Abstract
Human E4B, also called UFD2a, is a U-box-containing protein that functions as an E3 ubiquitin ligase and an E4 polyubiquitin chain elongation factor. E4B is thought to participate in the proteasomal degradation of misfolded or damaged proteins through association with chaperones. The U-box domain is an anchor site for E2 ubiquitin-conjugating enzymes but little is known of the binding mechanism. Using X-ray crystallography and NMR spectroscopy, we determined the structures of E4B U-box free and bound to UbcH5c and Ubc4 E2s. While previously characterized U-box domains are homodimeric, we show that E4B U-box is a monomer stabilized by a network of hydrogen bonds identified from scalar coupling measurements. These structural studies, complemented by calorimetry- and NMR-based binding assays, suggest an allosteric regulation of UbcH5c and Ubc4 by E4B U-Box and provide a molecular basis to understand how the ubiquitylation machinery involving E4B assembles.
INTRODUCTION
The covalent modification of proteins with ubiquitin or polyubiquitin chains regulates many cellular processes including protein degradation, cellular trafficking, DNA repair and DNA lesion bypass (Weissman, 2001; Passmore and Barford, 2004; Pickart and Eddins, 2004). Ubiquitylation involves a cascade of enzymatic reactions (Pickart, 2001) where ubiquitin is transferred from an E1 ubiquitin-activating enzyme to an E2 ubiquitin-conjugating enzyme forming a thioester conjugate with a cysteine of E2. E2 associates with an E3 ubiquitin ligase, which recognizes a protein substrate and facilitates the transfer of ubiquitin from E2 to the ζ-amino group of a lysine residue in the substrate (Ye and Rape, 2009). In some instances, a chain elongation factor E4 catalyzes polyubiquitin chain assembly in place of or after E3 ubiquitylation (Kuhlbrodt et al., 2005).
There are three classes of E3 ubiquitin ligases defined by the presence of a HECT, RING or U-box domain. The RING and U-box domain have the same fold but unlike the RING domain the U-box does not coordinate Zn2+ ions (Aravind and Koonin, 2000; Ohi et al., 2003). HECT E3s form a catalytic ubiquitin thioester intermediate before transferring ubiquitin to an acceptor protein (Huang et al., 1999) whereas RING and U-box E3s function as adaptors that bring in close proximity the ubiquitin-linked E2 and a target protein, with the RING or U-box domain being the anchor site for E2s. Binding of the RING domain was shown to activate the release of ubiquitin from E2 and this likely applies to the U-box domain as well but the activation mechanism is not known (Ozkan et al., 2005; Deshaies and Joazeiro, 2009). Overall, our understanding of the ubiquitin transfer mechanism from E2–E3/E4 to substrate is rudimentary and this is particularly true for U-box proteins.
S. cerevisiae Ufd2 (ubiquitin fusion degradation protein 2), the first chain elongation factor E4 to be described, has been shown to promote polyubiquitylation of artificial fusion proteins and is the prototype U-box protein (Koegl et al., 1999). Ufd2 binds the chaperone cdc48 component of the Ufd1–Npl4–cdc48 complex that facilitates the translocation of polyubiquitinated proteins from the endoplasmic reticulum into the cytosol for subsequent degradation by the 26S proteasome (Richly et al., 2005). Several other members of the U-box family of ubiquitin ligases interact with chaperones or chaperone binding proteins, suggesting a general function of U-box-containing ubiquitin ligases in the proteosomal degradation of misfolded or damaged proteins (Hatakeyama et al., 2004). The mammalian homologue of Ufd2, called UFD2a or E4B, is also a U-box-containing E4 enzyme that was shown to promote the degradation of a pathological form of ataxin-3, the gene product implicated in the Machado-Joseph disease (Matsumoto et al., 2004). Human E4B functions as an E3 ubiquitin ligase as well in vitro when provided with the human E2 conjugating enzymes Ubc4 or UbcH5c (Hatakeyama et al., 2001).
The mechanism by which U-box ubiquitin ligases function together with E2 conjugating enzymes is poorly understood. A molecular understanding of these enzymes is limited with only one U-box protein – CHIP (C-terminal Hsp70 interacting protein) – that has been characterized structurally in complex with different E2s (Xu et al., 2008; Zhang et al., 2005). Here we show that unlike CHIP and all other isolated U-box domains characterized to date, human E4B U-box is a monomer, both in the free state and in complex with human UbcH5c or Ubc4 E2 enzymes. We present the nuclear magnetic resonance (NMR) solution structure of E4B U-box, including an NMR-based characterization of a hydrogen bond network in the U-box domain. We also present the crystal structures of human E4B U-box, Ubc4 and E4B U-box in complex with UbcH5c. Data from NMR chemical shift mapping validate the crystal structure of E4B U-box–UbcH5c in solution and suggest a possible allosteric regulation of UbcH5c and Ubc4 by E4B U-Box.
Our structural studies of the interactions of human E4B with UbcH5c and Ubc4, combined with previously published structures of budding yeast Ufd2 (Tu et al., 2007) and human UbcH5c bound to ubiquitin (Brzovic et al., 2006) provide a molecular framework to better understand the mechanism by which U-box-containing ubiquitin ligases function.
RESULTS
Solution Structure of the U-box Domain of E4B
As a prelude to structure determination, we evaluated the oligomerization state of a region of human E4B, amino acids (aa) 1208 to 1302, that encompasses the U-box domain. The strong signals observed for this protein construct and the absence of changes in chemical shifts in the 1H-15N heteronuclear single quantum coherence (HSQC) NMR spectrum of E4B as a function of protein concentration in the ~0.4 – 2.0 mM range suggested that E4B U-box was a monomer. This is unlike other U-box domains characterized to date, which dimerize in this concentration range (Andersen et al., 2004; Vander Kooi et al., 2006). Consistent with a monomeric state, as explained below in the analysis of isothermal titration calorimetry (ITC) data, a control ITC dilution experiment that we performed with an initial high concentration (i.e. 3 mM) of E4B U-box did not reveal any marked change in heat signal that could be attributed to oligomer dissociation in the calorimeter cell. Dynamic light scattering measurements also suggested an apparent molecular weight of 11 kDa, which coincides with the molecular weight of a monomeric E4B U-box domain (data not shown). We note with interest that the full-length Ufd2 protein, thought to be the homologue of E4B in budding yeast, is also a monomer in solution (Tu et al., 2007).
