Abstract
Aldehyde dehydrogenase-1A1 (ALDH1A1) expression characterizes a subpopulation of cells with tumor initiating or cancer stem cell properties in several malignancies. Our goal was to characterize the phenotype of ALDH1A1-positive ovarian cancer cells and examine the biological effects of ALDH1A1 gene silencing. In our analysis of multiple ovarian cancer cell lines, we found that ALDH1A1 expression and activity was significantly higher in taxane and platinum-resistant cell lines. In patient samples, 72.9% of ovarian cancers had ALDH1A1 expression, in whom the percent of ALDH1A1-positive cells correlated negatively with progression-free survival (6.05 v 13.81 months, p<0.035). Subpopulations of A2780cp20 cells with ALDH1A1 activity were isolated for orthotopic tumor initiating studies, where tumorigenicity was approximately 50-fold higher with ALDH1A1-positive cells. Interestingly, tumors derived from ALDH1A1-positive cells gave rise to both ALDH1A1-positive and ALDH1A1-negative populations, but ALDH1A1-negative cells could not generate ALDH1A1-positive cells. In an in vivo orthotopic mouse model of ovarian cancer, ALDH1A1 silencing using nanoliposomal siRNA sensitized both taxane- and platinum-resistant cell lines to chemotherapy, significantly reducing tumor growth in mice compared to chemotherapy alone (a 74–90% reduction, p<0.015). These data demonstrate that the ALDH1A1 subpopulation is associated with chemoresistance and outcome in ovarian cancer patients, and targeting ALDH1A1 sensitizes resistant cells to chemotherapy. ALDH1A1-positive cells have enhanced, but not absolute, tumorigenicity, but do have differentiation capacity lacking in ALDH1A1-negative cells. This enzyme may be important for identification and targeting of chemoresistant cell populations in ovarian cancer.
Keywords: aldehyde dehydrogenase, chemotherapy resistance, cancer stem cell, small interfering RNA, ovarian cancer
INTRODUCTION
Ovarian cancer was expected to be diagnosed in 21,550 women in 2009, and take the lives of 14,600 women (1). Although ovarian cancer is among the most chemosensitive malignancies at the time of initial treatment (surgery and taxane/platinum-based chemotherapy), most patients will develop tumor recurrence and succumb to chemoresistant disease (2). An understanding of the mechanisms mediating survival of subpopulations of ovarian cancer cells is necessary to significantly improve outcomes in this disease.
In many malignancies, a subpopulation of malignant cells termed “cancer stem cells” (CSCs) or “tumor initiating cells” (TICs) has been hypothesized to represent the most tumorigenic and treatment-resistant cells within a heterogeneous tumor mass. Defined by their enhanced ability to generate murine xenografts and give rise to heterogeneous tumors that are composed of both TIC and non-TIC populations, these cells may also be more chemoresistant, and depend on unique biological processes compared to the majority of tumor cells (3–4). In ovarian cancer, many of these properties have been identified in populations of CD44/c-kit positive cells (5), CD133-positive cells (6–8), and Hoechst-excluding cells (the “side population”) (9).
Among several markers that have been used to identify cancer stem cells, aldehyde dehydrogenase-1A1 (ALDH1A1) has been a valid marker among several malignant and nonmalignant tissues (10–20). It holds the attractive distinction of not only being a potential marker of “stemness,” but potentially playing a role in the biology of TICs as well (10). ALDH1A1, one of 17 ALDH isoforms, is an intracellular enzyme that oxidizes aldehydes, serving a detoxifying role, and converts retinol to retinoic acid, mediating control on differentiation pathways. The ALDH1A1 population defines normal hematopoietic stem cells, being used to isolate cells for stem cell transplants in patients. Using the ALDEFLUOR assay, a functional flow cytometric assay that identifies cells with active ALDH1A1, TIC-enriched populations have been identified in multiple malignancies (20), including breast (11–14), colon (15–16), pancreas (17), lung (18), and liver (19). Whether or not the ALDH1A1-active population is enriched for TIC’s has not been demonstrated for ovarian cancer. More importantly, while ALDH1A1 is implicated in chemoresistance pathways, it is not known whether targeting ALDH1A1 can sensitize resistant cells to chemotherapy, and therefore represent a potential target for cancer stem cell-directed therapy. We sought to characterize expression of ALDH1A1 in ovarian cancer cell lines and patient samples, determine if it contains TIC properties, and examine whether targeting ALDH1A1 sensitizes cells to chemotherapy in both in vitro and in vivo ovarian cancer models.
METHODS
Cell lines and culture
The ovarian cancer cell lines SKOV3ip1, SKOV3TRip2, HeyA8, HeyA8MDR, A2780ip2, A2780cp20, IGROV-AF1, and IGROV-cp20 (21–22) were maintained in RPMI-1640 medium supplemented with 15% fetal bovine serum (Hyclone, Logan, UT). SKOV3TRip2 (taxane-resistant, a kind gift of Dr. Michael Seiden (23)) and HeyA8MDR were maintained with the addition of 150 nM of paclitaxel. The HIO-180 SV40-immortalized non-tumorigenic cell line derived from normal ovarian surface epithelium was a kind gift of Dr. Andrew Godwin. All cell lines were routinely screened for Mycoplasma species (GenProbe detection kit; Fisher, Itasca, IL) with experiments performed at 70–80% confluent cultures. Purity of cell lines were confirmed with STR genomic analysis, and cells used were always less than 20 passages from the stocks tested for purity.
Whole Genomic analysis
RNA was extracted from three independent collections of SKOV3ip1 and SKOV3TRip2 cells at 80% confluence with the RNeasy Mini Kit (Qiagen, Hilden, Germany). RNA was subjected to microarray analysis using the Illumina HumanRef-8 Expression BeadChip, which targets ~24,500 well-annotated transcripts. Microarray data were normalized by the cubic-spline method (24) using the Illumina BeadStudio software. The significance of differentially expressed genes was determined by student’s t-test followed by correction for false discovery (25). A heat map was generated using Cluster 3.0 and Java TreeView software. The array data has been registered with GEO (accession #GSE23779) for public access.
Western blot analysis
Cultured cell lysates were collected in modified radioimmunoprecipitation assay (RIPA) lysis buffer with protease inhibitor cocktail (Roche, Manheim, Germany) and subjected to immunoblot analysis by standard techniques (26) using anti-ALDH1A1 antibody (BD Biosciences, San Jose, CA) at 1:1000 dilution overnight at 4°C, or anti-β-actin antibody (Sigma Chemical, St. Louis, MO) at 1:2000.
Immunohistochemical staining and clinical correlations
Immunohistochemical (IHC) analysis was performed on formalin-fixed paraffin-embedded (FFPE) samples using standard techniques (26). For ALDH1A1, antigen retrieval was in citrate buffer for 45 mins in an atmospheric-pressure steamer, using anti-ALDH1A1 antibody (BD Biosciences) at 1:500 dilution in Cyto-Q reagent (Innovex Biosciences, Richmond, CA) overnight at 4°C. Primary antibody detection was with Mach 4 HRP polymer (Biocare Medical, Concord, CA) for 20 mins at RT, followed by DAB incubation. After IHC staining, the number of tumor cells positive for ALDH1A1 were counted and expressed as a percentage of all tumor cells by an examiner blinded to clinical outcome. Patient samples were categorized as having low (less than 1%), intermediate (1–20%), or high (21–100%) ALDH1A1 expression. IHC was performed on samples collected at primary debulking surgery on 65 untreated patients with stage III–IV, high grade papillary serous adenocarcinoma, and with IRB approval, clinical information was collected. Progression-free and overall survival were plotted with the Kaplan-Meier method for patients in each group of ALDH1A1 expression, and compared with the log rank statistic using PASW 17.0.
For dual staining of ALDH1A1 and CD68 (for macrophages), staining for ALHD1A1 was performed first as above, followed by exposure to anti-CD68 antibody (1:4,000, Dako, Cambridgeshire, UK) and goat anti-mouse-AP (Jackson Immunoresearch, West Grove, PA). AP was developed with Ferangi Blue chromagen kit (Biocare Medical). For dual staining of ALDH1A1 and hypoxic tumor regions, mice bearing SKOV3TRip2 xenografts were injected with 60mg/kg Hypoxyprobe-1 reagent (HPI, Inc, Burlington, MA). Tumor sections in FFPE were subjected to antigen retrieval as above, followed by exposure to FITC-conjugated anti-Hypoxyprobe-1 mouse antibody (1:50) overnight at 4°C. This was detected with HRP-conjugated anti-FITC antibody (1:500, Jackson Immunoresearch) and DAB resolution. Endogenous murine IgG was then blocked with anti-mouse IgG F(ab’)2 fragments (Jackson ImmunoResearch), and ALDH1A1 stained as above using AP-conjugated anti-mouse IgG and Ferangi Blue chromagen.
