Abstract
The cytidine deaminase AID hypermutates immunoglobulin genes but can also target oncogenes, leading to tumorigenesis. The extent of AID’s promiscuity and its predilection for immunoglobulin genes are unknown. We report here that AID interacted broadly with promoter-proximal sequences associated with stalled polymerases and chromatin-activating marks. In contrast, genomic occupancy of replication protein A (RPA), an AID cofactor, was restricted to immunoglobulin genes. The recruitment of RPA to the immunoglobulin loci was facilitated by phosphorylation of AID at Ser38 and Thr140. We propose that stalled polymerases recruit AID, thereby resulting in low frequencies of hypermutation across the B cell genome. Efficient hypermutation and switch recombination required AID phosphorylation and correlated with recruitment of RPA. Our findings provide a rationale for the oncogenic role of AID in B cell malignancy.
Somatic hypermutation (SHM) introduces non-templated point mutations in genes encoding immunoglobulin variable (V) domains at a frequency of about one mutation per kilobase (kb) per cell per generation. In germinal centers, this activity generates antibody variants that are selected on the basis of their affinity for antigen. In addition to altering antibodies via hypermutation, the germinal center response also shapes the effector function of antibody genes through class-switch recombination (CSR). This reaction replaces the μ-chain immunoglobulin heavy-chain locus constant (C) region (Igh-C) for one of a set of downstream Igh-C exons1,2.
AID initiates both CSR and SHM3,4 by deaminating cytidine residues in single-stranded DNA (ssDNA), which is exposed during immunoglobulin transcription by RNA polymerase II (PolII)5,6. The resulting U:G mismatches are processed by base excision–repair and mismatch-repair pathways that produce mutations or double-strand DNA breaks, which are obligate intermediates for CSR5,7. During SHM, AID seems to act mainly in a region 1.5 kb downstream of the transcription start sites (TSSs) of genes encoding immunoglobulin V domains8, whereas CSR-related double-strand DNA breaks map to switch (S) regions 1–12 kb in length that precede the participating Igh-C exons.
AID can also deaminate non-immunoglobulin genes, including CD79A, CD79B, MYC, RHOH, PIM1, PAX5, BCL6 and MIR142 and their mouse homologs9–13. Mutations occur at a lower frequency at such off-target sites than at immunoglobulin genes, in part because the initial lesions are repaired with high accuracy by physiological base excision–repair and mismatch-repair pathways12. Nevertheless, lesions in off-target sites for AID can cause deleterious mutations and large-scale chromosomal abnormalities that result in B cell lymphomas14. AID activity has also been linked to blast crisis progression in chronic myeloid leukemia15, prostate malignancies16 and gastric tumors17.
SHM outside the immunoglobulin loci may not be entirely pathological. AID is expressed in pluripotent tissues and has been shown to deaminate 5-methylcytosines in vitro18, which has led to the suggestion that it might mediate DNA demethylation in vertebrates19–21. Promiscuous AID activity, therefore, may promote developmental reprogramming in the embryo and potentially in activated B lymphocytes.
Several AID cofactors have been identified so far, including the ssDNA-binding protein RPA and protein kinase Ar1α22–26. RPA is of particular interest because of its established role in DNA recombination and repair27. In biochemical assays, RPA promotes the deamination of transcribed substrates by AID by stabilizing its interaction with ssDNA22, which suggests that the role of RPA in SHM and CSR is to provide access of AID to target DNA. In vivo, the formation of RPA-AID complexes is facilitated by phosphorylation of AID at Ser38 by protein kinase Ar1α23,26,28, and AID, RPA and protein kinase Ar1α all associate with sites of switch recombination, as determined by chromatin immunoprecipitation (ChIP)24,29.
Despite the importance of AID in shaping the antibody response and in promoting malignancy, there is little understanding of how immunoglobulin genes are preferentially hypermutated, the extent of AID off-target activity or how AID finds its ssDNA substrate near TSSs. To address these issues, we have defined the genome-wide association of AID and RPA in the context of the activated B cell epigenome, transcriptome and PolII. We found enrichment for AID across the genome at pausing sites for PolII, with the greatest abundance at the immunoglobulin μ-chain gene (Igh-6). Thus, our data support and extend the observation that Spt5, a factor required for polymerase stalling and CSR, is required for the association of AID with the transcribing holoenzyme30. In contrast, however, RPA associated mainly with the immunoglobulin locus, and this interaction was dependent on phosphorylation AID at Ser38 and Thr140. We propose that interaction of AID with Spt5-PolII complexes results in deamination of DNA across the genome in a manner that is proportional to the amount of AID recruited. However, optimal SHM and CSR may also require the recruitment of additional AID cofactors.