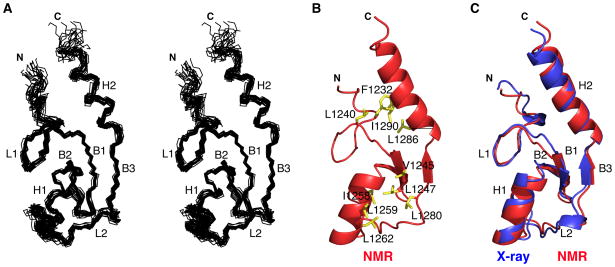
The structure of E4B U-box (aa 1208-1302) was determined using NMR spectroscopy (Figures 1A and 1B). The average pairwise root mean square deviation (r.m.s.d.) among the 20 best calculated structures superimposed on the backbone and all heavy atoms from residues 1228 to 1295 are 0.62 Å and 1.17 Å, respectively. The structural statistics are summarized in Table 1. The first 20 N-terminal amino acids of the E4B construct used for structure determination (aa 1208-1302) lack a defined conformation and connect to a long loop (L1: Ala1228 - Asp1243) that extends to a first β-strand (B1: Pro1244 - Leu1247). B1 forms a β-hairpin with β-strand 2 (B2: Thr1251 - Asp1254), and together with β-strand 3 (B3: Glu1281 - Pro1282) constitute a central three-stranded antiparallel β-sheet. Following B2 is a central α-helix (H1: Arg1255 - His1261) that connects to a second loop (L2: Leu1262 - Leu1280), and then to β-strand B3. B3 leads to a second α-helix (H2: Pro1284 -Arg1295) and finally to a seven-residue unstructured C-terminal segment (Glu1296 - His1302). Two hydrophobic cores are present in E4B U-box. The first hydrophobic core holds together the central α-helix H1 and the region surrounding the central β-sheet while the second core involves the C-terminal α-helix H2 packing against loop L1 (Figure 1B). The overall fold of E4B is similar to that of other U-box domains (Andersen et al., 2004; Ohi et al., 2003; Xu et al., 2006; Zhang et al., 2005).
Figure 1. Solution and Crystal Structures of E4B U-box.
(A) Superposition of the 20 lowest energy NMR structures of E4B U-box in stereoview showing only the N, Cα , and C’ trace for aa 1228 to 1300. The α-helices (H), β-strands (B) and loops (L) are indicated.
(B) Ribbon representation of the NMR structure of E4B U-box showing only residues 1226 to 1302 and amino acid side chains (yellow) forming two hydrophobic cores.
(C) Overlay of the crystal (aa 1226 to 1300) and NMR (aa 1226 to 1300) structures of E4B U-box where α-helices (H), β-strands (B) and loops (L) are indicated.
See also Figure S1. Structure representations were prepared using MOLMOL (Koradi et al., 1996) or PyMOL (http://pymol.sourceforge.net/).
Table 1.
NMR and Structure Refinement Statistics for E4B (aa 1208-1302)
| Number of restraints | |
| Total NOE restraints | 1368 |
| Intraresidue | 64 |
| Sequential (|i–j| = 1) | 384 |
| Medium-range (1 < |i–j| < 4) | 409 |
| Long-range (|i–j| >4) | 511 |
| Hydrogen bond restraints | 36 |
| Dihedral angle restraints | |
| Φ | 23 |
| Ψ | 23 |
| Structure statisticsa | |
| Violations (mean ± standard deviation) | |
| NOE restraints (Å) | 0.005 ± 0.001 |
| Dihedral angle restraints ( ) | 0.059 ± 0.045 |
| R.m.s. deviations from idealized geometry | |
| Bond lengths (Å) | 0.0011 |
| Bond angles (°) | 0.396 |
| Improper torsions (°) | 0.251 |
| Final energies (kcal mol−1) | |
| Total | 85.0 ± 2.7 |
| Bonds | 1.9 ± 0.2 |
| Angles | 70.3 ± 1.5 |
| van der Waals | 3.2 ± 0.7 |
| NOEs | 1.7 ± 0.6 |
| Coordinate precision (Å)b | |
| R.m.s. deviations | |
| Backbone atoms (aa 1228-1295) | 0.62 |
| Backbone in secondary structures | 0.48 |
| Heavy atoms (aa 1228-1295) | 1.17 |
| Heavy atoms in secondary structures | 1.07 |
| Ramachandran plot | |
| Preferred regions (%) | 59.1 ± 5.9 |
| Allowed regions (%) | 40.0 ± 5.5 |
| Outliers (%) | 0.9 ± 1.2 |
Structure statistics refer to an ensemble of 20 structures with lowest energies from 100 calculated structures.
Average pairwise r.m.s. deviation for the ensemble of 20 structures.
Crystal Structure of the E4B U-box Domain
To complement the NMR spectroscopy study, we determined the crystal structure of the U-box domain of E4B (aa 1208-1302) at a resolution of 2.6 Å (Figure 1C). The crystallography statistics are provided in Table 2. Crystals grew in the space group P432, with one molecule of E4B per asymmetric unit, consistent with a monomeric structure. Phasing was done by molecular replacement (MR) using the NMR-derived structure of E4B. Final R-factors are Rwork = 18.5% and Rfree = 25.4%. The electron density is clear for most of the U-box domain, the only disordered parts being the N- terminal residues 1208–1225 and C-terminal residues 1301–1302. These are the same regions of the NMR structure that lacked a defined conformation. The 2 Fo − Fc σA-weighted electron density map for E4B is shown in Figure S1A.
Table 2.
Crystallography Statistics
| E4B U-box | Ubc4 | E4B U-box–UbcH5c | |
|---|---|---|---|
| Data collection | |||
| Space group | P 4 3 2 | P 1 21 1 | P 6 2 2 |
| Cell dimensions | |||
| a, b, c (Å) | 83.40, 83.40, 83.40 | 28.23, 59.12, 45.42 | 142.71, 142.71, 83.13 |
| α β γ °) | 90, 90, 90 | 90, 106.73, 90 | 90, 90, 120 |
| Resolution (Å) | 24.07–2.60 (2.64–2.60) | 43.31–1.60 (1.64–1.60) | 32.79–3.17 (3.23–3.17) |
| Rsym or Rmerge | 0.139 (0.655) | 0.061 (0.123) | 0.192 (0.777) |
| I/σI | 46.2 (7.3) | 17.1 (13.2) | 18.1 (6.2) |
| Completeness (%) | 98.6 (99) | 99.9 (99.5) | 92.2 (96.0) |
| Redundancy | 37.0 (36.7) | 3.7 (3.6) | 58.8 (57.6) |
| Refinement | |||
| Resolution (Å) | 24.07–2.60 | 43.41–1.60 | 32.79–3.17 |
| No. reflections | 3328 | 17905 | 8196 |
| Rwork/Rfree | 0.185/0.254 | 0.189/0.229 | 0.236/0.279 |
| No. of atoms | 657 | 1449 | 1865 |
| Protein | 624 | 1246 | 1850 |
| Ligand/ion | N/A | N/A | N/A |
| Water | 33 | 203 | 15 |
| B-factors | |||
| Protein | 42.58 | 5.158 | 93.63 |
| Ligand/ion | N/A | N/A | N/A |
| Water | 41.52 | 18.27 | 65.53 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.002 | 0.023 | 0.003 |
| Bond angles (°) | 0.588 | 2.018 | 0.689 |
| Ramachandran plot | |||
| Preferred regions (%) | 94.3 | 94.9 | 88.5 |
| Allowed regions (%) | 5.7 | 4.4 | 6.6 |
| Outliers (%) | 0 | 0.7 | 4.9 |
Data for the highest resolution shell are shown in parentheses.
