Abstract
We applied the newly developed Fourier transform light scattering (FTLS) to study dynamic light scattering in single live cells, at a temporal scale of seconds to hours. The nanoscale cell fluctuations were measured with and without the active actin contribution. We found experimentally that the spatio-temporal signals rendered by FTLS reveal interesting cytoskeleton dynamics in glial cells (the predominant cell type in the nervous system). The active contribution of actin cytoskeleton was obtained by modulating its dynamic properties via Cytochalasin-D, a drug that inhibits actin polymerization/depolymerization.
OCIS codes: (180.0180) Microscopy, (170.0170) Medical optics and biotechnology, (120.5820) Scattering Measurements
1. Introduction
Recently, dynamic properties of cytoskeleton have been the subject of intense scientific interest [1–12]. In particular, it has been shown that actin filaments play an important role in various aspects of cell dynamics, including cell motility [2–4,12]. Actin filament polymerization has been studied in real time using total internal reflection fluorescence microscopy [5,6]. By observing the fluorescence signal arising from labeled filaments, the rate of rearward actin transport and that of forward cell movement have been quantified [8,9]. Particle tracking microrheology [13] has revealed unexpected behavior in live cell mechanics both by using passive (thermal) [14] and active (driven) [15] probing beads. This approach also revealed violation of the fluctuation-dissipation theorem in bacterial suspensions [16].
We have developed Fourier transform light scattering (FTLS) as a novel optical method that combines the high spatial resolution associated with optical microscopy and statistical averaging of light scattering techniques [17]. Because the measurement is performed in the image plane, where the optical field is uniform in amplitude, FTLS utilizes efficiently the dynamic range of the recording detector array and, thus, provides very high sensitivity to weakly scattering media such as thin tissue slices [18] and single cells [17]. Within a single measurement, FTLS renders dynamic information over a broad angular range, limited only by the numerical aperture of the microscope. Here we use FTLS to measure the spatio-temporal behavior of active (ATP consuming) dynamics due to F-actin in single glial cells. This activity mediated by motor protein Myosin II underlies diverse cellular processes, including cell division, developmental polarity, cell migration, filopodial extension, and intracellular transport. Extracellular signals mediate experience-induced changes of actin dynamics within synaptic microdomains of neurons [19,20]. These actin-based changes can transform cell state, locally and globally. In this paper we quantify the dynamics associated with the cell membrane and intrinsic cell structure motions. We used probing beads attached to the cell membrane as a control experiment to demonstrate the Cytochalasin-D efficacy in disrupting actin polymerization and, thus, prove that FTLS is sensitive to actin-driven dynamics.
2. Methods
2.1. Fourier transform light scattering (FTLS)
FTLS requires accurate optical phase retrieval for elastic light scattering measurements and, in addition, high phase stability for dynamic light scattering studies. The system diagram was depicted in our earlier publication [21] and it satisfies the requirements above by incorporating a common path interferometer with a commercial computer-controlled microscope. The second harmonic of a Nd:YAG laser (λ = 532 nm) is used to illuminate the sample in transmission. To ensure full spatial coherence, the laser beam is coupled into a single mode fiber and further collimated by a fiber collimator. The light scattered by the sample is collected by the objective lens of the microscope (Axio Observer Z1, Zeiss) and imaged at the side port of the microscope. A diffraction grating is placed at the image plane, thus generating multiple diffraction orders containing full spatial information about the image. In order to establish a common-path Mach-Zehnder interferometer, a standard spatial filtering lens system is used to select the two diffraction orders and generate the final interferogram at the CCD plane. The 0th order beam is low-pass filtered using a spatial filter positioned in the Fourier plane of the first lens, such that at the CCD plane it approaches a uniform field. Simultaneously, the spatial filter allows passing the entire frequency content