Abstract
The BUZ/Znf-UBP domain is a protein module found in the cytoplasmic deacetylase HDAC6, the E3 ubiquitin ligase BRAP2/IMP, and a subfamily of ubiquitin-specific proteases. Although several BUZ domains have been shown to bind ubiquitin with high affinity by recognizing its C-terminal sequence (RLRGG-COOH), it is currently unknown whether the interaction is sequence specific or whether the BUZ domains are capable of binding to proteins other than ubiquitin. In this work, the BUZ domains of HDAC6 and Ubp-M were subjected to screening against a one-bead-one-compound (OBOC) peptide library which displayed random peptide sequences with free C-termini. Sequence analysis of the selected binding peptides as well as alanine scanning studies revealed that the BUZ domains require a C-terminal Gly-Gly motif for binding. At the more N-terminal positions, the two BUZ domains have distinct sequence specificities, allowing them to bind to different peptides/proteins. A database search of the human proteome on the basis of the BUZ domain specificities identified 12 and 22 potential partner proteins for Ubp-M and HDAC6 BUZ domains, respectively. Peptides corresponding to the C-terminal sequences of four of the predicted binding partners (FBXO11, Histone H4, PTOV1, and FAT10) were synthesized and tested for binding to the BUZ domains by fluorescence polarization. All four peptides bound to the HDAC6 BUZ domain with low μM KD values and less tightly to the Ubp-M BUZ domain. Finally, in vitro pull-down assays showed that the Ubp-M BUZ domain was capable of binding to the histone H3-H4 tetramer protein complex. Our results suggest that BUZ domains are sequence-specific protein-binding modules, with each BUZ domain potentially binding to a different subset of proteins.
The conjugation of ubiquitin, a highly conserved 76-amino acid protein, to proteins is a key signaling event in cellular processes such as the proteolysis of cellular proteins (1), cell cycle control (2), and transcriptional regulation (3, 4). Several proteins involved in these processes need to specifically recognize ubiquitin to perform their function, such as deubiquitylating enzymes (DUBs) (5, 6), certain trafficking proteins (7, 8), and ubiquitin conjugating enzymes (9). In order to recognize ubiquitin, many proteins utilize ubiquitin-binding domains, such as UBA, UIM, MIU, DUIM, CUE, GAT, NZF, A20 ZnF, BUZ, UBZ, Ubc, UEV, UBM, GLUE, Jab1/MPN, and PFU domains (10). Most of these domains bind ubiquitin at a hydrophobic patch surrounding Ile-44 of ubiquitin (11, 12). The BUZ domain, however, has been found to interact with ubiquitin by binding to its free C-terminus (13, 14).
The BUZ domain, which is also known as the Znf-UBP (zinc finger-ubiquitin-specific processing protease) domain, DAUP (deacetylase/ubiquitin-specific protease) domain, or PAZ (polyubiquitin-associated zinc finger) domain, consists of approximately 100 residues in length and has the general structure of being organized around a central 5-stranded twisted β-sheet with a nearby α-helix (13–15). BUZ domains require one to three zinc ions to maintain structural integrity (13, 14, 16). The BUZ domain is present in at least 10 human ubiquitin-specific processing proteases (USPs) (6), which are the largest class of deubiquitylating enzymes. It is also found in BRCA1-associated protein 2 (BRAP2) (17), an E3 ubiquitin ligase, and in the cytoplasmic histone deacetylase 6 (HDAC6) (18, 19). Although the precise roles of BUZ domains in these proteins are not completely understood, it appears that one major function is to regulate the activity of BUZ-containing proteins by binding to the C-terminus of ubiquitin (6). In the case of BUZ-containing USPs, binding of ubiquitin has been shown to increase their catalytic activity (14, 20, 21), while binding of ubiquitin by the BUZ domain of HDAC6 may prevent HDAC6 from transporting polyubiquitinated proteins to aggresomes (8, 13).
