Abstract
Brain infection by the human immunodeficiency virus type 1 (HIV-1) has been investigated in many reports with a variety of conclusions concerning the time of entry and degree of viral compartmentalization. To address these diverse findings, we sequenced HIV-1 gp120 clones from a wide range of brain, peripheral and meningeal tissues from five patients who died from several HIV-1 associated disease pathologies. High-resolution phylogenetic analysis confirmed previous studies that showed a significant degree of compartmentalization in brain and peripheral tissue subpopulations. Some intermixing between the HIV-1 subpopulations was evident, especially in patients that died from pathologies other than HIV-associated dementia. Interestingly, the major tissue harboring virus from both the brain and peripheral tissues was the meninges. These results show that 1) HIV-1 is clearly capable of migrating out of the brain, 2) the meninges are the most likely primary transport tissues, and 3) infected brain macrophages comprise an important HIV reservoir during highly active antiretroviral therapy.
Keywords: HIV, Brain, Dementia, Meninges, Viral migration, Macrophages, HIV-associated disease pathologies
INTRODUCTION
One of the primary HIV-associated diseases caused by infection with the human immunodeficiency virus type 1 (HIV-1) is HIV-associated dementia (HAD). Since the institution of HAART therapy, the incidence of HAD has decreased, but the prevalence of HAD and neurocognitive dysfunction now exceeds 20% and may be increasing (McArthur, 2004). While some patients develop a slower, progressive form of HAD, others only develop neurological complications at advanced stages of AIDS and near the time of death. In the latter case, neurological complications are often secondary to other AIDS-associated diseases. In the brain, HIV infects macrophages that release cytokines that present a toxic environment for neurons. In the presence of HIV, astrocytes also synthesize complimentary factors that contribute to neuronal degeneration (Speth et al., 2002). Infected macrophages may also release sphingolipids (Campbell et al., 2002), which alter the functional plasticity of neurons in patients with Alzheimer’s disease (Haughey et al., 2010), a disease with similarities to post-HAART HIV-associated neurocognitive dysfunction.
Despite what is known about the effect of HIV in the brain, some debate exists concerning the timing of HIV entry into the central nervous system. Phylogenetic analysis has shown that viral variants in the brain are more compartmentalized than in peripheral tissues (Haggerty and Stevenson, 1991; Salemi et al., 2007; van't Wout et al., 1998; Wong et al., 1997). Other phylogenetic studies have identified a subset of brain viruses within the periphery (Liu et al., 2000; Wang et al., 2001). Early biological studies suggested that HIV enters the brain via monocyte trafficking in the early stages of infection (Kim et al., 2003) and begins to evolve independently from the virus in the periphery (i.e. compartmentalization) with subsequent viral entry into the brain inhibited by the establishment of an immune barrier (Williams and Hickey, 2002). Later studies suggested HIV enters the brain much later in infection due to general immune failure during the onset of AIDS (Fischer-Smith et al., 2008). A phylogenetic analysis of patients viruses with different disease pathologies showed that both models are possible (Lamers et al., 2010). An additional study by Zhao et al. (Zhao et al., 2009) showed that 46% of 13 patients were qPCR-negative for HIV within the brain at the time of death, indicating that many patients never develop an active infection in the brain. The biological mechanism(s) responsible for the wide spectrum of potential outcomes remains unclear. However, a recent study by Sacktor et al. (Sacktor et al., 2009) showed that specific HIV-1 subtypes are associated with a higher incidence of HAD than infection with other subtypes in the same geographic region. This finding suggests that particular genetic variants have increased neuropathogenesis. A better understanding of brain and periphery viral transport is fundamental to the identification of the tissue or tissues involved in viral gene flow between the two compartments. In this study, we examined brain and peripheral tissues from five patients who died due to varied disease pathologies to determine the degree of compartmentalization within the brain and to follow up on our earlier finding that showed evidence of the meninges acting as a transit route between brain compartments in one patient (Salemi et al., 2005).
MATERIALS AND METHODS
Samples
Frozen autopsy tissues from five patients and accompanying pathology records were obtained from the University of California, San Francisco AIDS and Cancer Specimen Resource (ACSR) (http://acsr.ucsf.edu). Tissues were obtained after appropriate consent and de-identification procedures were applied. Patient designations used throughout this study were generated randomly as shorthand used by technicians who performed the studies and names do not correlate to patient information. The ACSR is recognized by the Office of Biorepositories and Biospecimen Research at the National Institutes of Health as HIPAA compliant and in accordance with the ethical standards of the Declaration of Helsinki. All material was obtained under approval from the UCSF Committee on Human Research.