of the 1st diffraction order beam and blocks all the other orders. The 1st order is thus the imaging field and the 0th order plays the role of the reference field. The two beams propagate along a common optical path, thus significantly reducing the longitudinal phase noise. The final quantitative phase image of the sample is retrieved via spatial Hilbert transform [21–23]. Thus, from a single CCD exposure, we obtain the spatially-resolved phase and amplitude associated with the image field. From this image field information , the complex field can be numerically propagated at arbitrary planes; in particular, the far-field angular scattering distribution can be obtained simply via a Fourier transformation [17],
| (1) |
2.2. Cell preparation of enteric glial cells (EGCs)
We applied FTLS to study the slow active dynamics of glial cytoskeleton. Enteric glial cells (EGC) of the enteric (i.e. intestinal) nervous system have long been considered a mechanical support. However, more recent findings provide insight to more complex homeostatic and inflammatory interactions with neurons, lymphocytes, epithelial cells and capillaries of the gut to modulate gastrointestinal motility and respond to inflammation [24]. The EGC cell line was obtained from ATCC (CRL-2690, designation: EGC/PK060399egfr), cultured in DMEM (Gibco product #31053) with glucose (4.5 g/L), and supplemented with 3.0mM L-glutamine and 10% fetal bovine serum. Confluent cultures were propagated by incubation in trypsin EDTA to remove cells from culture flasks. The cell suspension in trypsin was diluted into culture media and gently triturated to break up aggregates. Cells were further diluted for culture passage or plating in glass-bottom culture Fluorodishes (catalogue number: FD-35; World Precision Instruments, Sarasota, FL) and were allowed to expand from initial plating for 1-2 days and imaged prior to reaching 90% confluence.
2.3. Blocking cell actin polymerization
During the FTLS measurement, the cells were maintained under constant temperature at 37°C via the incubation system that equips the microscope (Axio Observer Z1, Zeiss). The sensitivity of FTLS to actin dynamics was tested by controlling its polymerization activity. In order to inhibit actin polymerization, Cytochalasin-D (Cyto-D), approximately 5 µM in Hibernate-A, was added to the sample dishes. Cyto-D is a naturally occurring fungal metabolite known to have potent inhibitory action on actin filaments by capping and preventing filament polymerization and depolymerization at the rapidly elongating end of the filament. By capping this “barbed” end, the increased dissociation at the pointed end continues to shorten the actin filament. In this way, Cyto-D alters cytoskeleton and membrane dynamics which was captured via our FTLS measurement. The working algorithm and controlling efficiency of applying Cyto-D to inhibit the actin polymerization can be found in detail in Ref [25,26].
2.4. Particle tracking
We performed two sets of experiments: in addition to the main FTLS studies on actin dynamics, we also performed a control test to demonstrate the Cyto-D effect on actin polymerization. Thus, as a control experiment, we first used the single particle-tracking method [27,28] to monitor the actin-induced cell motions, with and without the Cyto-D inhibitor. The dynamic property of the cell membrane was investigated using micron-sized beads attached as probes to the cell membrane. Solutions of 1 micron diameter polystyrene beads (Polysciences Inc.) were diluted by mixing 2 µL with 1 mL Hibernate-A media (BrainBits LLC, Springfield, IL) and centrifuged at 4,400 rpm for 5 minutes to remove the liquid stock solution. After centrifugation, the media was aspirated almost entirely, down to approximately 50 µL Hibernate with concentrated beads. Hibernate-A (1 mL) was again added back to the vial and the media-bead solution was pipetted vigorously to resuspend beads in media. The bead suspension was heated to 37°C, and exchanged with culture media in the dish. Enteroglia culture dishes were sealed with parafilm and returned to the incubator. Cultures were left undisturbed for approximately 2 hours to enable beads to attach to enteroglia and for cells to adjust to the serum-free Hibernate media. Immediately prior to imaging, cultures were rinsed with 6-8 mL of Hibernate-A (37°C) to remove loosely attached and free-floating beads. Hibernate-A was used for all imaging of EGC dynamics, since it is buffered to retain optimal pH in ambient carbon-dioxide levels.
