasites. Arrowheads indicate the very apical end of the parasite, double arrowheads the apical cap, single arrow the very basal end, double-headed arrows the early bud and double arrowheads the 6 centrin2 foci on the apical cap. Inset is of boxed area. (D) YFP-IMC15 (green) expressing parasites co-stained with centrin antibody (red) in mature parasites. Arrows mark IMC15 localization to the duplicated centrosomes. Inset is of boxed area. (E) YFP-IMC15 (green) expressing parasites co-transfected with CherryRFP-MORN1 (red) and co-stained with anti-human centrin antibody (blue) in very early budding parasites (pre-mitotic as judged from the single, centrally located MORN1 highlighting the spindle pole). The parasite is outlined with a dotted line in the first panel. Insets are of the central region around MORN1. (F) S-phase wild-type parasites stained with anti-IMC15 serum (red) and co-stained with centrin antibody (green) and DAPI (blue). All insets are 2X enlargments.
The co-localization of IMC15 with the centrosomes is shown by the use of a centrin antibody (Fig. 4D–F). It appears that IMC15 localizes to the centrosomes at the time of or immediately following their duplication. These spots become very intense before the signal transitions into the early buds, arching over the apical side of the centrosomes (Fig. 4C,E,F). Previous to this study the localization of MORN1 above the centrosomes was the earliest known marker of budding (Gubbels et al., 2006). However, relative to the weak MORN1 signal in very early buds, already two very intense IMC15 bud structures are visible (Fig. 4E). IMC15 localization to the centrosomes and early buds was confirmed with a specific antiserum raised against the N-terminus of IMC15 (Fig. 4F and Fig. S3D,E). The antiserum does not recognize the apical accumulation possibly due to post-translational modifications or to the YFP tag blocking the antibody recognition site. Consistent with potential post-translational modification is the slower than expected migration of YFP-IMC15 fusion protein (Fig. S6).
A group of IMC proteins is part of the basal complex
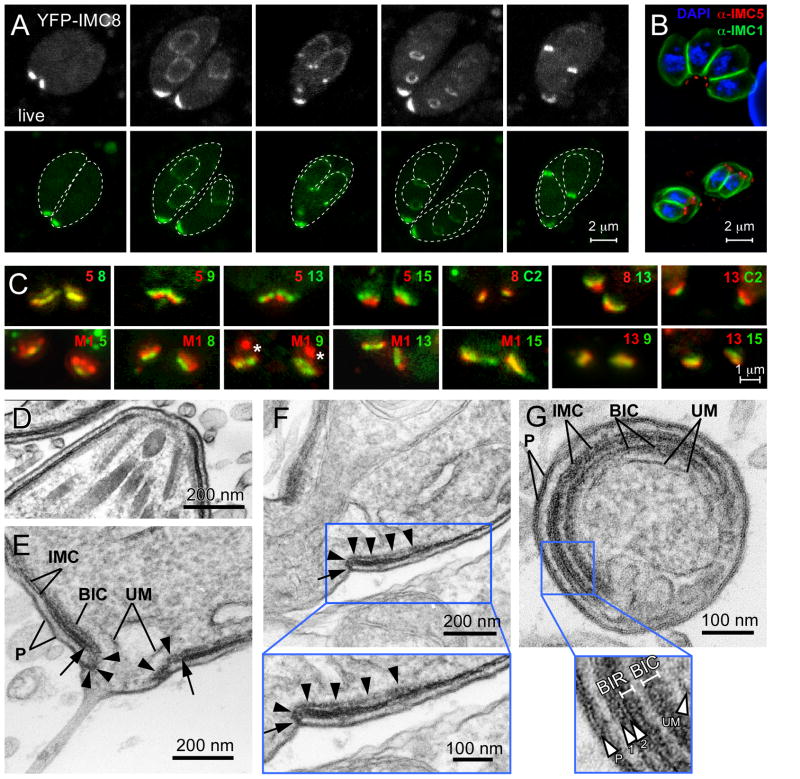
As shown in Figures 4 and S9, YFP-IMC15 and YFP-IMC11 co-localize with MORN1 at the basal end of the mature parasite. Surprisingly, we identified several IMC proteins that exclusively localize to the basal complex in mature parasites: IMC5, 8, 9, and 13 (Fig. 5). This group localizes to the whole daughter bud in the first half of budding and then halfway through budding they shift toward the basal complex (Fig. 5A). An N-terminal YFP fusion protein of IMC13 displays the same localization pattern as the C-terminal YFP fusion (Fig. S5F). Furthermore, we generated a specific antiserum against the N-terminus of IMC5, which confirmed its localization to the basal complex (Fig. 5B and Fig. S3C).