RESULTS
AID ChIP-seq in activated B cells
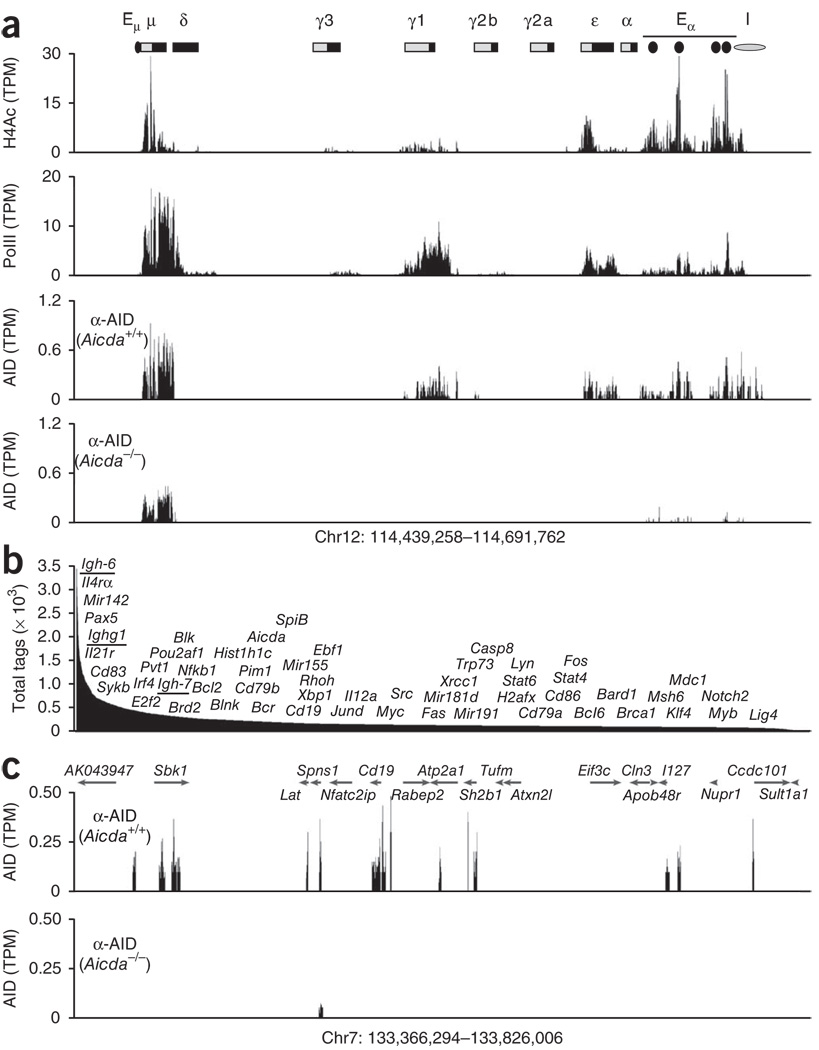
Although AID seems to target many non-immunoglobulin genes9,10,12,31, the extent of its off-target activity is not known. To define the genome-wide occupancy of AID in B cells, we stimulated AID-sufficient (Aicda+/+) B cells and AID-deficient (Aicda−/−) control B cells with lipopolysaccharide (LPS) and interleukin 4 (IL-4) and analyzed the cells by ChIP coupled with deep sequencing (ChIP-seq) using antibody to AID (anti-AID). These culture conditions induce AID expression and CSR to IgG1 (Ighg1) or IgE (Igh-7). As expected, AID was immunoprecipitated from the Igh μ-chain (Igh-6), γ1-chain (Ighg1) and ε-chain (Igh-7) (Fig. 1a), which are recombining, actively transcribed (as determined by the association of PolII with chromatin) and acetylated at histone H4 (Fig. 1a). Unexpectedly, outside the Igh locus, AID was also associated with as many as 5,910 genes (or 12,200 islands; Fig. 1b and Supplementary Table 1). Analogous to published ChIP-seq studies32, 396 genes (6.7%) also showed background signals in Aicda−/− cells (Supplementary Fig. 1a), probably because of the presence of sonication-hypersensitive chromatin in these sites (such as Spns1 (Fig. 1c) or Igh-6 (Fig. 1a); Supplementary Text). Nevertheless, with few exceptions, AID islands were distinguishable from background noise (P < 0.0005 (Wilcoxon rank-sum test); Supplementary Fig. 1b), and saturation studies confirmed the specificity of the AID islands observed in Aicda+/+ cells (Supplementary Fig. 2a).
Figure 1.
Extent of AID recruitment in activated B cells. (a) ChIP-seq analysis of the Igh locus (chromosome 12 (Chr12)) in B cells stimulated with LPS and IL-4, showing acetylation of histone H4 (H4Ac), as well as recruitment of PolII and AID; results are presented as sequence tags per million sequences (TPM) in 100-bp windows. Bottom row, specificity of AID immunoprecipitation, assessed in Aicda−/− cells with an AID-specific antibody (α-AID)22. Above, Igh locus, showing the S domains (light gray boxes) and C domains (black boxes) as well as the 5′ μ-chain enhancer (Eμ), 3′ α-chain enhancer (Eα) and insulator (I). (b) Total sequence tags of AID-recruiting genes (n = 5,910), showing the approximate positions of a subset of AID targets, including immunoglobulin genes (underlined). (c) Recruitment of AID at a domain of chromosome 7 ~500 kb in length containing sequence encoding the B cell–specific CD19 antigen, assessed in Aicda+/+ and Aicda−/− (control) cells. Data are representative of four independent experiments.
Ranking of genes associated with AID based on the absolute number of sequence tags positioned Igh-6 (encoding IgM) as the gene with the greatest recruitment of AID (Fig. 1b). This was followed by other previously characterized AID targets, including Pax5, Il21r, Cd83, Sykb, Pou2af1, Pim1, Cd79a, Cd79b, Aicda, Ebf1, Myc and H2afx, among others (complete list, Supplementary Table 1). Notably, Mir142, a Myc translocation partner in mouse and human B cell tumors14, was among the 15 genes with the greatest recruitment of AID (Supplementary Table 1). AID was also immunoprecipitated from Bcr and SpiB (Fig. 1b), presumed AID targets associated with chronic myelogenous leukemia15 and a subset of diffuse large B cell lymphomas33, respectively. Among the previously unknown targets identified, we found genes encoding microRNAs expressed in activated lymphocytes34 (Mir155, Mir181d and Mir21), transcription factors involved in cell differentiation (Stat6, Xbp1 and Nfkb1) and DNA-repair proteins (Brca1, Mdc1 and Lig4; Fig. 1b and Supplementary Table 1). We conclude that AID is recruited to a large number of genes in activated B cells.
AID recruitment indicates hypermutation
To confirm the ChIP-seq results for AID reported above, we obtained B cells from mice transgenic for expression of AID under the control of a promoter-enhancer cassette from the immunoglobulin κ-chain gene (Igk) and deficient in uracil DNA glycosylase (Igk-AID Ung−/− mice)9, then stimulated the cells with LPS and IL-4 and measured hypermutation. On the basis of results from a sample of nearly two dozen genes, these B cell cultures have been shown to achieve hypermutation frequencies similar to those measured in germinal centers and to maintain target selection specificity9,30. In agreement with the published data, mutation frequencies at five documented AID targets (Pim1, Myc, Pax5, Cd83 and H2afx) in Igk-AID Ung−/− cells were similar to those reported before for Ung−/− Peyer’s patch B cells also deficient in mutS homolog 2 (Msh2−/−)12 (Supplementary Table 2). In comparison, Igk-AID Ung−/− cultures had four times more hypermutation and more transition mutations than did Igk-AID Ung+/+ cultures (Supplementary Fig. 3).
