Abstract
Genome-wide association studies have led to a large number of single-SNP association findings, but there has been, so far, no investigation resulting in the discovery of a replicable gene-gene interaction. In this paper, we examine some of the possible explanations for the lack of findings, and argue that coverage of causal variation not only has a large effect on the loss in power, but that the effect is larger than in the single-SNP analyses. We show that the product of linkage disequilibrium measures, r2, between causal and tested SNPs offers a good approximation to the loss in efficiency as defined by the ratio of sample sizes that lead to similar power. We also demonstrate that, in addition to the huge search space, the loss in power due to coverage when using commercially available platforms makes the search for gene-gene interactions daunting.
1 Introduction
Recent research in the genetics of complex human traits has been fueled by the advances in genotyping technology that have permitted the simultaneous genotyping of hundreds of thousands (up to a million) of Single Nucleotide Polymorphisms (SNPs). This has led to a large number of genome-wide association studies (GWAS) (e.g. WTCCC, 2007) that have yielded an impressive number of discoveries. The vast majority of these discoveries have come from scanning the genome one marker at a time, and searching for relatively strong main effects. Although the field of complex traits genetics has been flooded with discoveries of genetic associations, they have failed to account for all the variation in phenotype (Manolio et al., 2009). Interactions such as gene-environment(GxE) and gene-gene (GxG) can explain some of the missing heritability, and have been the focus of many investigations.
It has been argued (Marchini et al., 2005) that testing strategies incorporating both single SNP and SNP-SNP interactions can be more powerful in the context of GWAS data than performing single-SNP analyses, even with a conservative penalty for multiple testing. The results from simulations provide convincing evidence for that claim, but the applications have not matched the theoretical expectations thus far. There have, as of yet, been no replicable G×G interactions discovered for a complex human disease. Some of the issues with published findings are: (i) there is some confusion on the difference between interaction versus marginal association; (ii) some papers describe interactions between loci in the same region, and in this situation it is difficult to assess the difference between interaction and main effects; (iii) some manuscripts report p-values calculated on sparse data using asymptotic approximations; (iv) some use incorrect corrections for multiple testing.
There are many methods for testing for GxG interaction (Cordell, 2009) and also many papers on the definition of interaction and on the difference between statistical and biological epistasis (Cordell, 2002, for example). An important aspect that has not yet enjoyed much attention is how to interpret the observed lack of replicable findings. The focus of this paper is on starting the discussion on study design and search planning which has a crucial effect on the discovery process. Note that these are directly related to power that tends to be much lower for GxG studies than single-SNP association. The obvious explanation for low power is the combination of sample sizes and underlying genetic models. It is possible that the deviation from statistical additivity in 2-SNP models is small, so only studies with very large sample sizes will detect it. It is also possible that the effects are concentrated in combinations of genotypes that have low population frequencies, and the sample size in these cells is relevant for determining the efficiency of the inference process. Moreover, in the case of a genome-wide search, one of the main hindrances is the huge search space (even in the case of 2-SNP models) that leads to stringent thresholds for declaring discovery.
In this paper we focus on a different reason behind the lack of findings, namely the coverage of causal variation. This issue has been explored extensively in the context of single-SNP analysis (see next section), but only partially in the context of GxG interactions; the effect of LD on power was indirectly quantified by measuring the effect of LD on odds ratios (Marchini et al., 2005). Here the questions we are trying to answer are: (i) what is the effect of using tag SNPs or platform SNPs on power? (ii) can we quantify that potential loss of power using measures of linkage disequilibrium (LD) as we do in the single-SNP case? Throughout, we measure power loss in terms of relative efficiency, which is the ratio of sample sizes of two statistical hypothesis tests that lead to the same level of power. In the GxG interaction testing context, coverage is the relative efficiency of these two scenarios: (i) test for interaction at the genotyped SNPs that best tag the causal SNPs; (ii) test for interaction at the causal SNPs. We argue below that this metric of coverage can be approximated using measures of LD. The accuracy of these metrics in the marginal association and interaction settings is also investigated.