ALDEFLUOR assay and tumorigenicity in limiting dilutions
Active ALDH1A1 was identified with the ALDEFLUOR assay according to manufacturer’s instructions (StemCell Technologies, Vancouver, BC). The ALDH1A1-positive population was defined by cells with increased FITC signal, with gates determined by DEAB-treated cells (DEAB being an inhibitor of ALDH1A1 activity). For tumorigenicity experiments, the ALDEFLUOR-positive population from A2780cp20 cells were sorted with a FACS Aria II flow cytometer (BD Biosciences), and reanalyzed to confirm at least 95% positivity. Collected cells were washed and resuspended in Ca2+- and Mg2+-free Hanks’ Balanced Salt Solution (HBSS, Gibco) and injected intraperitoneally (IP) into NOD-Scid mice in limiting dilutions, as shown in Table 2. Mice were followed for 1 year or until tumors formed, then sacrificed and tumor confirmed histologically. For flow cytometric analysis of these tumors, xenografts were dissociated mechanically with a scalpel, passed through a 70mm filter to collect single-cell suspensions, with the remaining clumped cells incubated in 0.5mg/mL collagenase and 0.0369mg/mL hyaluronidase (Calbiochem, Los Angeles, CA) for 30min at 37°C. These chemically digested cells were again filtered through a 70µm filter, added to the initial collection, and subjected to the ALDEFLUOR assay. ALDEFLUOR-positive cells or negative cells were then injected into additional mice (n=5) to examine maintenance of tumorigenicity.
Table 2.
Tumorigenicity of ALDEFLUOR-positive and negative cells
| A2780cp20 cells injected IP |
1 mil | 250k | 100k | 25k | 5k | 1k | Serial transplantation rate |
|---|---|---|---|---|---|---|---|
| ALDEFLUOR-negative | 5/5 | 4/5 | 2/5 | 2/5 | 0/5 | 0/5 | 0/5 |
| ALDEFLUOR-positive | 5/5 | 5/5 | 5/5 | 1/5 | 5/5 |
Primary xenograft development
With institutional IRB and IACUC approval, excess freshly-collected omental metastases from advanced-stage ovarian cancer patients were acquired after tissue required for diagnosis and management had been sequestered. 3–4mm3 sections were cut and implanted subcutaneously on the dorsal aspect of NOD-SCID mice. Adjacent sections were submitted for histologic analysis to confirm tumor. Tumors were measured in two dimensions twice per week. After progressive growth was noted, mice with formed tumors were treated with vehicle or cisplatin (7.5mg/kg weekly by intraperitoneal injection). Mice were treated for 8 weeks, then sacrificed and tumors harvested.
SiRNA downregulation in vitro
To examine downregulation of ALDH1A1 with siRNA, cells were exposed to 2.5µg/mL control siRNA (target sequence 5'-AATTCTCCGAACGTGTCACGT-3', Sigma, St. Louis, MO), or one of 3 tested ALDH1A1-targeting constructs (SASI_Hs01_00244055, 00244056, or 00303091, Sigma), at a 1:3 siRNA (µg) to Lipofectamine 2000 (µL) ratio. Lipofectamine and siRNA were incubated for 20min at RT, added to cells in serum-free RPMI to incubate for 6 hours, followed by 15% FBS/RMPI thereafter. Transfected cells were grown at 37°C for 48–72 hours and then harvested for Western Blot.
Assessment of cell viability with chemotherapy IC50 and cell cycle analysis
To a 96-well plate, 2000 cells/well were exposed to increasing concentrations of docetaxel or cisplatin in triplicate. Viability was assessed by 2-hour incubation with 0.15% MTT (Sigma) and spectrophotometer analysis at OD450. For effects of siRNA on IC50, cells were incubated with siRNA for 24 hours in 6-well plates, then replated in 96-well plates and chemotherapy added after 12 hours to allow attachment. IC50 was determined by finding the dose at which the drug had 50% of its effect, calculated by the equation [(OD450MAX−OD450MIN)/2) + OD450MIN]. Test of synergy was by the Loewe additivity model,(27) calculated by the equation CI = [D1/Dx1] + [D2/Dx2], where a CI (Combination Index) of 1 suggests an additive effect, <1 suggests synergy, and >1 suggests antagonism. For cell cycle analysis, cells were transfected with siRNA as above for 72 hours, trypsinized, washed in PBS, and fixed in 75% ethanol overnight. Cells were then centrifuged, washed x2 in PBS, and reconstituted in PBS with 50µg/mL propidium iodide. PI fluorescence was assessed by flow cytometry and percent cells in each cycle calculated by the cell cycle analysis module for FlowJo.
Orthotopic ovarian cancer model and in vivo delivery of siRNA
For orthotopic therapy experiments using ovarian cancer cell lines, female athymic nude mice (NCr-nu) were purchased from the National Cancer Institute (Frederick, MD) and cared for in accordance with guidelines of the American Association for Accreditation of Laboratory Animal Care. For all in vivo experiments, trypsinized cells were suspended in HBSS and 106 cells injected IP into 40 mice per experiment. After 1 week, mice were randomized to a) control siRNA/DOPC; b) control siRNA/DOPC plus chemotherapy; c) ALDH1A1-targeting siRNA/DOPC; or d) chemotherapy plus ALDH1A1-targeting siRNA/DOPC. SiRNA/DOPC dose was 5µg twice per week in a volume of 100µL IP. Chemotherapy doses were docetaxel 35µg IP weekly for SKOV3TRip2, or cisplatin 160 µg IP weekly for A2780cp20. Mice were treated for 4 weeks before sacrifice and tumor collection. SiRNA was incorporated into 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC) neutral nanoliposomes as previously described (28), lyophilized, and reconstituted in 0.9% saline for administration.
Statistical analysis
Comparisons between treatment groups of tumor weight was made with the two-tailed Student’s t test, if tests of data normality were met. Those represented by alternate distribution were examined by Mann-Whitney U statistic. Differences between groups were considered statistically significant at p<0.05. Number of mice per group (n=10) was chosen as directed by a power analysis to detect a 50% decrease in tumor growth with beta error of 0.2. Progression-free and overall survival in patients with three categories of ALDH1A1 staining were compared by plotting with the Kaplan Meier method and assessing for statistical differences with the log rank statistic using PASW 17.0 software.