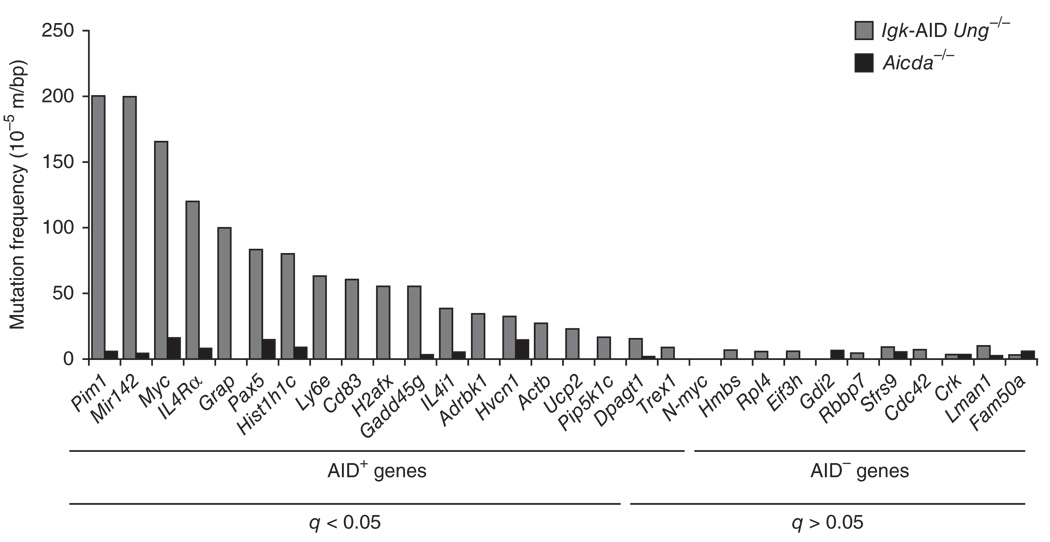
To examine additional predicted AID target genes, we amplified ~750 base pairs (bp) of DNA downstream of TSSs of genes associated with AID (n = 13) or not associated with AID (n = 11) and analyzed the sequences for point mutations. We analyzed a total of 1,496,058 bp. Most of the genes that recruited AID had significantly more mutations in Igk-AID Ung−/− B cells than in the background control Aicda−/− B cells (Fig. 2 and Supplementary Table 2). Among the genes with the most mutations (beyond the known targets of AID) were Il4ra, Grap, Hist1h1c, Ly6e, Gadd45g and Il4i1 (Fig. 2 and Supplementary Table 2). Notably, more than 86% of all mutations were C:G to T:A transitions and showed a strong bias toward RGYW-WRCY hot spots (Supplementary Table 2), which confirmed that they were indeed the result of AID deamination activity in the absence of Ung. In contrast, the mutation frequency of genes associated with AID that were highly transcribed was not distinguishable from background (q > 0.05; Fig. 2 and Supplementary Table 2). These results support the view that ChIP-seq for AID can be used to reliably predict the presence of AID activity.
Figure 2.
AID targets are somatically hypermutated. Mutation frequencies of genes that recruited AID (AID+; n = 9) or did not recruit AID (AID−; n = 11) in activated Igk-AID Ung−/− B cells and Aicda−/− B cells. Bottom, adjusted P values (q), calculated to control for false-discovery rate (Fisher’s exact test). Data are representative of one experiment.
AID preferentially targets genes in open chromatin
Transcription is required for AID activity in vitro and in vivo1,5,6,35. However, the relationship between transcription and AID targeting has been difficult to define because only a small number of genes have been assayed and this has been done in a limited way. To address this issue comprehensively, we used mRNA deep sequencing and ChIP-seq to compare AID, transcription and epigenetic marks on a genome-wide basis.
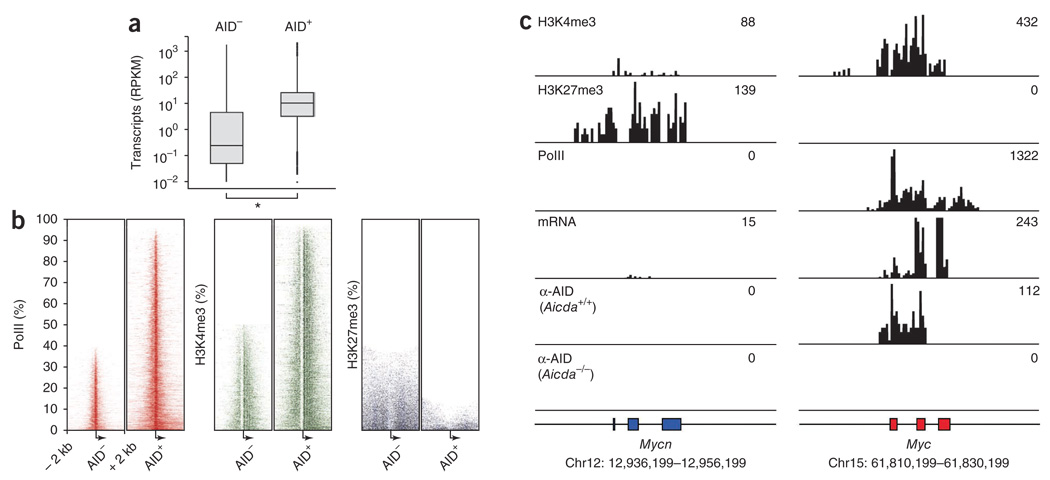
The genes that recruited AID (n = 5,910) had a median mRNA abundance 40 times greater than that of genes that did not recruit AID (n = 12,775; Fig. 3a and Supplementary Table 1). Consistent with that result, we detected PolII and trimethylated histone H3 Lys4 (H3K4me3), which are associated with gene activation36,37, in essentially all (>95%) genes associated with AID, compared with <50% of genes not associated with AID (Fig. 3b). Conversely, genes that recruited AID showed depletion of the polycomb group–inhibitory mark H3K27me3 (<12%), whereas it was associated with nearly 40% of those that did not recruit AID (Fig. 3b). These features were particularly evident at the Myc locus and Mycn locus (encoding N-Myc). Myc is expressed and preferentially translocated to Igh in mature B cells, whereas Mycn is not. Consistent with those observations, H3K4me3 marks, the presence of PolII and mRNA synthesis correlated with substantial association of AID at the Myc locus (Fig. 3c), whereas Mycn, which is not expressed in activated B cells, showed silencing by H3K27me3, had little H3K4me3 and lacked AID (Fig. 3c). Furthermore, we did not detect hypermutation at Mycn, whereas Myc intron 1 carried about 1 × 103 mutations per bp (Supplementary Table 2). Notably, the association of AID with Myc coincided with the mapped location of most AID-induced DNA breaks and canonical translocation breakpoints (at Myc exon 1–intron 1). Thus, we conclude that the interaction of AID with genes across the genome is biased toward genes associated with an open chromatin configuration. This feature provides a rationale for the greater incidence of Myc translocations than Mycn translocations in B lymphoid tumors.
Figure 3.
AID recruitment is biased toward actively transcribed genes associated with an open chromatin configuration. (a) Deep-sequencing analysis of mRNA for genes that recruited or did not recruit AID. Transcript abundance is presented as total mRNA sequences per gene length (in kb) per million aligned reads (RPKM), including the median (small horizontal lines), quartiles (vertical lines) and outliers (bolding). *P < 0.0005 (Wilcoxon rank-sum test). (b) Heat maps of ChIP-seq results for PolII, H3K4me3 and H3K27me3 at all (100%) genes that recruited or did not recruit AID, rank-ordered from least (top) to most (bottom) in terms of their association with PolII or chromatin modifications and centered on the TSS (arrow) of each gene ± 2 kb. P < 0.0005 (χ2 test). (c) Aligned ‘read’ density for H3K4me3, H3K27me3, PolII, mRNA and AID at Mycn and Myc loci; numbers in plots (top right) indicate total sequenced tags aligned in each genomic domain. Data are representative of one experiment.