The five patients in the study died from AIDS-related illnesses that were previously described in detail (Lamers et al., 2009). Briefly, patients CX and GA died from long-term progressive dementia followed by organ failure. The latest CD4 counts were unavailable for patient CX. The latest CD4 count for patient GA was 400. BW died from lymphoma, with meningeal involvement and with a CD4 count of 4. DY died from recurrent Mycobacterium avium complex (MAC) and with a CD4 count of 2. Both BW and DY showed signs of dementia near death; however, DY showed none of the typical histopathological signs of dementia within the brain. Patient AZ died from widespread atherosclerosis and with a CD4 count of 110. In Table 1 we present the histopathology of the brain of each patient at autopsy and the tissues used for generation of sequences. For all frozen tissues, parallel fixed tissues were available for immunohistochemical staining. Tissues from the meninges of all patients were stained with anti-HIVp24. All antibodies were obtained from DAKO and were used as suggested in the accompanying product insert and as previously described (Mack et al., 2003).
Table 1.
Patient information and samples used for study.
| Study ID |
Brain Histopathogology |
HAART? | Brain Tissues |
# of Brain Sequence |
Peripheral Tissues |
# of peripheral sequences |
|---|---|---|---|---|---|---|
| BW | Encephalitis | Yes | Basal ganglia Frontal lobe white Frontal lobe grey Temporal cortex |
81 | Meninges Spleen |
41 |
| AZ | Widespread atherosclerosis |
Yes | Frontal lobe | 22 | Meninges Liver Lymph node Spleen |
63 |
| DY | Slight encephalitis, normal brain tissue |
Yes | Basal ganglia Frontal lobe white Frontal lobe grey Temporal lobe |
87 | Meninges Liver Lymph node Spleen |
79 |
| GA | Considerable inflammation and macrophage infiltration throughout brain, Fibrotic meninges |
Yes | Frontal lobe white Temporal lobe |
37 | Meninges Lymph node Spleen |
95 |
| CX | Considerable inflammation and macrophage infiltration throughout brain |
No | Basal ganglia Occipital lobe grey Occipital lobe white Periventricular Choroid plexus Temporal lobe |
117 | Meninges Colon Periaortic lymph node |
70 |
Sequence generation, processing, and tropism prediction
The full gp120-nef region was amplified using primers and conditions previously described (Lamers et al., 2009). The sequences were deposited in Genbank (accession # HM001362-HM002482). HIV-1 envelope gp120 sequences from all individuals were aligned with the Clustal algorithm (Thompson et al., 1994) implemented in MEGA (Tamura et al., 2007) and manipulated by hand to optimize the alignment. An initial phylogenetic tree was calculated to verify the integrity of the data. Putative recombinant sequences identified in a previous study were removed from the data set and their analysis is described in detail in another publication (Lamers et al., 2009). The resulting data set only contained sequences that were functionally intact. The final number of non-recombinant sequences used for the current study is listed in Table 1. Sequence tropism was predicted using the charge of the V3 loop, where a charge of +3 or less is likely to utilize the CCR5 co-receptor and a charge of +5 or more is likely to utilize the CXCR4 co-receptor (Briggs et al., 2000).
Phylogenetic analysis of non-recombinant data sets
Phylogenies were inferred under a Bayesian framework assuming a relaxed molecular clock and a constant size population (Drummond et al., 2006; Drummond et al., 2005). Each aligned data set was partitioned in 1st+2nd and 3rd codon positions and the parameters of the nucleotide substitution (GTR+G+I) and demographic model were estimated independently for each partition. The Bayesian calculation consisted of 50,000,000 generations Markov Chain Monte Carlo (MCMC) with sampling every 5000th generation using the BEAST software package version 1.5.4 (Drummond and Rambaut, 2007). The convergence of the MCMC was calculated using the effective sampling size (ESS) of the combined runs. All parameter estimates showed significant ESS (>250). The maximum clade credibility tree was obtained for each patient after a 50% burn-in using the program TreeAnnotator within BEAST.
Maximum-likelihood (ML) trees were inferred using the best fitting nucleotide substitution model, as determined by a hierarchical likelihood ratio test and ML-estimated substitution parameters (Salemi et al., 2009). The heuristic search for the best tree was performed using a neighbor-joining (NJ) tree as the starting tree and the tree bisection and reconnection (TBR) branch-swapping algorithm. Calculations were performed with PHYML (Guindon and Gascuel, 2003). Statistical support for internal branches of each tree was obtained by bootstrapping (500 replicates). Each tree was mid-point rooted. ML and Bayesian methods were compared and in all cases the trees inferred the same topology.
