Abstract
Interleukin-13 is a Th2-associated cytokine responsible for many pathological responses in allergic asthma including mucus production, inflammation and extracellular matrix remodeling. In addition, IL-13 is required for immunity to many helminth infections. IL-13 signals via the type-II IL-4 receptor, a heterodimeric receptor of IL-13Rα1 and IL-4Rα, which is also utilized by IL-4. IL-13 also binds to IL-13Rα2, but with much higher affinity than the type-II IL-4 receptor. Binding of IL-13 to IL-13Rα2 has been shown to attenuate IL-13 signaling through the type-II IL-4 receptor. However, molecular determinants that dictate the specificity and affinity of mouse IL-13 for the different receptors are largely unknown. Here, we used high-density overlapping peptide arrays, structural modeling and molecular docking methods to map IL-13 binding sequences on its receptors. Predicted binding sequences on mouse IL-13Rα1 and IL-13Rα2 were in agreement with the reported human IL-13 receptor complex structures and site-directed mutational analysis. Novel structural differences were identified between IL-13 receptors, particularly at the IL-13 binding interface. Notably, additional binding sites were observed for IL-13 on IL-13Rα2. In addition, the identification of peptide sequences that are unique to IL-13Rα1 allowed us to generate a monoclonal antibody that selectively binds IL-13Rα1. Thus, high-density peptide arrays combined with molecular docking studies provide a novel, rapid, and reliable method to map cytokine- receptor interactions that may be used to generate signaling and decoy receptor-specific antagonists.
Keywords: Cytokine receptors, peptide arrays, protein-protein interactions, structure and protein modeling
INTRODUCTION
Among the Th2 cytokines, IL-13 is required for immunity against parasitic helminths yet is also responsible for pathological tissue remodeling of extracellular matrix1. Experimental models have revealed that IL-13 regulates hepatic fibrosis and chronic inflammatory diseases of the lung such as asthma, idiopathic pulmonary fibrosis, and COPD 1–4. IL-13 signals via the type-II IL-4 receptor comprised of IL-13Rα1 and IL-4Rα. This heterodimeric receptor is formed upon binding of IL-13 to IL-13Rα15. IL-4 can also signal through the same receptor complex. However, IL-4 binds to the IL-4Rα subunit first and then recruits IL-13Rα1 1,5. IL-4 can also signal via the type-I IL-4 receptor, a heterodimeric receptor formed of IL-4Rα and the IL-2 receptor gamma chain (IL-2γC)5. The type-II IL-4 receptor complex, expressed predominantly on non-hematopoietic cells is believed to be largely responsible for IL-13-dependent actions in vivo, including mucus production by epithelial cells and functional alterations in smooth muscle cells6–8. Notably, some of the pathological responses observed in vivo were shown to be independent of IL-4 yet dependent on IL-136,9. The recently solved crystal structures for the type-I and type-II IL-4 receptor complexes revealed important structural insights and provided some possible explanations for why IL-4 and IL-13 often exhibit distinct biological activities in vivo5. For example, alanine-scanning mutagenesis studies on residues W65A to A79Y within the N-terminal Ig-like domain of IL-13Rα1 showed loss of IL-13 binding but not IL-410, while Alanine substitutions at positions L35, L38 and V42 within domain D1 led to reduced IL-13 binding to IL-13Rα110. Interestingly, according to the recently reported crystal structure, these residues are located far away from the IL-13 binding interface5. This suggests multiple binding events or binding dynamics take place when IL-13 binds to its receptor.
Studies from our laboratory and others have demonstrated that IL13Rα2 primarily functions as a decoy receptor for IL-13. It prevents IL-13 from binding to the type II IL-4R complex, thus attenuating IL-13-mediated STAT6 phosphorylation11 6. Garcia and colleagues noted that the interaction between IL-13 and IL-13Rα2 is several orders of magnitude greater than the interaction between IL-13 and IL-13Rα1/IL-4Rα, confirming that the primary function of 13Rα2 is to prevent IL-13 from binding to the signaling receptor12–14. Nevertheless, a few recent studies have suggested that IL-13Rα2 might also signal under certain biological conditions15. While the exact signaling mechanism remains uncertain, it is clear that it does not induce the phosphorylation of STAT61,15–16. Because IL-13Rα1 and IL-13Rα2 exhibit opposing activities in a variety disease models7,17, a complete and detailed map of the IL-13 binding sites that are distinct and shared between IL-13Rα1 and IL-13Rα2 is needed, as this information may help explain their unique functional activities in vivo. A detailed map would also aid in the development of reagents that can selectively bind and inhibit the interaction of IL-13 with each receptor. Because membrane receptors are often difficult to express, purify, and crystallize, simplified methods are needed to map important protein-protein interactions. In the current study we used a novel high-density scanning peptide array platform to map IL-13 binding sites on IL-13Rα1 or IL-13Rα2 and show that this new methodology is fast, reliable, and highly efficient for mapping protein-protein interactions. We combined the peptide array data with protein structure modeling and molecular docking and identify specific IL-13 binding sites on IL-13Rα1 and IL-13Rα2.
MATERIALS AND METHODS
Peptide arrays
PepStar™ peptide microarrays prepared on glass slides were obtained from JPT Peptide Technologies, GmbH. In brief, 15-residue long overlapping peptides derived from IL-13Rα1 and IL-13Rα2 sequences were synthesized on modified cellulose membranes using SPOT technology18. The synthesized peptides were cleaved from the cellulose support using aqueous triethylamine (0.5% by vol.) and printed per spot on epoxy functionalized glass slides (spot size, 250 µM in diameter) using non-contact printing (JPT Ppetide Technologies GmbH). Slides were blocked using 1 mg/ml BSA in 1.5 mM SSC buffer pH 7.0 and 250 mM NaCl for 1 hr and washed extensively with 1.5 mM SSC buffer pH 7.0. Mouse IL-13 conjugated with alexa488 (label-per-protein molar ratio of 0.5) was added to a final concentration of 1 nM in PBST containing 1% BSA. After 6 h incubation in a hybridization chamber slides were washed five times with 1.5 mM SSC, followed by washings with water and then spun dry. The arrays were then imaged using a GenepixX microarray scanner and analyzed using Genepix Array list (GAL) files supplied by JPT18. The foreground intensity of the spots (three replicates for the individual spot) at the 595nm wavelength was used for analysis18.
Sequence alignment and homology modeling
Human and mouse primary protein sequences for IL-13Rα1 and IL-13Rα2 were aligned and homologous regions defined using Biology WorkBench (UCSD, California, USA). Sequences for mouse IL-13Rα1 (Genbank NP_598751.3), IL-13Rα2 (Genbank NP_032382.1) and IL-13 (Genbank NP_032381.1) were submitted to the LOMETS19 homology modeling server. The top scoring model in terms of length and sequence identity was chosen for further processing. Templates on which chosen models were constructed were PDB ID 3BPO chain A for IL-13 (62.5% sequence identity) and PDB ID 3BPN chain C for both IL-13Rα1 and PDB ID 3LB6 chain D for IL-13Rα2 (76% and 22% sequence identity, respectively). Models were checked for incorrect geometry and evaluated with PROCHECK20. Each model was minimized with the Powell conjugate gradient method using a non-bonded cutoff of 12 Å in implicit solvent with the AMBER7 ff99 force field in SYBYL 8.1 (Tripos) until a termination gradient of 0.05 kcal/mol·Å was reached.