AID recruitment follows PolII distribution
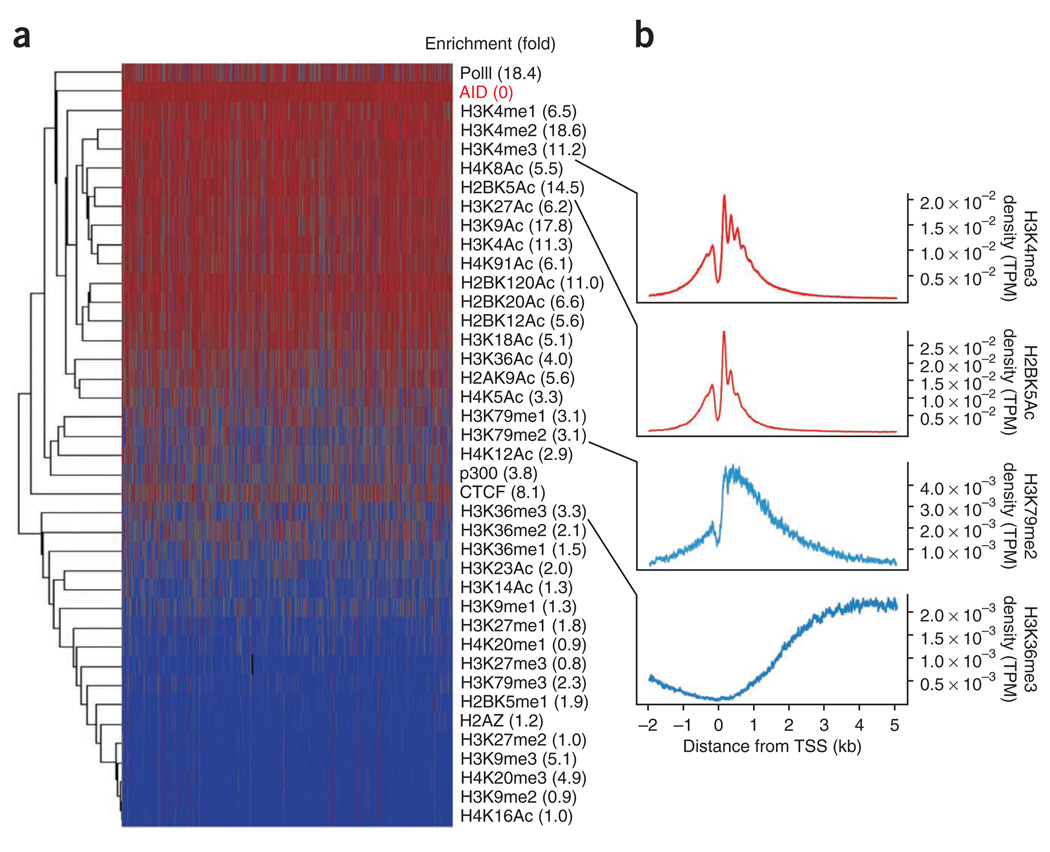
Despite the general correlation between mRNA accumulation and AID, our findings also indicated that transcription itself cannot be used to predict genome-wide recruitment of AID. Instead, a considerable fraction of genes with no detectable AID were abundantly transcribed (Fig. 3a, and Supplementary Table 1), including most of the genes assayed above that did not recruit AID (Fig. 2 and Supplementary Table 2). In addition, there was a wide variation in mRNA accumulation among genes targeted by similar amounts of AID (Supplementary Table 1). To clarify the precise nature of the interaction of AID with genes across the B cell genome, we determined the overlap between AID islands and histone modifications (n = 36), the insulator protein CTCF, the enhancer-binding acetyltransferase p300 and PolII in activated B cells (650,005 islands analyzed in total). For each variable, we calculated both its normalized Euclidean distance (Fig. 4a, dendrogram, and Supplementary Table 3) and its enrichment (Fig. 4a, right margin) relative to those in a random background model. These analyses confirmed the overall link between AID and active chromatin. For example, in addition to H3K27me3 (Fig. 3b), the inhibitory marks H4K20me1, H3K9me2 and H3K27me2 were rarely associated with AID islands (0.77 ≤ enrichment (fold) ≤ 1.01, Fig. 4a and Supplementary Table 3). Furthermore, most histone acetylation modifications, which as a group promote transcriptional activation, were closely associated with AID (Fig. 4a). Notably, hierarchical clustering of the distance matrix also indicated a preference of AID for promoter-proximal sequences (Supplementary Table 3). Methylation and acetylation marks that typically demarcate promoters of active genes (H3K4me1-H3K4me2-H3K4me3, H2BK5Ac, H3K27Ac and H3K9Ac) overlapped physically better with AID (6.17 ≤ enrichment (fold) ≤ 18.6) than did those showing enrichment at transcribed regions (H3K36me3, H3K79me2, H3K14Ac, H4K5Ac and H4K12Ac (1.26 ≤ enrichment (fold) ≤ 3.35); Fig. 4b and Supplementary Table 3).
Figure 4.
Epigenetic signature of AID recruitment. (a) Hierarchical clustering of AID, PolII, CTCF, p300 and 36 chromatin modifications on AID islands, presented as log2-transformed data and normalized Euclidean distances: red, high density; blue, low density. Numbers in parenthesis (right) indicate enrichment distance for each variable relative to that of AID. (b) Composite (metagene) profiles of ChIP-seq for H3K4me3, H2BK5Ac, H3K79me2 and H3K36me3, presented as tags per million aligned sequences per gene per nucleotide (density) versus position relative to the TSS (−2 kb to +5 kb). Data are representative of one experiment.