The x and y coordinates of the tracked beads were recorded as a function of time and the trajectories were used to calculate the mean squared displacement (MSD) [29],
| (2) |
where <…> indicates the average over time and also over all the tracked particles. Assuming thermal equilibrium, from Eq. (2) the conventional microrheology can be inferred via the generalized Stokes-Einstein equation [27,29–31].
3. Results
3.1 Bead tracking
Typically, a small region of interest surrounding a single tracked bead is captured and we record displacements of the attached bead before and after treatment with cyto-D. Figures 1a , 1b show examples of the bead trajectories before adding drug. These trajectories are broader, with larger displacements than those associated with the cells under cyto-D treatment (Figs. 1c, 1d). The trajectories of the tracked beads are further used to calculate the MSD as a function of time [Eq. (2)]. The results are summarized in Fig. 1e. Each curve is the result of an average over all the beads tracked under the same experimental condition. The data exhibits a power law trend over two distinct temporal regions. We first fit the data with a power law equation, tα, over the initial 150 seconds. Interestingly, the exponents obtained from the fit are below 1: α = 0.6 and 0.82 for before and after treatment, respectively, which is an indication of sub-diffusion. The change in slope at t~200 s is related to the actin polymerization lifetime, which is known to be in the minute range [12]. The fit at the t>150 s gives α = 0.91 and 0.99, respectively, for before and after treatment actin inhibition. These results, of exponents becoming closer to unity after drug treatment, indicate that blocking actin polymerization favors Brownian motion (for which α = 1), as expected [32]. The MSD before treatment is larger than that of after, which is expected, as the active contributions of the cytoskeleton contribute to cell motions. These findings indicate that our control for actin polymerization functions well.
Fig. 1.
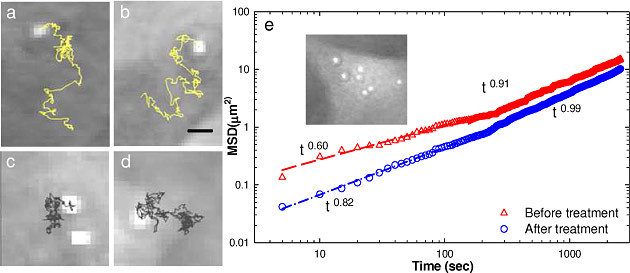
Tracking beads attached to the cell membrane: a)-b) bead trajectories before cyto-D treatment and c)-d) after drug treatment. e) Corresponding mean square displacement of the tracked beads before (red) and after (blue) treatment. The fitting with a power law function over two different time windows is indicated. The inset shows a quantitative phase image of a EGC with 1μm beads attached to the membrane.
3.2. Temporal fluctuations
To determine how the actin cytoskeleton contributes to the dynamic light scattering of cells alone, cells were imaged via FTLS by acquiring 512 frames, at 0.2 frames per second, over ~45 min prior to and after the Cyto-D application, respectively (Figs. 2a , 2b). The displacement was calculated with respect to the average for each point in the quantitative phase images. Figure 2c shows a comparison between the membrane displacement histograms of a cell before and after the actin inhibitor. It is evident from this result that the polymerization phenomenon is a significant contributor to the overall cell dynamics, as indicated by the broader histogram distribution. Further, both curves exhibit non-Gaussian shapes at displacements larger than 10 nm, which suggests that the cell motions both before and after actin inhibition are characterized by non-equilibrium dynamics, consistent with the short-time behavior in Fig. 1e. Note that these path-length fluctuations can have two main causes: membrane fluctuations, which modify the local cell thickness and intracellular mass transport, which produce refractive index fluctuations. The homogeneous appearance of the quantitative phase images and also the control experiments with the membrane-tagged beads seem to indicate that the membrane motions are dominant. It is known that actin is tethered to the membrane bilayer and keeps it under tension. This picture explains the significant effect of blocking actin activity, as resulting into a less tensed membrane and, thus, higher displacements.