Protein-protein docking
The RosettaDock program (v.3.1)21 was used to perform all-atom, perturbation protein-protein docking. Initially, a set of Rosetta run conditions were obtained using the mouse IL-13 and IL-13Rα1 models that reproduced the binding orientation of human IL-13 and IL-13α1 (PDB ID 3BPO). The run conditions consisted of holding the backbone rigid with the exception of IL-13Rα1 loop residues K189-N198. Backbone atoms for loop residues K189-N198 were “flexed” by using loop conformations generated from the Rosetta program. All side chains were allowed to move. Prior to docking, mouse IL-13 was placed 25Å from the side of the receptor that is analogous to the binding interface in human IL13α1. During the run, IL-13 was allowed to randomize its orientation relative to IL-13Rα1 before docking. Solutions were kept if they met a set of site constraints where certain IL13Rα1 residues were required to be at or near the interface (Supplementary Table 1). Ten thousand decoys were generated and the top 100 scoring (lowest energy) solutions were clustered based on an all-atom root mean square (rms) deviation cutoff of 5Å. The highest scoring (lowest energy) decoy from each cluster was retained and of these, the lowest energy solution was chosen for further analysis. Protein-protein docking was performed in the same way for mouse IL-13 and IL-13Rα2 with loop modeling performed on IL-13Rα2 residues S194-F204. All calculations were performed utilizing the high-performance computational resources of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, Md. (http://biowulf.nih.gov). Electrostatic potential was calculated using the Adaptive Poisson-Boltzmann Solver (APBS) program22 and mapped to a surface using the PyMol program (DeLano Scientific, Palo Alto, CA, USA). Residues were defined as being at a protein-protein interface if they were with 3.5Å of their partner.
Generation of monoclonal antibody
Mouse monoclonal antibodies were generated by using the peptides that correspond to IL-13Rα1 sequences. The peptides were conjugated to KLH and mice were immunized at Precision Antibody™ (Columbia, MD) using their proprietary protocols. We screened clones based on their positive reactivity to the individual peptides that are conjugated to BSA and also ability to bind the native IL-13Rα1 on a nitrocellulose membrane. The selected positive clone, 3D2 was slowly adapted to low serum media (DMEM containing 3% FCS) and cultured in large volumes. 3D2 culture supernatants were used to purify 3D2 antibody using a proteinA Q sepharose matrix (GE Healthcare, Piscataway, NJ). Antibody concentration was measured using absorbance at 280 nm and an extinction coefficient of 14 for mouse IgG.
Surface Plasmon Resonance
Surface Plasmon resonance experiments were performed with a Biacore 2000 (GE Healthcare, Piscataway, NJ). About 1000 resonance units (RU) of IL-13Rα1-Fc or IL-13Rα2-Fc (R&D Systems, MN, USA) were immobilized by amine coupling to one of the flow cells onto research grade Biacore CM5 biosensor chip (GE Healthcare, Piscataway, NJ). One of flow cells that lack immobilized protein was used as negative control for each binding. Purified monoclonal antibody, 3D2 (400 nM and 200 nM) was flown at a flow rate of 30µl/min over the above immobilized chip and signal obtained from negative control (empty cell) was subtracted to obtain final RU.
RESULTS AND DISCUSSION
Sequence analysis and structure homology modeling of IL-13 receptors
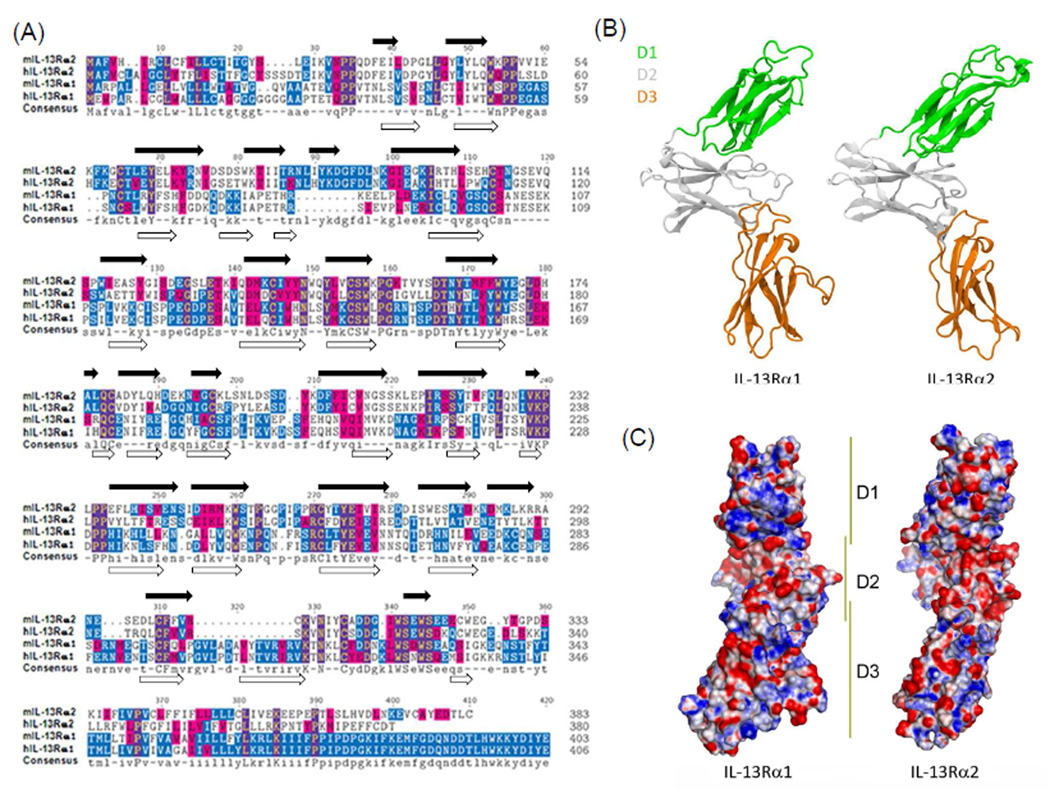
The biological functions of IL-13 are dependent on affinity driven binding of IL-13 to IL-13Rα1 or IL-13Rα2. Because the two receptors exhibit strikingly different biological effects when they engage IL-13, a complete understanding of the unique IL-13 binding sites used by each receptor may aid in the development receptor specific antagonists. Human and mouse sequences for IL-13Rα1 or IL-13Rα2 were aligned (Figure 1A) and found to be highly conserved with the mouse and human IL-13Rα1 subunit displaying an homology score of 73% and the mouse and human IL-13Rα2 chain a score of 56%. The relatively high sequence identities between both species suggest similar binding characteristics for the receptors in humans and mice. The low sequence identity observed between IL-13Rα1 and IL-13Rα2 (~20%), however, suggests different roles for each receptor.
Figure 1. Comparison of primary sequences and structural homology between IL-13Rα1 and IL-13Rα2.
(A) ClustalW multiple sequence alignment was performed between IL-13Rα1 and IL-13Rα2 sequences from human (hIL-13Rα1 and hIL-13Rα2) and mouse (mIL-13Rα1 and mIL-13Rα2). Residue backgrounds are highlighted as follows: blue, identical across sequences; magenta, similar across sequences. A consensus sequence is indicated at the bottom of the panel. The residue numbers for each sequence are indicated on the right side of the panel. Predicted β-sheets are indicated on top and bottom of aligned sequences for IL-13Rα2 and IL-13Rα1, respectively. (B) Ribbon diagram of mIL-13Rα1 and mIL-13Rα2 with domains colored. (C) Electrostatic potential at IL-13 binding surface of mIL-13Rα1 and mIL-13Rα2 (blue, positive; red, negative; white, neutral).
The sequence alignments of IL-13Rα1 and IL-13Rα2 suggest that differences in the secondary structure, particularly several loop regions, could cause these receptors to bind IL-13 with different affinity, specificity, or both. For example, in contrast to IL-13Rα1, the D1 domain of mouse and human IL-13Rα2 contains a 10-residue insertion (NLIYKDGFDL82–91) that appears to modulate receptor conformation and/or flexibility as this sequence is not in direct contact with IL-13 yet it strongly regulates its affinity for substrate. Similarly, a novel 13 residue insertion in domain D3 (GVLADAVYTVRVR298–310, mouse IL-13Rα1 sequence) is located outside the IL-13 binding interface in both human and mouse IL-13Rα1, however, it is not observed in IL-13Rα2.