Gene flow tests and migration counts
The hypothesis of compartmentalization, i.e. the existence of distinct HIV-1 sub-populations in different tissues, was tested using the Slatkin and Maddison test (Slatkin and Maddison, 1989) for gene flow in MacClade version 4 (Copyright © 2008 by David R. Maddison and Wayne P. Maddison, Sinauer Associates, Sunderland, MA). A one-character data matrix was obtained from the original data set by assigning each taxon in the tree a one-letter code that indicated its tissue of origin. The phylogenetic tree obtained from the nucleotide alignment was imported into MacClade and the putative origin of each ancestral sequence (i.e. internal node) in the tree was inferred by finding the most parsimonious reconstruction (MPR) of the ancestral character with the Fitch algorithm. The Hudson test was used to trace specific migrations among different compartments for each dataset (Ingvarsson, 2004). Because multiple MPRs were present in our data, the algorithm calculated the average migration count over all possible MPRs for each pair. Statistical significance was assessed by calculating the average number of migrations for all trees in the posterior distribution less a 50% burn-in and by calculating the standard deviation. If the 95% range was two standard deviations from the mean and did not include zero, the migration was determined as significantly greater than zero. We also estimated gene flow between brain, meninges and peripheral tissues using LAMARC, which allows a ML estimate of the rate and direction of gene flow between sub-populations.
RESULTS
P24 staining of HAD brain tissue
The tissue stains of the meninges for patient CX showed a background of HIV p24 positivity with intense staining localized to perivascular macrophages. The section shown in Figure 1 is typical of HIV p24 large vessel staining of a HAD brain section. P24 staining of the meninges for the other patients was very low, presumably due to the late stage of disease; however, as previously reported (Lamers et al., 2010), frontal lobe tissues for all patients stained positive for p24 with varying degrees of intensity.
Figure 1. HIV p24 stain of meninges: localization to perivascular and parenchyma macrophages in patient CX.
Three different magnifications of tissue are shown (A and B = 100X, C = 400X). In Panel A, SA indicates the subarachnoid space. HIV positive cells stain brown. Intermittent HIV-infected cells are found throughout the tissues (Panels A and B); however a large number of HIV positive cells are found lining up against the blood vessel (arrows in Panels A and C).
Phylogenetic analysis of HIV-1 in post-mortem brain and peripheral tissues
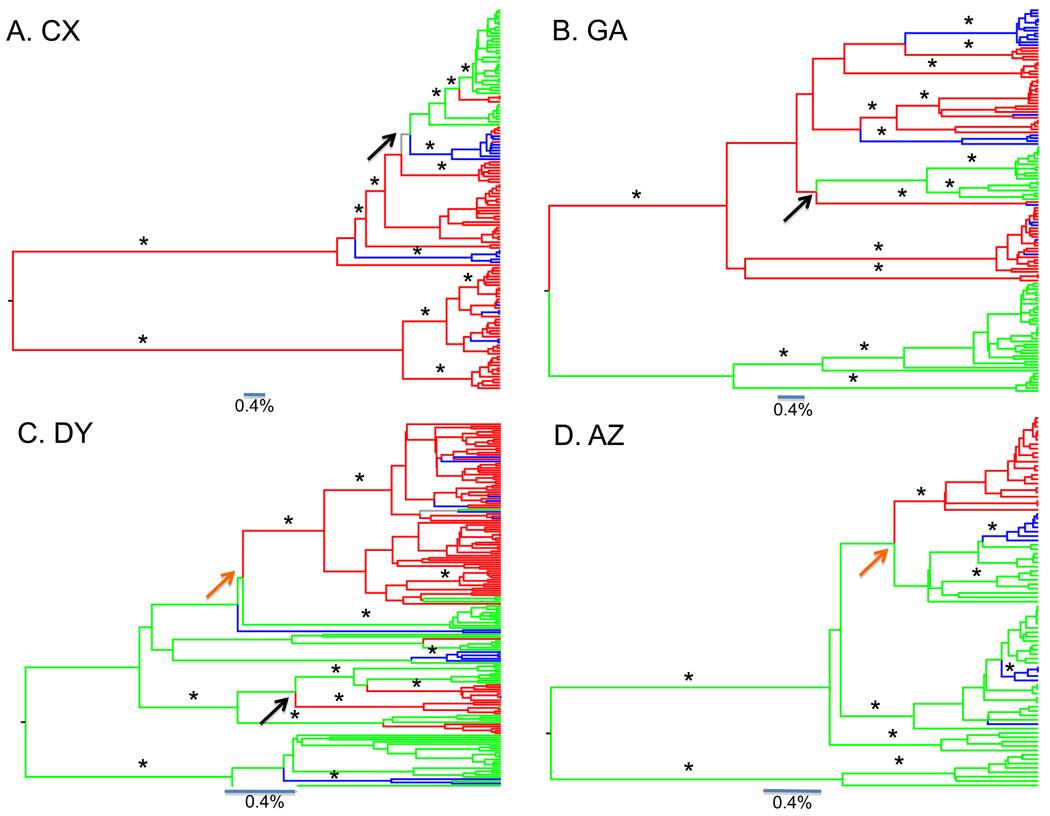
The maximum clade credibility (MCC) tree was obtained for each patient under a Bayesian framework. In patient CX (Figure 2A), the MCC tree showed two well-supported clades of brain sequences (basal ganglia, occipital lobe/grey, occipital lobe white matter, periventricular, choroid plexus, temporal lobe) that shared a common origin at the root of the tree, separated by very long branches. The V3 loop was predicted to utilize the CCR5 co-receptor with a charge of 3+ or less. Results from the Hudson test indicated there was no population structure evident among brain tissues (p>0.05) except for the temporal lobe (p<0.05). Interestingly, the peripheral sequences (colon and lymph node) emerged from one of the major brain clades. Another back migration from the periphery occurred to the basal ganglia. The meninges were again dispersed throughout the brain clade. Patient CX displayed signs of dementia over several years and was the only patient in the study that was not on HAART therapy. Although only two peripheral tissues were sampled in this patient, the virus in these tissues appeared to evolve directly from the brain.