Fig. 5.
IMC5, 8, 9 and 13 localize to the cortical IMC in early buds and then transition to and partially co-localize at the basal complex upon constriction. (A) Live ptub promoter driven YFP-IMC8 expressing parasites at different stages of tachyzoite development (independent parasites are shown in each panel). Mother and daughter parasites are traced by dotted lines in the lower series. (B) Wild type parasites co-stained with anti-IMC5 serum (red), IMC1 antibodies (green), and DAPI (blue). (C) Pair-wise comparisons of the members of the basal complex using co-transfected YFP and CherryRFP constructs. The numbers represent the tagged IMC protein, M1 is MORN1, C2 is centrin2, and the colors correspond with the fluorescent protein fusion. The asterisks in the MORN1 + IM9 panel mark the spindle pole localization of MORN1 (D–G) Electron micrographs identifying an inner collar at the basal end of the cytoskeleton. (D) Cross section through the apical complex demonstrating the absence of a comparable complex at the apical end. (E) Longitudinal section through the posterior end of a parasite displaying the basal inner collar (BIC) and the fold over the alveoli marked with arrowheads. The arrows mark the end of the alveolar vesicles. A unit membrane (UM) of unknown origin with an electron dense coating that is limited to the basal cup region is visible as a clear vesicle sitting within the very basal opening. (F) Longitudinal section through the basal complex. Arrowheads mark the BIC, which bends over the end of the alveolar membrane and connects with the plasma membrane as indicated by the arrow. The area marked by the blue box is enlarged. (G) Cross section through the basal complex displaying the continuity of the BIC and basal inner ring (BIR), which are visible in the enlarged area marked by the blue box. In addition, the two closely apposed UMs can be discerned. P is plasma membrane.
Subtle differences in IMC protein localizations within the basal complex were observed and we compared these pair wise with each other as well as with MORN1 and centrin2 (Fig. 5C). These co-localization studies resulted in the identification of three substructures in the basal complex: the widest structure contains MORN1, IMC9, 13, and 15, followed by a structure composed of IMC5 and 8 that is finally followed at the very basal end by the smallest structure containing centrin2, which partially overlaps with the IMC5 and 8 structure. Because definitive validation of these spatial relationships is limited by the resolution of light microscopy we sought to test whether the co-localizing proteins were interacting with each other and/or with their neighbors by yeast two-hybrid analysis. Unfortunately, these results were largely inconclusive (Table S2).
Since these distinctive basal structures have not been described at the ultrastructural level, we performed electron microscopy (EM) on the basal complex (Fig. 5D–G). Two distinct electron-dense structures were observed in the basal complex: one directly attached to the cytoplasmic side of the basal end of the IMC (the basal inner ring or BIR; Fig. 5G) and a second structure separated from the IMC and the inner ring, but extending over the same length of the basal IMC and bending toward the plasma membrane (the basal inner collar or BIC) (Fig. 5E–G). Though it is difficult to obtain a large number of properly sectioned parasites, among 30 parasites with properly oriented basal complexes the majority displayed this folding back of the BIC to contact the plasmalemma. Both the BIR and BIC are continuous (Fig. 5G) and absent from the apical end of the IMC (Fig. 5D).
The alveolin domain is the main determinant for localization to either the cortical or the basal cytoskeleton
We identified two major classes of IMC proteins: those that localize to the cortical cytoskeleton and those that localize to the basal cytoskeleton. Based on other classes of intermediate filament proteins their localization might be controlled by the N- or C-terminus since the central conserved domain is likely required for filament formation (Herrmann et al., 2004, Parry et al., 2007). To test this, we designed a series of deletion and chimeric constructs of IMC3 and IMC8, neither of which contain predicted palmitoylation sites that could potentially overrule amino acid motifs governing their localization (Fig. 1A). For IMC3 the expression of the alveolin domain alone showed the same localization pattern as its full-length counterpart (Fig. 6A). The results were the same for IMC8, except for the lack of pronounced basal localization in the buds when the alveolin domain is expressed alone (Fig. 6B). Additional deletion and chimeric constructs suggest that sequence is required on both ends of the alveolin domain to obtain the basal localization in the buds but the sequence does not have to be IMC8 specific (Fig. 6B, panels 5 and 6). These results suggest that the alveolin domain is necessary for IMC3 and IMC8 localization and sufficient for IMC3 localization in a wild-type background.