2 Measuring coverage in single-SNP association via r2
In the case of single-SNP analysis, the LD measure r2 has been used to characterize the loss of power due to using tag-SNPs in place of causal variants. The motivation for using it comes from one of the interpretations of this LD metric: the r2 between the causal and tested SNPs is approximately equal to the ratio of sample sizes (for testing the causal versus the genotyped SNP) that leads to the same power (Kruglyak, 1999; Pritchard and Przeworski, 2001) when testing for association with a particular phenotype. In other words, the sample size when using a proxy SNP needs to be larger by a factor of 1/r2 to maintain power similar to when using the causal variant.
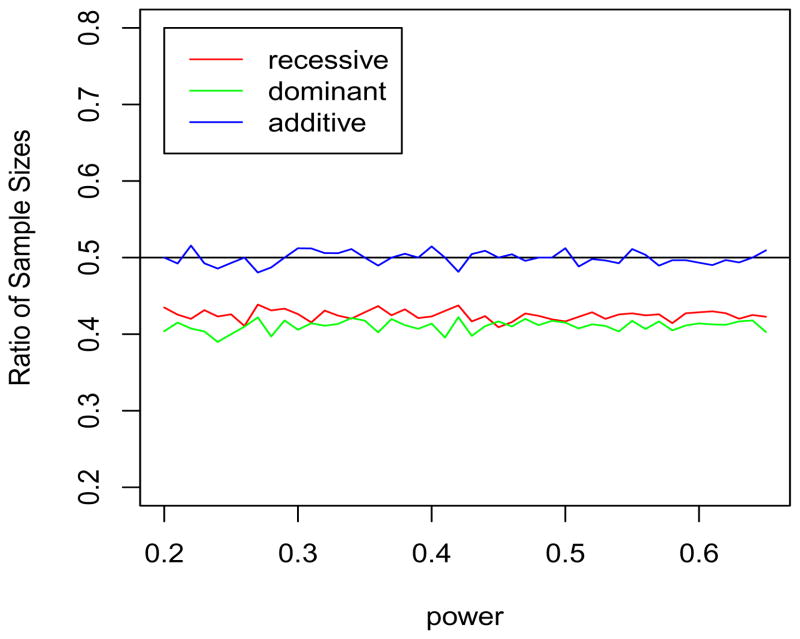
There are several papers (Nicolae et al., 2006; Barrett et al., 2006; Eberle et al., 2007) that use this interpretation of r2, or measures of multi-locus LD developed based on similar ideas (Nicolae, 2006), for comparing and characterizing GWAS platforms. It is not widely known that the r2 approximation of the relative efficiency offers, in many situations, an optimistic view on coverage. To illustrate, we simulated genotype data, G, for a SNP that is in LD, r2 = 0.5, with a causal SNP with data denoted by X. For simplicity we fixed the population allele frequency at 0.5 for both SNPs. We generated data under three different genetic models where the non-risk genotype penetrance was 10%: (i) recessive; (ii) dominant; and (iii) additive. We simulated genotypes for nX cases and nX population controls at the causal locus, and nG cases and nG population controls at the SNP in LD with the causal variant, with n’s between 1000 and 8000. For each setting, we performed 10000 repetitions, and estimated the power for a χ2 test at the 0.01 significance level. Figure 1 shows the ratio of sample sizes, nX/nG, that leads to similar power for the causal and proxy variants. Note that r2 offers a very good approximation under an additive model, but it is overly optimistic under recessive and dominant models. For these models, it is not sufficient to double the sample size in order to obtain similar power, but it is necessary to increase the sample size by a factor of 1/0.4 which is 2.5.
Figure 1.