RESULTS
Expression profiling of chemoresistant ovarian cancer cell lines
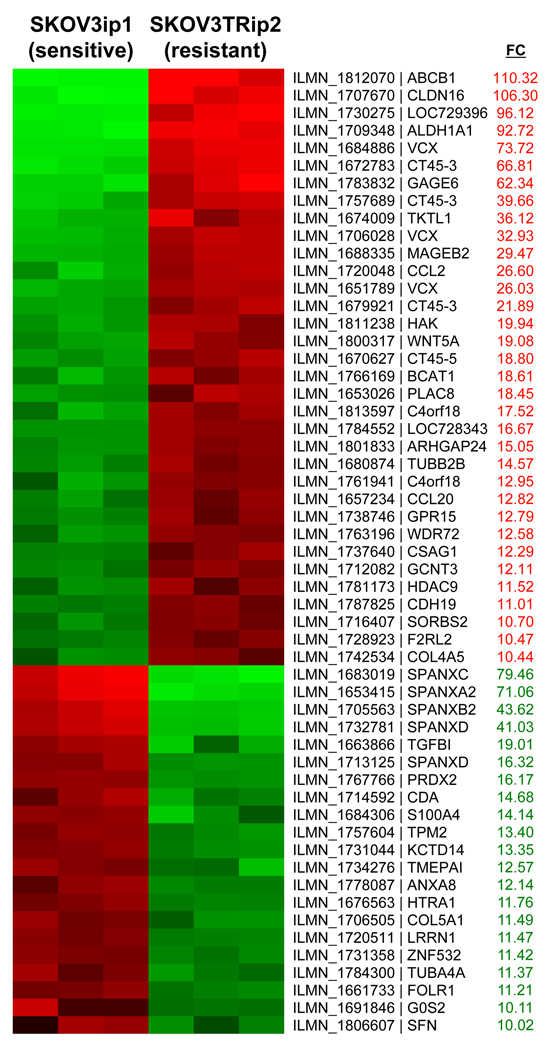
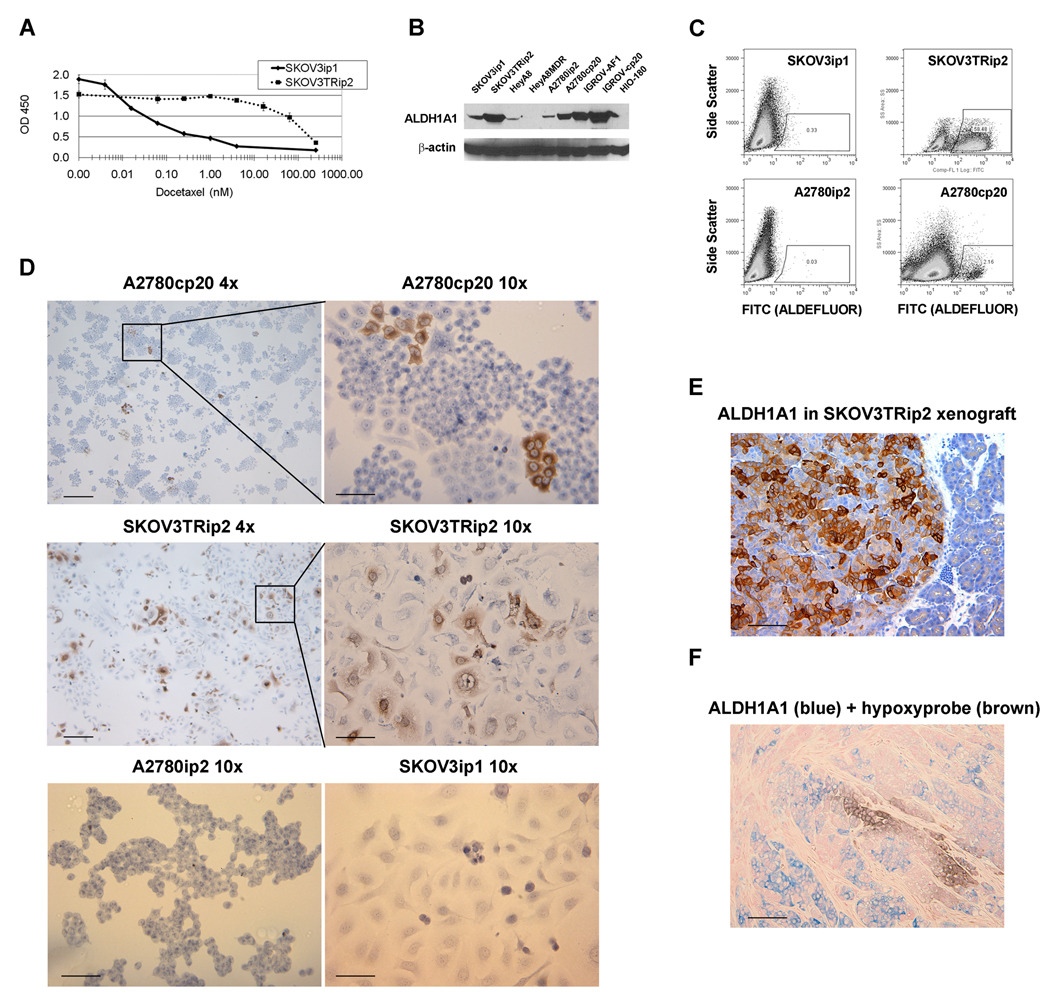
To discover genes mediating taxane resistance, expression profiling of parental SKOV3ip1 and taxane resistant SKOV3TRip2 cells was performed with microarray analysis using the Illumina HumanRef-8 Expression BeadChip. The SKOV3TRip2 cell line was previously generated through progressive exposure to paclitaxel (designated SKOV3TR) (23) and then passaged intraperitoneally (IP) in mice for two generations to select populations with enhanced tumorigenicity. Similarly, SKOV3ip1 were derived from SKOV3 parental cells to select for cells with enhanced tumorigenicity. We found 34 genes to be upregulated more than 10-fold in SKOV3TRip2 (Figure 1), among which was ALDH1A1, with a 92.7-fold increase (p=0.0025). 20 genes were more than 10-fold increased in SKOV3ip1. SKOV3TRip2 cells were confirmed to have approximately 3000-fold increased resistance to docetaxel, as measured by MTT IC50 (62.5 nM v. 0.02 nM, Figure 2A).
Figure 1. Comparison of whole genome expression profiling between SKOV3TRip2 and SKOV3ip1 cell lines.
Total RNA from the SKOV3TRip2 and SKOV3ip1 cell lines were subjected to whole genome expression profiling using the Illumina platform. The genes with a greater than 10-fold increase (FC-fold change) in SKOV3TRip2 are shown in red, those with a greater than 10-fold increase in SKOV3ip1 are shown in green.
Figure 2. ALDH1A1 expression in ovarian cancer cell lines.
A) As measured with the MTT viability assay, the SKOV3TRip2 ovarian cancer cell line has a docetaxel IC50 approximately 3,000-fold higher than its parental SKOV3ip1 cell line. B) Expression of ALDH1A1 by Western blot in four pairs of chemosensitive and chemoresistant ovarian cancer cell lines, and the nontransformed HIO-180 normal ovarian surface epithelium line. In all cases except HeyA8/HeyA8MDR, in which both lines had minimal expression, the chemoresistant line had increased ALDH1A1 expression. C) As measured by the ALDEFLUOR assay, the A2780cp20 (cisplatin resistant) and SKOV3TRip2 (taxane resistant) also contained a higher percentage of cells with functional ALDH1A1. D) This was confirmed by immunohistochemistry (IHC) for ALDH1A1 on these cell lines in vitro, where individual cells appeared either negative or strongly positive, demonstrating heterogeneity of ALDH1A1 expression in the cell line population. A low-power (4x) view gives an appreciation for the distinct colonies of ALDH1A1-positive cells, while examination at high power (10x) shows the definitive ALDH1A1-positive or negative nature of individual A2780cp20 and SKOV3TRip2 cells, but an absence of ALDH1A1 in parental A2780ip2 and SKOV3ip1 lines. E) This heterogeneity is also present in tumor xenografts, as seen with IHC for ALDH1A1 in SKOV3TRip2 tumors grown in mice (intraperitoneal location is confirmed by presence of normal pancreatic tissue on the right side of the slide) F) ALDH1A1 expression is not limited to hypoxic cells, as demonstrated in xenografts collected from mice given the hypoxyprobe reagent and subjected to co-IHC for ALDH1A1 (in blue) and the hypoxyprobe byproduct (in brown). Scale bars represent 50µm in 10x views, 100µm in 4x views and (E, F).
ALDH1A1 expression in ovarian cancer cell lines
To confirm an increase in ALDH1A1 expression/activity in SKOV3TRip2, and examine expression in other ovarian cancer cell lines, 4 pairs of parental and chemoresistant cell lines were examined: SKOV3ip1 / SKOV3TRip2; HeyA8 / HeyA8MDR (multi-drug resistant); A2780ip2 / A2780cp20 (10-fold increased cisplatin resistance); and IGROV-AF1 / IGROVcp20 (5-fold increased cisplatin resistance). Additionally, an immortalized non-transformed cell line derived from normal ovarian surface epithelium, HIO-180, was examined. We found that expression of total ALDH1A1, as measured by Western blot analysis, was in each case higher in the chemoresistant cell line, with the exception of HeyA8/HeyA8MDR, in which ALDH1A1 was low to absent in both (Figure 2B). To examine whether ALDH1A1 was not only present, but also active, we subjected cells to flow cytometric analysis using the ALDEFLUOR assay. This functional assay predominantly identifies active ALDH1A1, by conversion of a chemical to a fluorochrome. Presence of a subpopulation of ALDH1A1-active cells could be readily identified in SKOV3TRip2 (58% of the total population) and A2780cp20 (2.2%), but not in their parental cell line (Figure 2C). Furthermore, the strong shift in fluorescent signal in some cells suggests that there was not simply a general increase in expression in all cells, but rather separate populations of ALDH1A1–positive and negative cells. This was confirmed by immunohistochemistry, which showed distinct populations of ALDH1A1-positive or negative cells in A2780cp20 and SKOV3TRip2 cells, but not in the parental A2780ip2 and SKOV3ip1 cells in culture (Figure 2D). Finally, we observed that this heterogeneous profile was maintained in tumors. After IP injection of SKOV3TRip2 cells into nude mice and collection of the resulting orthotopic tumor implants, immunohistochemical staining of for ALDH1A1 showed both positive and negative ALDH1A1 subpopulations (Figure 2E). To examine whether this heterogeneity in expression was due to differential expression in hypoxic regions, a tumor-bearing mouse was injected with hypoxyprobe reagent and sacrificed after 30 minutes. The tumor was co-stained with ALDH1A1 and anti-hypoxyprobe antibody. We found that the ALDH1A1-positive cells were not preferentially localized to hypoxic regions in the tumor, with only 1.5% of ALDH1A1-positive cells concurrently positive for hypoxyprobe, and only 3.3% of hypoxyprobe-positive cells also positive for ALDH1A1 (p<0.01, Figure 2F).