Fig. 2.
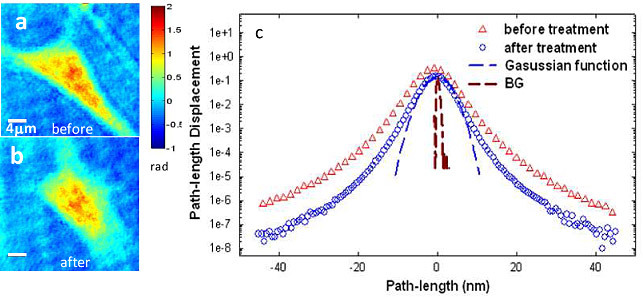
a-b) Quantitative phase images of glial cell before and after Cyto-D treatment. c) Histogram of the path-length displacements of a glial cell before and after drug treatment, as indicated. The blue dash line indicates the fit with a Gaussian function and brown dash line shows the background fluctuation.
Figure 3a presents the comparison of the spatially-averaged power spectra associated with the FTLS signal for glial cells averaged over three individual measurements which were taken at different days but following the same sample preparation procedures and experimental condition, before and after treatment with the actin blocking drug. The broader power spectrum of the untreated cell membrane motions (Fig. 3a) is consistent with the histogram distribution in Fig. 2c. The intact cell exhibits a power spectrum that departs from a Lorentzian shape at frequencies of the order 4-6x10−3 rad/s, which indicates that actin affects the dynamics predominantly at short times (i.e. minute scale). Further, both frequency-averaged (statics) curves shown in Fig. 3b indicates similar functional dependence on the wavenumber q, but with enhanced fluctuations for the normal cell, by a factor of ~3.4.
Fig. 3.
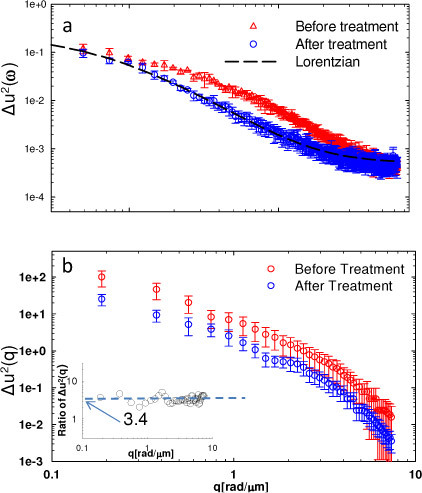
a) Spatially-averaged power spectrum of glial cells before and after drug treatment, as indicated. Error bars show the standard deviation (same for all the followed figures) and dash lines show the fit with Lorentzian equations. b) Temporally-averaged power spectrum before and after drug treatment. The inset shows the ratio of the two spectra.
3.3. Spatio-temporal fluctuations
One key feature of FTLS is its ability to render simultaneously angular scattering from an entire range of angles. Figures 4a –4d show the power spectrum of the fluctuations for the same cells before and after the actin inhibition, as function of both frequency ω (at two particular q-values) and wavenumber q (at two particular frequencies ω). Thus, after actin inhibition, the functional dependence of Δu2(ω) assumes a Lorentzian shape and does not change notably with varying q, which contrasts with the situation where the cell cytoskeleton is intact. This interesting behavior can be seen in Figs. 4a, 4b, where the temporal power spectra at 2 particular wavenumbers are shown. These findings suggest that, as the actin is disrupted, the dynamic scattering signal is most likely due to Brownian-like fluctuations. To better capture dynamic activity within our experimental temporal resolution, the power spectra vs. q at two frequencies divided far enough are shown in Figs. 4c, 4d. At frequencies above certain threshold ω0 (Fig. 4c), there is a significant mismatch in shape between the “before” and “after” Δu2(q) curves. On the other hand, for lower frequencies (Fig. 4d), the two dependencies look similar, with the normal cell exhibiting consistently higher fluctuations. The results in Fig. 3 and 4 suggest that the actin contribution is characterized by a certain lifetime, which is consistent with previous findings of polymerization kinetics [12].