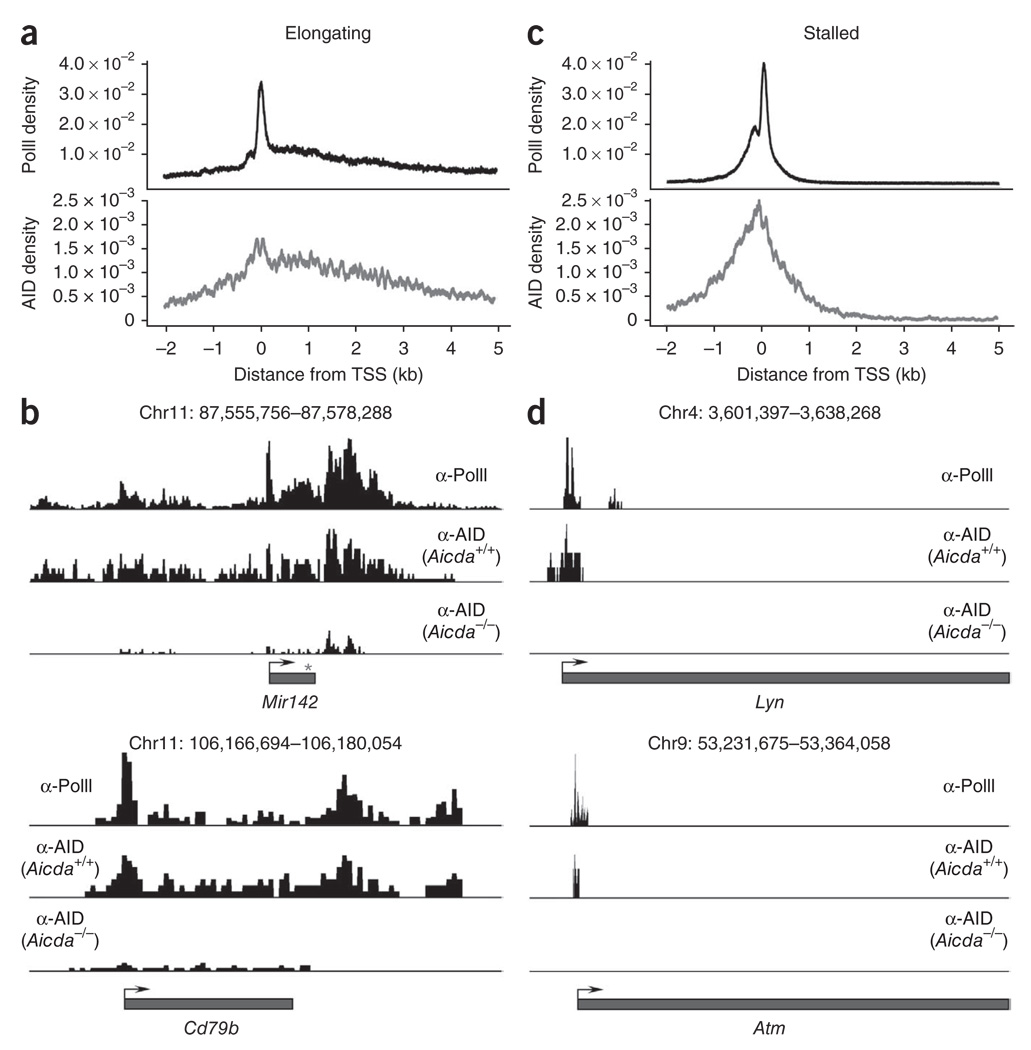
To confirm that AID has high occupancy of promoter regions, we generated composite profiles of AID and PolII around TSSs (from −2 kb to +5 kb relative to the TSS). On the basis of PolII-binding activity38, we classified AID-recruiting genes as either stalled (stalling index > 3 for n = 4,756 genes (80%)) or elongating (1 ≤ stalling index ≤ 3 for n = 352 genes (6%)). The stalling index (also known as the traveling or pausing index) is a measure of the ratio of the density of PolII at the promoter (±1 kb relative to the TSS) versus its density in the gene body. Rather than indicating the presence or absence of transcription, the stalling index reflects the dynamics of PolII assembly and promoter clearance38. In stalled genes, for example, the rate of promoter clearance is lower than that of holoenzyme assembly; as a result, PolII disproportionately accumulates (pauses) at promoter-proximal sequences. In contrast, in elongating genes, the rates of promoter assembly and clearance are more equivalent and thus PolII is more easily detected in gene bodies by ChIP-seq analysis38. We found that AID density closely matched the overall PolII profiles. In elongating genes, the density of AID and PolII peaked at the TSS and decreased thereafter to background density 4–5 kb downstream (Fig. 5a). Two illustrative examples of this correlation were Mir142 and Cd79b, for which AID density quantitatively mirrored, nearly peak by peak, the PolII-association profiles at the gene body and transcription termination sites (Fig. 5b). In stalled genes, both PolII and AID were immunoprecipitated mainly from a 2-kb area centered on the TSS that included the basal promoter (Fig. 5c). Lyn and Atm were both in this promoter-stalled category (Fig. 5d). On the basis of these findings we conclude that genome-wide AID occupancy mirrors PolII density. Furthermore, the overall bias of AID toward promoter-proximal sequences is explained by the fact that, as in embryonic stem cells38, most genes in activated B lymphocytes accumulate paused polymerases at promoter areas. These conclusions were further supported by the observation that the overall AID-recruitment profiles correlated better with PolII phosphorylated at Ser5 than with PolII phosphorylated at Ser2 (Supplementary Fig. 4). Of note, the former has been shown to associate mainly with transcriptional initiation and pausing, whereas sites of elongation and transcriptional termination show enrichment for the latter39.
Figure 5.
Genome-wide correlation between AID and PolII occupancy. (a) Composite profiles of the density of PolII and AID at elongating genes (n = 352) that recruit AID (presented as in Fig. 4b). (b) Quantitative correlation of PolII and AID at the Mir142 and Cd79b loci. Bottom row, microsequencing background in Aicda−/− cells. (c) Composite profiles of the density of PolII and AID at stalled genes (n = 4,756) that recruit AID (presented as in Fig. 4b). (d) Quantitative correlation of PolII and AID at the Lyn and Atm loci (stalled genes). Data are representative of one experiment.
In addition to interacting with gene domains, a fraction of AID islands (34%) were intergenic, with three fourths of these localizing together with PolII, p300 and/or CTCF (Supplementary Fig. 5a). A particularly good example of this was the 3′ Eα enhancer, where both PolII and AID distinctly immunoprecipitated from each of the enhancer elements (Fig. 1a; other examples, Supplementary Fig. 5b). As a group, these sites may represent promoters of unknown genes, true intergenic AID-binding sites and/or regulatory elements at which AID is crosslinked as a result of long-range interactions with promoters.
Direct deamination of basal promoters by AID
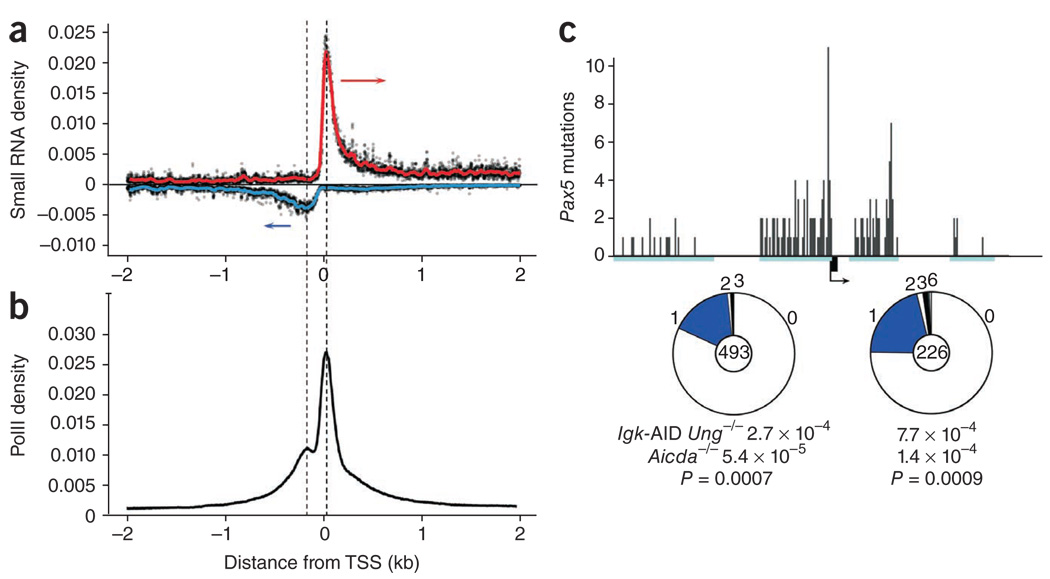
In addition to producing conventional elongation of gene bodies, mammalian RNA polymerases also transcribe promoters in an orientation opposite that of the annotated gene40. Because of efficient polymerase stalling38, the resulting antisense transcripts do not typically extend very far beyond the basal promoter40. In agreement with such studies, analysis of deep-sequenced RNA isolated from activated B cells34 showed that PolII-stalling profiles at promoters of B cell genes coincided precisely with the TSSs of both sense and antisense RNA (Fig. 6a,b).