Recent biochemical and structural analysis of the isopeptidase T (isoT) BUZ domain revealed a distinct ubiquitin-binding mode, in which the C-terminal RGG motif of ubiquitin inserts in to a deep pocket on the BUZ domain (14). The more N-terminal residues including Arg-72 and Leu-73 also make specific contacts with the BUZ domain, likely contributing to both specificity and overall affinity. In addition, in the case of isoT, Phe-224 form the L2A loop of the BUZ domain interacts with a small hydrophobic patch on the ubiquitin surface (formed by Leu-8 and Ile-36). Consistent with this observation, any modification that disrupts the free C-terminal Gly-Gly motif abolished the binding of the ubiquitin peptide to BUZ domains (13). Since ubiquitin is the only currently known ligand of BUZ domains, it is not clear whether the BUZ domains can bind to other peptides/proteins. One approach to addressing this question is to screen the BUZ domains against a combinatorial peptide library (22). We have previously developed a methodology for synthesizing and screening one-bead-one-compound (OBOC) peptide libraries that display support-bound peptides with free C-termini and applied it to profiling the sequence specificity of postsynaptic density-95/discs large/zona occluden-1 (PDZ) domains (23). Herein, we extended this approach to systematically profile the sequence specificity of the BUZ domains of mutant ubiquitin-processing protease (Ubp-M, also known as USP16) and HDAC6. Our results demonstrate that the BUZ domain is a sequence-specific protein-binding module, with each domain recognizing a specific subset of C-terminal sequences. This suggests that the BUZ domains may bind to other cellular proteins in addition to ubiquitin.
MATERIALS AND METHODS
Materials
Fmoc L-amino acids and coupling reagents for peptide synthesis were purchased from Advanced ChemTech (Louisville, KY) and NovaBiochem (La Jolla, CA). TentaGel resin was purchased from Peptides International (Louisville, KY), while Wang resin was from Advanced ChemTech. Streptavidin-alkaline phosphatase (SA-AP) conjugate was purchased from Prozyme (San Leandro, CA). Phenyl isothiocyanate (PITC) was purchased in 1-mL sealed ampoules from Sigma (St. Louis, MO). 5-Bromo-4-chloro-3-indolyl phosphate (BCIP) was from Sigma. Boc-Glu(OFm)-OH was purchased from Chem-Impex International Inc. (Wood Dale, IL). Thrombin (from bovine plasma) was purchased from Fisher Scientific (Pittsburgh, PA). DNA plasmids for GST-Ubp-M BUZ (BUZ residues 10–143) and GST-HDAC6 BUZ (residues 1059–1215) (both pGEX-2T vectors) and (His)6-tagged Ubp-M BUZ (in pET-15b vector, residues 22–143) have previously been described (13).
Purification and Labeling of BUZ Domains
E. coli Rosetta BL21(DE3) cells were transformed with either GST-HDAC6 BUZ, GST-Ubp-M BUZ, or (His)6-Ubp-M BUZ plasmids and grown in Luria-Bertani media (containing 500 μM ZnSO4) at 37 °C until OD600 reached 0.6. For the production of GST-Ubp-M BUZ fusion protein, the cells were induced by addition of 90 μM isopropyl-β-D-thiogalactoside (IPTG) for 5 h at 30 °C. For (His)6-Ubp-M BUZ and GST-HDAC6 BUZ proteins, the cells were induced with 200 μM IPTG for 15 h at 20 °C. The cells were collected by centrifugation at 5000 RPM for 20 min in a Sorvall RC-5C Plus rotor and lysed by sonication in either 50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 5 mM imidazole [for (His)6-Ubp-M BUZ] or 20 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM β-mercaptoethanol (for GST fusion proteins) containing protease inhibitors phenylmethylsulfonyl fluoride (35 mg/L), trypsin inhibitor (20 mg/L), and pepstatin (1 mg/L). The GST fusion proteins were purified on a glutathione-agarose column according to the manufacturer’s instructions. Free glutathione was removed by size exclusion chromatography in 30 mM HEPES, pH 7.4, 150 mM NaCl. For library screening, the GST fusion proteins (≥2 mg/mL) were biotinylated by treatment with 2 equivalents of (+)-biotin N-hydroxysuccinimide (NHS) ester (a 10 mg/mL biotin-NHS stock solution was prepared in DMSO). The pH of the reaction solution was adjusted to ~8 by the addition of 1 M NaHCO3 (pH 8.4) and the reaction was allowed to proceed for 1 h at 4°C. Any unreacted biotin-NHS was quenched by the addition of 1 M Tris buffer (pH 8.3) to a final concentration of 50 mM. Free biotin was then removed by size exclusion chromatography in 30 mM HEPES, pH 7.4, 150 mM NaCl. The protein concentration was determined by the Bradford method, using bovine serum albumin as the standard. The proteins were flash frozen in 33% glycerol using dry ice/isopropyl alcohol and stored at −80 °C. (His)6-tagged Ubp-M BUZ domain was purified by metal affinity chromatography (Ni-NTA column) and ion exchange chromatography (Q-Sepharose). For fluorescence polarization experiments, the proteins were exchanged into a buffer containing 20 mM sodium phosphate, pH 7.0, and 100 mM NaCl by size exclusion chromatography after affinity purification. For fluorescence polarization studies with the HDAC6 BUZ domain, the GST tag was removed by treatment of the fusion protein still bound to the glutathione resin with thrombin (GE Healthcare) for 16 h at 4 °C. The GST-free protein was eluted from the resin with a buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 2.5 mM CaCl2.