The crystal structure and average NMR structure superimpose well with an r.m.s.d of 1.04 Å for the backbone atoms N, Cα , C’ of residues 1228 to 1299 (Figure 1C). In the crystal lattice there is no pairing of U-box domains corresponding to the previously identified interface in homodimeric U-box proteins, a result also consistent with the monomeric nature of E4B U-box (Figure S1B).
Hydrogen Bond Network in E4B Determined Experimentally from Scalar Couplings
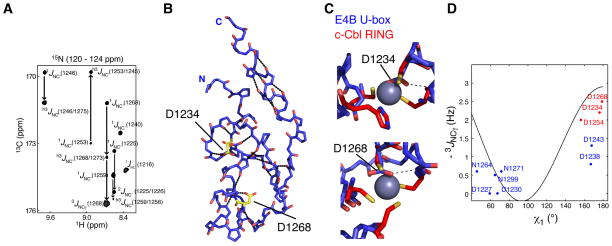
An extensive hydrogen bond (H-bond) network is predicted from the solution and crystal structures of E4B U-box. Canonical H–N · · ·O=C H-bonds can be detected experimentally via long-range h3JNC’ scalar couplings that connect the 15N nucleus of the donor amino acid to the carbonyl 13C nucleus of the acceptor (Cordier and Grzesiek, 1999a). From such measurements in E4B, a total of nineteen H-bonds were unambiguously identified. Twelve are backbone H-bonds that link inter-strand residues in the β-sheet as well as i and i + 4 residues within the central and C-terminal helices (Figure 2A and 2B, and Table S1). Seven H-bonds were identified in loops regions (Table S1).
Figure 2. Identification of Hydrogen Bonds in E4B U-box by NMR Spectroscopy.
(A) A region from the constant time 3D J-HNCO NMR spectrum of E4B U-box showing peaks correlated through 1JNC’, 2JNC’, 3JNCγ and h3JNC’ scalar couplings.
(B) The nineteen H-bonds in E4B U-box that are listed in Table S3 were identified through h3JNC’ scalar coupling measurements. The dotted lines illustrate these H-bonds. Residues Asp1232 and Asp1268 for which side chain h3JNC’ scalar couplings were measured are shown. Asp1232 and Asp1268 correspond to the zinc binding regions of RING domains.
(C) Asp1232 and Asp1268 of E4B U-box approximately occupy the zinc centers (purple spheres) in c-Cbl RING domain (PDB entry 1FBV) when the U-box and RING domains are superimposed. H-bonds involving the two aspartate side chains were determined from h3JNC’ scalar coupling measurements and are shown by the dashed lines.
(D) Calibration curve for the dihedral angle (χ1) dependence of 3JNCγ scalar couplings in aspartate and asparagine residues. Blue and red points correlate experimental 3JNCγ and corresponding χ1 angle values from the crystal structure of E4B U-box. The red points correspond to residues for which a side chain carbonyl participates in a H-bond as determined from h3JNC scalar couplings.
See also Table S1 for the values of measured coupling constants.
From comparison of the structures of E4B U-box and C3HC4-type zinc-binding RING finger motif of c-Cbl (Zheng et al., 2000), E4B residues Asp1234, Met1237, Asp1254 and Ser1256 are found to approximate the histidine and cysteine residues in the first zinc binding site of the RING motif. Similarly, Ser1249, Thr1251, Asp1268 and Asn1271 are the residues corresponding to the second zinc binding site (Figure 2C). Noticeably, the carboxylate groups of Asp1234 and Asp1268 roughly occupy the place of cationic zinc metal centers in the first and second zinc binding sites in the RING motif, respectively. Supporting this observation, the side chain carbonyls of Asp1234, Asp1254 and Asp1268 were identified through h3JNC’ scalar couplings to be involved in H-bonds (Figure 2B and 2C, and Table S1). The dihedral angles χ1 (defined by atoms N-Cα-Cβ-Cγ ) measured for Asp1234, Asp1254 and Asp1268 in the crystal structure of E4B U-box and the corresponding experimental 3JNCγ scalar couplings are in good agreement with the predicted dependence of 3JNCγ on χ1 values (Juranić et al., 2005) (Figure 2D). This is consistent with the aspartate side chains adopting a defined conformation in solution. Notice that 3JNCγ scalar couplings for E4B surface Asp1238 and Asp1243 are not predictive of the angles measured in the crystal structure (angle difference ≥ 30°), an indication that these side chains are in a state of free rotation around χ1 in solution. 3JNCγ values for other aspartate and asparagine residues of E4B are consistent with the χ1 dihedral angles measured in the crystal structure (Figure 2D). Taken together, our data show that the U-box motif is stabilized by a network of H-bonds with some H-bonds substituting for Zn+2 chelation in RING domains.
Comparison of E4B U-box Monomer to Dimeric U-box Domains
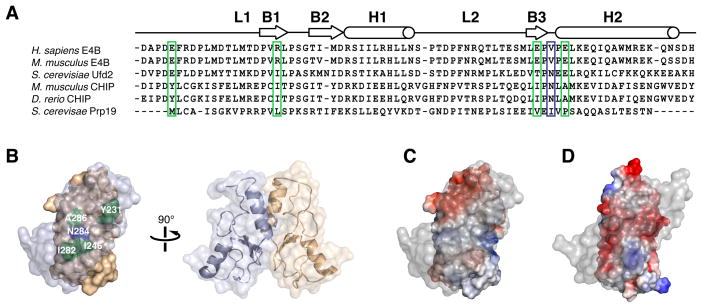
E4B U-box is monomeric unlike all other U-box domains characterized previously, which have a conserved dimeric topology (Vander Kooi et al., 2006; Xu et al., 2006; Zhang et al., 2005). This difference in oligomerization state can be understood from comparison of the structure and amino acid sequence of E4B U-box to those of dimeric U-box proteins such as CHIP or Prp19 (Figure 3A and 3B). In mouse CHIP, homodimerization is mediated by the hydrophobic residues Tyr231, Ile246, Ile282 and Ala286, and an asparagine (Asn284) that forms H-bonds with the same residue of the complementary CHIP molecule. The corresponding first four residues in human E4B are all charged: Glu1231, Arg1246, Glu1281 and Glu1285, and two are conserved in Saccharomyces cerevisiae Ufd2 (Figure 3A). The hydrophobic nature of the interface in CHIP is in stark contrast with the corresponding negatively charged surface of a hypothetical E4B dimer, likely explaining the monomeric state of human E4B U-box (Figure 3C and 3D). This also applies to mouse E4B U-box whose NMR structure was reported recently (Nordquist et al., 2010). Despite the dissimilarities in oligomerization states of E4B and CHIP U-boxes, we note that full-length mouse CHIP has an asymmetric conformation in which the E2 binding site of one of the U-box domains is blocked (Zhang et al., 2005). Thus, a CHIP dimer, like the E4B monomer, only binds one E2 ubiquitin-conjugating enzyme.