Figure 6.
AID hypermutates basal promoters. (a) Alignment of sequences from small RNA cDNA libraries (n = 14.7 × 106) relative to gene TSS (± 2 kb) in activated B cells. Arrows indicate sense of transcription. (b) PolII density at all genes in B cells activated with LPS and IL-4. Dashed lines indicate the coincidence of the two stalling peaks with the TSSs of both sense and antisense RNA in a. (c) AID-mediated hypermutation of the Pax5 promoter-proximal area in Igk-AID Ung−/− B cells (top) detected from four PCR products (cyan rectangles); black box indicates Pax5 exon 1, and arrow indicates TSS and direction of transcription. Middle, mutation frequency: segment size indicates the proportion of sequences with the number of mutations noted along circle margin; center, total sequences. Bottom, mutation frequency (calculated as total mutations per total base pairs sequenced) and P value (relative to Aicda−/− control). Data are representative of one experiment.
Although our observations indicated that SHM is associated with PolII stalling, published studies have indicated that the mutation track is unidirectional, occurring mainly 3′ of TSSs8. To determine whether divergent PolII stalling also renders basal promoters susceptible to SHM, we analyzed six AID-recruiting genes (Pax5, Myc, Grap, Ighg1, Pim1 and Il4ra) for point mutations 5′ of their respective TSSs. Notably, except for Il4ra and Grap, all promoters had mutation frequencies above background in an AID-dependent manner (Fig. 6c and Supplementary Table 4). Mutation frequencies, which ranged between 1.7 × 10−4 and 4.7 × 10−4, were equivalent to those measured at gene bodies of other AID targets, such as Pax5 (7.0 × 10−4) or H2afx (5.5 × 10−4; Supplementary Table 2 and Supplementary Fig. 6). Consistent with the PolII and small RNA profiles at basal promoters (Fig. 6a,b), promoter mutations were abundant near the TSS and became increasingly rare at upstream sequences (Fig. 6c). Notably, the overall mutation spectrum, consisting almost entirely (>94%) of transition mutations, was biased toward RGYW-WRCY hotspots, showing the distinctive footprint of AID (Supplementary Table 4). Finally, we also found hypermutation of the Pax5 basal promoter in Peyer’s patches from Ung−/−Msh2−/− germinal centers (Supplementary Table 4), which demonstrated that hypermutation of this promoter also occurs as part of physiological immune responses. We conclude that AID can deaminate basal promoters in activated B cells in a manner consistent with divergent polymerase profiles. Together with the observation that PolII paused at immunoglobulin S core domains41,42 (Fig. 1a), our results further support the idea that SHM is targeted across the genome to sites of polymerase stalling30.
RPA is restricted to Igh loci
Co-occupancy of AID and PolII across the genome provides a mechanistic explanation for the widespread hypermutation observed in primary and transformed B lymphocytes. However, the disparity in the magnitude of the mutation load at immunoglobulin and off-target sites cannot be fully explained on the basis of AID-binding density. For example, whereas the association of AID with chromatin was ~1.5-fold greater at Igh-6 than at Mir142 (Supplementary Table 1), the mutation frequency at the 5′ end of the Igh-6 S domain (Sμ) was 10 times that measured at Mir142 (Supplementary Table 2). Furthermore, hypermutation at the Sμ core is probably orders of magnitude higher43.
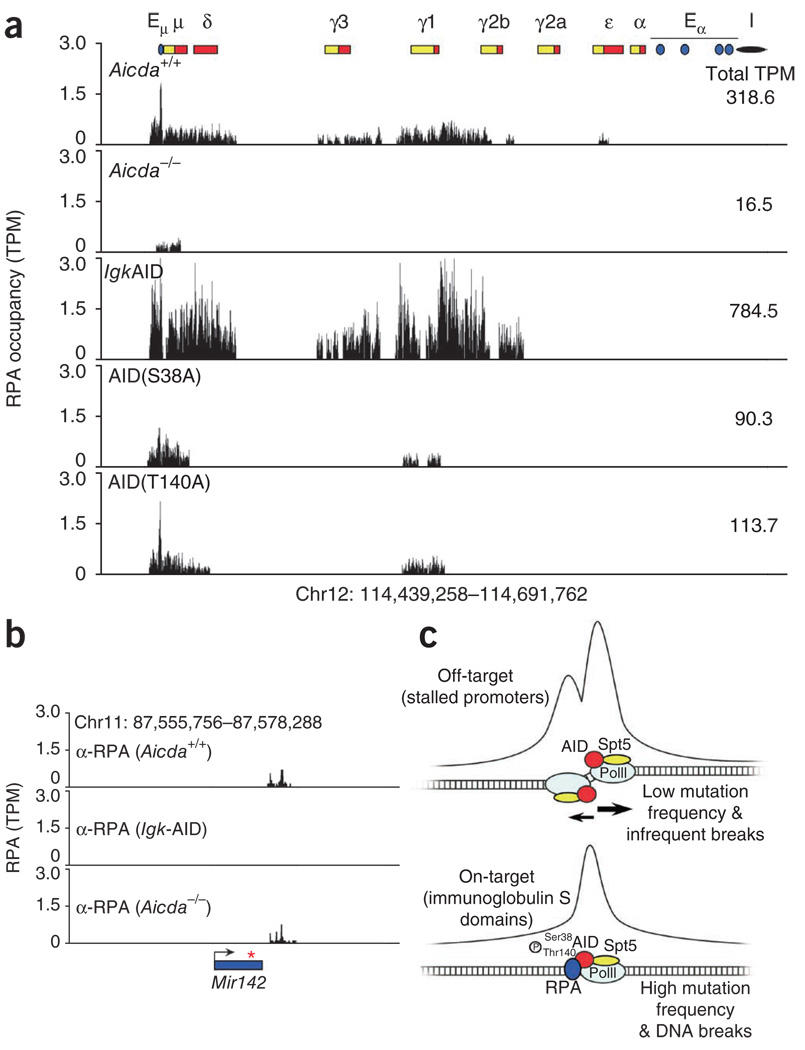
Several lines of evidence indicate that the ssDNA-binding protein RPA may have a role in hypermutation. In biochemical assays, RPA enhances the deamination of transcribed DNA by AID22,23, and RPA interacts in an AID-dependent manner with S regions in B cells undergoing CSR24. However, whether recruitment of RPA correlates with AID activity across the B cell genome or whether it is specific for immunoglobulin genes has not been fully explored. To determine the genomic occupancy by RPA in AID-expressing cells, we activated B cells with LPS and IL-4 and assayed the cells by ChIP-seq with RPA-specific antibodies. Two RPA islands were reproducibly immunoprecipitated from Igh; one spanned the entire Igh-6 μ-chain and one centered on the γ1-chain (Fig. 7a, top row). In contrast, only a very small number of sequence ‘reads’ aligned near Sμ in the absence of AID24 (Fig. 7a, top and second rows). Notably, Igk-AID B cells, which have higher AID expression9, had RPA signals 2.5 times more abundant than those measured in Aicda+/+ cells (784.5 versus 318.6 tags per million sequences; Fig. 7a, top and third rows). In cells undergoing CSR to the γ3-chain (cells treated with LPS plus anti-δ-dextran), as expected, RPA was associated with both gene segments encoding the μ-chain and γ3-chain (data not shown). Thus, RPA is recruited to Igh in a manner that is dependent on and directly proportional to AID expression.