Synthesis of Nα-Boc-Glu(δ-N-hydroxysuccinimidyl)-O-CH2-CH=CH2
Boc-Glu(OFm)-OH (0.426 g, 1 mmol) was dissolved in 1.4 mL of DCM, followed by the addition of NaHCO3 (0.168 g, 2 mmol) and H2O (1.7 mL). Allyl bromide (0.363 g, 3 mmol) was then added at 0 °C, followed by Aliquate-336 (0.388 g, 0.96 mmol). The reaction mixture was stirred at 35 °C for 16 h. After that, the organic and aqueous phases were separated and the aqueous fraction was extracted with DCM (2 × 1 mL) and the organic fractions were combined and dried over MgSO4. The solvent was removed by evaporation under vacuum and the crude product was purified by silica gel column chromatography (2:1 hexane to ethyl acetate) to give a white solid after being dried under vacuum overnight (0.37 g, 80%). The product was dissolved in 10% (v/v) piperidine in DCM (16 mL) and stirred for 2 h at room temperature. The solvent was removed under reduced pressure. The product was dissolved in 10% NaHCO3 and extracted by diethyl ether (20 mL). The aqueous layer was then acidified to pH ~4 with 1 M HCl. The desired product was extracted with ethyl acetate (3 × 20 mL) and dried over Na2SO4. After removal of the solvent under reduced pressure, the product (0.174 g, 76.3%) was dissolved in 20 mL of DCM containing 1.2 equivalents (0.72 mmol, 0.083 g) of N-hydroxysuccinimide, and the mixture was stirred vigorously for 30 min at room temperature. Next, diisopropylcarbodiimide (0.72 mmol, 0.091 g) was added to the mixture and the solution was stirred overnight at room temperature. The solvent was removed under reduced pressure and the reaction mixture was extracted with ethyl acetate (3 × 30 mL), and water (20 mL). The organic portions were then combined and washed with brine (40 mL), and dried with Na2SO4. The solvent was evaporated under reduced pressure and the product was purified by silica gel column chromatography (1:2 hexane to ethyl acetate) to give a white solid (0.18 g, 78%). 1H NMR (250 MHz, CDCl3) δ 1.28 (s, 9H), δ 1.84–1.99 (m, 1H), δ 2.08–2.23 (m, 1H), δ 2.53–2.62 (m, 2H), δ 2.68 (s, 4H), δ 4.15–4.29 (m, 1H), δ 4.49 (d, J = 5.7 Hz, 2H), δ 5.11 (dd, J = 1.2 Hz, 10.3 Hz, 2H), δ 5.22 (d, J = 1.4 Hz, 1H), δ 5.68–5.83 (m, 1H). HRESI-MS: C17H24N2O8Na+ ([M + Na]+), calcd 407.1425, found 407.1421.
Synthesis of Peptide Library
The library was synthesized on 2.0 g of TentaGel S NH2 resin (90 μm, 0.28 mmol/g) by modifying a previously reported procedure (23). The BBLLM linker was synthesized using 4 equiv Fmoc-amino acids using HBTU/HOBt/NMM as the coupling agents. Each amino acid was coupled for 1 h, followed by exhaustive washing the resin with DMF and DCM. Before the coupling of the next amino acid, the N-terminal Fmoc group was removed by treatment of the resin with 20% piperidine in DMF (5 + 15 min), followed by exhaustive washing of the resin with DMF, DCM, and DMF again. To segregate the beads into outer and inner layers, the resin (after removing the N-terminal Fmoc group) was soaked in 75% DMF in water (5 min), 50% DMF in water (5 min), 25% DMF in water (5 min), and washed with water. The resulting resin was incubated in water overnight. The following day the water was drained and the resin was quickly resuspended in 30 mL of 55:45 (v/v) DCM/diethyl ether containing 0.5 equivalent of Nα-Boc-Glu(δ-N-hydroxysuccinimidyl)-O-CH2CH=CH2 (0.28 mmol) and incubated for 30 min on a rotary shaker. The resin was washed with 55:45 (v/v) DCM/diethyl ether (3 × 50 mL) and DMF (8 × 50 mL) and treated with 4 equiv of Fmoc-Gly-OH plus HBTU/HOBt/NMM (in 30 mL DMF) for 1 h. Next, the Nα-Boc group was removed by treatment with a solution containing 90% trifluoroacetic acid (TFA), 2.5% triisopropylsilane, 2.5% ethanedithiol, and 5% DCM for 30 min. The resin was drained and was neutralized with 10% triethylamine in DMF for 