Figure 3. Electrostatic Surface Potentials of E4B and CHIP U-box Domains Affect Oligomerization States.
(A) Sequence alignment of U-box domains from different E3 ubiquitin ligases indicating locations of secondary structure elements. CHIP and Prp19 are dimers while Ufd2 and E4B are monomers. Key CHIP and Prp19 residues mediating homodimerization are highlighted. Hydrophobic residues in CHIP and Prp19 (green box) correspond to charged residues in E4B while an asparagine residue (Asn284) in mouse CHIP (blue box) is replaced by a valine residue (Val1283) in human E4B.
(B) Molecular representation of mouse CHIP homodimer showing the important dimerization interface residues described in A.
(C) Electrostatic surface potential of mouse CHIP U-box homodimer (PDB entry 2C2V) showing predominantly non-polar character at the dimerization interface. Foremost protomer is rendered transparent for clarity.
(D) Electrostatic surface potential of a hypothetical homodimer of E4B U-box showing a negatively charged dimer interface.
Structure of the Complex of Human E4B U-box and UbcH5c
The ability of the human ubiquitin ligase E4B to participate in its own ubiquitylation and that of bacterial substrates was shown previously (Hatakeyama et al., 2001). In these in vitro assays, E4B was mixed with E1, different E2 enzymes, ubiquitin and bacterial lysate, and then immunoblotted with antibodies against ubiquitin to detect the ubiquitylation that occurred (Hatakeyama et al., 2001). These experiments showed that ubiquitylation required the U-box domain of E4B and that the enzymatic reaction was most efficient with the E2 conjugating enzymes UbcH5c and Ubc4. Little enzymatic activity was detected when other E2s including Ubc2a, Ubc2b, Ubc3, UbcH6, UbcH7 and UbcH8 were present.
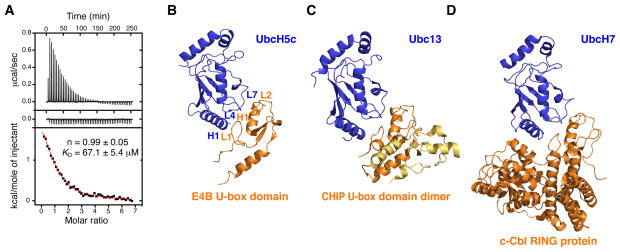
Using ITC, we confirmed the interaction of E4B U-box and full-length UbcH5c (aa 1-147) and Ubc4 (aa 1-147). A dissociation constant (KD) of 67.1 ± 5.4 μM at 22 °C was determined for the E4B U-box–UbcH5c interaction with a stochiometry close to unity (n = 0.99 ± 0.05) (Figure 4A). The interaction is endothermic with an unfavorable observed enthalpy change (Δ Hobs = 2.9 kcal·mol−1) but is entropically favored (−TΔ Sobs = −8.5 kcal·mol−1). Similar thermodynamic parameters were obtained for the calorimetric titration of Ubc4 with E4B U-box (data not shown). The control ITC dilution experiment in which E4B U-box is injected into a buffer solution does not show any marked change in heat of dilution as the concentration of E4B U-box increases in the calorimeter cell, consistent with a monomeric state of E4B U-box (Figure 4A).
Figure 4. Interaction of E4B U-box with UbcH5c and Ubc4 E2 Conjugating Enzymes.
(A) Isothermal titration calorimetry of UbcH5c with E4B U-box. Shown are the integrated heat measurements from injecting 3 mM E4B U-box into the calorimeter cell containing UbcH5c at an initial concentration of 100 μM (top panel) or buffer solution (middle panel). A standard one-site model was used for curve fitting (bottom panel) in the determination of KD and stoichiometry (n), the values of which are shown.
(B) Crystal structure of human E4B U-box in complex with UbcH5c.
(C) Crystal structure of mouse CHIP U-box in complex with Ubc13 (PDB entry 2C2V).
(D) Crystal structure of the human c-Cbl RING E3 ligase in complex with UbcH7 (PDB entry 1FBV).
See also Figure S2.
To gain additional insight into these interactions we attempted crystallization of E4B U-box in complex with Ubc4 and UbcH5c. From these trials, we initially obtained crystals of Ubc4 (aa 1-147) alone and determined its structure to a resolution of 1.6 Å by MR using the atomic coordinates of UbcH5c (Protein Data Bank entry 1X23) (Figure S2A). The final R-factors are Rwork = 18.9% and Rfree = 22.9%. Refinement statistics are summarized in Table 2. The crystals have a space group of P21 and contain one molecule of Ubc4 in the asymmetric unit. The electron density is well defined as illustrated by the 2Fo − Fc σA-weighted map shown in Figure S2B.
We were able to determine the crystal structure of UbcH5c (aa 1-147) in complex with the U-box domain of human E4B (aa 1208-1302) to a resolution of 3.17 Å (Figure 4B). The crystals have one molecule each of UbcH5c and E4B U-box per asymmetric unit in the space group P622. Phasing was done by MR using the crystal structures of free human Ubc4 and E4B U-box, both from this study, as starting models. The final R-factors are Rwork = 23.6% and Rfree = 27.9% and refinement statistics can be found in Table 2. The resulting electron density map has a good fit for most regions of E4B and UbcH5c, except for the first eighteen N-terminal residues (aa 1208-1225) and last two C-terminal residues (1301 and 1302) of E4B, which are not visible in the electron density.
These regions of the protein also lack a stable conformation in the solution and crystal structures of E4B (vide supra). Ten surface residues of UbcH5c and nine of E4B with long side chains–arginine, lysine or methionine–are poorly defined with incomplete electron density. In the protein cores and at the interface of the two proteins in the crystal all residues have electron density guiding their position. A 2Fo − Fc σA-weighted electron density map is shown in Figure S3. Noticeably, of the three crystals used to elucidate the E4B U-box–UbcH5c structure, all had a missing layer of electron density equal to one-third of the unit cell. This is likely due either to one-dimensional twinning or other crystalline disorder(s). The lack of electron density could originate from the N- and C-terminal flexible segments of E4B, which point toward the disordered region of the crystals. No crystals of the complex could be obtained with shorter versions of E4B. Further remarks on this subject are presented in the Supplemental Discussion.