Estimated ratios of samples sizes for marginal association testing, nX/nG, are shown for three genetic models. The sample sizes, nX for the causal SNP and nG for the tag SNP, are chosen such that the power to detect association at the two markers is similar. The tag and causal SNPs have an LD r2 = 0.5.
3 The coverage of SNP-pairs
In this section we investigate the coverage of SNP pairs when testing for gene-gene interactions. Here we argue that not genotyping the causal variant results in a higher loss of efficiency in the case of gene-gene interaction tests than in the case of single-SNP analysis. This is a consequence of the loss of information at both loci, loss that accumulates in a multiplicative fashion.
We use notation similar to that used in the previous section. We assume that the two causal SNPs genotypes are denoted by X1 and X2, and that the sample size corresponding to a study where these variants are typed is nXX. The SNPs that are genotyped in the study of interest have genotypes denoted by G1 and G2, where Xi and Gi (i=1,2) are correlated (in linkage disequilibrium). The sample size corresponding to a study on these SNPs is denoted by nGG. We denote with the LD measure between Xi and Gi. The goal is to quantify the loss in power due to not typing the causal variants. We do this by evaluating the ratio of sample sizes, nXX/nGG, that leads to similar power under the two scenarios.
We argue here that for the case only gene-gene interaction test, the product of the two LD measures, offers a good approximation to relative efficiency. The argument is clear from the following diagram,
where we show three testing scenarios (each scenario corresponds to testing one SNP pair). We assume that the sample sizes (nXX, nXG, nGG) are such that we have approximately equal power under the three scenarios. We are trying to quantify
Note that nXX/nXG corresponds to the loss in power when having a proxy at the second locus, and thus depends on the accuracy of the proxy (or the linkage disequilibrium between X2 and G2) and on the underlying genetic risk model. In the case-only interaction test, the sufficient statistics consist of the cases counts for the 3×3 tables of genotype combinations. In its simplest form, the interaction is tested using a chi-square statistic, and it is equivalent to a genotype association test on X2 when the “case-control” status is given by the genotypes at X1. Therefore, the loss in power for not typing X2 (the first part of the above diagram) can be quantified using , with the same caveats discussed in the previous section: r2 offers, for many genetic models, an optimistic view on coverage. In other words, is an approximation of nXX/nXG in the case-only GxG test. Similarly, the loss in power corresponding to the second part of the diagram can be approximated by . Overall, coverage can be quantified using
This approximation of the effect of coverage does not restrict to case-only inference. We show in the next section, using simulations, that the product of r2’s can be also used when the test is based on logistic regression using both cases and controls.
4 Empirical Study
In this section we present two sets of simulations to study the difference in power, in testing for interactions, between the complete (testing the causal SNPs) and incomplete (using tag SNPs for causal variation) data settings. Our main goal is to compare the power loss in several genetic models to the product of the r2. We do not attempt to completely describe the nature of the power loss, as that is beyond the scope of a small scale simulations section, but merely to explore enough to gain an understanding of how coverage could impact the results of a GWAS for gene-gene interactions.
In our first set of simulations, we consider four genetic models labeled A–D. The penetrance tables, assuming 5% disease prevalence, are shown in Table 1. We assume that all allele frequencies are 50%. In model A there is only one nonzero interaction term, while each consecutive model has one additional interaction term from its predecessor. In each case, the signal is divided evenly amongst the interaction terms. We use samples of half cases and half controls throughout. For various r2 combinations we simulate the power curves (α = 0.01 and 1000 repetitions for each point on each curve), as a function of the sample size, when testing for a significant interaction term in a full logistic regression model for the complete and incomplete data settings. We then compare the curves to see what ratio of incomplete/complete sample sizes lead to the same power.
Table 1.