Fig. 4.
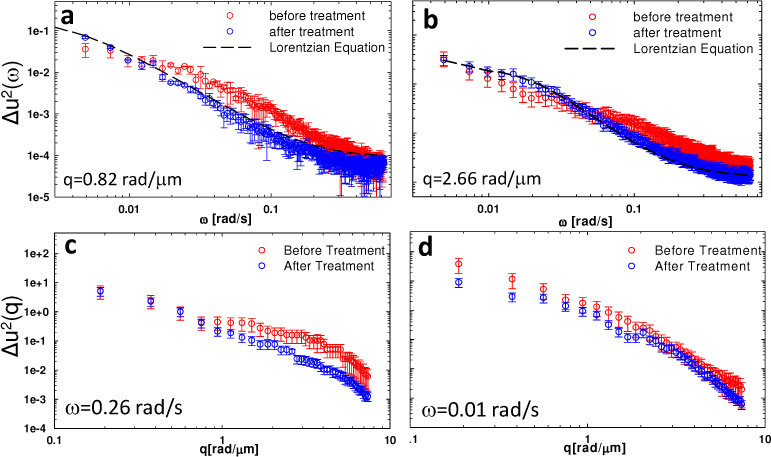
a-b) Temporal power spectra of glial cells before and after drug treatment at two different spatial frequencies, as indicated. Dash lines show fits with Lorentzian equations for the power spectra after drug treatment; c-d) Spatial power spectra at two different frequencies, as indicated. The error bars indicate the standard deviation associated 3 different measurements.
4. Conclusion
In summary, we applied FTLS to measure the effects of actin cytoskeleton on the spatio-temporal fluctuations of cell dynamics. We believe that these label-free results are extremely promising for studying cytoskeleton dynamics and will complement very well the existing fluorescence studies. Retrieving the membrane tension and actin lifetime without physical contact will likely provide a useful approach for studying fundamental cell biology phenomena, such as mitosis, phagocytosis, or motility [33]. With further theoretical developments, the FTLS data will be connected with the cell microrheology obtained via one- and two- particle tracking [27,28].
Acknowledgements
This research was supported in part by the National Science Foundation (CAREER 08-46660) and the Grainger Foundation. L.J.M. was supported by the National Institute of Health (HD007333) Developmental Psychobiology and Neurobiology Training Grant. M.U.G. was supported by National Institute of Health (HL086070). For more information, visit http://light.ece.uiuc.edu/.
References and links
- 1.Gardel M. L., Shin J. H., MacKintosh F. C., Mahadevan L., Matsudaira P., Weitz D. A., “Elastic behavior of cross-linked and bundled actin networks,” Science 304(5675), 1301–1305 (2004). 10.1126/science.1095087 [DOI] [PubMed] [Google Scholar]
- 2.Pollard T. D., Borisy G. G., “Cellular motility driven by assembly and disassembly of actin filaments,” Cell 112(4), 453–465 (2003). 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- 3.Cooper J. A., Schafer D. A., “Control of actin assembly and disassembly at filament ends,” Curr. Opin. Cell Biol. 12(1), 97–103 (2000). 10.1016/S0955-0674(99)00062-9 [DOI] [PubMed] [Google Scholar]
- 4.Mitchison T. J., Cramer L. P., “Actin-based cell motility and cell locomotion,” Cell 84(3), 371–379 (1996). 10.1016/S0092-8674(00)81281-7 [DOI] [PubMed] [Google Scholar]
- 5.Kuhn J. R., Pollard T. D., “Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy,” Biophys. J. 88(2), 1387–1402 (2005). 10.1529/biophysj.104.047399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollard T. D., “The cytoskeleton, cellular motility and the reductionist agenda,” Nature 422(6933), 741–745 (2003). 10.1038/nature01598 [DOI] [PubMed] [Google Scholar]
- 7.Amann K. J., Pollard T. D., “Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy,” Proc. Natl. Acad. Sci. U.S.A. 98(26), 15009–15013 (2001). 10.1073/pnas.211556398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Theriot J. A., Mitchison T. J., “Actin microfilament dynamics in locomoting cells,” Nature 352(6331), 126–131 (1991). 10.1038/352126a0 [DOI] [PubMed] [Google Scholar]