ALDH1A1 expression in human ovarian cancer specimens
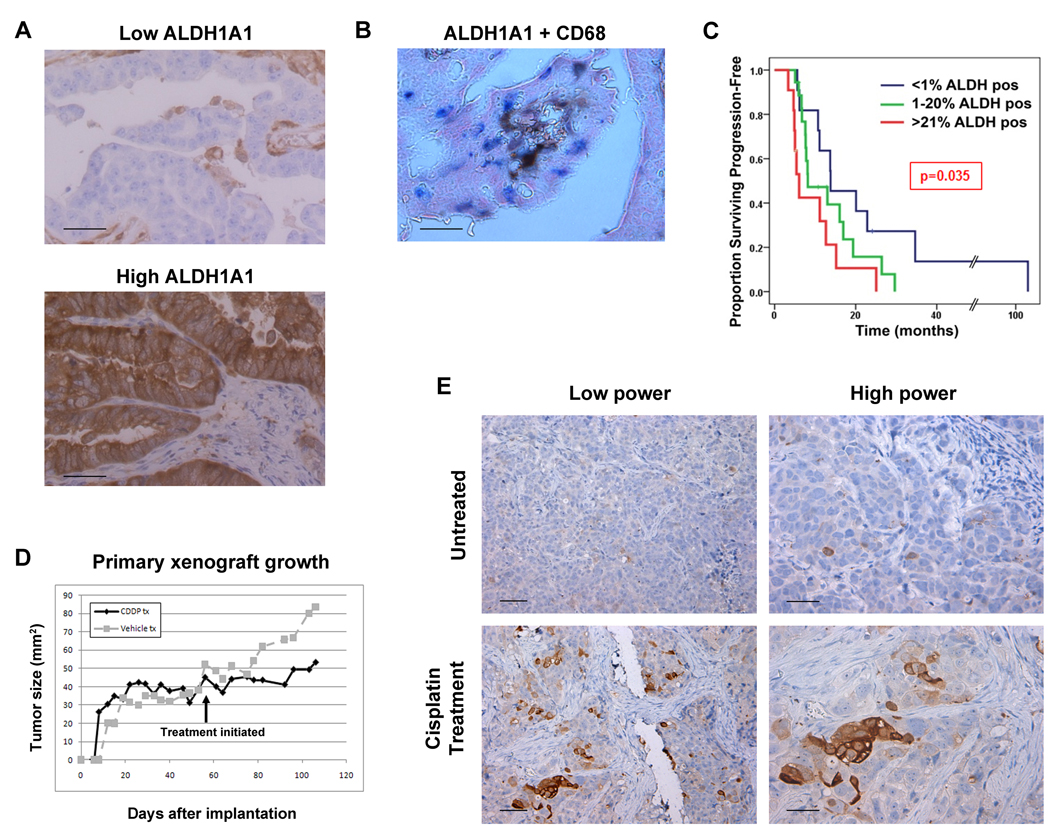
To determine the pattern of ALDH1A1 expression and possible correlations with chemoresistance in patients, we next examined ALDH1A1 expression in 65 untreated high-grade papillary serous stage III–IV ovarian cancer patient specimens (patient characteristics in Table 1). We found a wide range of expression patterns (Figure 3A). There was no ALDH1A1 in tumor cells in 27.1% of samples. ALDH1A1 expression was noted in 1–20% of cells in 44% of tumors, representing the largest cohort of expression patterns. As in xenografts from cell lines, expression was typically strong in some cells and negative in others, signifying distinct heterogeneity in the tumor. There was no distinct histologic pattern to the location of the positive cells (such as around vasculature, or on the leading edge of the tumor), but positive cells did tend to cluster together. The remaining tumors (28.9%) all had between 21 and 100% staining, with 10% of all patients having strong ALDH1A1 expression in nearly 100% of their tumor cells. To confirm that ALDH1A1 expression was not being mistakenly identified in tumor-infiltrating macrophages, dual staining for ALDH1A1 and CD68 was performed on several snap-frozen samples. Although images are not as detailed as those from paraffin-embedded samples, dual staining clearly shows that the majority of macrophages (blue) are ALDH1A1-negative, and therefore the heterogeneous ALDH1A1 positivity in tumors is not simply due to detection of macrophage infiltration (Figure 3B).
Table 1.
Characteristics of patients tested for ALHD1A1 expression (n=65)
| Characteristic | Percent or Average (range) |
|---|---|
| Age at diagnosis | 62.2 (34–89) |
| Caucasian Race | 71% |
| Pretreated with chemo | 0% |
| Stage | |
| Stage III | 74% |
| Stage IV | 26% |
| Ca125 | 3,071 (161–9600) |
| Ascites | 87% |
| Optimal Debulking | 74% |
| Papillary serous histology | 100% |
| Platinum/taxane primary therapy | 96% |
| Progression-free survival | 14.2 months (1.7–108) |
| Overall survival | 2.5 years (0.2–11.8) |
| ALDH1A1 staining | |
| Absent | 27.1% |
| 1–20% of cells | 44.0% |
| 21–100% of cells | 28.9% |
Figure 3. ALDH1A1 expression in ovarian cancer patients.
ALDH1A1 was assessed by IHC in 65 high-grade stage III–IV papillary serous ovarian cancer patients. A) Several expression patterns were seen, including absent, spotty (e.g. “Low ALDH”), and diffuse (“High ALDH”) staining. Consistent with staining in cell lines, both strongly positive and negative populations were noted. B) To confirm the “spotty” ALDH1A1 pattern was not identifying infiltrating macrophages, co-IHC on frozen tissue for ALDH1A1 (brown) and CD68 (a pan-macrophage marker, blue) was performed. C) Patients were stratified into less than 1%, 1–20% and greater than 20% ALDH1A1 expression, and progression-free and overall survival plotted by the Kaplan-Meier method and tested for statistical significance with the log rank test. There was a significantly shorter progression-free survival in patients with increasing ALDH1A1 expression. D) Mice with established primary subcutaneous xenografts were treated with vehicle or cisplatin for 5 weeks. E) Tumors from these mice were harvested and subjected to IHC for ALDH1A1. Tumors treated with cisplatin demonstrated a significant increase in the number of ALDH1A1-positive tumors cells. Magnification at low and high power are shown; scale bars represent 50µm in (A, B), and high power (E), 100µm in low power (E).
Correlation of ALDH1A1 expression with clinical outcomes
To determine if ALDH1A1 expression correlated with clinical outcomes, we compared progression-free survival and overall survival from patient samples described above (and in Table 1) in cohorts with no ALDH1A1 expression, 1–20% expression, and greater than 20% expression, as this grouping allowed similar numbers between groups,. Patients with >20% ALDH1A1-positive cells had a shorter median progression-free survival (6.1 months) compared to those with 1–20% ALDH1A1-positive cells (8.2 months) or those with no ALDH1A1-positive cells (13.8 months), which was statistically significant by the log rank test (p=0.035, Figure 3C). Overall survival, which will reflect resistance to multiple chemotherapeutic agents used in the recurrent setting, showed a trend towards a poor outcome with increasing ALDH1A1 expression (median OS 1.09 v. 1.84 v. 2.32 years) but the trend was not statistically significant (p=0.33, Suppl. Fig 1).
Preferential survival of ALDH1A1-positive cells with cisplatin treatment
To determine if the ALDH1A1-positive cells have preferential survival in the tumor microenvironment with platinum treatment, we established mouse xenografts from primary patient samples by subcutaneously implanting a freshly-collected tumor specimen into NOD-Scid mice. A subcutaneous rather than orthotopic model was utilized so that tumor growth and response could be accurately measured. Once tumors were established and growing, and achieved a size of approximately 1cm3, treatment with 7.5µg/kg of cisplatin IP weekly was initiated, while only vehicle was administered to controls (Figure 3D). When tumors grew to 2 cm3 in controls, having remained stable with cisplatin-treatment, tumors were harvested and sections stained for ALDH1A1 expression. Baseline expression of ALDH1A1 in the implanted tumor was seen in approximately 1% of cancer cells, and similar levels were found in growing xenografts in untreated mice (Figure 3E). A significant increase in the percentage of ALDH1A1-positive cells was, however, noted in cisplatin-treated xenografts to 38% (p<0.001, Figure 3E). Consistent with this, the ALDEFLUOR assay on the dissociated tumor showed that 0.6% of cells from untreated tumors were ALDEFLUOR-positive, while 17.6% of cells from cisplatin-treated tumors were ALDEFLUOR-positive. Because the treated xenograft in this case did not regress, but rather remained stable in size, cisplatin exposure may have induced ALDH1A1 expression in surviving cells, in addition to preferential killing of ALDH1A1-negative cells.