Penetrances for the models used in simulations. For each of the models, shown are P(D|X1, X2), where X1 and X2 are genotypes at two causal interacting SNPs.
| Model A | Model B | ||||||
|---|---|---|---|---|---|---|---|
| X1/X2 | 0 | 1 | 2 | X1/X2 | 0 | 1 | 2 |
| 0 | 0.045 | 0.045 | 0.045 | 0 | 0.036 | 0.036 | 0.036 |
| 1 | 0.045 | 0.045 | 0.045 | 1 | 0.036 | 0.036 | 0.108 |
| 2 | 0.045 | 0.045 | 0.116 | 2 | 0.036 | 0.036 | 0.108 |
| Model C | Model D | ||||||
| X1/X2 | 0 | 1 | 2 | X1/X2 | 0 | 1 | 2 |
| 0 | 0.036 | 0.036 | 0.036 | 0 | 0.036 | 0.036 | 0.036 |
| 1 | 0.036 | 0.036 | 0.080 | 1 | 0.036 | 0.061 | 0.061 |
| 2 | 0.036 | 0.080 | 0.080 | 2 | 0.036 | 0.061 | 0.061 |
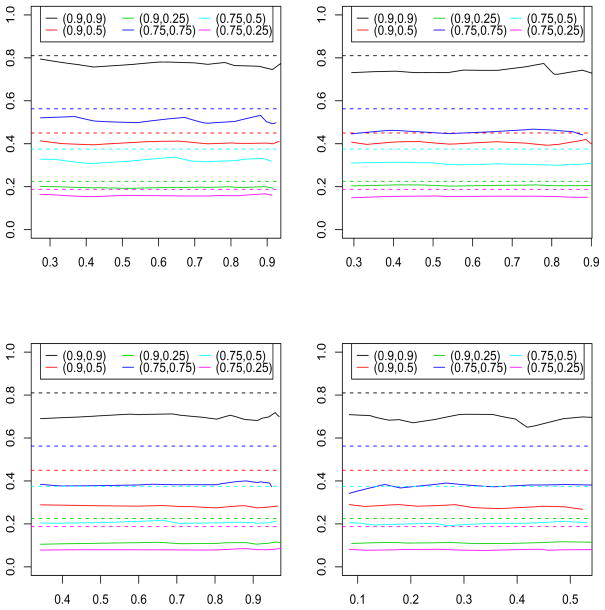
The top left panel of Figure 2 is a plot of the ratio of sample sizes (y-axis) that lead to the same power (x-axis) for the various r2 combinations in model A. We also include horizontal lines corresponding to the product of the two r2 values. As we can see, the product of the r2 slightly underestimates the power loss, but is still a fairly good approximation in this setting. The top right panel of Figure 2 shows the corresponding results for model (B). The product of the r2 is still a lower bound on the power loss, but does not approximate it well. The bottom left and bottom right panels of Figure 2 shows the corresponding results for models (C) and (D) respectively. As in model (B), the product of the r2 is a lower bound on the power loss, but is far from the true power loss.
Figure 2.
The solid line is the ratio of sample sizes (y-axis) that lead to a particular power (x-axis). The dashed line is the product of the r2. The top left, top right, bottom left, and bottom right panels correspond to models A, B, C, and D respectively. All results are based on 1000 repetitions.
Interestingly, the power loss seems to depend heavily on the number of significant parameters in the model. The higher the number of parameters, the higher the power loss. The product of the r2 is a lower bound, but things could be much worse.
For our second set of simulations we examine the effect allele frequency has on power loss. We consider Model A, but for varying allele frequencies. For each linked pair of SNPs (causal and tag), we assume that the minor allele frequencies are the same, but that the frequencies differ between the pairs; we consider allele frequencies of 0.5, 0.4, 0.3, and 0.2. For these simulations we used 2000 repetitions. Table 2 gives the relative efficiency for power levels 0.3, 0.6, and 0.9 and we consider r2 values of 0.9, 0.7, and 0.5. Each entry of the table is around or a bit below the products of the r2 values, which is what we would expect from the discussion in the previous section. This suggests the allele frequencies do not play a large role in the power loss, supporting the viability of the products of the r2 as a prescreening metric.