- 9.Theriot J. A., Mitchison T. J., Tilney L. G., Portnoy D. A., “The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization,” Nature 357(6375), 257–260 (1992). 10.1038/357257a0 [DOI] [PubMed] [Google Scholar]
- 10.Uttenweiler D., Veigel C., Steubing R., Götz C., Mann S., Haussecker H., Jähne B., Fink R. H. A., “Motion determination in actin filament fluorescence images with a spatio-temporal orientation analysis method,” Biophys. J. 78(5), 2709–2715 (2000). 10.1016/S0006-3495(00)76815-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C. C., Lin J. Y., Chen H. C., Lee C. H., “Dynamics of cell membranes and the underlying cytoskeletons observed by noninterferometric widefield optical profilometry and fluorescence microscopy,” Opt. Lett. 31(19), 2873–2875 (2006). 10.1364/OL.31.002873 [DOI] [PubMed] [Google Scholar]
- 12.Watanabe N., Mitchison T. J., “Single-molecule speckle analysis of actin filament turnover in lamellipodia,” Science 295(5557), 1083–1086 (2002). 10.1126/science.1067470 [DOI] [PubMed] [Google Scholar]
- 13.Mason T. G., Ganesan K., vanZanten J. H., Wirtz D., Kuo S. C., “Particle tracking microrheology of complex fluids,” Phys. Rev. Lett. 79(17), 3282–3285 (1997). 10.1103/PhysRevLett.79.3282 [DOI] [Google Scholar]
- 14.Caspi A., Granek R., Elbaum M., “Diffusion and directed motion in cellular transport,” Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 66(1), 011916 (2002). 10.1103/PhysRevE.66.011916 [DOI] [PubMed] [Google Scholar]
- 15.Deng L. H., Trepat X., Butler J. P., Millet E., Morgan K. G., Weitz D. A., Fredberg J. J., “Fast and slow dynamics of the cytoskeleton,” Nat. Mater. 5(8), 636–640 (2006). 10.1038/nmat1685 [DOI] [PubMed] [Google Scholar]
- 16.Chen D. T. N., Lau A. W. C., Hough L. A., Islam M. F., Goulian M., Lubensky T. C., Yodh A. G., “Fluctuations and rheology in active bacterial suspensions,” Phys. Rev. Lett. 99(14), 148302 (2007). 10.1103/PhysRevLett.99.148302 [DOI] [PubMed] [Google Scholar]
- 17.Ding H. F., Wang Z., Nguyen F., Boppart S. A., Popescu G., “Fourier transform light scattering of inhomogeneous and dynamic structures,” Phys. Rev. Lett. 101(23), 238102 (2008). 10.1103/PhysRevLett.101.238102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding H., Nguyen F., Boppart S. A., Popescu G., “Optical properties of tissues quantified by Fourier-transform light scattering,” Opt. Lett. 34(9), 1372–1374 (2009). 10.1364/OL.34.001372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matus A., “Actin-based plasticity in dendritic spines,” Science 290(5492), 754–758 (2000). 10.1126/science.290.5492.754 [DOI] [PubMed] [Google Scholar]
- 20.Zito K., Knott G., Shepherd G. M. G., Shenolikar S., Svoboda K., “Induction of spine growth and synapse formation by regulation of the spine actin cytoskeleton,” Neuron 44(2), 321–334 (2004). 10.1016/j.neuron.2004.09.022 [DOI] [PubMed] [Google Scholar]
- 21.Popescu G., Ikeda T., Dasari R. R., Feld M. S., “Diffraction phase microscopy for quantifying cell structure and dynamics,” Opt. Lett. 31(6), 775–777 (2006). 10.1364/OL.31.000775 [DOI] [PubMed] [Google Scholar]