10 min. 4-Hydroxymethylphenoxyacetic acid (HMPA, 2 equiv) was then coupled to the surface layer of the beads using HBTU. The N-terminal Fmoc group of the peptides in the inner layer was removed by piperidine and Fmoc-Arg(Pbf)-OH (0.7 equiv) was coupled to the free N-terminus using HBTU/HOBt/NMM (45 min). Upon removal of the Fmoc protecting group, the resin was split into 20 equal portions (100 mg each) and each was coupled to a different Fmoc-amino acid (4 equiv). The addition of the first random residue employed diisopropylcarbodiimide (4 equiv) and 4-dimethylaminopyridine (0.1 equiv) in DCM as the coupling reagents (6 h), while the other random residues were coupled using the standard HBTU/HOBt/NMM chemistry (1 h). Each coupling reaction was repeated once to ensure complete reaction. To differentiate isobaric amino acids by MS sequencing, 5% (mol/mol) CD3CO2D was added to the coupling reactions of Leu and Lys while 5% (mol/mol) CH3CD2CO2D was added to the coupling reactions of Nle (24, 25). After synthesis of the random region of the library, an Ala-Ala dipeptide was added to the N-terminus of all peptides. The resin was treated overnight with tetrakis(triphenylphosphine)palladium (1 equiv), triphenylphosphine (3 equiv), formic acid (10 equiv), and diethylamine (10 equiv) in anhydrous THF. The resin was washed with 1% diisopropylethylamine in DMF, 1% sodium dimethyldithiocarbamate hydrate in DMF, DMF, DCM, and DMF. The Fmoc group was removed with piperidine and the resin was washed with DMF, 1 M HOBt in DMF, DMF, and DCM. The surface peptides were cyclized by incubating the resin in a solution of PyBOP/HOBt/NMM (5/5/10 equiv, respectively) in DMF for 3 h. The resin was washed with DMF and DCM and treated with 50 mL of a modified Reagent K (7.5% phenol, 5% water, 5% thioanisole, 2.5% ethanedithiol, 1% anisole in TFA) for 2 h. The resulting resin was washed with TFA and DCM, dried under vacuum, and stored at −20 °C.
Library Screening
A typical screening reaction involved 10–50 mg of the peptide library in a MicroBio-Spin column (0.8 mL, Bio-Rad). The resin was swelled in DCM for 10–15 min, thoroughly washed with DMF and water, and incubated in 1 mL of HBST-gelatin buffer (30 mM Hepes, 150 mM NaCl, 0.05% Tween-20, 0.1% gelatin, pH 7.4) for 4 h. Afterwards, the gelatin buffer was drained and the resin was resuspended in 1 mL of HBST-gelatin buffer containing 1 μM biotinylated BUZ domain protein and 1 mM tris(2-carboxyethyl)phosphine (TCEP). The resin was incubated overnight at 4 °C with gentle mixing. The protein solution was gently drained and the resin was resuspended in 1 mL of SA-AP binding buffer (30 mM Tris-HCl, pH 7.4, 250 mM NaCl, 10 mM MgCl2, 70 μM ZnCl2, 20 mM imidazole) containing 1 μg/mL SA-AP. The resin was incubated for 10 min at 4 °C. The SA-AP buffer was drained and the resin was washed with the SA-AP binding buffer (2 × 1 mL) and SA-AP reaction buffer (30 mM Tris-HCl, pH 8.5, 100 mM NaCl, 5 mM MgCl2, 20 μM ZnCl2, 20 mM imidazole, 0.01% Tween-20) (2 × 1 mL). The resin was then transferred into a single well of a 12-well plate (BD Falcon) by using the SA-AP reaction buffer (3 × 300 μL). After the addition of 100 μL of a BCIP solution (5 mg/mL in SA-AP reaction buffer), the resin was incubated at room temperature on a rotary shaker and monitored under a dissecting microscope for the development of turquoise color on positive beads. Once the desired color intensity was developed on the positive beads (typically ~1 h), the staining reaction was quenched by the addition of 100 μL of 1 M HCl. The colored beads were manually removed by using a micropipette under a dissecting microscope. The peptides on the positive beads were individually sequenced by partial Edman degradation-mass spectrometry (PED-MS) as previously described (24, 25).