Figure 7.
Recruitment of RPA to on-target sites of AID. (a) RPA occupancy at the immunoglobulin gene locus in B cells (genotype, top left) stimulated for 72 h with LPS and IL-4. AID(S38A) or AID(T140A), replacement of AID Ser38 or AID Thr140, respectively, with alanine. Numbers in plots (top right) indicate total sequence tags per million for the entire locus. (b) Background signals at the Mir142 locus after ChIP-seq analysis of RPA. Red asterisk indicates position of the microRNA in the microRNA transcript (arrow). (c) AID recruitment and activity in B lymphocytes. Top: AID accumulates predominantly at PolII pausing sites across the genome by interacting with the stalling factor Spt5. At proximal promoter areas, RPA is either absent or fails to accumulate in sufficient amounts to fully engage AID activity. This results in little hypermutation both upstream and downstream of TSSs. With few exceptions, hypermutation at these sites is efficiently erased by base excision–repair and mismatch-repair activity. Bottom: at on-target immunoglobulin sites, RPA is recruited as a result of site-specific phosphorylation (P) of AID at Ser38 and Thr140. This activity enhances DNA deamination and the formation of double-stranded DNA breaks, particularly in the core of immunoglobulin S domains, where PolII frequently stalls.
AID is phosphorylated at Ser38 and Thr140 in activated B cells23,25,26,44. Substitution of either residue does not affect AID deamination activity but interferes with SHM and CSR in vivo44,45. Furthermore, phosphorylation of Ser38 is required for the association of AID with RPA both in vitro23 and in vivo24. To assess the role of AID phosphorylation in genome-wide RPA recruitment, we did ChIP-seq for RPA with mutant B cells in which AID Ser38 or AID Thr140 is replaced with an alanine residue44. The association of RPA with chromatin was much less (about one third as many sequence reads) in both AID-mutant samples than in Aicda+/+ samples (Fig. 7a, fourth and fifth rows versus top row). These results indicate that phosphorylation of AID contributes to the interaction of RPA with Igh.
In contrast to the ChIP-seq results for AID, which identified nearly 6,000 genomic targets of AID (>12,000 islands), we identified only a small number of islands outside Igh by ChIP-seq for RPA (~200 per sample; Supplementary Table 5 and Supplementary Fig. 7). We detected most of these islands in only one of the four independent ChIP-seq experiments with Aicda+/+ cells (including Igk-AID), and those that we reproducibly detected in all samples (16 in total) were also present in the absence of AID (data not shown). Notably, a substantial fraction of non-immunoglobulin islands localized together with background noise obtained with Aicda−/− lymphocytes after ChIP-seq for AID (for example, the Mir142 locus; Figs. 5b and 7b). Our failure to detect RPA-specific targets outside Igh was not due to lack of sequence tag saturation, because a combined analysis of Aicda+/+ samples (n = 3) and Igk-AID samples (n = 2) analyzed by ChIP-seq for RPA (44,764,916 total tags) did not show substantial association of RPA with AID-recruiting genes (Supplementary Fig. 8 and Supplementary Table 6). We conclude that in contrast to genomic occupancy by AID itself, genomic occupancy by its cofactor RPA is restricted to immunoglobulin genes (Fig. 7c).
DISCUSSION
By means of deep sequencing, we have defined the global occupancy by AID and its cofactor RPA in the B cell genome. To better characterize the data, we further annotated the B cell genome by comprehensively mapping 36 epigenetic marks, the mRNA transcriptome, PolII, p300 and CTCF binding. We found that the association of AID with genes across the genome was widespread and correlated with activating chromatin marks. Among those we found acetylation of H3, H4 and H2B, as well as all three methylated forms of H3K4. By extension, genes with an inhibitory chromatin configuration (such as H3K27me3 and H3K20me3) failed to recruit AID or hypermutate. One example of this was Mycn, which is epigenetically and transcriptionally silent, does not bind AID and is rarely involved in chromosomal translocations in the mature B cell compartment46,47.
Our data have established a tight correlation between AID and PolII, as predicted by the coimmunoprecipitation of AID and PolII from ex vivo–activated B cells29. The pausing factor Spt5 is required for the linkage of AID to the transcription apparatus30. In agreement with those data, we have now shown that AID associated mainly with paused polymerases at promoter-proximal sequences across the genome. Published observations have suggested a link between AID activity and pausing of gene transcription. For example, stalling of PolII at tandem repeats of the immunoglobulin S domain has been associated with AID activity during CSR41,42. One interpretation of those results was that pausing of the transcription machinery might promote DNA deamination by facilitating the interaction of AID with ssDNA substrates41,42. Consistent with that hypothesis, we found that AID occupancy and hypermutation coincided with stalling of divergent polymerases upstream of TSSs, a feature not previously appreciated. In addition, deep-sequencing studies have shown tight correlation between genome-wide recruitment of AID and sites of ssDNA (A.Y. and R.C., unpublished data). On the basis of these findings, we postulate that Spt5-stalled polymerases recruit AID across the genome, thus explaining the degree of AID’s promiscuity in B cells.
The large number of AID targets explains the broad genomic instability observed in primary and premalignant cells after sustained or aberrant AID expression14. The data also underscore the decisive role of base-excision repair and mismatch repair in safeguarding the genome from promiscuous SHM. A pertinent example is the Myc proto-oncogene, which accumulates substantial hypermutation in Ung−/− or Ung−/−Msh2−/− cells but is fully protected in wild-type germinal center or activated B cells9,12, as shown here. Despite efficient repair, however, the Myc locus often participates in large-scale chromosomal alterations and translocations that are dependent on AID14,46,48,49. Thus, high-fidelity repair at off-target sites is not sufficient to prevent (and perhaps even promotes) DNA breaks, chromosomal translocations and B cell malignancy. In this context, we have shown that AID occupancy at Myc coincided precisely with mapped sites for canonical translocation breakpoints. We anticipate that the AID ChIP-seq data will help identify new tumor-inducing targets of AID.