- 22.Popescu G., Ikeda T., Goda K., Best-Popescu C. A., Laposata M., Manley S., Dasari R. R., Badizadegan K., Feld M. S., “Optical measurement of cell membrane tension,” Phys. Rev. Lett. 97(21), 218101 (2006). 10.1103/PhysRevLett.97.218101 [DOI] [PubMed] [Google Scholar]
- 23.Ikeda T., Popescu G., Dasari R. R., Feld M. S., “Hilbert phase microscopy for investigating fast dynamics in transparent systems,” Opt. Lett. 30(10), 1165–1167 (2005). 10.1364/OL.30.001165 [DOI] [PubMed] [Google Scholar]
- 24.Bassotti G., Villanacci V., Fisogni S., Rossi E., Baronio P., Clerici C., Maurer C. A., Cathomas G., Antonelli E., “Enteric glial cells and their role in gastrointestinal motor abnormalities: introducing the neuro-gliopathies,” World J. Gastroenterol. 13(30), 4035–4041 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casella J. F., Flanagan M. D., Lin S., “Cytochalasin D inhibits actin polymerization and induces depolymerization of actin filaments formed during platelet shape change,” Nature 293(5830), 302–305 (1981). 10.1038/293302a0 [DOI] [PubMed] [Google Scholar]
- 26.Flanagan M. D., Lin S., “Cytochalasins block actin filament elongation by binding to high affinity sites associated with F-actin,” J. Biol. Chem. 255(3), 835–838 (1980). [PubMed] [Google Scholar]
- 27.Levine A. J., Lubensky T. C., “One- and two-particle microrheology,” Phys. Rev. Lett. 85(8), 1774–1777 (2000). 10.1103/PhysRevLett.85.1774 [DOI] [PubMed] [Google Scholar]
- 28.Lau A. W. C., Hoffman B. D., Davies A., Crocker J. C., Lubensky T. C., “Microrheology, stress fluctuations, and active behavior of living cells,” Phys. Rev. Lett. 91(19), 198101 (2003). 10.1103/PhysRevLett.91.198101 [DOI] [PubMed] [Google Scholar]
- 29.Dieterich P., Klages R., Preuss R., Schwab A., “Anomalous dynamics of cell migration,” Proc. Natl. Acad. Sci. U.S.A. 105(2), 459–463 (2008). 10.1073/pnas.0707603105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crocker J. C., Valentine M. T., Weeks E. R., Gisler T., Kaplan P. D., Yodh A. G., Weitz D. A., “Two-point microrheology of inhomogeneous soft materials,” Phys. Rev. Lett. 85(4), 888–891 (2000). 10.1103/PhysRevLett.85.888 [DOI] [PubMed] [Google Scholar]
- 31.Mason T. G., Weitz D. A., “Optical measurements of frequency-dependent linear viscoelastic moduli of complex fluids,” Phys. Rev. Lett. 74(7), 1250–1253 (1995). 10.1103/PhysRevLett.74.1250 [DOI] [PubMed] [Google Scholar]
- 32.Feder T. J., Brust-Mascher I., Slattery J. P., Baird B., Webb W. W., “Constrained diffusion or immobile fraction on cell surfaces: a new interpretation,” Biophys. J. 70(6), 2767–2773 (1996). 10.1016/S0006-3495(96)79846-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai J., Ting-Beall H. P., Sheetz M. P., “The secretion-coupled endocytosis correlates with membrane tension changes in RBL 2H3 cells,” J. Gen. Physiol. 110(1), 1–10 (1997). 10.1085/jgp.110.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]