Figure 2. Bayesian genealogies of HIV-1.
Bayesian maximum clade credibility trees assuming a relaxed molecular clock and constant population size coalescent prior generated from the posterior distribution of trees less a 50% burn-in. Branch lengths are shown according to the scale bar at the bottom of each panel, in relative units of time. Posterior probabilities greater than 85% are indicated with an asterisk. Internal branches are colored according to the maximum parsimony reconstruction of the ancestral tissue origin. Brain to periphery migrations are indicated with black arrows. Periphery to brain migrations are indicated with orange arrows. Branches are colored according to tissue from which virus was isolated: red branches indicate brain tissue, green branches indicate peripheral tissue, blue branches indicate meninges, gray branches indicate multiple equally parsimonious (i.e. uncertain) reconstructions. Phylogenies are shown for patient CX (A), GA (B), AZ (C), DY (D) and BW (E).
For patient GA, the MCC tree (Figure 2B) was also consistent with an early viral infection model. Similar to the sequence of patient CX, all of GA’s sequences were predicted to utilize the CCR5 co-receptor, with a V3 loop charge of 3+ or less. A well-supported peripheral and brain clade shared a common ancestor at the root of the tree. Results from the Hudson test showed that the peripheral sequences (lymph and spleen) did not have structure between them (p>0.05). The temporal and frontal lobe/white matter were structured with respect to each other (p<0.05). Also similar to patient CX, the meninges were interspersed within the brain clade only. Again one well-supported clade consisting of peripheral sequences from both lymph and spleen emerged from a larger brain clade. This result indicated re-seeding of the peripheral tissues from the brain.
The MCC tree for patient DY (Figure 2C) provided support for late viral invasion into brain tissues. In addition, all sequences from patient DY had a charge of +6 or greater, indicating infection of the brain with CXCR4 viruses, a phenotype typically associated with T-cell tropic viruses. One primary well-supported brain clade emerged from the peripheral tissues (liver, lymph, spleen). Again, results from the Hudson test indicated no population structure among the peripheral tissues (p>0.05). Sequences from the meninges were interspersed within both brain and peripheral tissue. Four additional introductions occurred from periphery to the brain. Sequences from specific regions of the brain lacked population structure (basal ganglia, frontal lobe/white matter, frontal lobe/grey matter, p>0.05), with the exception of the temporal lobe (p<0.05). Furthermore, a few sequences from peripheral tissues emerged from the brain clade, suggesting a back-migration event(s).
The MCC tree for patient AZ (Figure 2D) provided another example of late viral invasion into the brain. The peripheral sequences (liver, lymph, and spleen) were basal to the brain clade and were not clustered by tissue of origin. A separate clade of peripheral sequences (liver and spleen) was separated by a long branch, all peripheral sequences potentially using the CXCR4 co-receptor with a V3 loop charge of +5 (all other AZ sequences had a v3 loop charge of 3+ or less). The Hudson test confirmed a lack of population subdivision among the three peripheral tissues (p>0.05). A well-supported monophyletic brain clade comprised of frontal lobe/white matter sequences emerged late in the tree. Again, the meninges are not monophyletic, but in this case were interspersed within the peripheral tissue and appear to be recently infected by peripheral tissues.
The MCC tree for patient BW (Figure 2E) was consistent with the classic pattern of compartmentalization: a well-supported peripheral clade and a brain clade shared a common ancestor at the root of the tree. The brain clade was comprised of several monophyletic clades of sequences from different regions (frontal lobe/white matter, frontal lobe/grey matter, basal ganglia, and temporal lobe). The Hudson test indicated strong evidence for population subdivision (p<0.001). The only exception was the meninges sequences, which were interspersed throughout the brain clades, indicating that the meninges were infected either by virus from different regions of the brain or by different peripheral isolates over the course of infection. The V3 loop charge for BW’s sequence were predominantly +4, a charge associated with both CCR5 and CXCR4 phenotypes.