Table 2.
Relative efficiencies for Model A under various r2 combinations and allele frequencies. The first column shows the r2 values between the causal and tag SNPs. Various combinations of allele frequencies are investigated. Each entry of the table gives the ratio of sample sizes that lead to the same power. All results are based on 2000 repetitions.
| Allele Frequencies | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| ( ) | Power | (0.5,0.4) | (0.5,0.3) | (0.5,0.2) | (0.4,0.3) | (0.4,0.2) | (0.3,0.2) | ||
| (0.9,0.9) | 0.81 | 0.3 | 0.771 | 0.762 | 0.760 | 0.775 | 0.772 | 0.761 | |
| 0.6 | 0.785 | 0.791 | 0.790 | 0.783 | 0.789 | 0.772 | |||
| 0.9 | 0.794 | 0.787 | 0.783 | 0.774 | 0.794 | 0.792 | |||
| (0.9,0.7) | 0.63 | 0.3 | 0.589 | 0.614 | 0.669 | 0.610 | 0.639 | 0.655 | |
| 0.6 | 0.602 | 0.603 | 0.642 | 0.617 | 0.649 | 0.668 | |||
| 0.9 | 0.581 | 0.615 | 0.661 | 0.620 | 0.67 | 0.668 | |||
| (0.9,0.5) | 0.45 | 0.3 | 0.411 | 0.465 | 0.500 | 0.432 | 0.505 | 0.535 | |
| 0.6 | 0.413 | 0.443 | 0.511 | 0.445 | 0.531 | 0.531 | |||
| 0.9 | 0.415 | 0.449 | 0.507 | 0.458 | 0.521 | 0.549 | |||
| (0.7,0.7) | 0.49 | 0.3 | 0.439 | 0.47 | 0.467 | 0.486 | 0.483 | 0.518 | |
| 0.6 | 0.452 | 0.443 | 0.473 | 0.478 | 0.486 | 0.523 | |||
| 0.9 | 0.440 | 0.466 | 0.474 | 0.464 | 0.503 | 0.503 | |||
| (0.7,0.5) | 0.35 | 0.3 | 0.310 | 0.316 | 0.363 | 0.353 | 0.389 | 0.410 | |
| 0.6 | 0.305 | 0.322 | 0.373 | 0.347 | 0.398 | 0.409 | |||
| 0.9 | 0.313 | 0.331 | 0.393 | 0.336 | 0.390 | 0.422 | |||
| (0.5,0.5) | 0.25 | 0.3 | 0.217 | 0.233 | 0.259 | 0.241 | 0.261 | 0.310 | |
| 0.6 | 0.216 | 0.233 | 0.262 | 0.241 | 0.274 | 0.297 | |||
| 0.9 | 0.218 | 0.235 | 0.266 | 0.241 | 0.269 | 0.300 | |||
5 The coverage of commonly used platforms
We explore here the coverage of the platforms that are commonly used for GWAS studies. Our interest is in testing for gene-gene interactions, and so the coverage is measured by defined in the previous sections. In an ideal setting, we would examine the platform coverage of the complete set of variants in the human genome as this would include all causal variation. Unfortunately this is not yet possible, although the 1000 Genome Project will soon yield a more complete picture of human variation. Instead we use the data in Phase II HapMap (The International HapMap Consortium, 2005), and pairwise LD measures we obtained from the database SCAN (SNP and Copy number ANnotation database; http://www.scandb.org) that contains SNP information such as LD patterns and eQTLs from expression studies in LCLs (Gamazon et al., 2010; Nicolae et al., 2010).