Tumor-initiating capacity of ALDH1A1-positive ovarian cancer cells
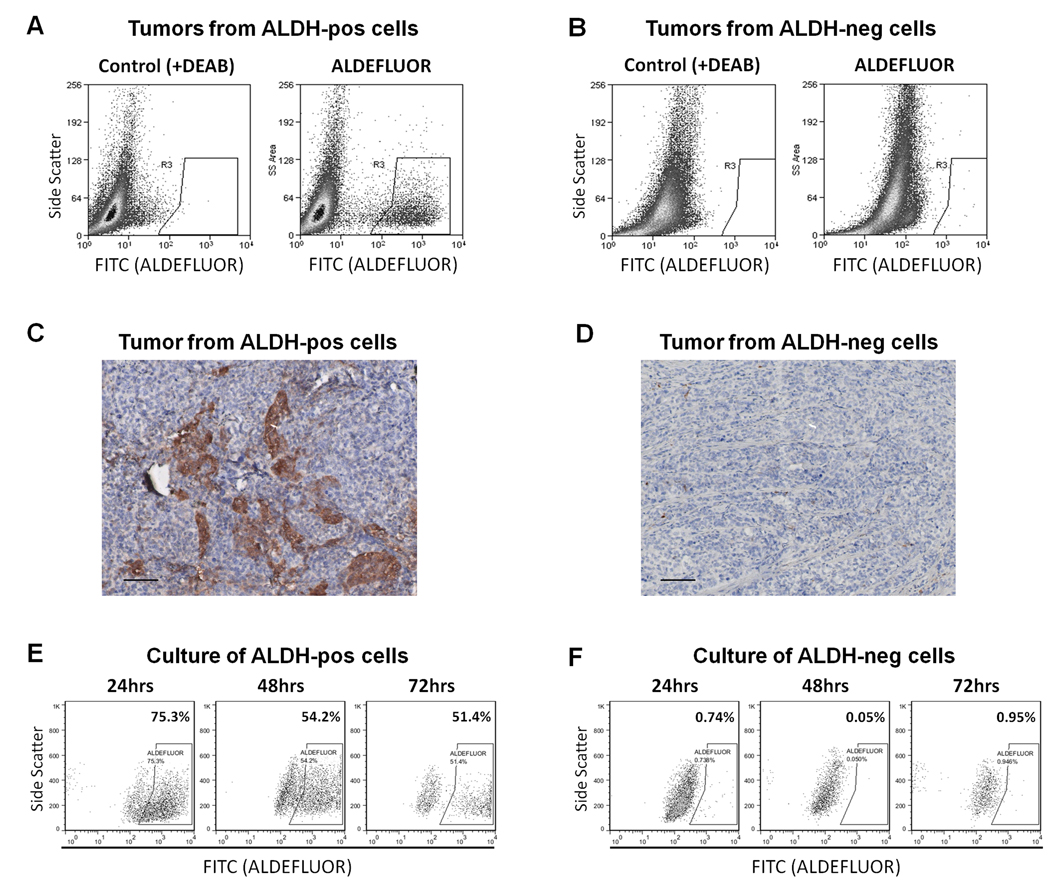
In breast and other cancers, the ALDH1A1-active cancer cells have been shown to represent a tumor initiating population (10–19). To determine if this were the case in ovarian cancer, we sorted ALDH1A1-positive and negative populations from the A2780cp20 cell line using the ALDEFLUOR assay and injected cells intraperitoneally into NOD-Scid mice in limiting dilutions to determine tumor initiating potential. As summarized in Table 2, ALDEFLUOR-positive cells exhibited increased tumorigenic potential, with 100% tumor initiation after injection of 100,000, 25,000, or 5,000 cells, and 1 tumor established after 1,000 cells injected. ALDEFLUOR-negative cells were also able to form tumors, although at a lower rate: two of 5 mice formed tumors after injection of 25,000 or 100,000 cells, and no tumors formed after injection of 5,000 or 1,000 cells. Mice were followed for 1 year after injection, and thorough necropsies were performed in remaining mice to confirm that tumors failed to develop. The TD50, or dose of cells required to permit tumor formation in 50% of animals, was 50-fold lower with ALDEFLUOR-positive cells. Perhaps more striking was the make-up of these tumors. One requirement of a tumor-initiating population is that they have the capacity to give rise to heterogeneous tumors, composed of both stem cell and non-stem cell populations, therefore demonstrating multipotent differentiating potential. This was noted in tumors that formed after injection of ALDEFLUOR-positive cells. In all 16 of these tumors, a strongly-positive ALDH1A1 population was noted in the minority of the sample, on average 4.7% of the tumor (range 2.4–6.1%, Figure 4A). However, no ALDEFLUOR-positive cells were found in the tumors that formed after injection of ALDH1A1-negative cells (Figure 4B). This was confirmed with IHC (Figure 4C,D). This argues against the idea that tumors formed because of contamination with ALDEFLUOR-positive cells, or that ALDH1A1 expression is simply induced by the tumor microenvironment regardless of the capacity of the cells. This difference in the capacity to generate ALDEFLUOR-positive cells was also noted in vitro. SKOV3TRip2 cells sorted into ALDEFLUOR-positive and negative populations were cultured separately, and the ALDEFLUOR assay performed on the different populations at 24, 48, and 72 hours (Figure 4E,F). Of the ALDEFLUOR-positive cells, the population gradually reverted to 75.3%, 54.2%, and 51.4% ALDEFLUOR-positive, respectively for each timepoint. However, the ALDEFLUOR-negative cells could not produce any ALDEFLUOR-positive cells.
Figure 4. ALDH1A1 populations in A2780cp20 xenografts.
Intraperitoneal tumors that developed after injection of ALDEFLUOR–positive or negative A2780cp20 cells were assessed for ALDH1A1 composition. A) Tumors that formed after injection of purely ALDEFLUOR-positive cells demonstrated both ALDEFLUOR-positive and negative populations, and recapitulated the TIC phenotype of having a small (2.4–6.1%) percentage of ALDEFLUOR-positive cells. B) Interestingly, tumors that formed after injection of purely ALDEFLUOR-negative cells contained only ALDEFLUOR-negative cells, showing an absence of capacity for differentiation, at least in terms of ALDEFLUOR positivity. C–D) This expression discrepancy was also noted on IHC for ALDH1A1 from these samples. Scale bars represent 100µm. E) Similarly, in vitro, SKOV3TRip2 ALDEFLUOR-positive cells give rise to both ALDEFLUOR-positive and ALDEFLUOR-negative cells, re-establishing baseline levels at 48 hours, whereas (F) ALDEFLUOR-negative cells cannot give rise to ALDEFLUOR-positive cells.
To confirm that the ALDEFLUOR-positive cells from tumors maintained tumorigenicity, these populations were sorted and re-injected intraperitoneally into mice, and continued to form tumors at 100% rate with 25,000 cells injected. However, ALDEFLUOR-negative cells from the tumors forming after ALDEFLUOR-negative cells were injected did not form tumors. Taken together, these studies show that ALDEFLUOR-positive cells have increased but not absolute tumorigenicity, but do have a differentiating capacity and a maintenance of the tumorigenic phenotype that is absent in ALDEFLUOR-negative cells.
In an effort to determine whether ALDEFLUOR-positive cells freshly collected from ovarian cancer patients has similar tumorigenicity, we have sorted ALDEFLUOR-positive and negative cells from five separate ovarian cancer patients, dissociating tumors metastatic to the omentum at the time of primary debulking surgery. In this cohort 1.5–17.8% of cells were ALDEFLUOR-positive. 25,000 ALDEFLUOR-positive cells, 100,000 ALDEFLUOR-negative cells, or 100,000 unsorted cells were injected IP into 5 mice per group per patient. Unfortunately, no tumors formed in any mice, highlighting the difficulty of tumorigenicity studies in primary ovarian cancer samples dissociated to single cell suspensions.
In order to preliminarily determine if there is overlap between the ALDEFLUOR-positive population and other markers of putative stem cells in ovarian cancer, these five samples were also profiled for CD44, c-kit, and CD133. We were not able to identify a convincing positive c-kit population from any sample. CD133-positive cells made up an average of 3.1% of total tumor cells (range 0.6–5.7%), and in all five samples were greater than 80% ALDEFLUOR positive (mean 86.7%, range 81.5–100%). CD44 was more commonly expressed, representing an average of 45.7% of tumors (but with a very broad range of 2.4–98.2%). Of the CD44-positive cells, 75.4% were also ALDEFLUOR-positive (range 46.6–88.8%). Similarly, the SKOV3TRip2 line has 82% CD44-positive cells, and of these 74% are ALDEFLUOR positive. Although a great number of samples will need to be examined to fully delineate if multiple-marker-positive cells can more accurately define the most pure tumorigenic cell, there is certainly overlap in marker expression. There are both double-positive CD44/ALDEFLUOR and CD133/ALDEFLUOR-positive populations that may prove more discerning as CSC populations, and ongoing studies could assess this distinction. Interestingly, the A2780cp20 cell line is completely negative for CD44, and the HeyA8 cell line is negative for ALDH1A1/ALDEFLUOR, despite the fact that both are highly tumorigenic. This highlights the fact that these cannot be the sole mediators of tumorigenicity in mice.