Since no structure is available for mouse IL-13Rα1 or IL-13Rα2, we modeled these structures based on the crystal structure of human IL-13Rα1 (PDB ID 3BPN)5 and human IL-13Rα2, respectively (PDB ID 3LB6)12. Likewise, no structure is currently available for mouse IL-13 and this was modeled using human IL-13 (PDB ID 3BPO chain A) as a template5. A secondary structure comparison was made between mouse IL-13Rα1 and mouse IL-13Rα2 and the differences noted (Figure 1A). Overall, both structures consist of an S-type Ig fold domain attached to two tandem Fibronectin-III-like domains with a WSXWS motif that together form the familiar, elbow-shaped cytokine binding region (CHR)23–24. The secondary structures of each domain (D1, D2, and D3) were similar between the models with each domain consisting of only β-strands (Figure 1) The relatively few amino acid changes coupled with a strong similarity in overall secondary and tertiary structure is analogous to the observations made between the human IL-13Rα1 and IL-13Rα2 receptors12.
It is likely that only a handful of residue changes lead to differences in affinity and specificity. The effects of amino acid composition differences between the receptors are better gauged by examining the differences in electrostatic potential across the domains, in particular at their IL-13 binding interface (Figure 1C). One key electrostatic difference is in the domain D1 c′ strand at positions Asp73, Lys74, and Lys75 (IL-13Rα1 numbering) where the analogous positions in IL-13Rα2 are Lys76, Thr77, and Ile78. Another difference occurs in the neighboring domain D1 c strand at position Arg63 wherein IL-13Rα2, there is a glutamate (Glu62). Another difference in charge occurs in domain D3 where Arg251 is replaced with a proline (Pro250). Both structures have a conserved arginine at position 253 (262 in IL-13Rα2). The addition of Arg251 next to Arg253 strengthens the overall positive character of this region.
Molecular docking studies between IL-13 and its binding receptors
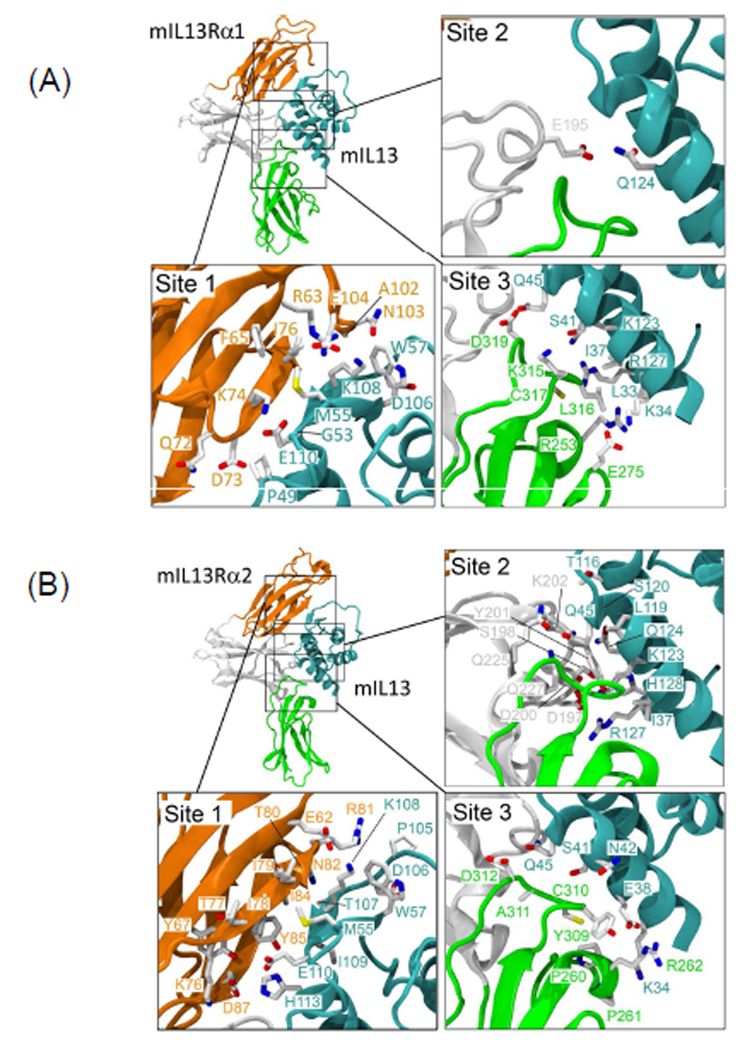
We further analyzed IL-13 and its receptor interactions using RosettaDock and obtained lowest energy structures for both the IL-13Rα1/IL-13 and IL-13Rα2/IL-13 complexes (Figure 2). The lowest energy docking solution for mouse IL-13 and IL-13α1 corresponded nicely to the human IL-13/IL-13α1 crystal structure (rmsd = 1.45Å using backbone atoms). Mirroring the methods used for the IL-13/IL-13Rα1 docking solution, we chose the lowest energy solution for IL-13/IL-13Rα2. Like IL-13/IL-13Rα1, this docking solution exhibited only a modest deviation from the human IL-13/IL-13Rα2 crystal structure. Overall the IL-13 footprint for the mouse receptors was very similar to their human counterparts. Both mouse receptors appeared to bind to IL-13 largely through their D1 and D3 domains like the configurations seen in the human crystal structures.
Figure 2. Computational structure prediction of mouse IL-13Rα1/IL-13 and IL-13Rα2/IL-13.
(A) Low-energy complex for mouse IL-13Rα1/IL-13 generated by RosettaDock. Enlarged view for the interactions of domain D1 (site 1, orange), domain D2 (site2, white), domain D3 (site3, green) of mouse IL-13Rα1 with IL-13 (Cyan) (B) Low-energy complex for mouse IL-13Rα2/IL-13 generated by RosettaDock. Enlarged view for the interactions of domain D1 (site 1, orange), domain D2 (site2, white), domain D3 (site3, green) of mouse IL-13Rα2 with IL-13 (Cyan)
For IL-13Rα1, direct interactions with IL-13 were contributed by side chains and also main-chain amide groups of the D1 c′ strand (Lys74 and Ile76) (Figure 2A and Supplementary Table 1). This is in agreement with the reported crystal structure for hIL-13Rα1. Deletion of the above three residues resulted in the loss of IL-13 binding10. As shown in Figure 2A and Table 1, Ile79, Lys108 and Glu110 of mIL-13 are directly involved in the interaction with the mIL-13Rα1 c′ strand. Similarly, domain D1 residues Glu62, Lys76, Ile78, Ile79, Thr80, Arg81, Asn82, Ile84, Tyr85 and Asp87 of mouse IL-13Rα2 interact directly with IL-13 (Figure 2B and Supplementary table 2). Interestingly, four of these residues lie within the ten residue insertion NLIYKDGFDL82–91 suggesting that this sequence plays a role in determining affinity. In addition, several other residues from domain D1 in IL-13Rα1 participate in binding to IL-13, namely Lys66 (c strand) and Glu105 and His106 (loop between f and g strands).
Docking solutions of IL-13/IL-13Rα1 revealed the Lys189-Asn198 region (domain D2 loop between strands e-f) as “helix-like.” This region was not solved in the crystal and in our model makes minimal contact with IL-13 through residue Glu195, forming an H-bond acceptor-donor pair with Gln124 of IL-13. We found that when other conformations for this region were used in docking, such as an extended loop, IL-13 would dock away from the expected binding site. The presence of a helix-like structure is also seen in the human IL-13Rα2 crystal structure and was incorporated into our model. The analogous residues in mouse IL-13Rα2 (positions 194–203) also form a helix-like conformation during docking. Like the IL-13Rα1 complex, IL-13 contacts with residues Asp197, Ser198 and Asp200 from IL-13Rα2. Like domain D1, the D3 domain plays an important role in IL-13 binding. In the IL-13/IL-13Rα1 model, IL-13 contacts receptor residues Arg253, Glu275, Lys315, Leu316, Cys317, and Asp319, making a combination of electrostatic and hydrophobic interactions. These residues lie in loop regions between the individual β-strands of domain D3. Analogously, IL-13 contacts IL-13Rα2 receptor residues Arg262, Tyr309, Cys310, Ala311 and Asp312 which are also located in loop regions. In both cases, these residues are located between β-strands b-c and f-g. Overall, there are differences in electrostatic interactions within D3, but the interactions with residues in domain D1, especially with the c′ strand, are likely modulating binding affinity more than other regions.