In the crystal, the most energetically favorable interface between E4B U-box and UbcH5c, as determined using PISA (Krissinel and Herrick, 2007), occurs along each of the crystallographic six-fold symmetry axes. As explained below, this also corresponds to the binding site identified in solution using NMR spectroscopy. The relative positioning of the two proteins is such that H1, L4, and L7 of UbcH5c make contacts with L1, H1, and L2 of E4B, respectively (Figure 4B). Polar interactions at the interface include a salt bridge between E4B U-box Asp1238 and UbcH5c Lys8 (Figure 5A). Based on (N)H···O(C) distances shorter than 3.8 Å, there likely are intermolecular H-bonds involving the following atoms: E4B-Leu1236 O and UbcH5c-Arg5 NH1, E4B-Pro1269 O and UbcH5c-Ser94 OG, E4B-Asn1264 ND2 and UbcH5c-Phe62 O, E4B-Asn1264 O and UbcH5c-Lys63 NZ and E4B-Arg1272 and Gln92 OG1 (Figure 5A). There are three hydrophobic clusters at the interface. The first, composed of Leu1236, Met1237, Ile1257, Arg1260, His1261 and Asn1264 of E4B, forms a deep pocket where Phe62 of UbcH5c is completely buried (Figure 5B). With Arg1260 close to Phe62 at 4.5 Å, there is a possible cation-π interaction between the guanidinium group and the phenyl ring. A similar cationπ interaction is present in mouse and zebrafish CHIP U-box complexes (Xu et al., 2008; Zhang et al., 2005). The second cluster composed of Leu1236, Met1253, His1261, Pro1269 and Phe1270 of E4B forms a channel in which Ser94, Pro95 and Ala96 of UbcH5c are located (Figure 5B). This UbcH5c motif, termed S-P-A, is conserved and is part of a similar interface in the E2–E3 structures of mouse and zebrafish CHIP in complex with Ubc13 and UbcH5a, respectively (Xu et al., 2008; Zhang et al., 2005), and in the RING domain protein TRAF2 bound to Ubc13 (Yin et al., 2009). The final hydrophobic cluster encompasses E4B Leu1236 and Met1237 which both contact the surface of UbcH5c made up of Ala2, Leu3, Lys4, Arg5 and Lys8 (Figure 5C). Overall, the E4B–UbcH5c interface resembles those of other E2–E3 complexes such as CHIP–Ubc13–Uev1a (Zhang et al., 2005) and c-Cbl–UbcH7 (Zheng et al., 2000) where the E3 is a U-box- or RING domain-containing protein (Figures 4C and 4D).
Figure 5. Interaction Interface Between E4B U-box and UbcH5c.
(A) E4B U-box and UbcH5c interface showing possible salt bridges and hydrogen bonds.
(B) E4B U-box and UbcH5c interface showing how F62 and S94-P95-A96 (S-P-A motif) of UbcH5c interact with E4B U-box.
(C) E4B U-box and UbcH5c interface illustrating how L1236 and M1237 of E4B U-box interact with UbcH5c. See also Figure S3 for the electron density map of the E4B U-box–UbcH5c complex.
Mapping the E4B Interaction Interface with UbcH5c and Ubc4 in Solution
To further probe how E4B U-box binds UbcH5c and Ubc4 in solution and validate the crystal structure of E4B U-box–UbcH5c complex, we acquired a series of 1H-15N HSQC NMR spectra on 15N-labeled E4B U-box, alone and titrated with nonlabeled Ubc4 (aa 1-147) or UbcH5c (aa 1-147) (Figures 6A and 6B). Only one set of E4B signals was observed during the entire titration with either UbcH5c or Ubc4. A subset of these signals shifted and broadened, indicating that the exchange between E4B in the free state and bound to Ubc4 or UbcH5c is fast to intermediate on the NMR chemical shift time scale. This exchange regime is consistent with the KD of 67.1 ± 5.4 μM for the E4B U-box–UbcH5c interaction derived from ITC measurements (Figure 4A). The E4B residues for which changes occurred in the 1H-15N HSQC spectrum during titration by Ubc4 or UbcH5c were mapped on the structure of E4B. As shown in Figure 6C, these residues are all in the vicinity of the most favorable binding interface identified in the crystal structure of the E4B U-box–UbcH5c complex.
Figure 6. NMR Titrations and Mapping of Interaction Interfaces in E4B U-box, UbcH5c and Ubc4.
(A, B) Superposition of the 1H-15N HSQC titration spectra of 15N-labeled E4B U-box from free (black) to the bound (red) states with nonlabeled Ubc4 (A) and UbcH5c (B). The arrows indicate the directions of the titrations.
(C) In the crystal structure of E4B U-box and UbcH5c complex, E4B residues affected by UbcH5c titration are colored red (signals with chemical shift changes ≥ 0.02 ppm) or blue (weakened or disappeared signals, namely those of Asp1234, Leu1236, Asp1238, Leu1247, Ile1258, Asn1271 and Arg1272). The active site Cys85 is shown in yellow.
(D, E) Superposition of the 1H-15N HSQC titration spectra of 15N-labeled Ubc4 (D) or UbcH5c (E) from the free (black) to the bound (red) states with nonlabeled E4B U-box. The arrows indicate the directions of the titrations.
(F) In the crystal structure of E4B U-box and UbcH5c complex, UbcH5c residues affected by E4B U-box titration are colored red (signals with chemical shift changes from 0.03 to 0.06 ppm) or blue (chemical shift changes from 0.07 to 0.16 ppm). The active site Cys85 is shown in yellow.
(G) UbcH5c residues distant from the binding interface with E4B U-box and for which changes in chemical shifts were detected are shown in gray. The active site Cys85 is shown in yellow.
To identify the binding site of the E2 conjugating enzymes, complementary NMR experiments were performed where 15N-labeled Ubc4 and UbcH5c were titrated with nonlabeled E4B U-box (Figures 6D and 6E). For these studies, we used the previously published resonance assignments of human Ubc4 (Farrow et al., 2000). Because of the 97% amino acid sequence identity between Ubc4 and UbcH5c and their similar structures, these assignments could also be used for UbcH5c (Jensen et al., 1995). As expected, the chemical shift changes observed for the two E2 enzymes were similar (Figures 6D and 6E). Residues exhibiting chemical shift perturbations were highlighted in the crystal structure of E4B U-box–UbcH5c. Perturbed resonances primarily come from residues in loops L4 and L7, and the N-terminal helix H1 of UbcH5c, and fit well the binding interface seen in the crystal structure of the E4B U-box–UbcH5c complex (Figure 6F). This demonstrates a close agreement between the binding interface in solution and in crystalline state.