The phylogenetic trees for patients CX and GA displayed more evolutionary distance between major clades than the trees from patients AZ, DY and BW (note the distance bar at the bottom of the phylogenies).
Viral gene flow among post-mortem tissues
The Slatkin-Maddison test was used to investigate the hypothesis of gene flow among brain, peripheral, and meninges sequences. The average number of observed migrations among the three tissues was calculated for the maximum clade credibility tree (Table 2). In four patients (BW, DY, GA, and CX) significant migration was detected from the brain to the meninges (p<0.0001). Migration was also detected from the meninges to the periphery in DY, and from the meninges to the brain in patient CX. Significant migration was found from the periphery to the brain in three patients (AZ, DY, and CX). A similar analysis with the program LAMARC, which allows estimating the direction and rate of gene flow among different sub-population with maximum likelihood, gave identical results (data not shown). In Table 3 we summarize the conclusions from the combined phylogenetic and migration analyses. Gene flow patterns among different lymphoid tissues, as well as different brain tissues, were also examined. In all cases, however, the number of observed migrations among strains infecting different tissues lied within the 95% confidence interval of a distribution of 10,000 trees where tissue assignments on the tips were randomly scrambled (data not show). Therefore, no statistically significant inferences could be made about preferential/asymmetric gene flow in this case.
Table 2.
Migration of the virus among tissues.
| Destination Tissue | ||||
|---|---|---|---|---|
| Study ID | Tissue of Origin | Periphery | Brain | Meninges |
| BW | Periphery | - | 0.5 | 0 |
| Brain | 0 | - | 4.5 | |
| Meninges | 0 | 0.5 | - | |
| AZ | Periphery | - | 1 | 3 |
| Brain | 0 | - | 0 | |
| Meninges | 0 | 0 | - | |
| DY | Periphery | - | 5 | 3.8 |
| Brain | 2.7 | - | 7.7 | |
| Meninges | 0.8 | 0 | - | |
| GA | Periphery | - | 0.5 | 0 |
| Brain | 1.5 | - | 7 | |
| Meninges | 0 | 0 | - | |
| CX | Periphery | - | 1 | 0.4 |
| Brain | 0.7 | - | 4.7 | |
| Meninges | 0.4 | 2 | - | |
For each patient, the number of migrations from a given tissue (indicated at the left) to another (on top) is shown, averaged over all reconstructions. The numbers shaded and in bold indicate values significantly greater than zero over all trees in the posterior distribution.
Table 3.
Summary of Phylogenetic Results.
| Study ID | Intra-Brain Population Structure |
Intra- Periphery Population Structure |
Meninges | Migration From Periphery To Brain |
Migration From Brain To Periphery |
Root Ancestor |
Model |
|---|---|---|---|---|---|---|---|
| BW | Yes | NA | Brain Clades | Yes | No | Indeterminate | Early Infection |
| AZ | NA | No | Peripheral | Yes | No | Peripheral | Late Invasion |
| DY | No (Except Temporal) |
No | Brain and Peripheral Clades |
Yes | Yes | Peripheral | Late Invasion |
| GA | Yes | No | Brain Clades | Yes | Yes | Indeterminate | Early Infection |
| CX | No (Except Temporal) |
No | Brain Clades | Yes | Yes | Brain | Early |
NA = analysis was not performed because only one tissue was sampled.
DISCUSSION
In the current study, we amplified virus from brain and peripheral tissues, including the meninges, from five patients. Two patients died primarily from progressive HAD, whereas the other three patients died from lymphoma, systemic infection or atherosclerosis. In every patient, sequences from the meninges were interspersed within the brain, the periphery, or both tissue types, thus identifying the meninges as the primary supplier of HIV to and from the brain. Although there has been much discussion as to how the virus enters the brain on a cellular level, this is the first study to our knowledge that used a bioinformatics approach in order to precisely identify the tissue responsible for viral transport between the brain and periphery. Another important finding from the study is that the brain is clearly capable of re-seeding the periphery with HIV-1. Three previous studies described complete compartmentalization of viruses in the brain (Haggerty and Stevenson, 1991; van't Wout et al., 1998; Wong et al., 1997); however, in these studies meninges were not sampled. In a later study Liu et al. (Liu et al., 2000) sequenced virus from meninges and found, similar to our results, that meningeal-derived sequences clustered with both brain and peripheral sequences although they could not establish directional gene-flow, as clearly shown by our data. Although only five patients were examined in this study, our results indicate that subsequent studies that attempt to identify the establishment and maintenance of CNS HIV-1 reservoirs, or examine the evolution and tracking of HIV-1 in the CNS, should also examine virus harbored in the meninges.