Downregulation of ALDH1A1 sensitizes ovarian cancer cells to chemotherapy
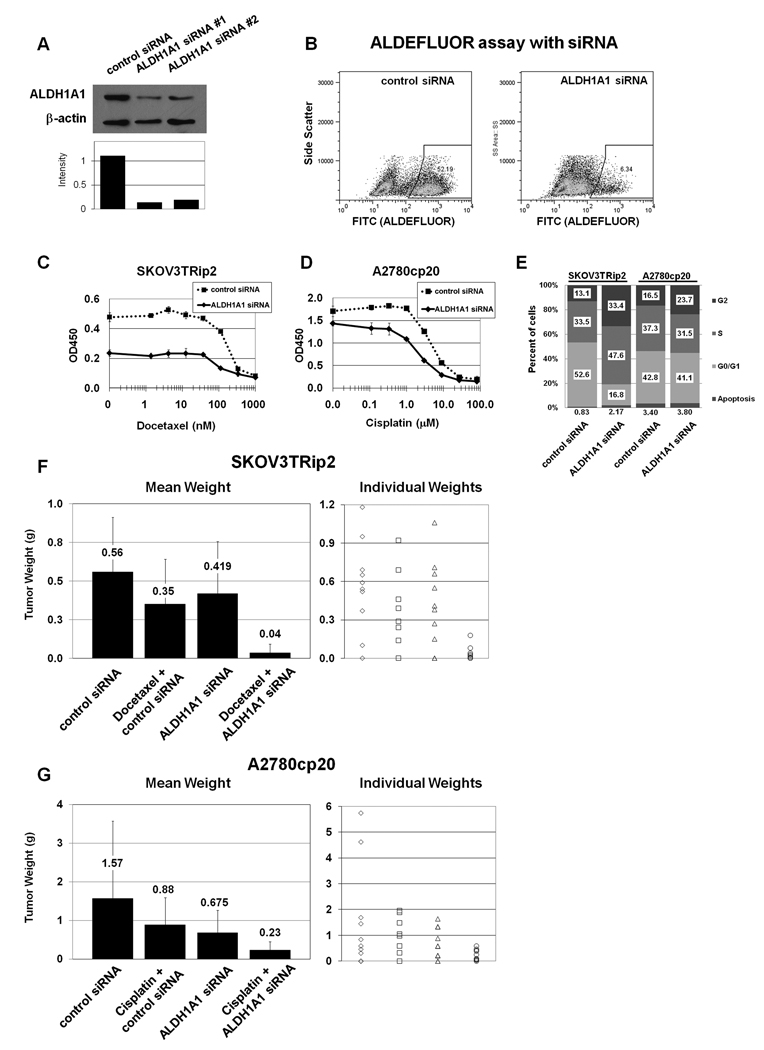
Given the association of ALDH1A1 expression with chemoresistant cell lines and a decreased progression-free survival in ovarian cancer patients, we asked whether downregulation of ALDH1A1 could sensitize resistant cells to chemotherapy. Two different siRNA constructs were identified that reduced ALDH1A1 expression by greater than 80% (Figure 5A). Reduction in the ALDEFLUOR population was confirmed (Figure 5B). SKOV3TRip2 or A2780cp20 cells were exposed to ALDH1A1-targeting siRNA (ALDH1A1 siRNA) or control siRNA for 24 hours before replating and adding increasing concentrations of docetaxel or cisplatin, respectively. Cell viability four days after addition of chemotherapy was assessed with the MTT assay. In SKOV3TRip2 cells, siRNA-ALDH1A1 alone reduced viability by 49% (Figure 5C, p<0.001). Downregulation of ALDH1A1 also reduced the docetaxel IC50 from 178 nM to 82 nM. In A2780cp20, the effects of ALDH1A1 downregulation alone were modest (Figure 5D, reduced viability by 15.9%, p=0.040), but sensitization to cisplatin was considerable, with a decrease in the IC50 from 5.1µM to 2.0µM. Tests for synergy suggest moderate synergy in each cell line (CI=0.82 for SKOV3TRip2, and 0.75 for A2780cp20). The contrasting effects of ALDH1A1-siRNA alone are consistent with the number of ALDH1A1-active cells in these cell lines, with SKOV3TRip2 cell lines having 50–60% ALDEFLUOR-positive cells, and A2780cp20 having just 2–3%. To determine how ALDH1A1 downregulation alone may affect cell growth, cell cycle analysis was performed in a separate experiment. We found that ALDH1A1 downregulation induced an accumulation of SKOV3TRip2 cells in S and G2 phases (p<0.001 compared to control siRNA), but had only minimal effects on the cell cycle of A2780cp20 cells (Figure 5E).
Figure 5. Efficacy of ALDH1A1 downregulation with siRNA in vitro and in vivo.
A) Identification of siRNA constructs that decrease ALDH1A1 expression were confirmed with Western blot, and B) by flow cytometry using the ALDEFLUOR assay in the SKOV3TRip2 cell line. C) Downregulation of ALDH1A1 with siRNA 48hrs prior to treatment of SKOV3TRip2 cells with increasing doses of docetaxel demonstrated a sensitization effect, decreasing the IC50 from 178nM to 82nM. siRNA alone also showed an effect, with decreased viability by 49%. D) In the A2780cp20 cell line, downregulation of ALDH1A1 alone had minimal effect, but sensitized cells to cisplatin, decreasing the IC50 from 5.1µM to 2.0µM. E) Cell cycle analysis shows that ALDH1A1 downregulation induces accumulation of cells in S and G2 phases in SKOV3TRip2, with little effect on A2780cp20. F) In vivo, mice injected intraperitoneally with SKOV3TRip2 cells were treated with ALDH1A1-siRNA incorporated in DOPC nanoparticles, docetaxel/control siRNA in DOPC, or the combination, and compared to mice treated with control siRNA in DOPC. Mice treated with either single agent had minimal effect, but the combination showed a significant reduction compared to treatment with control siRNA (94% reduction in tumor growth, p<0.001) or either single agent (90–91% reduction, p<0.005). G) Similarly, mice injected with A2780cp20 cells showed a minimal non-significant reduction in growth with cisplatin or ALDH1A1-siRNA in DOPC, but combination therapy was statistically superior to either single agent (65–73% reduction, p<0.04), and control siRNA (85% reduction, p=0.048). Mean tumor weight and individual tumor sizes are presented.
There are no known inhibitors of ALDH1A1 for in vivo studies. Therefore we utilized a method for delivery of siRNA in vivo using DOPC nanoparticles. We and others (28–32) have previously demonstrated delivery of siRNA incorporated into DOPC nanoliposomes to the tumor parenchyma with subsequent target downregulation. In this study nude mice were injected intraperitoneally with either SKOV3TRip2 or A2780cp20 cells and randomized to four treatment groups to begin 1 week after cell injection: 1) control siRNA in DOPC, delivered IP twice per week; 2) docetaxel 35 mg, delivered IP weekly (for SKOV3TRip2 model) or cisplatin 160 µg, delivered IP weekly (for A2780cp20 model); 3) ALDH1A1-siRNA in DOPC, IP twice per week; or 4) ALDH1A1-siRNA in DOPC plus docetaxel (for SKOV3TRip2) or cisplatin (for A2780cp20). After four weeks of treatment, mice were sacrificed and total tumor weight recorded. Immunohistochemical analysis confirmed reduced ALDH1A1 expression with ALDH1A1-siRNA/DOPC treatment compared to controls but not with chemotherapy alone (Suppl Figure 2; too little tissue was available to examine with the ALDEFLUOR assay). In SKOV3TRip2 xenografts (Figure 5F) there was a non-significant reduction in tumor growth with docetaxel treatment of 37.0% (p=0.17) and with ALDH1A1 siRNA treatment of 25.0% (p=0.38) compared to control-DOPC. The observation that ALDH1A1 downregulation alone significantly decreased SKOV3TRip2 growth in vitro but was less pronounced in vivo suggests that tumor microenvironment factors such as supporting stromal cells may be able to protect cells from ALDH1A1 depletion. However, the combination of ALDH1A1 siRNA and docetaxel resulted in significantly reduced growth, by 93.6% compared to control siRNA (p<0.001), by 89.8% compared to docetaxel plus control siRNA (p=0.003), and by 91.4% compared to ALDH1A1 siRNA (p=0.002). In A2780cp20 (Figure 5G), there was a similar non-significant reduction in tumor weight with cisplatin alone of 43.9% (p=0.32) and with ALDH1A1 siRNA treatment of 57.0% (p=0.19). These effects may be even less significant than the mean tumor weights suggest, given the presence of two especially large tumors in the control siRNA group. However, again combined therapy showed a sensitization to chemotherapy with ALDH1A1 siRNA, with combination therapy reducing growth by 85.0% compared to control siRNA (p=0.048), by 73.4% compared to cisplatin plus control siRNA (p=0.013), and by 65.3% compared to ALDH1A1 siRNA alone (p=0.039). Given the minimal effects of either single agent and the consistent finding of significant improvement with combined therapy, these data suggest a synergy between ALDH1A1 downregulation and both taxane and platinum chemotherapeutic agents, though formal dose-finding experiments would be required to definitively prove synergy.