Interestingly, six residues conserved in Ubc4 and UbcH5c that are distant from the binding interface also experience shifts in their NMR signals upon interaction with E4B U-box (Figure 6G). As discussed next in light of previous work on the interaction of UbcH5b and the RING domain of E3 ubiquitin ligase CNOT4 (Dominguez et al., 2004; Ozkan et al., 2005), the observed changes in chemical shifts suggest that E4B U-box could regulate UbcH5c and Ubc4 through an allosteric mechanism.
DISCUSSION
The question of whether U-box and related RING E3 ubiquitin ligases are only adaptor proteins that bring substrate and E2 in close proximity or contribute also to an induced activating conformational change in their cognate E2 enzymes has been subject to debate (Deshaies and Joazeiro, 2009). The crystal structure of human E4B U-box–UbcH5c complex does not reveal any change in conformation when compared to the structures of the two proteins in their unbound state. NMR spectroscopy is a very sensitive technique to probe changes in conformations. As mentioned above, upon complex formation, shifts in 1H and 15N signals of surface residues of E4B U-box and UbcH5c map to the binding interface identified in the crystal structure, therefore validating this interface in solution. Importantly, after addition of E4B U-box we also observed changes in chemical shifts for six resonances associated with amino acids in UbcH5c and Ubc4 that are distant from the interface: Thr36, Ile37, Asp87, Ser91, Gln92, Ile106 (Figures 6D, 6E, 6F and 6G). This is consistent with subtle changes in flexibility or conformation, or both, that propagate away from the E4B binding site. From their location in UbcH5c and Ubc4, these residues may connect the binding interface to the active site cysteine (Cys85) (Figure 6G), suggesting a possible allosteric regulation of the enzymatic activity by E4B through alteration of the E2 active site.
It was previously shown that binding of the RING domain of human E3 ubiquitin ligase CNOT4 to the E2 enzyme UbcH5b allosterically enhances the release of ubiquitin from UbcH5b–ubiquitin thioester (Ozkan et al., 2005). Because UbcH5b is highly similar to UbcH5c and Ubc4 (97.3% and 99.3% amino acid identity, respectively) and the U-box and RING domains bind the same surface of the E2 enzyme, it is likely that E4B U-box contributes to the activation of UbcH5c and Ubc4. Supporting this possibility, the UbcH5c and Ubc4 residues not involved in E3 binding and for which changes in chemical shifts were observed upon interaction with the U-box domain are the same or in the vicinity of residues in UbcH5b (Ile37, Ile88, Leu89, Ile106 and Asn114) which when mutated inhibited the RING domain-stimulated release of ubiquitin from the active site cysteine (Ozkan et al., 2005). More work will be needed to test this allosteric activation hypothesis and to understand how ubiquitin released from E2 is transferred to the substrate.
To examine how E4B U-box bound to UbcH5c is positioned in the context of full-length E4B, we generated a model by superimposing the structure of human E4B U-box–UbcH5c complex onto the structures of budding yeast Ufd2 (Tu et al., 2007) and human UbcH5c bound non-covalently to ubiquitin (Brzovic et al., 2006) (Figure 7). When its first 187 N-terminal amino acids are excluded, Ufd2 has high sequence similarity (i.e. 56% similarity and 30% identity) with the C-terminal half of human E4B (aa 560-1302), therefore making Ufd2 (aa 188-961) a reasonable template for this segment of E4B. The region of similarity encompasses a large N-terminal core domain made of five repeating units that resemble tandem Armadillo repeats and also includes the C-terminal U-box domain (Figure 7).
Figure 7. Model Structure of E4B, UbcH5c and Ubiquitin Ternary Complex.
The structures of budding yeast Ufd2 (aa 188-961 from PDB entry 2QIZ), human UbcH5c–ubiquitin (PDB entry 2FUH) and human E4B U-box–UbcH5c (this study) were superimposed to model the complex. Ubiquitin (yellow), UbcH5c (gray) and Ufd2 (body in green and U-box in blue) fit together in the model with very few steric clashes and leave the active site cysteine of UbcH5c (red) accessible to react with a ubiquitin molecule.
When the U-box in the E4B U-box–UbcH5c complex is overlaid to the U-box of Ufd2, the positioning of UbcH5c has few steric or surface charge clashes with the core region of Ufd2. Moreover, the small clashes are likely insignificant since structures of Ufd2 from two crystal forms revealed flexibility in the hinge region connecting the U-box to the rest of the protein (Tu et al., 2007). Normal mode calculations performed with NOMAD (Lindahl et al., 2006) on Ufd2 also support a hinge-bending motion of the U-box relative to the protein core (data not shown). In the overlaid structures, the orientation of UbcH5c is such that its non-covalent ubiquitin binding site can be occupied by ubiquitin. Also unhindered is the active site cysteine of UbcH5c to which a ubiquitin molecule can be covalently linked without creating any steric clash with the rest of the complex.
Presumably, the large E4B domain made of tandem Armadillo repeats or residues N-terminal from this domain and lacking in our model, or both, are involved in the recognition of substrate proteins. Work is in progress to characterize full-length E4B and its interaction with a protein substrate, a requirement to understand the ubiquitin transfer mechanism.
EXPERIMENTAL PROCEDURES
Protein Preparation
The cDNAs of full-length Ubc4 and UbcH5c were cloned in a pT7.7 vector encoding an N-terminal non-cleavable (His)6 tag while a DNA segment corresponding to aa 1203-1302 of E4B was inserted in a pET28b vector encoding an N-terminal (His)6 tag preceding a thrombin cleavage site. The proteins were produced in E. coli BL21(DE3) cells (Novagen) grown in LB media at 37 °C until an A600 of about 0.8, transferred to 15 °C and then induced with 1 mM final concentration of IPTG 45 min later. Cells were harvested 12–16 h afterwards.
Cell pellets were resuspended in 50 mM sodium phosphate, pH 7.5, 300 mM NaCl (buffer 1) supplemented with 5 mM imidazole and lysed using a high-pressure homogenizer Emulsiflex C-5 (Avestin Inc.). The soluble and insoluble fractions were separated by centrifugation and the lysate loaded onto a Ni2+-NTA column (Qiagen). Buffer 1 with 20 mM and 500 mM imidazole was used to wash the column and elute the protein, respectively. Where applicable, the (His)6 tag was then cleaved with thrombin at room temperature overnight, leaving the sequence GSHKF at the N-terminus of E4B U-box. All proteins were passed through a Superdex 75 size-exclusion chromatography column (GE Healthcare), equilibrated with buffer 1.