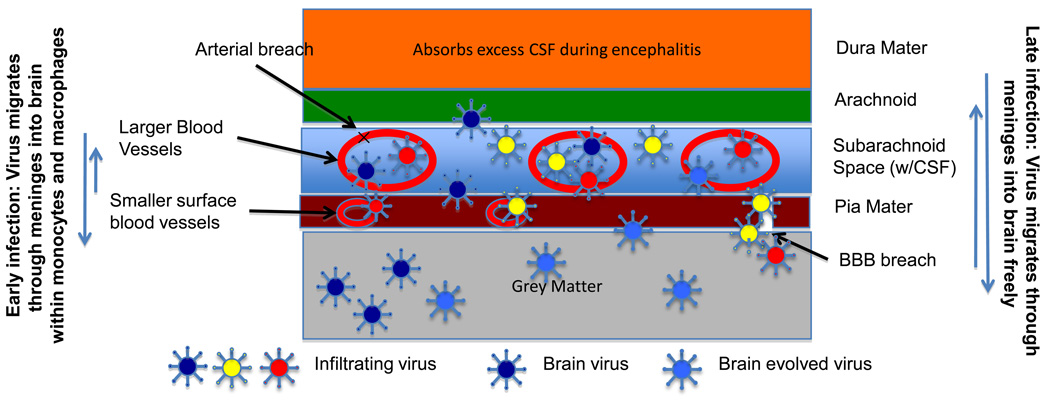
The meninges are the protective layer that covers the entire central nervous system and provides an interface between the periphery and brain. Although distinct from the brain, they are interweaved within the folds of the outer tissues of the brain. The meninges contain three layers of tissue: 1) the Dura mater, which is closest to the skull, is a tough leather-like layer that protects the brain and is fused to the skull at several points, 2) the Arachnoid layer, which is a spider-like tissue, laced throughout with large blood vessels and is in contact with the Dura Mater, and, 3) the Pia Mater, which is separated from the Arachnoid layer by the subarachnoid space, adheres to the brain and contains finer blood vessels and capillary beds. Cerebrospinal fluid flows above the Pia Mater in the subarachnoid space, which contains the spider-like fibers of the Arachnoid. The cells within the meninges include fibroblasts, mast cells, dendritic cells and macrophages. The meninges do not mount an immune response, however, they have been shown to absorb cerebral-spinal fluid via Arachnoid cisterns that drain into the Dura vasculature (Zenker et al., 1994). This represents yet another way they could harbor HIV viruses due to early encephalitis Additionally, Mercier and Hatton (Mercier and Hatton, 2000) have challenged the current model of brain architecture with confocal 3D images that show meningeal projections directly into the brain and components of the meninges in the choroid plexus, through the parenchyma of the ventricles and in contact with astrocytes and joining with blood vessels. They suggest that previously unsuspected connections between the meninges and the brain are considerable. Although it is known that HIV-infected macrophages and monocytes are the most likely carriers of the virus into the brain during early infection, late viral infection of the brain has also been suggested due to overall immune breakdown and breach of the blood-brain barrier. In Figure 1 we show the p24 stain of a blood vessel within meningeal tissue. Although there is only slight p24 positivity in the overall tissue sample, p24 positive cells surround the perivascular surface of the blood vessels, indicating a high possibility of viral transfer at the site.
Figure 3 provides a cartoon representation of the three layers of the meninges and proposes a model of viral transport to the brain that combines findings in this paper as well as several others (Fischer-Smith et al., 2008; Herbein and Varin, 2010; Lamers et al., 2010; Salemi et al., 2005; Williams and Hickey, 2002): 1) The blood vessels within the Subarachnoid and Pia Mater of the meninges provide a mechanism for HIV to come in close contact with the brain; 2) Infection of the brain is likely during onset of early HIV-induced encephalitis, when a markedly increased amount of leukocytes are present within the cerebral spinal fluid; 3) Due to specific HIV genetic signatures, some viruses may be more capable of evolving within brain tissues than other viruses. Viruses that are not allowed to replicate in brain tissues do not persist. This hypothesis explains why some patients develop a slower progressive dementia while others present only with late-stage dementia.
Figure 3. The layers of the meninges and proposed models for movement of virus to and from brain.