The broad recruitment of AID in the B cell genome raises the question of whether AID has additional functions beyond diversification of immunoglobulin genes. Studies have linked cytidine deamination and AID to the elusive mechanism of DNA demethylation. In zebrafish, AID and Apobec2 seem to be required for the demethylation of exogenous DNA21, and AID deficiency is reported to result in genome-wide hypermethylation of mouse primordial germ cells20. The reprogramming of mouse-human heterokaryons, which requires the demethylation of promoters of genes encoding key transcription factors, is also facilitated by AID19. On the basis of those observations, it has been proposed that high AID expression in germinal center B cells might also engage in active demethylation of the B cell genome19,20. Our observation that AID deaminated basal promoters would be consistent with that hypothesis.
A prominent feature of our results is that whereas AID was highly promiscuous, its cofactor RPA seemed to be specific for immunoglobulin genes under the conditions tested. However, our data do not exclude the possibility that RPA is recruited to any given off-target site in only a fraction of the cells being assayed. A signal present in only some cells and in different genomic locations in subpopulations of cells would not be detected above background. Heterogeneity among dividing cells is probably the reason we did not detect interaction of RPA with the DNA-replication machinery in our nonsynchronized cultures. As expected from published work24, the localization of RPA to Igh requires phosphorylation of AID. The disparity between the genome-wide recruitment of AID and RPA is also consistent with the idea that RPA may function as an amplifier of AID activity on the immunoglobulin locus22,23. In biochemical assays, RPA seems to stabilize ssDNA displaced by the transcribing holoenzyme22,23,50, thus providing AID with a window of opportunity to initiate cytidine deamination. Given the established role of RPA in DNA repair27, an additional, not mutually exclusive possibility is that RPA functions downstream of the initial DNA damage, for example, by stabilizing ssDNA exposed during the repair phase of an AID lesion. In conclusion, we have identified here the broad range of genes targeted by AID, and the epigenetic and PolII stalling signature associated with targeting. We have demonstrated that AID recruitment alone is insufficient to explain the difference in mutator activity at immunoglobulin and off-target genes.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/natureimmunology/.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Schatz for comments on the manuscript; J. Chaudhuri (Memorial Sloan-Kettering Cancer Center) and F. Alt (Harvard University) for antibodies to AID; J. Simone for cell sorting; G. Gutierrez for technical assistance with the genome analyzer; and C. Ansarah-Sobrinho and S. Nelson for help with sequencing. Supported by the National Institutes of Health (Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases; and AI037526 to M.C.N.) and the Howard Hughes Medical Institute (M.C.N.).
Footnotes
Note: Supplementary information is available on the Nature Immunology website.
AUTHOR CONTRIBUTIONS
A.Y. did deep sequencing, cloning and conventional sequencing experiments; W.R. and H.-w.S. analyzed data; N.K. contributed data; Z.L. maintained the mouse colonies and cultured cells; D.F.R. contributed the Igk-AID mice; M.C.N. made suggestions for experiments and reviewed and wrote sections of the manuscript; R.C. designed the experiments and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu. Rev. Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 3.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 5.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 6.Peled JU, et al. The biochemistry of somatic hypermutation. Annu. Rev. Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 7.Delker RK, Fugmann SD, Papavasiliou FN. A coming-of-age story: activation-induced cytidine deaminase turns 10. Nat. Immunol. 2009;10:1147–1153. doi: 10.1038/ni.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 9.Robbiani DF, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol. Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 11.Pasqualucci L, et al. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc. Natl. Acad. Sci. USA. 1998;95:11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu M, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 13.Gordon MS, Kanegai CM, Doerr JR, Wall R. Somatic hypermutation of the B cell receptor genes B29 (Igβ, CD79b) and mb1 (Igα, CD79a) Proc. Natl. Acad. Sci. USA. 2003;100:4126–4131. doi: 10.1073/pnas.0735266100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klemm L, et al. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer Cell. 2009;16:232–245. doi: 10.1016/j.ccr.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin C, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto Y, et al. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat. Med. 2007;13:470–476. doi: 10.1038/nm1566. [DOI] [PubMed] [Google Scholar]
- 18.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 19.Bhutani N, et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010 doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popp C, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rai K, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 23.Basu U, et al. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438:508–511. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- 24.Vuong BQ, et al. Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nat. Immunol. 2009;10:420–426. doi: 10.1038/ni.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBride KM, et al. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc. Natl. Acad. Sci. USA. 2006;103:8798–8803. doi: 10.1073/pnas.0603272103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasqualucci L, Kitaura Y, Gu H, Dalla-Favera R. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc. Natl. Acad. Sci. USA. 2006;103:395–400. doi: 10.1073/pnas.0509969103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 28.McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J. Exp. Med. 2004;199:1235–1244. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nambu Y, et al. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- 30.Pavri R, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasqualucci L, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 32.Ji Y, et al. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lenz G, et al. Aberrant immunoglobulin class switch recombination and switch translocations in activated B cell-like diffuse large B cell lymphoma. J. Exp. Med. 2007;204:633–643. doi: 10.1084/jem.20062041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuchen S, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Storb U, et al. Targeting of AID to immunoglobulin genes. Adv. Exp. Med. Biol. 2007;596:83–91. doi: 10.1007/0-387-46530-8_8. [DOI] [PubMed] [Google Scholar]
- 36.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buratowski S. Transcription. Gene expression–where to start? Science. 2008;322:1804–1805. doi: 10.1126/science.1168805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Wuerffel R, Feldman S, Khamlichi AA, Kenter AL. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J. Exp. Med. 2009;206:1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajagopal D, et al. Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J. Exp. Med. 2009;206:1237–1244. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue K, Rada C, Neuberger MS. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J. Exp. Med. 2006;203:2085–2094. doi: 10.1084/jem.20061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McBride KM, et al. Regulation of class switch recombination and somatic mutation by AID phosphorylation. J. Exp. Med. 2008;205:2585–2594. doi: 10.1084/jem.20081319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng HL, et al. Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice. Proc. Natl. Acad. Sci. USA. 2009;106:2717–2722. doi: 10.1073/pnas.0812304106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kovalchuk AL, et al. AID-deficient Bcl-xL transgenic mice develop delayed atypical plasma cell tumors with unusual Ig/Myc chromosomal rearrangements. J. Exp. Med. 2007;204:2989–3001. doi: 10.1084/jem.20070882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malynn BA, et al. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000;14:1390–1399. [PMC free article] [PubMed] [Google Scholar]
- 48.Takizawa M, et al. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J. Exp. Med. 2008;205:1949–1957. doi: 10.1084/jem.20081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robbiani DF, et al. Activation induced deaminase is required for the chromosomal translocations in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.