Fig. 6.
The alveolin domain contains the localization signal differentiating cortical from basal IMCs. IMC3 (A) and IMC8 (B) are dissected to determine whether the N-terminus (“N”), alveolin domain (“A”), or C-terminus (“C”) is/are responsible for their localization patterns. The domains indicated above or below each panel are fused to an N-terminal YFP and all constructs were driven by the ptub promoter.
Discussion
Our data show that the 14-member sub-family of alveolin motif containing intermediate filament-like proteins has distinct dynamics and localization patterns throughout tachyzoite development, which include several phenomena not previously described. Based on our findings we established a development time line outlined in Figures 7A. Formation of the daughter cytoskeleton begins with the localization of IMC15 to the centrosome at approximately the time of duplication. All other IMC proteins except IMC7, 12, and 14 assemble and grow with the daughter buds, with IMC3, 6, and 10 exhibiting significant abundance in the daughter buds compared to the mature mother IMC. Halfway through daughter formation IMC5, 8, 9, and 13 shift their localization to the basal complex and the basal complex starts to constrict (Fig. 5). Completion of cytokinesis coincides with the appearance of three distinct regions within the basal complex (Fig. 7B). At this point IMC1, 3, 4, 6, 10, and 15 are present along the full length of the cortical cytoskeleton. IMC11 and 15 occupy the apical cap subdomain of the IMC, and IMC15 is additionally observed at the very apical and basal ends of the IMC. Now in G1, IMC7, 12, and 14 begin to appear at the cortical IMC of the mature parasites. This series of events completes a full development cycle.
Fig. 7.
Summarizing schematics of the IMC protein dynamics throughout tachyzoite development and the structure of the basal cytoskeleton. (A) Groups of IMC proteins with a similar behavior are shown in the same color and the groups are introduced at the stage of their defining role; among the yellow, cortical IMC proteins, the ones with a preference for the immature buds are outlined (IMC3, 6, and 10). IMC11 is not included. (B) The tentative structure of the basal complex in mature parasites is composed of three layers. The top layer (green) is composed of MORN1, IMC9, IMC13, and IMC15, the middle layer of IMC5 and 8 and the very basal tip contains centrin2, which overlaps with the middle layer. Data in Figure 5C do not include clear candidates for the bend of the inner collar toward the plasma membrane seen by EM (Fig. 5D–H). Interpretation of the posterior cup is based on data presented in (Mann et al., 2001).
The behavior of the various IMC proteins appears to be controlled at three different levels: expression patterns, primary protein sequences, and post-translational modifications. At the transcriptional level three different expression patterns were defined (Fig. 1B). At the primary protein sequence level we showed that the alveolin motif is a determinant for localization to either the cortical or basal sections of the cytoskeleton (Fig. 6). However, it is possible that the IMC proteins first assemble into homo-dimers or -oligomers before being targeted to cytoskeleton as in other intermediate filament systems (Herrmann et al., 2004, Parry et al., 2007). The alveolin domain deletion constructs could oligomerize with the untagged full-length native IMCs and be carried along to the appropriate location. Until the alveolin domains alone are shown to control the spatio-temporal patterns of the IMC proteins in gene-specific knockout backgrounds it must be considered that localization information for the IMC proteins could be contained in the N- or C-terminal domains as well. At the post-translational level protein modifications can influence the function and assembly of the IMC proteins. IMC1 undergoes proteolytic cleavage coinciding with a transition in filament skeleton rigidity (Mann et al., 2002). The shorter than expected bands for several IMC YFP fusion proteins in the western blots could be indicative of such proteolytic processing; however, at this point we can not exclude the possibility that these sizes could originate in degradation during sample preparation (Fig. S5). In other intermediate protein filament systems assembly is controlled by post-translational modifications such as acylation and phosphorylation (Gruenbaum et al., 2005, Herrmann et al., 2004). Several of the IMC proteins contain predicted palmitoylation sites (Fig. 1A) and numerous potential phosphorylation sites (not shown). Our future work will focus on dissecting the contribution of post-translational modifications to IMC protein function and behavior.