DISCUSSION
We have found that ALDH1A1 expression and activity are increased in chemoresistant ovarian cancer cell lines, and in in situ primary ovarian cancer xenografts treated with cisplatin. Expression of ALDH1A1 is frequent in ovarian tumors, and patients with low ALDH1A1 expression have a more favorable outcome than those with more ALDH1A1-positive cells. ALDEFLUOR-positive cells have increased (but not absolute) tumorigenicity compared to ALDEFLUOR-negative cells, and have a differentiating capacity not present in the ALDEFLUOR-negative population. Most importantly, downregulation of ALDH1A1 expression sensitized normally chemoresistant tumors to both docetaxel and cisplatin both in vitro and in an orthotopic mouse model of ovarian cancer.
The search for tumor initiating cells in ovarian cancer has resulted in observations that the CD44+/c-kit+ population has an approximately 5,000-fold increase in tumorigenicity, with tumors forming after injection of as few as 100 cells from primary tumor, xenograft, or spheroid heterogeneous populations (5), and that the CD133+ population has approximately 20 fold increased tumorigenicity, with tumor formation with as few as 100–500 cells from murine xenografts, and tumor formation 4 times faster with CD133+ cells (7). Furthermore, the increased tumorigenicity of CD133+ cells can be inhibited by interfering with binding between CD44 and its ligand hyaluronic acid (6). Other investigators have found equal rates of tumor formation among CD133+ and CD133− cells from the A2780 cell line, but a faster growth rate in CD133+ cells (8). The side population (SP) cells from the MOVCAR cell line also formed tumors more frequently and appeared 3–4 weeks sooner than tumors derived from non-SP cells (9). In all of these studies, as in ours, the tumors resulting from the putative TIC population contained both TIC and non-TIC populations, demonstrating multipotentiality. Interestingly, we have seen that cells comprising tumors formed from ALDH1A1-negative cells lack the capacity to generate ALDH1A1-positive cells, and do not continue to propagate tumors over multiple generations, suggesting that their multipotentiality is limited. This lack of differentiating capacity has also been noted in ALDEFLUOR-negative cells from breast cancer cell lines (33).
The most appropriate source of tumor cells for tumorigenicity experiments is of some debate. While it is desirable to use samples freshly collected from primary tumors, sorting these samples and establishing primary xenografts has proven problematic. Ovarian cancer xenografts and cells lines have traditionally been challenging to establish from primary samples. All previously reported studies of ovarian tumor-initiating cells have used selected cells of some sort, either from xenografts of varying generations or cells grown in differentiation-inhibiting media (to form tumorspheres), to serve as a compromise between freshly collected specimens and cell lines. However, those cells that form tumors in mice even in the first generation almost certainly represent some select portion of the original tumor. That these xenografts still contain only a small percentage of TIC’s speaks to either the appropriateness of this approach, or to the testament that the tumor-forming cells are multipotent, give rise to TIC-negative populations, and remain relatively rare. Use of cell lines is often discouraged due to their homogenous nature. But clearly, even within cell lines there is heterogeneity in ALDH1A1 expression, demonstrated by the detection of distinct populations with flow cytometric and immunohistochemical analysis (Figure 2). Distinct ALDEFLUOR-positive and negative populations have also been found in several breast cancer cell lines, with ALDEFLUOR-positive cells having increased tumorigenicity and differing molecular signatures (33). Therefore, our finding that the ALDEFLUOR-positive population in cell lines has increased tumorigenicity may reflect the more aggressive phenotype of ALDH1A1-active cells, but does not represent proof that this population is important to in situ ovarian cancers. Evidence that patients with increasing ALDH1A1 expression have poor outcomes suggests this association, but additional tumorigenicity experiments from freshly-collected tumors would more appropriately define the ALDEFLUOR population as a clinically significant TIC.
The importance of tumorigenicity in defining cancer stem cells has also been debated. While tumor formation with 100–500 ALDEFLUOR-positive cells and a lack of tumor formation with injection of 105 ALDEFLUOR-negative cells definitely reflects an aggressive phenotype, the biologic processes required for xenograft formation – survival under stressful experimental conditions, adhesion, time to proliferation, and variations in host immunocompetence – may not reflect the true population that cancer stem cell research seeks to identify. Our ultimate goal should be to identify the subpopulations in parent tumors that survive chemotherapy and therefore are more likely to cause recurrence. Stem cells that survive chemotherapy should exhibit chemoresistance to be clinically relevant. In breast cancer, for example, the CD44+/CD24− population is highly tumorigenic. However, Tanei et al studied tissue obtained prior to and after neoadjuvant chemotherapy and found that despite a positive response to treatment, the proportion of CD44+/CD24− negative cells was unchanged. In these samples, however, the ALDH1A1-positive population was significantly increased (34).
ALDH1A1 has previously been proposed to play a role in chemoresistance, having been noted to be higher in proteomic profiling of IGROV platinum resistant ovarian cancer cells (35), in genomic profiling of multidrug resistant gastric carcinoma (36), and in cells resistant to cyclophosphamide (37–38), oxazaphosphorines (39), and now docetaxel and cisplatin. ALDH1A1 oxidizes many intracellular aldehydes into carboxylic acids (40), detoxifying many of the free oxygen radicals generated by chemotherapeutic agents. It stands to reason that a stem cell population should be resistant to multiple chemotherapeutic agents, rather than being specific to one class. This also follows clinically, in that most ovarian cancer patients that develop resistance to platinum agents have resistance to multiple agents (2). ALDH1A1 has been shown to be associated with BRCA1 in breast cancer, in that knockdown of BRCA1 increases the ALDEFLUOR population, and ALDEFLUOR-positive cells preferentially contain BRCA1 loss of heterozygocity (41). These findings could be important to BRCA-mediated ovarian cancer as well. Despite this body of evidence for the importance of ALDH1A1, it is not fully understood if any of the additional ALDH1 isoforms are important to stem cell biology. In our study, ALDH1A1 can be specifically identified with isotype-specific antibodies (as used for IHC and WB). However, the more important and consistently-used identifier of a stem cell population is the ALDEFLUOR assay, which, while primarily dependent on ALDH1A1, may also identify ALDH1A2 and ALDH1A3 isotypes ((42) and unpublished data by Stem Cell Technologies). As a therapeutic, we have seen positive effects by targeting ALDH1A1 with siRNA, but to maximize the efficacy of therapeutics, the contribution of these additional isotypes will need to be defined with additional studies.
Although our finding of a poor outcome in patients with high ALDH1A1 expression agrees with similar investigations in breast cancer (12–13) and ovarian cancer (20), one interesting report found that high ALDH1A1 expression actually confers a positive prognosis in ovarian cancer (43). This cohort also contained patients with absent, scattered, and diffuse staining. However, this cohort included patients with stage I and II disease and low-grade tumors, and ALDH1A1 expression was higher in these patients (confirming findings from a previous report (44)). Furthermore, with multivariate analysis only stage correlated with survival – ALDH1A1 expression no longer predicted outcomes. In ovarian cancer there is a well-recognized dichotomy in carcinogenesis and pathobiology (45), whereby low-grade tumors (which are more often diagnosed at stage I or II) are paradoxically more chemoresistant but have prolonged survival due to slow growth. Given these collective data, and the several mechanisms by which ALDH1A1 has been shown to contribute to chemoresistance, it may be that ALDH1A1 is more frequently expressed in, low-grade tumors, but participates in chemoresistance to both high-grade and low-grade subtypes.
We have demonstrated that the ALDH1A1-positive population has properties of cancer stem cells, is associated with both taxane and platinum resistance, and can be resensitized to chemotherapy with downregulation of ALDH1A1 in vitro and in vivo. Therefore ALDH1A1 is not just a marker of an aggressive population but a mediator of the phenotype and a viable target for therapy. As better models are developed to more purely define the true chemoresistant population in de novo patient tumors, the ALDH1A1 population, either alone or in combination with other markers and mediators of resistance, may represent a population that must be targeted in order to achieve increased response rates and survival in ovarian cancer patients.