The three major layers of the meninges are shown along with the subarachnoid space. Blood vessels in the CSF and Pia Mater are indicated. In early infection, viruses may enter the brain due to the elevated number of HIV infected macrophages in the CSF due to early encephalitis. Certain viral variants may be more suited to the brain environment and set up a slower long-term dementia process in which viruses evolve within brain tissue resident macrophages. In other cases, patients may develop a late-stage HAD when the patient is again at risk for encephalitis. This, in combination with complete immune breakdown and other disease processes such as lymphoma, atherosclerosis or lipid disorders, may damage both the vessels of the Pia Mater and subarachnoid space, as well as cause breaches across the BBB and allow more rapid movement of virus in and out of the brain.
This hypothesis would also explain the findings of Sacktor et al. (Sacktor et al., 2009) showing that a specific subtype of HIV is associated with a significantly higher degree of dementia rate than another subtype in the same geographic area. In addition, this hypothesis explains the observed semi-compartmentalization of brain viruses; 4) Due to the lack of selective constraints within the meninges, virus can constantly move from the brain and back into the meninges, but this movement is likely reduced during non-encephalitic stages; 5) During late-stage disease, especially when complicated with other disease pathologies that could damage blood vessels (atherosclerosis) or the BBB (lymphoma and systemic infection), virus can leak at a higher rate from the blood vessels of the meninges and accumulate in the tissues surrounding the brain and may increase the likelihood for the migration of viruses that utilize the CXCR4 co-receptor; 6) Accumulation of virus in the meninges and surface of the brain increases the likelihood of viral transport to and from the brain; 7) A large amount of virus and virus by-products cause late stage encephalitis and dementia.
The successful reduction in the occurrence of HAD will depend on the development of drugs that target HIV-infected macrophages. Additionally, the identification of neurotropic strains of HIV could assist in alternative approaches to identify those that develop HAD. Furthermore, approaches that reduce AIDS-associated diseases such as lymphoma and lipid disorders may also reduce the occurrence of HAD.
Acknowledgements
The project was funded by NIH grants U01 CA066529 and U19 MH081835. MS is supported by NIH grant R01 NS063897-01A2. RRG was supported by T32 NIH training grant_CA-09126. The authors would like to thank those who assisted in generation and proofing of the data: Derek Galligan, Li Zhao and Tulio de Oliveira.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Briggs DR, Tuttle DL, Sleasman JW, Goodenow MM. Envelope V3 amino acid sequence predicts HIV-1 phenotype (co-receptor usage and tropism for macrophages) AIDS. 2000;14:2937–2939. doi: 10.1097/00002030-200012220-00016. [DOI] [PubMed] [Google Scholar]
- Campbell SM, Crowe SM, Mak J. Virion-associated cholesterol is critical for the maintenance of HIV-1 structure and infectivity. AIDS. 2002;16:2253–2261. doi: 10.1097/00002030-200211220-00004. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- Fischer-Smith T, Bell C, Croul S, Lewis M, Rappaport J. Monocyte/macrophage trafficking in acquired immunodeficiency syndrome encephalitis: lessons from human and nonhuman primate studies. J Neurovirol. 2008;14:318–326. doi: 10.1080/13550280802132857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Haggerty S, Stevenson M. Predominance of distinct viral genotypes in brain and lymph node compartments of HIV-1-infected individuals. Viral Immunol. 1991;4:123–131. doi: 10.1089/vim.1991.4.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughey NJ, Bandaru VV, Bae M, Mattson MP. Roles for dysfunctional sphingolipid metabolism in Alzheimer's disease neuropathogenesis. Biochim Biophys Acta. 2010;1801:878–886. doi: 10.1016/j.bbalip.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbein G, Varin A. The macrophage in HIV-1 infection: from activation to deactivation? Retrovirology. 2010;7:33. doi: 10.1186/1742-4690-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson PK. Population subdivision and the Hudson-Kreitman-Aguade test: testing for deviations from the neutral model in organelle genomes. Genet Res. 2004;83:31–39. doi: 10.1017/s0016672303006529. [DOI] [PubMed] [Google Scholar]
- Kim WK, Corey S, Alvarez X, Williams K. Monocyte/macrophage traffic in HIV and SIV encephalitis. J Leukoc Biol. 2003;74:650–656. doi: 10.1189/jlb.0503207. [DOI] [PubMed] [Google Scholar]