IMC15 colocalizes with the centrosomes upon division and currently is the earliest known marker for the new daughter buds, appearing earlier than MORN1 (Fig. 4). A potential model is that the centrosomal association of IMC15 provides a cue for the start of daughter budding by recruiting MORN1 and the other IMC proteins. Regardless of the exact mechanism, IMC15 highlights a key step in the connection between the cell cycle and mitosis. Elucidating this connection will be the focus of our future work.
The timing of the shift of IMC5, 8, 9, and 13 from the small buds to the basal complex is reminiscent of the timing of the previously described assembly of centrin2 on the basal complex, which marks the start of basal complex constriction to establish the tapered basal end of the cytoskeleton (Figs. 5,7A). Centrin2 has been suggested to provide the constrictive force (Hu, 2008). Daughter maturation and emergence coincides with relative localization shifts within the basal complex to generate three discernable regions that could coincide with the BIR and BIC revealed by EM (Fig. 7B, 5E–G). Moreover, the BIC appears to bend over the alveoli and connect with the plasma membrane, providing the only visible connection between the IMC and plasma membrane. However this bend is not observed at all times and could either be transient or be a transitional architecture in development. A possible function besides the constriction of the forming daughters for this complicated basal structure is the maintenance of cellular integrity. Host cell invasion is accompanied by significant forces on the cytoskeleton when the parasite squeezes through a narrow aperture. Where the conoid reinforces the apical end, the basal complex could reinforce the basal end. A function for the basal IMC proteins in the basal complex would predict that the basal IMC proteins would be conserved across the Apicomplexa, especially since the role of major basal complex component MORN1 is conserved across division modes in the Apicomplexa (Ferguson et al., 2008). A posterior cup as observed for Toxoplasma tachyzoites (Mann et al., 2001) is also present in E. tenella merozoites (Russell et al., 1984). However, the basal structure of Plasmodium sporozoites contains a branching ER with a posterior polar ring, but this is not an as clearly defined a structure as in the Toxoplasma tachyzoite (Kudryashev et al., 2010). Although it is possible that different development stages have different posterior structures, the basal IMC proteins are very poorly conserved across the Apicomplexa (see Table S3), which could support that the structure of the posterior end or basal complex is not necessarily well conserved across the Apicomplexa. Lastly, potential stage specific roles for IMC proteins have been suggested by differential expression profiles across Plasmodium berghei life stages (Tremp et al., 2008).
An unexpected event is the appearance of IMC7, 12, and 14 in the G1 phase of the cell cycle (Fig. 3). At this point the cytoskeleton is fully formed so the need for these additional components is puzzling. One possible mechanism is that the G1 phase separates two subsequent cytokinetic events and these IMC proteins serve as markers to distinguish the mother cytoskeleton from future budding daughters. This model complements one of the most remarkable features of internal budding by Toxoplasma: it takes place in the presence of a mature mother cytoskeleton that at some point must be disassembled concurrently with the maturation of the daughters. Although this is a speculative model, it would resolve several issues presented by differential stability of mother and daughter during cytokinesis. Most Apicomplexa divide through schizogony wherein the mother’s IMC is disassembled long before new daughters are being assembled (Ferguson et al., 2008) and, thus, do not require features to differentiate mother and daughter. The possibility of IMC7, 12, and 14 playing the role of maturity marker is supported by the lack of orthologs in other Apicomplexa. For IMC14 no robust direct orthologs were identified, but hits with significant e-values were present (Table S3). Unlike IMC7, 12, and 14, and the basal IMC proteins, IMC1, 3, 4, 6, and to a lesser extent 10 are well conserved across the Apicomplexa (Table S3). Next to Neospora, convincing IMC15 orthologs were only identified in the Plasmodia. It should be noted that IMC proteins not orthologous to IMC1, 3, 4, 6 or 10 are present in other Apicomplexa (Gould et al., 2008).
Taken together, several IMC proteins are conserved, likely playing key roles in cytoskeleton assembly. However, expansion of the IMC protein family in Toxoplasma appears to have created new functions for several IMC proteins potentially not shared across the Apicomplexa. In our future work we will address the function of the various IMC proteins by creating direct and conditional knockouts. Given the potential redundancy of IMC proteins with similar behavior, multi-gene knockouts might be required to better define their function. Furthermore, gene knockouts will provide us with a model to directly study the domain requirements for targeting IMC proteins.