High-density scanning peptide array analysis
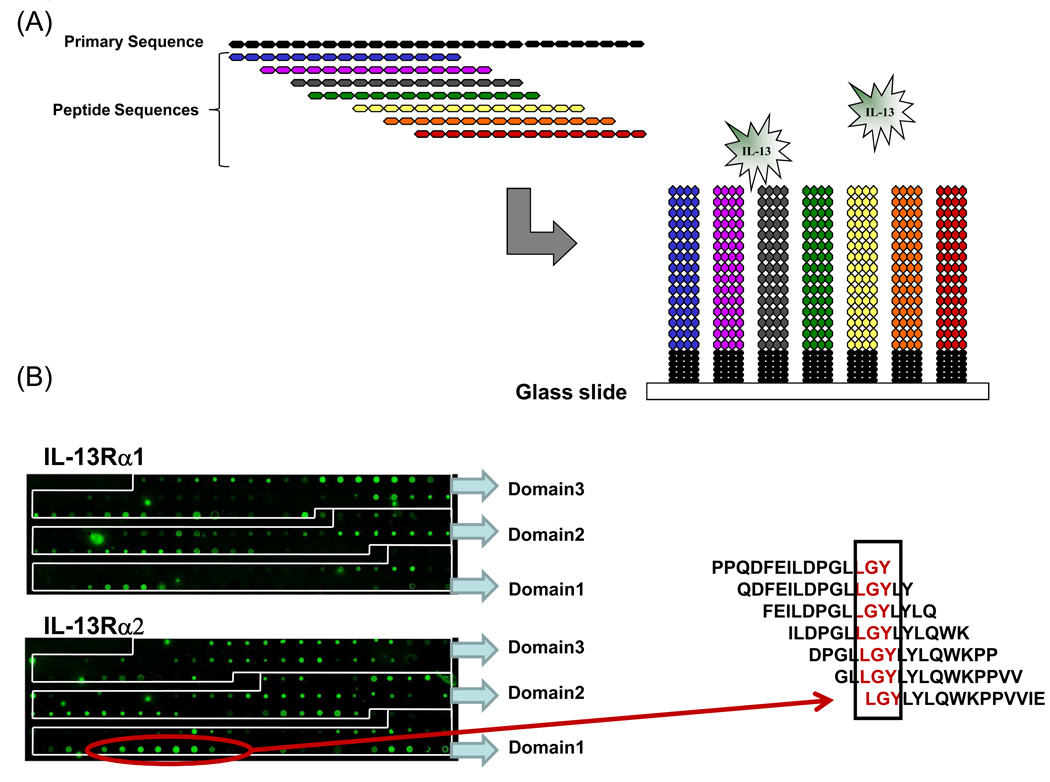
We next used peptide array technology to further interrogate the molecular docking studies and define molecular determinants at their binding interfaces. For peptide arrays, we synthesized libraries of overlapping 15-mer peptides, each shifted by three amino acids, spanning the complete sequence of the extracellular domains of mouse IL-13Rα1 and IL-13Rα2 (Figure 3A). The peptides were printed on a glass surface at a high density, providing a highly effective method to detect and map binding interactions between IL-13 and its receptor sequences. IL-13 receptor peptides and control spots were printed in triplicate on the slide (data not shown). Sequences for controls peptides were derived from immunoglobulin heavy chains of mouse, goat and human and these peptides were hydrophobic, hydrophilic and amphipathic in nature. Fluorescence intensity signal from these control peptide spots due to IL-13 binding was below background signal (data not shown). With these arrays, binding patterns were highly reproducible between two independent experiments and we observed low background signals, supporting previous validations of this technology18. As shown in figure 3B, several positive spots were identified in the array, revealing potential IL-13/ receptor sequence interactions.
Figure 3. Probing IL-13 binding sequences on mIL-13Rα1 and mIL-13Rα2 using peptide arrays.
(A) Schematic representation of overlapping peptide array design. (B) Captured images of fluorescence signals from IL-13 bound to mIL-13Rα1 and mIL-13Rα2 specific peptide microarray slides. The red arrow illustrates a positive signal on the mouse IL-13Rα2 array and the corresponding alignment to the right, which identifies the critical residues within the binding sequence.
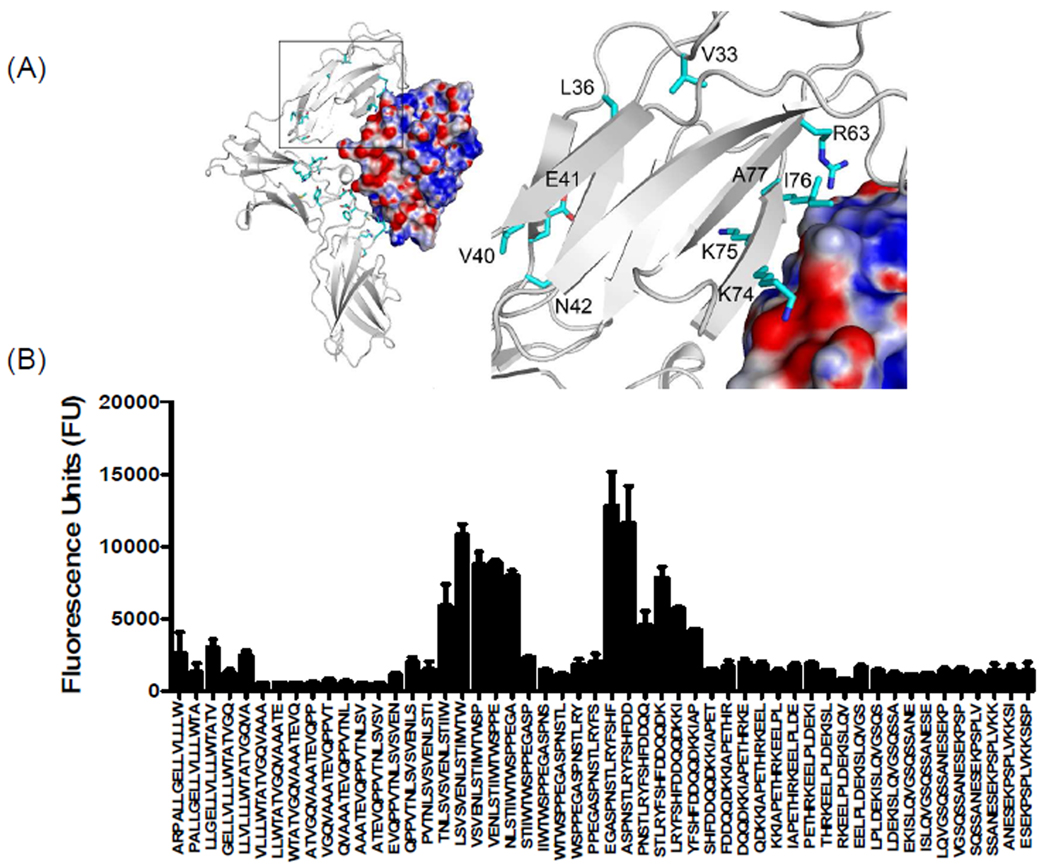
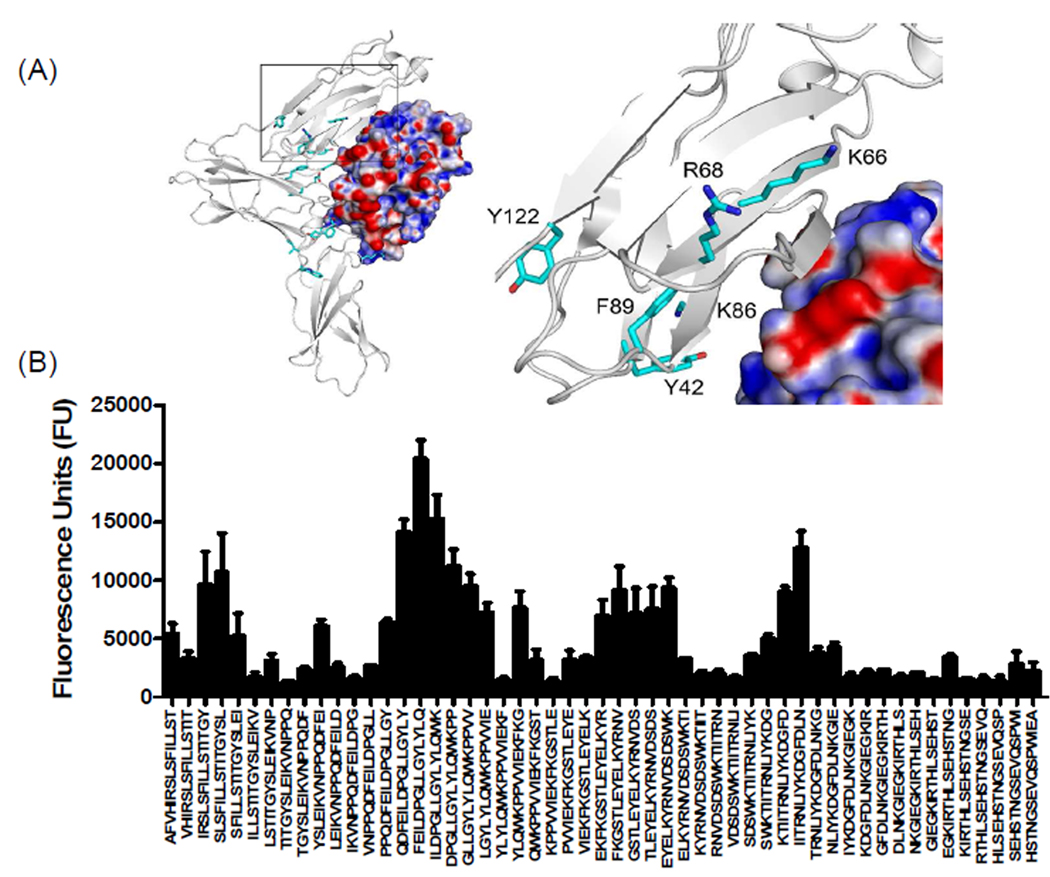
As shown in figure 4B, we observed two dominant binding regions in domain D1 of mouse IL-13Rα1. One of the binding regions was found in the c′ strand of the D1 domain of IL-13Rα1, agreeing with the molecular docking studies (Figure 4A). This data also agreed nicely with the recent crystal structure of human IL-13Rα1 complex5. The importance of the c′ strand of the D1 domain of IL-13Rα1 is further supported by mutational analyses that suggest three amino acids, Lys76, Lys77 and Ile78 contribute to the binding of IL-1310. Data obtained from our peptide arrays and molecular docking studies suggest that similar conserved hydrophobic interactions are involved in the binding of mouse IL-13 to IL-13Rα1. Ito and colleagues showed that N-terminal deletion or Ala-substitutions at Val35, Leu38 and Val42 of the N-terminal D1 domain of human IL-13Rα1 results in a loss of IL-13 binding10. Because these residues are buried between β-sheets, we speculate that these residues help stabilize the domain rather than mediating the direct binding of IL-13. However, data from our peptide array