Isotopically-labeled proteins were produced using similar methods but replacing LB media with M9 media containing 1 g/L 15N-NH4Cl, 4 g/L unlabeled glucose or 2 g/L of 13C-enriched glucose, 1 g/L Isogro or 10% (v/v) Silantes OD2 media that are 15N- or 15N/13C-enriched and H2O or D2O (Botuyan et al., 2004). Isotopes were purchased from Isotec and CIL.
NMR Spectroscopy
All NMR spectra were recorded at 25 °C using Bruker Avance spectrometers operating at proton frequencies of 500 MHz and 600 MHz (with cryoprobe). Multiple samples of nonlabeled, 15N-labeled, 15N/13C-labeled and 15N/13C/2H-labeled E4B U-box were prepared at concentrations of 0.5 to 2 mM in 20 mM sodium phosphate buffer, pH 7.0, 50 mM NaCl. Backbone and side chain resonance assignments were carried out using standard NMR experiments including HNCO, HNCACO, HNCA, HNCOCA, HNCACB, HNCOCACB, CCONH-TOCSY, HCCONH-TOCSY, HBHACONH, HCCH-TOCSY, 1H-15N TOCSY-HSQC, and 2D TOCSY (Ferentz and Wagner, 2000). Interproton distance restraints were derived from 15N-edited NOESY (mixing time of 100 ms) and 13C-edited NOESY (mixing time of 100 ms) spectra of 15N- and 15N/13C-labeled E4B, respectively, as well as from a series of 2D NOESY spectra of nonlabeled E4B U-box recorded with mixing times of 20, 30, 50, 60 and 100 ms. All data were processed with NMRPipe/NMRDraw (Delaglio et al., 1995) and analyzed with NMRView (Johnson and Blevins, 1994). Assignment of NOEs was facilitated by using the noeassign module of CYANA 2.1 (Güntert, 2004).
Identification of Hydrogen Bonds in E4B U-box
The H-bond h3JNC’, peptide bond 1JNC’ and intra-residue 3JNC’ scalar coupling constants were measured using constant time 3D J-HNCO experiments (Cordier et al., 1999b; Cordier et al., 1999c; Cornilescu et al., 1999). The J-HNCO spectra were recorded with constant times of 35 ms and 70 ms and with evolution times of 16, 20, 24, 28, 32, 33, 34 ms; and 40, 50, 54, 65, 66, 66.5 ms. The coupling constants were determined by fitting the time evolution of all observed NC’ couplings. H-bond dHO distances [(N)H···O(C)] and H-bond acceptor angles [(N)H···O=C] listed in Table S1 were estimated from the magnitudes of the couplings (Juranić and Macura, 2001; Juranić et al., 2006).
NMR Structure Calculations
NMR structure calculations were performed with CYANA 2.1 (Güntert, 2004) using a total of 1368 NOE-derived interproton distances, 36 H-bond distances identified from slow exchanging amide protons and from measurement of scalar couplings as explained above, and 46 dihedral φ and ψ angle restraints derived from TALOS analysis (Cornilescu et al., 1999). The structures were then refined by simulated annealing with XPLOR (Schwieters et al., 2003). For further validation of the structures, 63 1H-15N residual dipolar couplings (RDC) restraints were included in a separate set of calculations using XPLOR. The RDCs were measured by weak alignment of E4B U-box in stretched polyacrylamide gel (Chou et al., 2001).
Isothermal Titration Calorimetry
All ITC measurements were recorded at 22 °C with a VP-ITC titration calorimeter (MicroCal). All protein samples were in 50 mM Tris-HCl, pH 7.5, 20 mM NaCl at concentrations of 100 μM for UbcH5c and Ubc4 and 3 mM for E4B U-box. The calorimeter syringe was used to deliver E4B U-box as 50 injections (1 x 3 μL followed by 49 x 6 μL) at 5 min intervals into the calorimetric cell containing 1.42 mL of UbcH5c or Ubc4 solution. Control experiments were performed under identical conditions to determine the heat signals that arise from injecting E4B U-box into the buffer solution. The initial data point (from the first 3 μL injection) was routinely deleted. Data were analyzed by Levenberg Marquardt nonlinear regression fitting of each ITC isotherm using a model corresponding to one independent binding event (Turnbull and Daranas, 2003).
Crystallization and X-ray Structure Determination
All crystals were obtained at 22 °C using the hanging drop vapor diffusion method by mixing 1 μL of 1 mM protein solution and 1 μL of precipitant solution. The proteins were in 50 mM Tris-HCl, pH 7.5, 50 mM NaCl. Crystals of free E4B U-box and E4B U-box–UbcH5c complex grew overnight in 2 M tacsimate and 4 M sodium formate, respectively, while crystals of Ubc4 formed in 2–4 days in 2.0 M sodium formate, 0.1 M sodium acetate trihydrate, pH 4.6. The crystals were cryoprotected by soaking in the respective mother liquor supplemented with 30% glucose and then flash frozen in liquid nitrogen.
Diffraction data were collected at the Advanced Photon Source (APS) 19-BM beamline, Argonne National Laboratory, for E4B U-box and E4B U-box–UbcH5c complex (wavelength was 0.97918 Å) and using a Rigaku/MSC CuKα Microfocus 007 diffractometer at a wavelength of 1.54 Å for Ubc4. Data were integrated, scaled, and merged using HKL2000 (Otwinowski et al., 1997). For all structures, phasing by molecular replacement was done using Phaser (McCoy et al., 2007). Initial model building was performed using Coot (Emsley and Cowtan, 2004). Refinement of the structures of E4B U-box in the free state and E4B U-box–UbcH5c complex was done using Phenix (Adams et al., 2002) while refinement of the structure of Ubc4 was carried out using Refmac 5 (Murshudov et al., 1997).
ACCESSION NUMBERS
Atomic coordinates and structure factors for the crystal structures of E4B U-box, E4B U-box–UbcH5c complex and Ubc4, and NMR structure of E4B U-box have been deposited in the Protein Data Bank under accession codes 1L1X, 3L1Z, 3L1Y and 2KRE, respectively.