Materials and methods
Parasites
RH strain parasites and transgenic derivatives were used throughout this study. Parasites were maintained in human foreskin fibroblasts (HFF) as previously described (Roos et al., 1994). Stable parasites expressing transgenes were selected under chloramphenicol or pyrimethamine pressure and cloned by limiting dilution.
RACE
For IMC genes 7, 9, and 12–15 we determined the 5′- and 3′-ends by RACE using the GeneRacer kit (Invitrogen) according to the manufacturer’s instruction. All primer sequences are provided in Supplementary Table S4.
Sequence analysis
The IMC gene family was identified through reciprocal BLASTP searches of the T. gondii genome of the ME49 strain in ToxoDB version 5.1 using a cut-off e-value of 10−3 (Gajria et al., 2008, Kissinger et al., 2003). The alveolin repeat domains of each IMC were determined by first identifying the VPV and EKIVEVP repeats as defined in (Gould et al., 2008) and then analyzing the protein sequences for novel repetitive regions with the REPRO program (George et al., 2000). The final determination of each alveolin domain’s boundaries was done manually based on the V/E/K/Q/R/P richness of the domain. Predictions of lipid modification (myristoylation, farnesylation, geranylgeranylation, palmitoylation) were performed using the suite available at http://mendel.imp.ac.at/mendeljsp/index.jsp, (Maurer-Stroh et al., 2005, Ren et al., 2008)).
Plasmids
All PCR primer sequences are provided in Supplementary Table S4. Plasmids ptub-YFP-IMCx/sagCAT and ptub-IMCx-YFP/sagCAT are based on the ptub-YFP2(MCS)/sagCAT plasmid (Gubbels et al., 2003) with an extra multiple cloning site containing EcoRV and XmaI/SmaI introduced behind the stop codon of the second YFP based on the ptub-CherryRFP2/sagCAT plasmid (kindly provided by Giel van Dooren and Boris Striepen, University of Georgia). Complete ORFs were amplified from cDNA generated from either Type I (RH strain) or Type II (Prugniaud strain; cDNA library kindly shared by Peter Bradley, UCLA) tachyzoites. Amplicons were BglII/AvrII cloned for C-terminal YFP fusions or AvrII/EcoRV or AvrII/XmaI for N-terminal YFP fusions. The same strategy was employed to generate cherryRFP fusions. 1500 bp endogenous promoters were PCR amplified from genomic DNA and cloned by PmeI/BglII into ptub-YFP-IMCx/sagCAT or ptub-IMCx-YFP/sagCAT. Deletion and chimeric proteins were cloned by PCR-fusion amplification of select domain inserts flanked with AvrII/EcoRV sites (Szewczyk et al., 2006) and cloned into ptubYFP2(MCS)/sagCAT.
Generation of antisera
The full length ORF of IMC11 and the 5′-end before the alveolin repeats from IMC3 (1-120 aa), IMC5 (1-350 aa), and IMC15 (1-350 aa) were amplified from cDNA and cloned into plasmid AVA0421 (Alexandrov et al., 2004) to generate a His6 N-terminal fusion. Fusion proteins were expressed in BL21 E. coli (Invitrogen) and purified over TALON Resin (Clontech). Polyclonal antisera were generated by rat immunizations (Covance). Antibodies were affinity purified against corresponding recombinant protein cross-linked to cyanogen bromide Sepharose 4B (Sigma) (Harlow et al., 1988).
Immunofluorescence assays
IFAs were performed as described (Gubbels et al., 2006). The following primary antibodies were used: rat anti-IMC3, rat anti-IMC5, rat anti-IMC15, rabbit anti-IMC3 (Gubbels et al., 2004), rabbit anti-MORN1 (Gubbels et al., 2006), MAb 45:36 IMC1 (kindly provided by Gary Ward, Univ. Vermont), rabbit anti-human centrin (kindly provided by Iain Cheeseman, Whitehead Institute), rabbit anti-GAP45 (Gaskins et al., 2004). Secondary antibodies conjugated to Alexa350, 488 or 594 were used (Invitrogen). Nuclear material was co-stained with 4′,6-diamidino-2-phenylindole (DAPI).
Fluorescence microscopy
A Zeiss Axiovert 200M wide-field fluorescence microscope equipped with a α-Plan-Fluar 100x/1.45 NA oil objective, and a Hamamatsu C4742-95 CCD camera was used. In addition, a Leica TCS SP5 inverted confocal microscope with a 100x/1.4 NA oil objective was used. Time-lapse microscopy was performed on the Zeiss microscope. Images were analyzed and processed using Openlab and Volocity (Improvision).