analysis revealed a direct interaction between mIL-13Rα1 sequence LSVSVENLS36–44 and IL-13. In addition, residues Val33, Leu36 and Val40 of mIL-13Rα1 and the analogous positions in hIL-13Rα1 (Val35, Leu38 and Val42) are absent in IL-13Rα2 (Figure 1A). The IL-13 binding signal observed on our peptide array corresponding to the N-terminal D1 domain of mouse IL-13Rα2 suggested that a large hydrophobic region LGYLY42–46 contributes to IL-13 binding (Figure 5). A binding region contributed by the c and c′ strand of mouse IL-13Rα2, in particular the Lys66 residue of c strand, contacts the Pro 105 present on the CD loop of IL-13 through a van der Waals interaction (Figure 5 and Supplementary table 2). The IL-13 binding sequence NLIYKDGFDL82–91 found on IL-13α2 also provides a unique IL-13 binding region that is absent in IL-13Rα1 (Figure 1A and 5B). This IL-13Rα2 site is highly conserved between human and mouse.
Figure 4. Characterization of binding interactions between mouse IL-13Rα1 domain D1 and IL-13 using peptide array analysis.
(A) Structure of the complex and enlarged view showing the interactions between domain D1 of mIL-13Rα1 (white) and IL-13. The electrostatic potential at the molecular surface of IL-13 is shown (blue, positive; red, negative; white, neutral) and residues identified using peptide arrays are highlighted on domain D1 of mouse IL-13Rα1. (B) The bar chart represents fluorescence reference units corresponding to overlapping peptide sequences from domain D1 of mIL-13Rα1.
Figure 5. Characterization of binding interactions between mouse IL-13Rα2 domain D1 and IL-13 using peptide array analysis.
(A) Structure of the complex and enlarged view showing the interactions between domain D1 of mIL-13Rα2 (white) and IL-13. The electrostatic potential at the molecular surface of IL-13 is shown (blue, positive; red, negative; white, neutral) and residues identified using peptide arrays are highlighted on domain D1 of mouse IL-13Rα2. (B) The bar chart represents fluorescence reference units corresponding to overlapping peptide sequences from domain D1 of mIL-13Rα2.
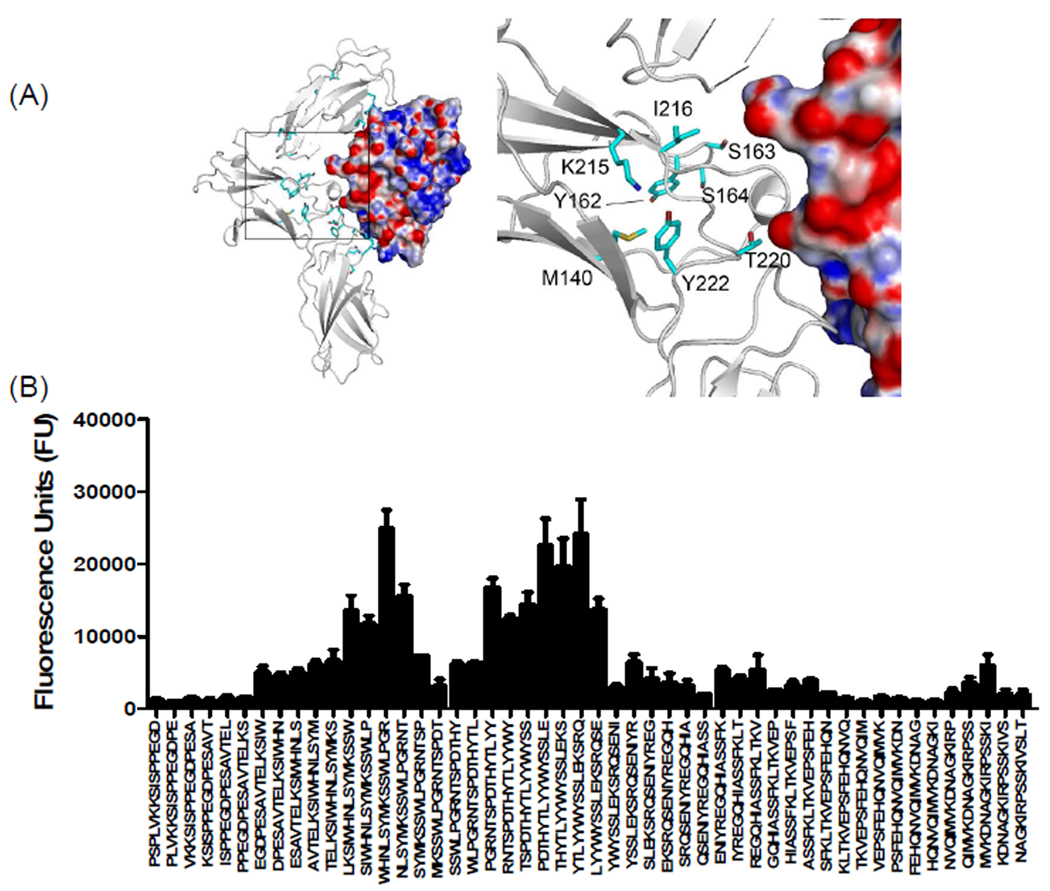
Peptide array analysis of domain D2 of mIL-13Rα1 revealed two distinct binding regions that were not evident in our docking studies or in the reported crystal structure of human IL-13Rα1 complex. These binding regions (SYMKCSW138–144 and YTLYYW156–161) are highly conserved and the observed binding signature was similar between IL-13Rα1 and IL-13Rα2 (Figure 6 and 7). Although, we have no additional evidence, it is tempting to speculate that this unique binding site directly interacts with IL-13. The peptide arrays also revealed a novel binding site on mIL-13Rα2 in sequence SDYKDFFIC199–207 (Figure 7). This sequence is also highly conserved in hIL-13Rα2 but is not found in mouse or human IL-13Rα1 (Figure 1A and 6). This sequence may be important, since replacing Tyr207 with Ala in loop EF of hIL-13Rα2 resulted in a significant loss of hIL-13 binding25.