Supplementary Material
Acknowledgements
Funding support provided by the Reproductive Scientist Development Program through the Ovarian Cancer Research Fund and the National Institutes of Health (K12 HD00849), the Department of Defense Ovarian Cancer Research Academy, a Career Development Award from the UT MD Anderson Cancer Center Specialized Program of Research Excellence in ovarian cancer (CA083639), and the Gynecologic Cancer Foundation to CNL; NIH (CA109298 and CA110793, P50 CA083639; CA128797, RC2GM092599), Program Project Development Grant from the Ovarian Cancer Research Fund, the Marcus Foundation, and the Betty Ann Ashe Murray Distinguished Professorship to AKS.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Hoskins W, Perez C, Young R, Barakat R, Markman M, Randall M. Principles and Practice of Gynecologic Oncology. 4th ed. Philadelphia: Lippincott Williams & Wilkins; 2005. [Google Scholar]
- 3.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 5.Zhang S, Balch C, Chan MW, et al. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68:4311–4320. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slomiany MG, Dai L, Tolliver LB, Grass GD, Zeng Y, Toole BP. Inhibition of Functional Hyaluronan-CD44 Interactions in CD133-positive Primary Human Ovarian Carcinoma Cells by Small Hyaluronan Oligosaccharides. Clin Cancer Res. 2009;15:7593–7601. doi: 10.1158/1078-0432.CCR-09-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curley MD, Therrien VA, Cummings CL, et al. CD133 Expression Defines a Tumor Initiating Cell Population in Primary Human Ovarian Cancer. Stem Cells. 2009 doi: 10.1002/stem.236. [DOI] [PubMed] [Google Scholar]
- 8.Baba T, Convery PA, Matsumura N, et al. Epigenetic regulation of CD133 and tumorigenicity of CD133+ ovarian cancer cells. Oncogene. 2008 doi: 10.1038/onc.2008.374. [DOI] [PubMed] [Google Scholar]
- 9.Szotek PP, Pieretti-Vanmarcke R, Masiakos PT, et al. Ovarian cancer side population defines cells with stem cell-like characteristics and Mullerian Inhibiting Substance responsiveness. Proc Natl Acad Sci U S A. 2006;103:11154–11159. doi: 10.1073/pnas.0603672103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moreb JS. Aldehyde dehydrogenase as a marker for stem cells. Curr Stem Cell Res Ther. 2008;3:237–246. doi: 10.2174/157488808786734006. [DOI] [PubMed] [Google Scholar]
- 11.Balicki D. Moving forward in human mammary stem cell biology and breast cancer prognostication using ALDH1. Cell Stem Cell. 2007;1:485–487. doi: 10.1016/j.stem.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charafe-Jauffret E, Ginestier C, Iovino F, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Croker AK, Goodale D, Chu J, et al. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med. 2008 doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang EH, Hynes MJ, Zhang T, et al. Aldehyde Dehydrogenase 1 Is a Marker for Normal and Malignant Human Colonic Stem Cells (SC) and Tracks SC Overpopulation during Colon Tumorigenesis. Cancer Research. 2009;69:3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carpentino JE, Hynes MJ, Appelman HD, et al. Aldehyde Dehydrogenase-Expressing Colon Stem Cells Contribute to Tumorigenesis in the Transition from Colitis to Cancer. Cancer Research. 2009;69:8208–8215. doi: 10.1158/0008-5472.CAN-09-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dembinski JL, Krauss S. Characterization and functional analysis of a slow cycling stem cell-like subpopulation in pancreas adenocarcinoma. Clin Exp Metastasis. 2009;26:611–623. doi: 10.1007/s10585-009-9260-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ucar D, Cogle CR, Zucali JR, et al. Aldehyde dehydrogenase activity as a functional marker for lung cancer. Chem Biol Interact. 2009;178:48–55. doi: 10.1016/j.cbi.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma S, Chan KW, Hu L, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 20.Deng S, Yang X, Lassus H, et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS One. 2010;5:e10277. doi: 10.1371/journal.pone.0010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buick RN, Pullano R, Trent JM. Comparative properties of five human ovarian adenocarcinoma cell lines. Cancer Res. 1985;45:3668–3676. [PubMed] [Google Scholar]
- 22.Yoneda J, Kuniyasu H, Crispens MA, Price JE, Bucana CD, Fidler IJ. Expression of angiogenesis-related genes and progression of human ovarian carcinomas in nude mice. J Natl Cancer Inst. 1998;90:447–454. doi: 10.1093/jnci/90.6.447. [DOI] [PubMed] [Google Scholar]
- 23.Duan Z, Feller AJ, Toh HC, Makastorsis T, Seiden MV. TRAG-3, a novel gene, isolated from a taxol-resistant ovarian carcinoma cell line. Gene. 1999;229:75–81. doi: 10.1016/s0378-1119(99)00042-6. [DOI] [PubMed] [Google Scholar]
- 24.Illumina I. BeadStudio Normalization Algorithms for Gene Expression Data. TECHNICAL NOTE: ILLUMINA® RNA ANALYSIS. 2007 cited; Available from: http://www.illumina.com/Documents/products/technotes/technote_beadstudio_normalization.pdf.
- 25.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 26.Landen CN, Jr, Lu C, Han LY, et al. Efficacy and antivascular effects of EphA2 reduction with an agonistic antibody in ovarian cancer. J Natl Cancer Inst. 2006;98:1558–1570. doi: 10.1093/jnci/djj414. [DOI] [PubMed] [Google Scholar]
- 27.Straetemans R, O'Brien T, Wouters L, et al. Design and analysis of drug combination experiments. Biom J. 2005;47:299–308. doi: 10.1002/bimj.200410124. [DOI] [PubMed] [Google Scholar]
- 28.Landen CN, Jr, Chavez-Reyes A, Bucana C, et al. Therapeutic EphA2 Gene Targeting In vivo Using Neutral Liposomal Small Interfering RNA Delivery. Cancer Res. 2005;65:6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 29.Gray MJ, Dallas NA, Van Buren G, et al. Therapeutic targeting of Id2 reduces growth of human colorectal carcinoma in the murine liver. Oncogene. 2008;27:7192–7200. doi: 10.1038/onc.2008.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halder J, Kamat AA, Landen CN, Jr, et al. Focal adhesion kinase targeting using in vivo short interfering RNA delivery in neutral liposomes for ovarian carcinoma therapy. Clin Cancer Res. 2006;12:4916–4924. doi: 10.1158/1078-0432.CCR-06-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamat AA, Feng S, Agoulnik IU, et al. The role of relaxin in endometrial cancer. Cancer Biol Ther. 2006;5:71–77. doi: 10.4161/cbt.5.1.2289. [DOI] [PubMed] [Google Scholar]
- 32.Villares GJ, Zigler M, Wang H, et al. Targeting melanoma growth and metastasis with systemic delivery of liposome-incorporated protease-activated receptor-1 small interfering RNA. Cancer Res. 2008;68:9078–9086. doi: 10.1158/0008-5472.CAN-08-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charafe-Jauffret E, Ginestier C, Iovino F, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanei T, Morimoto K, Shimazu K, et al. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin Cancer Res. 2009;15:4234–4241. doi: 10.1158/1078-0432.CCR-08-1479. [DOI] [PubMed] [Google Scholar]
- 35.Le Moguen K, Lincet H, Marcelo P, et al. A proteomic kinetic analysis of IGROV1 ovarian carcinoma cell line response to cisplatin treatment. Proteomics. 2007;7:4090–4101. doi: 10.1002/pmic.200700231. [DOI] [PubMed] [Google Scholar]
- 36.Ludwig A, Dietel M, Lage H. Identification of differentially expressed genes in classical and atypical multidrug-resistant gastric carcinoma cells. Anticancer Res. 2002;22:3213–3221. [PubMed] [Google Scholar]
- 37.Russo JE, Hilton J. Characterization of cytosolic aldehyde dehydrogenase from cyclophosphamide resistant L1210 cells. Cancer Res. 1988;48:2963–2968. [PubMed] [Google Scholar]
- 38.Yoshida A, Dave V, Han H, Scanlon KJ. Enhanced transcription of the cytosolic ALDH gene in cyclophosphamide resistant human carcinoma cells. Adv Exp Med Biol. 1993;328:63–72. doi: 10.1007/978-1-4615-2904-0_8. [DOI] [PubMed] [Google Scholar]
- 39.Kohn FR, Landkamer GJ, Manthey CL, Ramsay NK, Sladek NE. Effect of aldehyde dehydrogenase inhibitors on the ex vivo sensitivity of human multipotent and committed hematopoietic progenitor cells and malignant blood cells to oxazaphosphorines. Cancer Res. 1987;47:3180–3185. [PubMed] [Google Scholar]
- 40.Riveros-Rosas H, Julian-Sanchez A, Pina E. Enzymology of ethanol and acetaldehyde metabolism in mammals. Arch Med Res. 1997;28:453–471. [PubMed] [Google Scholar]
- 41.Liu S, Ginestier C, Charafe-Jauffret E, et al. BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci U S A. 2008;105:1680–1685. doi: 10.1073/pnas.0711613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yokota A, Takeuchi H, Maeda N, et al. GM-CSF and IL-4 synergistically trigger dendritic cells to acquire retinoic acid-producing capacity. Int Immunol. 2009;21:361–377. doi: 10.1093/intimm/dxp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang B, Liu G, Xue F, et al. ALDH1 expression correlates with favorable prognosis in ovarian cancers. Mod Pathol. 2009;22:817–823. doi: 10.1038/modpathol.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanner B, Hengstler JG, Dietrich B, et al. Glutathione, glutathione S-transferase alpha and pi, and aldehyde dehydrogenase content in relationship to drug resistance in ovarian cancer. Gynecol Oncol. 1997;65:54–62. doi: 10.1006/gyno.1996.4593. [DOI] [PubMed] [Google Scholar]
- 45.Landen CN, Jr, Birrer MJ, Sood AK. Early events in the pathogenesis of epithelial ovarian cancer. J Clin Oncol. 2008;26:995–1005. doi: 10.1200/JCO.2006.07.9970. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.