Electron microscopy
Intracellular parasites were fixed in 4% glutaraldehyde in 0.1 M phosphate buffer pH 7.4 and processed for routine electron microscopy (Ferguson et al., 1999). In summary, cells were post-fixed in OsO4 and treated with uranyl acetate prior to dehydration in ethanol, treatment with propylene oxide, and embedding in Spurr’s epoxy resin. Thin sections were stained with uranyl acetate and lead citrate and examinaned with a JEOL 1200EX electron microscope.
Supplementary Material
Acknowledgments
We thank Chris Kvaal, Giel van Dooren, Boris Striepen, Gary Ward, Iain Cheeseman, Peter Bradley, Nicole van Poppel, Dick Schaap, and John Murray for generously sharing reagents and Michael White and Michael Behnke for providing genome wide expression profiles as well as reagents. We also thank Marisha Collins and Kathleen Moorehouse for cloning assistance.
This work was supported by a Smith Family Foundation New Investigator grant, a Knights Templar Eye Foundation Research grant, and a National Institutes of Health grant (AI081924) awarded to MJG.
References
- Agop-Nersesian C, Naissant B, Ben Rached F, Rauch M, Kretzschmar A, Thiberge S, et al. Rab11A-controlled assembly of the inner membrane complex is required for completion of apicomplexan cytokinesis. PLoS Pathog. 2009;5:e1000270. doi: 10.1371/journal.ppat.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov A, Vignali M, LaCount DJ, Quartley E, de Vries C, De Rosa D, et al. A facile method for high-throughput co-expression of protein pairs. Mol Cell Proteomics. 2004;3:934–938. doi: 10.1074/mcp.T400008-MCP200. [DOI] [PubMed] [Google Scholar]
- Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson DJ, Cesbron-Delauw MF, Dubremetz JF, Sibley LD, Joiner KA, Wright S. The expression and distribution of dense granule proteins in the enteric (Coccidian) forms of Toxoplasma gondii in the small intestine of the cat. Exp Parasitol. 1999;91:203–211. doi: 10.1006/expr.1998.4384. [DOI] [PubMed] [Google Scholar]
- Ferguson DJ, Sahoo N, Pinches RA, Bumstead JM, Tomley FM, Gubbels MJ. MORN1 has a conserved role in asexual and sexual development across the Apicomplexa. Eukaryot Cell. 2008 doi: 10.1128/EC.00021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajria B, Bahl A, Brestelli J, Dommer J, Fischer S, Gao X, et al. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 2008;36:D553–556. doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskins E, Gilk S, DeVore N, Mann T, Ward G, Beckers C. Identification of the membrane receptor of a class XIV myosin in Toxoplasma gondii. J Cell Biol. 2004;165:383–393. doi: 10.1083/jcb.200311137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George RA, Heringa J. The REPRO server: finding protein internal sequence repeats through the Web. Trends Biochem Sci. 2000;25:515–517. doi: 10.1016/s0968-0004(00)01643-1. [DOI] [PubMed] [Google Scholar]
- Gilk SD, Raviv Y, Hu K, Murray JM, Beckers CJ, Ward GE. Identification of PhIL1, a novel cytoskeletal protein of the Toxoplasma gondii pellicle, through photosensitized labeling with 5-[125I]iodonaphthalene-1-azide. Eukaryot Cell. 2006;5:1622–1634. doi: 10.1128/EC.00114-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SB, Tham WH, Cowman AF, McFadden GI, Waller RF. Alveolins, a new family of cortical proteins that define the protist infrakingdom Alveolata. Molecular biology and evolution. 2008;25:1219–1230. doi: 10.1093/molbev/msn070. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nature reviews. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- Gubbels MJ, Li C, Striepen B. High-throughput growth assay for Toxoplasma gondii using yellow fluorescent protein. Antimicrob Agents Chemother. 2003;47:309–316. doi: 10.1128/AAC.47.1.309-316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbels MJ, Vaishnava S, Boot N, Dubremetz JF, Striepen B. A MORN-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. J Cell Sci. 2006;119:2236–2245. doi: 10.1242/jcs.02949. [DOI] [PubMed] [Google Scholar]