Figure 6. Characterization of binding interactions between mouse IL-13Rα1 domain D2 and IL-13 using peptide array analysis.
(A) Structure of the complex and enlarged view showing the interactions between domain D2 of mIL-13Rα1 (white) and IL-13. The electrostatic potential at the molecular surface of IL-13 is shown (blue, positive; red, negative; white, neutral) and residues identified using peptide arrays are highlighted on domain D2 of mouse IL-13Rα1. (B) The bar chart represents fluorescence reference units corresponding to overlapping peptide sequences from domain D2 of mIL-13Rα1.
Figure 7. Characterization of binding interactions between mouse IL-13Rα2 domain D2 and IL-13 using peptide array analysis.
(A) Structure of the complex and enlarged view showing the interactions between domain D2 of mIL-13Rα2 (white) and IL-13. The electrostatic potential at the molecular surface of IL-13 is shown (blue, positive; red, negative; white, neutral) and residues identified using peptide arrays are highlighted on domain D2 of mouse IL-13Rα2. (B) The bar chart represents fluorescence reference units corresponding to overlapping peptide sequences from domain D2 of mIL-13Rα2.
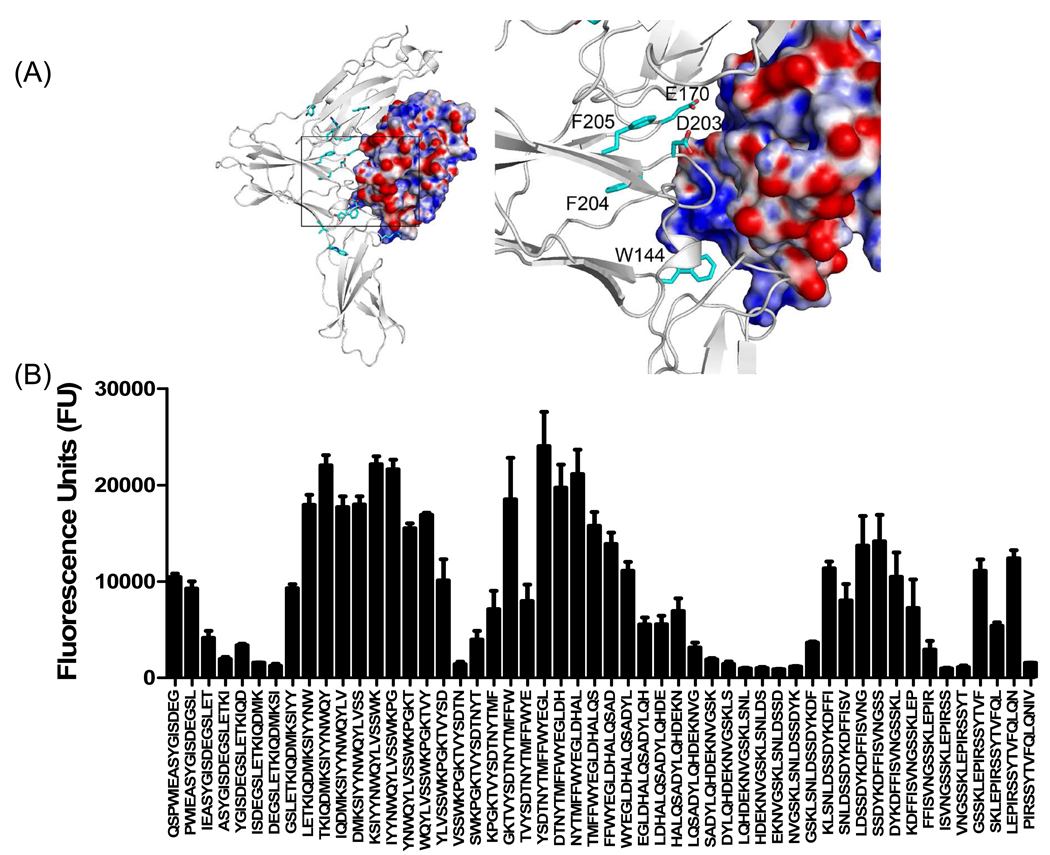
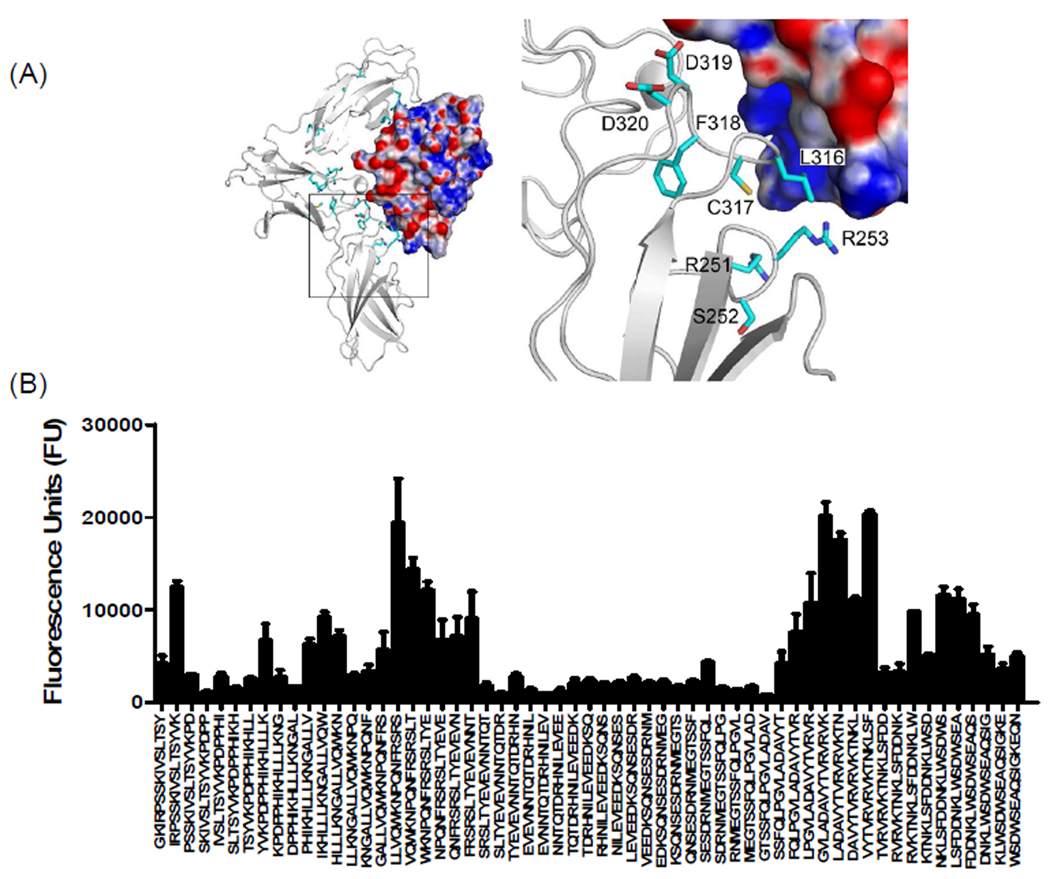
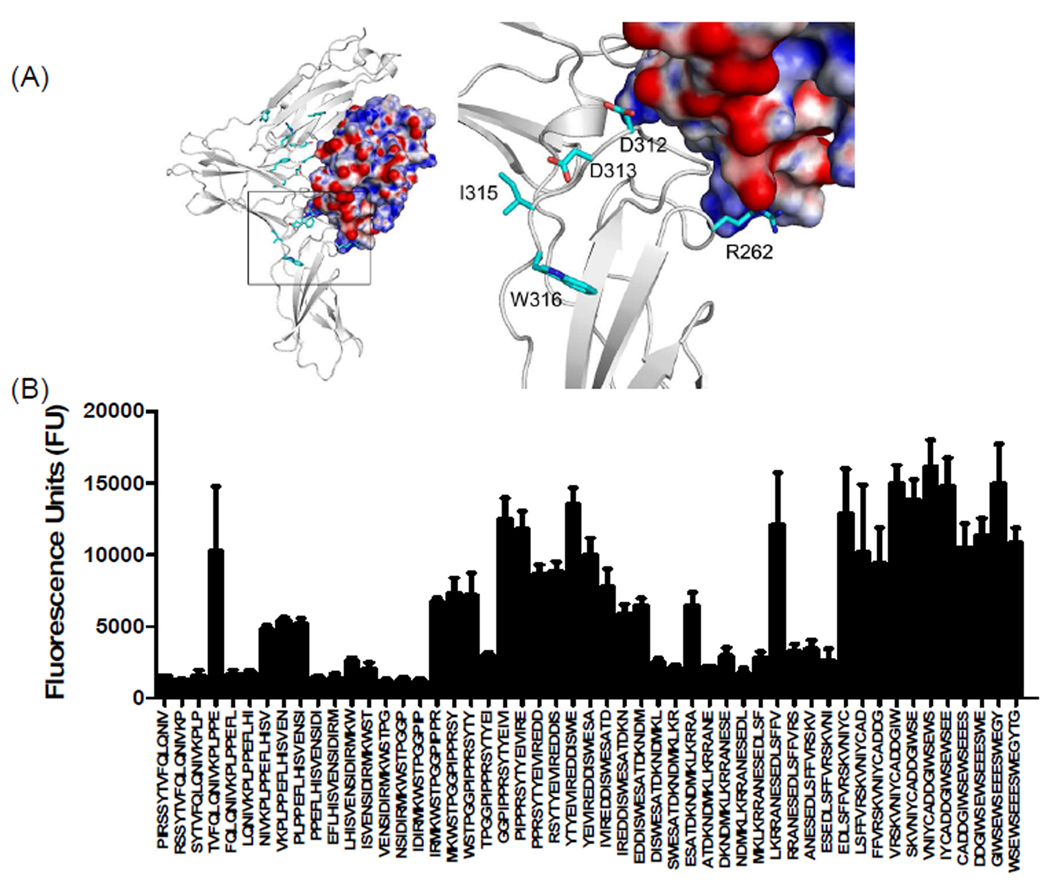
Molecular docking studies and peptide array analysis of domain D3 of mIL-13Rα1 are also in agreement with the recently reported crystal structure of hIL-13Rα1/IL-13 complex. In particular, residues in the BC loop of domain D3 (FRSR250–253) and FG loop (KLCFDD315–321) provide a landing pad for IL-13 that involves both hydrophobic and electrostatic interactions with residues found in helix A and helix D of IL-13 (Figure 8, 9 and supplementary table 2). The overall importance of the FG loops of IL-13Rα1 and IL-13Rα2 in IL-13 binding is further supported by prior site-directed mutagenesis studies. Replacement of Asp318 with Gln in IL-13Rα2 or Asp324 of IL-13Rα1 with Ala resulted in a significant loss of IL-13 binding25. An additional novel binding region in mIL-13Rα1, TVRVRV306–311 was also discovered with our peptide array analysis, suggesting this region plays an important role in IL-13 binding. Notably, this sequence is not present in IL-13Rα2 (Figure 1A and 9).