We used LD patterns in both the Caucasian (CEU) part of HapMap as well as in the samples of African descent (YRI). The YRI LD data capture patterns we would see in a GWAS on African-American cohorts, for example. SCAN contains data on 2,543,887 SNPs in the CEU dataset and 2,852,184 in the YRI. We focus the array comparison to three platforms for which there are data in SCAN: Affymetrix 6.0, Illumina HumanHap 650K and Illumina 1M. For each of these platforms, and for each SNP in HapMap, the database contains the pairwise r2 between the SNP and the best proxy on the platform (e.g. the maximum r2 over all SNPs in the same region). For each platform and each pair of SNPs in Phase II HapMap, we calculated the product of these r2 and summarized the results in Figure 3.
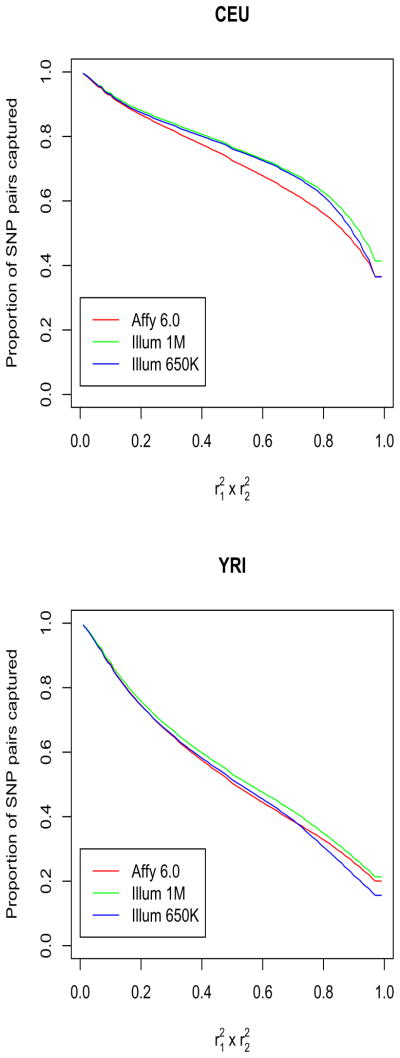
Figure 3.
The GxG coverage for three major genome-wide association platforms. We show the proportion of HapMap SNP pairs that are covered by pairs of SNPs on the genotyping platform at a level at least as large as the corresponding value on the x-axis. The linkage disequilibrium measures were calculated from HapMap data on a Caucasian population (top plot) and an African population (bottom plot).
Figure 3 shows, for each of the platforms, the fraction of pairs of Phase II HapMap SNPs (on the y-axis) that are captured by the best tagging pair of SNPs on the platform at a given efficiency level. For example, in the YRI panel less than 40% of the pairs of SNPs are captured at more than 80% efficiency (for any of the platforms), and around 20% of the pairs of SNPs are captured at full efficiency. As expected from the population LD structure, the coverage is better in samples of European descent than in samples of African ancestry. The platforms have similar coverage, with Illumina 1M slightly edging out in both CEU and YRI samples.
6 Discussion
In single SNP analyses the LD measure r2 is often used to measure the power loss, at least approximately, when using a tag SNP as a proxy for a causal SNP. We argue here that it is reasonable to use the product of the r2 as an approximation to the order of power loss when using proxies for gene-gene interactions. However, our simulations indicate that the product is in many cases an upper bound and that the power loss could be significantly higher. This suggests that coverage of causal variation is crucial when searching for gene-gene interactions as the power loss can be quite high even for larger r2 values.
The plots in Figure 3 provide an overly optimistic view not only because the metric we use is a lower bound for the loss in power, but also because we assume that Phase II HapMap (our reference set) contains the causal variants. Single-SNP coverage of the SeattleSNPs obtained from a resequencing project is around 20% lower than those in HapMap (Bhangale et al., 2008). For example, at r2 ≥ 0.8, SeattleSNPs coverage in the Illumina 1M is 55% which is much lower compared to the 80% coverage of HapMap variation (Bhangale et al., 2008). Also the coverage estimates we show for the various genotyping platforms are optimistic because of the design of these arrays: SNPs were chosen to capture HapMap variation and will perform well under this type of calculation.