Synthesis of Individual Peptides
Each peptide was synthesized on 50 mg of Wang resin (0.8 mmol/g). The first amino acid was coupled to the resin using diisopropylcarbodiimide as the coupling reagent and dimethylaminopyridine as a catalyst. The remaining amino acids were coupled using standard Fmoc/HBTU chemistry. To generate fluorescently labeled peptides, 100 μL of an FITC solution (20 mg/mL FITC in anhydrous DMSO) was added to ~5 mg of the resin in a 0.5 mL microcentrifuge tube, along with 3.2% (v/v) diisopropylethylamine. The resin was incubated with FITC at room temperature for 4 h in the dark. The resin was then transferred into a 0.8-mL Bio-Rad Micro-Spin column and washed with dichloromethane. Side-chain deprotection and peptide cleavage from the resin were achieved by treatment with 1 mL of the modified reagent K for 2 h. The crude peptide was triturated three times with cold diethyl ether and purified on a semi-preparative HPLC column (C18 column). The identity of each peptide was confirmed by MALDI-TOF mass spectrometry. Peptide concentration was determined by the absorbance of FITC at 495 nm.
Determination of Dissociation Constants by Fluorescence Polarization
The binding affinities of the BUZ domains to individual peptides were determined by fluorescence polarization at 25 °C on a SpectraMax M5 Multi-Mode Microplate Reader (Molecular Devices, Sunnyvale, CA). A fluorescently labeled peptide was dissolved in ddH2O containing 1% (w/v) bovine serum albumin and added to solutions containing increasing concentrations of GST-HDAC6 BUZ or (His)6-Ubp-M BUZ protein (in 20 mM sodium phosphate, pH 7.0, 100 mM NaCl) to give a final concentration of 90 nM peptide and 0.1% (w/v) bovine serum albumin. For binding studies with HDAC6 BUZ (no GST), the buffer contained 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 2.5 mM CaCl2. Fluorescence anisotropy (A) values were measured in triplicates and the dissociation constant (KD) was determined by nonlinear regression fitting of the anisotropy data against the protein concentration using KaleidaGraph software (version 3.6, Synergy Software, Reading, PA):
where A is experimentally measured anisotropy, Af is the anisotropy of the unbound peptide, Ab is the anisotropy of the peptide-protein complex, Qf is the fluorescent intensity of the free peptide, Qb is the fluorescence intensity of the bound peptide, [LT] is the total concentration of labeled peptide, and [PT] is the total concentration of protein.
GST Pull-Down Assay
DNA constructs encoding Xenopus laevis histone proteins H3 and H4 were kindly provided by Dr. Karolin Luger (Colorado State University). The (H3 + H4)2 tetrameric protein complex was overexpressed and purified as described previously (26). A small column filled with ~200 μL of glutathione-agarose (Sigma) was washed with PBS buffer (25 mM sodium phosphate, 100 mM NaCl, pH 7). Either the GST-BUZ domain protein (4.0 nmol) or GST alone (negative control) was loaded to the column, which was exhaustively washed with PBS until no GST-BUZ or GST protein was detected in the flow-through fractions. An equal amount of the (H3 + H4)2 tetrameric complex (4.0 nmol) was added to the column and incubated on ice for 10 min with rotary mixing. The column was then washed with 10 mL of PBS containing 100 mM NaCl and eluted with PBS containing 100 mM NaCl (3 × 200 μL), 300 mM NaCl (3 × 200 μL), and 700 mM NaCl (3 × 200 μL). All of the fractions were analyzed by SDS-PAGE and visualized by Coomassie blue staining.