Supplementary Material
Acknowledgments
We acknowledge the use of beamline 19-BM at Argonne National Laboratory, Structural Biology Center at the Advance Photon Source (APS). Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. We thank Dr. Younchang Kim at APS for assistance with X-ray data collection. We thank Ema Stokasimov for her early contribution to this project. Partial support from the National Institutes of Health grant CA109449 and a March of Dimes Basil O’Connor scholarship to G.M. is gratefully acknowledged. J.R.T. acknowledges support from the Minnesota Partnership for Biotechnology and Medical Genomics. Y.N. was recipient of a Kendall-Mayo postdoctoral fellowship in biochemistry.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data include Supplemental Discussion, one table and three figures and can be found with this article online at http://www.cell.com/structure/supplemental.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr Sect D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Andersen P, Kragelund BB, Olsen AN, Larsen FH, Chua NH, Poulsen FM, Skriver K. Structure and biochemical function of a prototypical Arabidopsis U-box domain. J Biol Chem. 2004;279:40053–40061. doi: 10.1074/jbc.M405057200. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. The U box is a modified RING finger - a common domain in ubiquitination. Curr Biol. 2000;10:132–134. doi: 10.1016/s0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- Botuyan MV, Nominé Y, Yu X, Juranić N, Macura S, Chen J, Mer G. Structural basis of BACH1 phosphopeptide recognition by BRCA1 tandem BRCT domains. Structure. 2004;12:1137–1146. doi: 10.1016/j.str.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Chou JJ, Gaemers S, Howder B, Louis JM, Bax A. A simple apparatus for generating stretched polyacrylamide gels, yielding uniform alignment of proteins and detergent micelles. J Biomol NMR. 2001;21:377–382. doi: 10.1023/a:1013336502594. [DOI] [PubMed] [Google Scholar]
- Cordier F, Grzesiek S. Direct observation of hydrogen bonds in proteins by interresidue 3hJNC’ scalar couplings. J Am Chem Soc. 1999a;121:1601–1602. doi: 10.1021/ja038616m. [DOI] [PubMed] [Google Scholar]
- Cordier F, Dingley AJ, Grzesiek S. A doublet-separated sensitivity-enhanced HSQC for the determination of scalar and dipolar one-bond J-couplings. J Biomol NMR. 1999b;13:175–180. doi: 10.1023/a:1008301415843. [DOI] [PubMed] [Google Scholar]
- Cordier F, Rogowski M, Grzesiek S, Bax A. Observation of through-hydrogen-bond 2hJHC' in a perdeuterated protein. J Magn Res. 1999c;140:510–512. doi: 10.1006/jmre.1999.1899. [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro AP. Ring domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dominguez C, Bonvin AM, Winkler GS, van Schaik FM, Timmers HT, Boelens R. Structural model of the UbcH5b/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure. 2004;12:633–644. doi: 10.1016/j.str.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Farrow NA, Archer SJ, Wu ZJ, Camac DM, Parsons T, Rolfe M, Domaille PJ. Backbone resonance assignment of human UBC4. J Biomol NMR. 2000;18:363–364. doi: 10.1023/a:1026525817687. [DOI] [PubMed] [Google Scholar]
- Ferentz AE, Wagner G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Q Rev Biophys. 2000;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Matsumoto M, Yada M, Nakayama KI. Interaction of U-box-type ubiquitin-protein ligases (E3s) with molecular chaperones. Genes Cells. 2004;9:533–548. doi: 10.1111/j.1356-9597.2004.00742.x. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP. Structure of an E6AP-UbcH7 Complex: Insights into Ubiquitination by the E2–E3 Enzyme Cascade. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- Jensen JP, Bates PW, Yang M, Vierstra RD, Weissman AM. Identification of a family of closely related human ubiquitin conjugating enzymes. J Biol Chem. 1995;270:30408–30414. doi: 10.1074/jbc.270.51.30408. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Juranić N, Macura S. Correlations among 1JNC' and h3JNC' coupling constants in the hydrogen-bonding network of human ubiquitin. J Am Chem Soc. 2001;123:4099–4100. doi: 10.1021/ja015647d. [DOI] [PubMed] [Google Scholar]
- Juranić N, Atanasova E, Moncrieffe MC, Prendergast FG, Macura S. Calcium-binding proteins afford calibration of dihedral-angle dependence of 3JNCγ coupling constant in aspartate and asparagine residues. J Magn Reson. 2005;175:222–225. doi: 10.1016/j.jmr.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Juranić N, Moncrieffe MC, Atanasova E, Macura S, Prendergast FG. NMR h3JNC’ couplings provide comprehensive geometrical constraints for protein H-bonds in solution. Croat Chem Acta. 2006;79:503–507. [Google Scholar]
- Krissinel E, Herrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K, Mouysset J, Hoppe T. Orchestra for assembly and fate of polyubiquitin chains. Essays Biochem. 2005;41:1–14. doi: 10.1042/EB0410001. [DOI] [PubMed] [Google Scholar]
- Lindahl E, Azuara C, Koehl P, Delarue M. NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids res. 2006;34:W52–W56. doi: 10.1093/nar/gkl082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Yada M, Hatakeyama S, Ishimoto H, Tanimura T, Tsuji S, Kakizuka A, Kitagawa M, Nakayama KI. Molecular clearance of ataxin-3 is regulated by a mammalian E4. EMBO J. 2004;23:659–669. doi: 10.1038/sj.emboj.7600081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum–likelihood method. Acta Crystallogr Sect D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nordquist KA, Dimitrova YN, Brzovic PS, Ridenour WB, Munro KA, Soss SE, Caprioli RM, Klevit RE, Chazin WJ. Structural and functional characterization of the monomeric U-box domain from E4B. Biochemistry. 2010;49:347–355. doi: 10.1021/bi901620v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol. 2003;10:250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W, Carter Charles W., Jr Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci U S A. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Barford D. Getting into position: the catalytic mechanisms of protein ubiquitylation. Biochem J. 2004;379:513–525. doi: 10.1042/BJ20040198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Marius Clore G. The Xplor-NIH NMR molecular structure determination package. J Magn Res. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Tu D, Li W, Ye Y, Brunger AT. Structure and function of the yeast U-box-containing ubiquitin ligase Ufd2p. Proc Natl Acad Sci U S A. 2007;104:15599–15606. doi: 10.1073/pnas.0701369104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull WB, Daranas AH. On the Value of c: Can low affinity systems be studied by isothermal titration calorimetry? J Am Chem Soc. 2003;125:14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- Vander Kooi CW, Ohi MD, Rosenberg JA, Oldham ML, Newcomer ME, Gould KL, Chazin WJ. The Prp19 U-box crystal structure suggests a common dimeric architecture for a class of oligomeric E3 ubiquitin ligases. Biochemistry. 2006;45:121–130. doi: 10.1021/bi051787e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Xu Z, Devlin KI, Ford MG, Nix JC, Qin J, Misra S. Structure and interactions of the helical and U-box domains of CHIP, the C terminus of HSP70 interacting protein. Biochemistry. 2006;45:4749–4759. doi: 10.1021/bi0601508. [DOI] [PubMed] [Google Scholar]
- Xu Z, Kohli E, Devlin KI, Bold M, Nix JC, Misra S. Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct Biol. 2008;8:26. doi: 10.1186/1472-6807-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Lin SC, Lamothe B, Lu M, Lo YC, Hura G, Zheng L, Rich RL, Campos AD, Myszka DG, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation–crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.