Moreover, the power loss is even more significant when moving from 2-SNP models to settings where several SNPs are studied simultaneously (higher order interactions). It is easy to see that the power loss is multiplicative in the number of proxy SNPs used in the analysis, and unless we have strong priors on which variants to use in analysis, a higher order interaction has an extremely low chance of detection using GWAS data.
For these reasons, a great deal of thought needs to be put in the design of a GxG interaction search. For example, a strategy for detecting interactions using all data from an African-American GWAS done with the Affymetrix 100K platform has an extremely low chance of success and it should likely not be attempted.
Thus the picture is that of a double edged sword. On one hand, if the coverage is low then the power loss is large when using SNP proxies. On the other hand, a larger coverage necessarily implies a larger search space, which is typically enormous, when searching for interactions. Therefore any search strategy that fails to appreciate these limitations is likely doomed to fail or requires significant luck.
There are many aspects that enter in the evaluation of power for gene-gene interaction testing, such as: (i) the underlying genetic model; (ii) the statistical test for interaction; (iii) the number of pairs investigated and the corresponding multiple testing correction (in a Bayesian setting, the prior on each pair); (iv) the coverage of causal variation. The goal of this paper is to investigate the latter issue, but obviously a full discussion of power should involve all of the above and it is beyond the goal of this manuscript. We will not discuss in detail the effect of (i) and (ii), and just state that our conclusion holds for the models we tested and the statistical inference (logistic regression) we used. It is worth noting that different strategies for prioritizing pairs of SNPs can have different effects on the power loss due to coverage. If the prioritization is done from external resources (e.g. testing pairs of SNPs in genes that are directly linked in a genetic network), our quantification holds because the measures of coverage are independent of power. If an alternative strategy is used, where we first test for marginal effects and then look only at associated loci, our global measures can be misleading because truly associated SNPs tend to be in higher LD with causal SNPs than randomly chosen causal-tag pairs of SNPs.
Acknowledgments
The authors are grateful to Nancy Cox for helpful comments, and to Eric Gamazon for providing the LD data on platform coverage. The research was supported in part by the NIH grants 1RC2-HL101651 and U01-HL084715.
References
- Bhangale TR, Rieder MJ, Nickerson DA. Estimating coverage and power for genetic association studies using near-complete variation data. Nature Genetics. 2008;40:841843. doi: 10.1038/ng.180. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Cardon LR. Evaluating coverage of genome-wide association studies. Nature Genetics. 2006;38:65962. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- Cordell HJ. Epistasis: what it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum Mol Genet. 2002;11:2463–2468. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- Cordell HJ. Detecting genegene interactions that underlie human diseases. Nature Rev Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle M, Ng P, Kuhn K, Zhou L, Peiffer D, et al. Power to detect risk alleles using genome-wide tag SNP panels. PLoS Genetics. 2007;3(10):e170. doi: 10.1371/journal.pgen.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Zhang W, Konkashbaev A, Duan S, Kistner EO, et al. SCAN: SNP and copy number annotation. Bioinformatics. 2010;26:259262. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A Haplotype Map of the Human Genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nature Genetics. 1999;22:139144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nature Genetics. 2005;37:413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- Nicolae DL, Wen X, Voight B, Cox NJ. Coverage and characteristics of the Affymetrix GeneChip Human Mapping 100K SNP Set. PLoS Genetics. 2006;2(5):e67. doi: 10.1371/journal.pgen.0020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL. Quantifying the amount of missing information in genetic association studies. Genetic Epidemiology. 2006;30(8):703–717. doi: 10.1002/gepi.20181. [DOI] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-Associated SNPs Are More Likely to Be eQTLs: Annotation to Enhance Discovery from GWAS. PLoS Genetics. 2010;6(4):e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J, Przeworski M. Linkage disequilibrium in humans: models and data. Am J Hum Geneta. 2001;69:114. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]