- Gubbels MJ, Wieffer M, Striepen B. Fluorescent protein tagging in Toxoplasma gondii: identification of a novel inner membrane complex component conserved among Apicomplexa. Mol Biochem Parasitol. 2004;137:99–110. doi: 10.1016/j.molbiopara.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: a laboratory manual Cold Spring Harbor. Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hartmann J, Hu K, He CY, Pelletier L, Roos DS, Warren G. Golgi and centrosome cycles in Toxoplasma gondii. Mol Biochem Parasitol. 2006;145:125–127. doi: 10.1016/j.molbiopara.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu Rev Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- Hu K. Organizational changes of the daughter basal complex during the parasite replication of Toxoplasma gondii. PLoS Pathog. 2008;4:e10. doi: 10.1371/journal.ppat.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Johnson J, Florens L, Fraunholz M, Suravajjala S, DiLullo C, et al. Cytoskeletal components of an invasion machine - the apical complex of Toxoplasma gondii. PLoS Pathog. 2006;2:121–138. doi: 10.1371/journal.ppat.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Mann T, Striepen B, Beckers CJ, Roos DS, Murray JM. Daughter cell assembly in the protozoan parasite Toxoplasma gondii. Mol Biol Cell. 2002;13:593–606. doi: 10.1091/mbc.01-06-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khater EI, Sinden RE, Dessens JT. A malaria membrane skeletal protein is essential for normal morphogenesis, motility, and infectivity of sporozoites. J Cell Biol. 2004;167:425–432. doi: 10.1083/jcb.200406068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger JC, Gajria B, Li L, Paulsen IT, Roos DS. ToxoDB: accessing the Toxoplasma gondii genome. Nucleic Acids Res. 2003;31:234–236. doi: 10.1093/nar/gkg072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryashev M, Lepper S, Stanway R, Bohn S, Baumeister W, Cyrklaff M, Frischknecht F. Positioning of large organelles by a membrane- associated cytoskeleton in Plasmodium sporozoites. Cell Microbiol. 2010;12:362–371. doi: 10.1111/j.1462-5822.2009.01399.x. [DOI] [PubMed] [Google Scholar]
- Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- Leander BS, Keeling PJ. Morphostasis in alveolate evolution. trends in Ecology and Evolution. 2003;18:395–402. [Google Scholar]
- Mann T, Beckers C. Characterization of the subpellicular network, a filamentous membrane skeletal component in the parasite Toxoplasma gondii. Mol Biochem Parasitol. 2001;115:257–268. doi: 10.1016/s0166-6851(01)00289-4. [DOI] [PubMed] [Google Scholar]
- Mann T, Gaskins E, Beckers C. Proteolytic Processing of TgIMC1 during Maturation of the Membrane Skeleton of Toxoplasma gondii. J Biol Chem. 2002;277:41240–41246. doi: 10.1074/jbc.M205056200. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Eisenhaber F. Refinement and prediction of protein prenylation motifs. Genome Biol. 2005;6:R55. doi: 10.1186/gb-2005-6-6-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissette NS, Sibley LD. Cytoskeleton of apicomplexan parasites. Microbiol Mol Biol Rev. 2002;66:21–38. doi: 10.1128/MMBR.66.1.21-38.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi M, Hu K, Murray JM, Roos DS. Organellar dynamics during the cell cycle of Toxoplasma gondii. J Cell Sci. 2008 doi: 10.1242/jcs.021089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DA, Strelkov SV, Burkhard P, Aebi U, Herrmann H. Towards a molecular description of intermediate filament structure and assembly. Experimental cell research. 2007;313:2204–2216. doi: 10.1016/j.yexcr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng Des Sel. 2008;21:639–644. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos DS, Donald RG, Morrissette NS, Moulton AL. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol. 1994;45:27–63. doi: 10.1016/s0091-679x(08)61845-2. [DOI] [PubMed] [Google Scholar]
- Russell DG, Burns RG. The polar ring of coccidian sporozoites: a unique microtubule-organizing centre. J Cell Sci. 1984;65:193–207. doi: 10.1242/jcs.65.1.193. [DOI] [PubMed] [Google Scholar]
- Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, et al. Fusion PCR and gene targeting in Aspergillus nidulans. Nature protocols. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Tremp AZ, Khater EI, Dessens JT. IMC1b is a putative membrane skeleton protein involved in cell shape, mechanical strength, motility, and infectivity of malaria ookinetes. J Biol Chem. 2008;283:27604–27611. doi: 10.1074/jbc.M801302200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.