Figure 8. Characterization of binding interactions between mouse IL-13Rα1 domain D3 and IL-13 using peptide array analysis.
(A) Structure of the complex and enlarged view showing the interactions between domain D3 of mIL-13Rα1 (white) and IL-13. The electrostatic potential at the molecular surface of IL-13 is shown (blue, positive; red, negative; white, neutral) and residues identified using peptide arrays are highlighted on domain D3 of mouse IL-13Rα1. (B) The bar chart represents fluorescence reference units corresponding to overlapping peptide sequences from domain D3 of mIL-13Rα1.
Figure 9. Characterization of binding interactions between mouse IL-13Rα2 domain D3 and IL-13 using peptide array analysis.
(A) Structure of the complex and enlarged view showing the interactions between domain D3 of mIL-13Rα2 (white) and IL-13. The electrostatic potential at the molecular surface of IL-13 is shown (blue, positive; red, negative; white, neutral) and residues identified using peptide arrays are highlighted on domain D3 of mouse IL-13Rα2. (B) The bar chart represents fluorescence reference units corresponding to overlapping peptide sequences from domain D3 of mIL-13Rα2.
Generation of receptor specific monoclonal antibodies
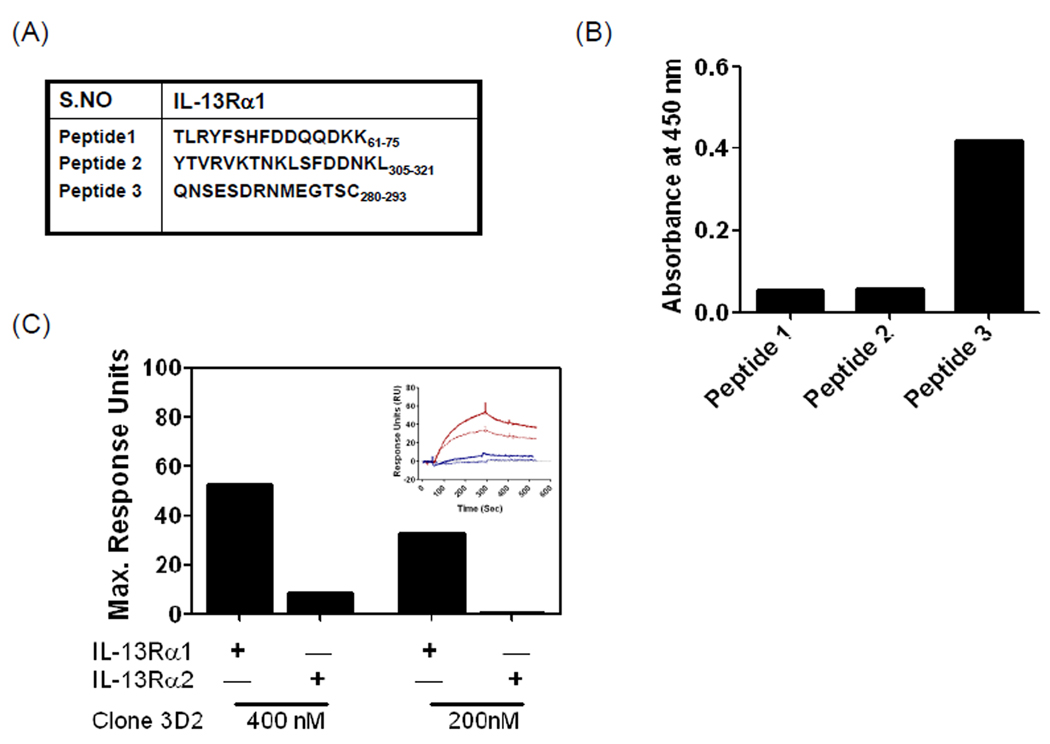
The above binding data were used to design three different peptides unique to IL-13Rα1 that are believed to fall in the region critical for IL-13 binding (Figure 10A). Mice were immunized with the peptides and monoclonal hybridomas were generated and selected based on their reactivity to the above peptides and IL-13Rα1 extracellular domain (data not shown). As shown in figure 10B, culture supernatant from clone 3D2 showed positive reactivity to peptide 3 that has a sequence derived from domain D3 of IL-13Rα1. Binding studies were conducted using surface plasmon resonance to compare the specificity of antibody 3D2. As shown in figure 10C, antibody 3D2 displayed a dose dependent increase in binding to IL-13Rα1. However, no significant binding was observed between 3D2 and IL-13Rα2 (Figure 10C). Thus, these data show the utility of using peptide arrays to generate highly specific monoclonal antibodies. Currently, studies are in progress to improve the binding affinity of the antibodies.