RESULTS
Synthesis and Screening of Peptide Library with Free C-Termini
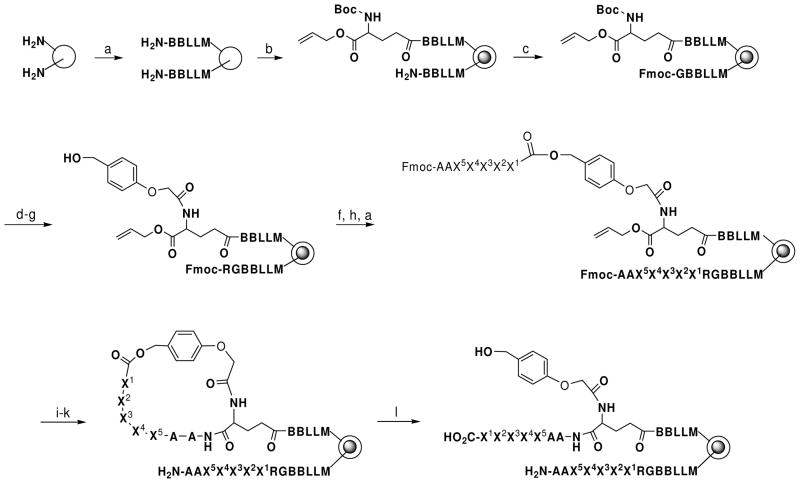
Earlier studies have shown that the BUZ domains of isoT and Ubp-M interact with ubiquitin by recognizing its extreme C-terminal sequence (13, 14). A peptide corresponding to the last five residues of ubiquitin bound to the Ubp-M BUZ domain with a KD value of 15.9 μM, similar to the binding affinity between the BUZ domain and full-length ubiquitin. However, it is currently unknown whether BUZ domains are capable of binding to other peptides/proteins. To answer this question, we decided to systematically profile the sequence specificity of BUZ domains by screening them against a combinatorial OBOC peptide library. Since the BUZ domains require a free C-terminus for binding (13), we designed a peptide library in the form of resin-MLLBBE’AAX5X4X3X2X1-CO2H [where B is β-alanine, E′ is a modified glutamic acid, and X1–X5 are L-α-aminobutyrate (Abu or U, used as a replacement of Cys), L-norleucine (Nle or M, used as Met replacement), or 18 proteinogenic amino acids except for Cys and Met], which featured five random residues near the free C-terminus (Figure 1). Because solid-phase peptide synthesis usually starts from the C-terminus, the desired peptide library cannot be prepared by conventional peptide synthesis chemistry. We have recently developed a novel strategy to overcome this technical difficulty (23). In this strategy, each resin bead was topologically segregated into two layers, with the surface layer displaying an inverted peptide containing a free C-terminus, while the inner core containing the same peptide sequence in the normal orientation (which is attached to the support via its C-terminus) as an encoding tag for later sequence analysis (Figure 1). The peptide library was synthesized on TentaGel microbeads (90 μm, 0.28 mmol/g, 2.86 × 106 beads/g) by a modification of our previously reported method (23) and has a theoretical diversity of 205 or 3.2 × 106. The library was screened against the biotinylated GST-BUZ domain proteins as previously described for other protein domains (27). Briefly, binding of a biotinylated BUZ domain to a resin bead recruits streptavidin-alkaline phosphatase to the bead surface. Subsequent incubation in the presence of 5-bromo-4-chloro-3-indolyl phosphate results in the formation of turquoise color on the positive beads (27). Note that during the library screening, the BUZ domains were too large to diffuse into the beads and therefore have access only to the inverted peptides on the bead surfaces.
Figure 1.
Synthesis of a C-Terminal One-Bead-One-Compound Peptide Library. Reagents and conditions: (a) standard Fmoc/HBTU chemistry; (b) soak in water and then 0.5 equiv Nα-Boc-Glu(δ-N-hydroxysuccinimidyl)-O-CH2CH=CH2; (c) Fmoc-Gly-OH/HBTU; (d) TFA; e) HMPA/HBTU; (f) piperidine; (g) Fmoc-Arg(Pbf)-OH/HBTU; (h) Fmoc-AA/DIC; (i) Pd(PPh3)4; (j) piperidine; (k) PyBOP, HOBt; (l) TFA. B, β-alanine, X, random residues.
Sequence Specificity Profiles of Ubp-M and HDAC6 BUZ Domains
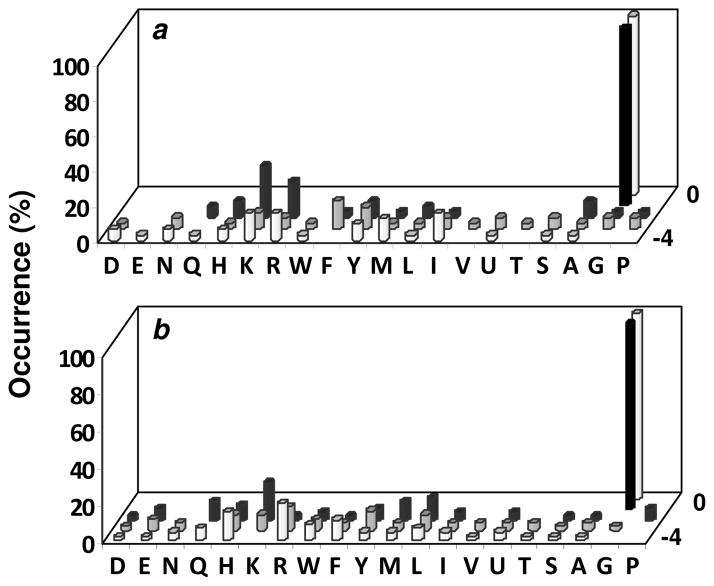