- Lamers SL, Salemi M, Galligan DC, de Oliveira T, Fogel GB, Granier SC, Zhao L, Brown JN, Morris A, Masliah E, McGrath MS. Extensive HIV-1 intra-host recombination is common in tissues with abnormal histopathology. PLoS One. 2009;4:e5065. doi: 10.1371/journal.pone.0005065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers SL, Salemi M, Galligan DC, Morris A, Gray R, Fogel G, Zhao L, McGrath MS. Human immunodeficiency virus-1 evolutionary patterns associated with pathogenic processes in the brain. J Neurovirol. 2010;16:230–241. doi: 10.3109/13550281003735709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tang XP, McArthur JC, Scott J, Gartner S. Analysis of human immunodeficiency virus type 1 gp160 sequences from a patient with HIV dementia: evidence for monocyte trafficking into brain. J Neurovirol. 2000;6 Suppl 1:S70–S81. [PubMed] [Google Scholar]
- Mack KD, Jin X, Yu S, Wei R, Kapp L, Green C, Herndier B, Abbey NW, Elbaggari A, Liu Y, McGrath MS. HIV insertions within and proximal to host cell genes are a common finding in tissues containing high levels of HIV DNA and macrophage-associated p24 antigen expression. J Acquir Immune Defic Syndr. 2003;33:308–320. doi: 10.1097/00126334-200307010-00004. [DOI] [PubMed] [Google Scholar]
- McArthur JC. HIV dementia: an evolving disease. J Neuroimmunol. 2004;157:3–10. doi: 10.1016/j.jneuroim.2004.08.042. [DOI] [PubMed] [Google Scholar]
- Mercier F, Hatton GI. Immunocytochemical basis for a meningeo-glial network. J Comp Neurol. 2000;420:445–465. doi: 10.1002/(sici)1096-9861(20000515)420:4<445::aid-cne4>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Sacktor N, Nakasujja N, Skolasky RL, Rezapour M, Robertson K, Musisi S, Katabira E, Ronald A, Clifford DB, Laeyendecker O, Quinn TC. HIV subtype D is associated with dementia, compared with subtype A, in immunosuppressed individuals at risk of cognitive impairment in Kampala, Uganda. Clin Infect Dis. 2009;49:780–786. doi: 10.1086/605284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemi M, Burkhardt BR, Gray RR, Ghaffari G, Sleasman JW, Goodenow MM. Phylodynamics of HIV-1 in lymphoid and non-lymphoid tissues reveals a central role for the thymus in emergence of CXCR4-using quasispecies. PLoS One. 2007;2:e950. doi: 10.1371/journal.pone.0000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemi M, Lamers SL, Yu S, de Oliveira T, Fitch WM, McGrath MS. Phylodynamic analysis of human immunodeficiency virus type 1 in distinct brain compartments provides a model for the neuropathogenesis of AIDS. J Virol. 2005;79:11343–11352. doi: 10.1128/JVI.79.17.11343-11352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemi M, Vandamme A-M, Lemey P. The phylogenetic handbook : a practical approach to phylogenetic analysis and hypothesis testing. 2nd ed. Cambridge, UK ; New York: Cambridge University Press; 2009. [Google Scholar]
- Slatkin M, Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123:603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth C, Schabetsberger T, Mohsenipour I, Stockl G, Wurzner R, Stoiber H, Lass-Florl C, Dierich MP. Mechanism of human immunodeficiency virus-induced complement expression in astrocytes and neurons. J Virol. 2002;76:3179–3188. doi: 10.1128/JVI.76.7.3179-3188.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Wout AB, Ran LJ, Kuiken CL, Kootstra NA, Pals ST, Schuitemaker H. Analysis of the temporal relationship between human immunodeficiency virus type 1 quasispecies in sequential blood samples and various organs obtained at autopsy. J Virol. 1998;72:488–496. doi: 10.1128/jvi.72.1.488-496.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TH, Donaldson YK, Brettle RP, Bell JE, Simmonds P. Identification of shared populations of human immunodeficiency virus type 1 infecting microglia and tissue macrophages outside the central nervous system. J Virol. 2001;75:11686–11699. doi: 10.1128/JVI.75.23.11686-11699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KC, Hickey WF. Central nervous system damage, monocytes and macrophages, and neurological disorders in AIDS. Annu Rev Neurosci. 2002;25:537–562. doi: 10.1146/annurev.neuro.25.112701.142822. [DOI] [PubMed] [Google Scholar]
- Wong JK, Ignacio CC, Torriani F, Havlir D, Fitch NJ, Richman DD. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J Virol. 1997;71:2059–2071. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenker W, Bankoul S, Braun JS. Morphological indications for considerable diffuse reabsorption of cerebrospinal fluid in spinal meninges particularly in the areas of meningeal funnels. An electronmicroscopical study including tracing experiments in rats. Anat Embryol (Berl) 1994;189:243–258. doi: 10.1007/BF00239012. [DOI] [PubMed] [Google Scholar]
- Zhao L, Galligan DC, Lamers SL, Yu S, Shagrun L, Salemi M, McGrath MS. High level HIV-1 DNA concentrations in brain tissues differentiate patients with post-HAART AIDS dementia complex or cardiovascular disease from those with AIDS. Sci China C Life Sci. 2009;52:651–656. doi: 10.1007/s11427-009-0085-5. [DOI] [PubMed] [Google Scholar]