Figure 10. Generation of mouse IL-13Rα1 specific antibodies.
(A) Three unique peptide sequences of mIL-13Rα1 were conjugated to KLH and mice were immunized with the peptides to generate peptide specific monoclonal antibodies. (B) ELISA absorbance values for the three peptides with culture supernatant obtained from hybridoma 3D2, showing specificity for peptide 3. (C) Bar graph showing maximum resonance units due to the binding of 400 nM or 200 nM of 3D2 antibody to mIL-13Rα1-Fc and mIL-13Rα2-Fc at room temperature. The inserted figure shows SPR sensograms for the binding of 400 nM (solid line) or 200 nM (dashed line) of 3D2 antibody to mIL-13Rα1-Fc (red) and mIL-13Rα2-Fc (blue).
In summary, peptide arrays and molecular docking studies identified IL-13 binding sequences on IL-13Rα1 and IL-13Rα2. The present findings support the recent crystal structures of the human receptor equivalents5,10,12,25. Use of peptide arrays revealed the binding elements that are distinct or similar between IL-13 binding receptors. These sequences will be useful in the future to generate receptor specific agonists or antagonists. They will also allow us to design molecules that can result in the therapeutic intervention of distinct IL-13 functions in vivo. This study clearly shows the utility of peptide arrays in mapping protein-protein interactions and may provide a more rapid, alternative, or complimentary approach to crystallography.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by the Intramural Research Program of the NIH, NIAID. We would like to thank Dr. Larry M. Lantz, NIAID Custom Antibodies Core, Flow Cytometry Section, Research Technologies Branch, NIAID, for his help with the purification of antibodies.
REFERENCES
- 1.Wynn TA. IL-13 effector functions. Annu Rev Immunol. 2003;21:425–456. doi: 10.1146/annurev.immunol.21.120601.141142. [DOI] [PubMed] [Google Scholar]
- 2.Wills-Karp M. Interleukin-13 in asthma pathogenesis. Immunol Rev. 2004;202:175–190. doi: 10.1111/j.0105-2896.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- 3.Wilson MS, Wynn TA. Pulmonary fibrosis: pathogenesis, etiology and regulation. Mucosal Immunol. 2009;2(2):103–121. doi: 10.1038/mi.2008.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beghe B, Hall IP, Parker SG, Moffatt MF, Wardlaw A, Connolly MJ, Fabbri LM, Ruse C, Sayers I. Polymorphisms in IL13 pathway genes in asthma and chronic obstructive pulmonary disease. Allergy. 2009 doi: 10.1111/j.1398-9995.2009.02167.x. [DOI] [PubMed] [Google Scholar]
- 5.LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, Keegan AD, Garcia KC. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008;132(2):259–272. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, Donaldson DD. Interleukin-13: central mediator of allergic asthma. Science. 1998;282(5397):2258–2261. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 7.Ramalingam TR, Pesce JT, Sheikh F, Cheever AW, Mentink-Kane MM, Wilson MS, Stevens S, Valenzuela DM, Murphy AJ, Yancopoulos GD, Urban JF, Jr, Donnelly RP, Wynn TA. Unique functions of the type II interleukin 4 receptor identified in mice lacking the interleukin 13 receptor alpha1 chain. Nat Immunol. 2008;9(1):25–33. doi: 10.1038/ni1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morimoto M, Zhao A, Sun R, Stiltz J, Madden KB, Mentink-Kane M, Ramalingam T, Wynn TA, Urban JF, Jr, Shea-Donohue T. IL-13 receptor alpha2 regulates the immune and functional response to Nippostrongylus brasiliensis infection. J Immunol. 2009;183(3):1934–1939. doi: 10.4049/jimmunol.0804299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grunig G, Warnock M, Wakil AE, Venkayya R, Brombacher F, Rennick DM, Sheppard D, Mohrs M, Donaldson DD, Locksley RM, Corry DB. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282(5397):2261–2263. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito T, Suzuki S, Kanaji S, Shiraishi H, Ohta S, Arima K, Tanaka G, Tamada T, Honjo E, Garcia KC, Kuroki R, Izuhara K. Distinct structural requirements for interleukin-4 (IL-4) and IL-13 binding to the shared IL-13 receptor facilitate cellular tuning of cytokine responsiveness. J Biol Chem. 2009;284(36):24289–24296. doi: 10.1074/jbc.M109.007286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiaramonte MG, Donaldson DD, Cheever AW, Wynn TA. An IL-13 inhibitor blocks the development of hepatic fibrosis during a T-helper type 2-dominated inflammatory response. J Clin Invest. 1999;104(6):777–785. doi: 10.1172/JCI7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lupardus PJ, Birnbaum ME, Garcia KC. Molecular basis for shared cytokine recognition revealed in the structure of an unusually high affinity complex between IL-13 and IL-13Ralpha2. Structure. 18(3):332–342. doi: 10.1016/j.str.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrews AL, Holloway JW, Puddicombe SM, Holgate ST, Davies DE. Kinetic analysis of the interleukin-13 receptor complex. J Biol Chem. 2002;277(48):46073–46078. doi: 10.1074/jbc.M209560200. [DOI] [PubMed] [Google Scholar]
- 14.Mentink-Kane MM, Wynn TA. Opposing roles for IL-13 and IL-13 receptor alpha 2 in health and disease. Immunol Rev. 2004;202:191–202. doi: 10.1111/j.0105-2896.2004.00210.x. [DOI] [PubMed] [Google Scholar]
- 15.Fichtner-Feigl S, Strober W, Kawakami K, Puri RK, Kitani A. IL-13 signaling through the IL-13alpha2 receptor is involved in induction of TGF-beta1 production and fibrosis. Nat Med. 2006;12(1):99–106. doi: 10.1038/nm1332. [DOI] [PubMed] [Google Scholar]
- 16.Andrews AL, Nasir T, Bucchieri F, Holloway JW, Holgate ST, Davies DE. IL-13 receptor alpha 2: a regulator of IL-13 and IL-4 signal transduction in primary human fibroblasts. J Allergy Clin Immunol. 2006;118(4):858–865. doi: 10.1016/j.jaci.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 17.Wilson MS, Elnekave E, Mentink-Kane MM, Hodges MG, Pesce JT, Ramalingam TR, Thompson RW, Kamanaka M, Flavell RA, Keane-Myers A, Cheever AW, Wynn TA. IL-13Ralpha2 and IL-10 coordinately suppress airway inflammation, airway-hyperreactivity, and fibrosis in mice. J Clin Invest. 2007;117(10):2941–2951. doi: 10.1172/JCI31546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nahtman T, Jernberg A, Mahdavifar S, Zerweck J, Schutkowski M, Maeurer M, Reilly M. Validation of peptide epitope microarray experiments and extraction of quality data. J Immunol Methods. 2007;328(1–2):1–13. doi: 10.1016/j.jim.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 19.Wu S, Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35(10):3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231(4):1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 21.Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. J Mol Biol. 2007;373(2):503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 22.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bazan JF. Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci U S A. 1990;87(18):6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Vos AM, Ultsch M, Kossiakoff AA. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science. 1992;225(5042):306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 25.Arima K, Sato K, Tanaka G, Kanaji S, Terada T, Honjo E, Kuroki R, Matsuo Y, Izuhara K. Characterization of the interaction between interleukin-13 and interleukin-13 receptors. J Biol Chem. 2005;280(26):24915–24922. doi: 10.1074/jbc.M502571200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.