The BUZ domain of Ubp-M was chosen for this study because it is a representative of a family of BUZ domain-containing ubiquitin-specific processing (USP) proteases (6). HDAC6, on the other hand, is one of only a few BUZ domain-containing proteins that are not in the USP family. For each BUZ domain, a total of ~300 mg of the library (~850,000 beads) was screened in several separate experiments. Although the amount of resin used did not cover the entire sequence space of the peptide library, we have previously demonstrated that screening only a fraction of the library (~10% or more of its theoretical diversity) is sufficient to define the sequence specificity of a binding domain (28, 29). For the Ubp-M BUZ domain, library screening resulted in a total of 128 hits, which can be divided into five different classes based on sequence similarity (Table 1). The class I peptides (35 sequences) all contained a C-terminal Gly-Gly motif, similar to the C-terminal sequence of ubiquitin (RLRGG-CO2H). It has broader but still significant specificity at the more N-terminal positions. For example, it strongly prefers a positively charged residue (Arg or Lys) at the -2 position (relative to the C-terminal residue, which is defined as position 0) (Figure 2). It tolerates a wide variety of residues at the -3 and -4 positions, with some preference for an aromatic hydrophobic residue at the -3 position and a basic or hydrophobic residue at the -4 position. The class II peptides (18 sequences) contained a consensus motif of DG(F/Y), often but not always at the C-terminus. These sequences were apparently selected because of their ability to bind to the GST tag (vide infra). The class III peptides (19 sequences) all contained an HPQ motif, which has previously been identified as a specific ligand of streptavidin (30). Apparently, beads containing the class III peptides directly bound and recruited SA-AP to their surfaces. The class IV peptides (6 sequences) contained an HXH or HXXH motif (where X represents any amino acid). We have previously shown that they were false positives during screening of inverted peptide libraries against GST-PDZ fusion proteins and the origin of their selection is not yet clear (23). Finally, a large number of peptides (50 total) with diverse sequences were selected and were collectively categorized as the class V peptides. The origin of their selection is not yet clear and we have not previously observed this type of sequences during our library screening against numerous other protein domains. Interestingly, some of the selected sequences were highly similar with each other (e.g., PSDHV and PSDYV, LRKMG and RLRMG), suggesting that they were positively selected against some component of the library screening system.
Table 1.
Peptide Sequences Selected from Peptide Library against Ubp-M BUZ Domaina
| Class I | Class II | HPQXX | FDAGK |
| WYAGG | LSDGF | HPQRY | VASGL |
| KFAGG | LQDGF* | WUHPQ | PSHHL |
| RRAGG | RWDGF | WTHPQ | IFISL |
| LHFGG | LYDGF | AEAAM | |
| RKGGG | MDGLF | Class IV | FIYDM |
| MYHGG | XDGMF | IYHQH | HRAGM |
| IRHGG | DGFWF | YVHTH | GMGLM |
| KVHGG | FDGFM* | HSHKT | YDDQM |
| RYIGG | DWDGM | HKHYT | HQKGQ |
| YSKGG | LDGYM | HHHXX | PULGQ |
| QDKGG | UDGYM | QHLTH | YFGKQ |
| IAKGG* | WDGYS | KIQLQ | |
| RWKGG | NDGFW | Class V | SIWUR |
| MFKGG | HDGFY | PSDHV* | DMUGR |
| MNKGG | FRDGY | PSDYV | UUKSS |
| KYKGG | LMDGY | YRFDA | QKKRT |
| EFKGG | YFDGY | QFLDA | HQVNW |
| NNKGG | LFDGY | KGGGA | FYEMW |
| HTKGG | VVVPU | YTHRV | |
| HRLGG | Class III | LDHRD | FRAGR |
| NPLGG | RHPQA | SVIKE | ESHGD |
| KSMGG | HPQNU | HGMSE | RNRNV |
| YGPGG | THPQF | UKFAF | KRYHH |
| YFQGG | HPQUG | DLVGF | YINGY |
| IIQGG | HPQWG | LHFLF | LFWMY |
| DIRGG | HPQWG | ISVQF | QRDTY |
| DKRGG | SHPQN | UGLVF | PDIWY |
| MGRGG | HPQRP | LRKMG | |
| SPRGG | WRHPQ | RLRMG | |
| KLRGG | WNHPQ | MITWG | |
| IMRGG | LYHPQ | YNSGH | |
| RURGG | GWHPQ | PFYMH | |
| UFYGG | RHPQQ | WGPYH | |
| IUYGG | WHPQR | FMUAI | |
| AKYGG | HPQXX | QLYQI |
M, norleucine; U, (S)-2-aminobutryic acid; X, amino acids whose identity could not be unambiguously determined.
sequences selected for resynthesis and binding analysis.
Figure 2.
Sequence specificity of Ubp-M (a) and HDAC6 BUZ domains (b). Displayed are the amino acids identified at each position from the C-terminus (position 0) to the -4 position (z axis). Occurrence on the y-axis represents the percentage of selected sequences that contained a particular amino acid at a certain position.
The HDAC6 BUZ domain also selected the same five classes of peptides (Table 2). Again, the class I peptides (50 sequences) all contained a C-terminal GG motif. Among the class I peptides, HDAC6 has weaker selectivity for basic residues at the -2 position as compared to Ubp-M; although lysine was frequently selected, Arg was not (Figure 2). On the other hand, hydrophobic residues Leu and Nle were also selected at this position. At the -4 position, HDAC6 BUZ domain prefers an Arg, His, or Phe residue. The four class II peptides all contained the DG(M/Y) motif and were selected against the GST tag. Screening against the HDAC6 BUZ domain also led to the same false positive sequences containing the HPQ (class III) and HXH motifs (class IV) as the Ubp-M domain. Again, the HDAC6 BUZ domain selected a large number of class V peptides (137 sequences), some of which are highly homologous to each other (e.g., MYRHF and MSRHF, QTVFL and QLVLL, YRRWQ and SRRWQ, SWRAR and SLRAR).
Table 2.
Peptide Sequences Selected from Peptide Library against HDAC6 BUZ Domaina
| Class I | Class II | MVRHH | KRYAR | RGYRR |
| HHQGG | DDGMF | WKWIH | NALER | WNGGY |
| FRAGG | FEDGM | LRPKH | IRIFR | RREHY |
| RYUGG | WADGY | HRWLH | VMPGR | RIHKY |
| FKUGG | ADGMY | UKQRH | SRUHR | WKALY |
| RKDGG | FLRSH | AWHHR | YGHPY | |
| QREGG | Class III | RHIWH | YYHHR | DIAYR |
| RCEGG | SHPQU | RTTWH | GYMHR | MIKRY |
| RSEGG | SHPQF | RFQYH | VIRHR | RFQRY |
| WUFGG | HPQXX | YIYAI | QVRHR | LIKYY |
| WYHGG | HPQXX | THMDI | FKWHR | MYYYY |
| HAHGG | HPQXX | WVYGI | RYDIR | |
| VKHGG | WHPQG | VLRHI | RQLLR | |
| WMHGG | VVGLI | RVTLR | ||
| HEIGG | Class IV | IWIMI | LRUMR | |
| YLIGG | HSHIH | LFFQI | VRSMR | |
| XXKGG | FRHYH | IFEYI | WSARR | |
| LHKGG | YHKHT | YRHFK | RYFRR | |
| HDKGG | HKHYW | URQKK | LKGRR | |
| TRKGG | HRHPY | PGNDL | LLHRR | |
| AFKGG | HRHXX | QTVFL | AGIRR | |
| LLKGG | HTHRR | QLVLL | RIQRR | |
| IKKGG | HIHRR | IMAQL | KFYRR | |
| XXKGG | HAHXX | IYFQL | LRATR | |
| RNKGG | YHDHV | WWHRL | YHNTR | |
| REKGG | RKQRL | GIRVR | ||
| NALGG* | Class V | LSMSL | QHPWR | |
| RVLGG | LSGPA | FHKVL | QRUYR | |
| HTLGG | ETWUU | MNWFM | FKFFS | |
| EGLGG | WRIFU | IRHHM | MLMGS | |
| DHLGG | WMQFU | YEALM | MUITS | |
| QELGG | RRWNU | FQULM | FLLAT | |
| FTMGG | WMEPD | KPMLM | HPVHT | |
| RRMGG | MFNMD | KGPQM | YRRQT | |
| MYMGG | RWHAF | RVSQM | LRLRT | |
| WWMGG | LELAF | KIQRM | NNMTT | |
| FNMGG | YRRFF | LURRM | IFLYT | |
| HWPGG | RRAGF | TLUSM | LUEHV | |
| NYPGG | MYRHF | DVHTM | UYRHV | |
| SWPGG | MSRHF | RMHVM | KIYKV | |
| MIQGG | UHHKF | IYWYM | FSIRV | |
| LLQGG | EYFLF | HIYYM | AKDWV | |
| IRQGG | RHGLF | VVDQP | RHGRW | |
| ULQGG | GIILF | GLRVP | HUUWW | |
| UMRGG | MPVLF | RQFRQ | RRUWW | |
| RFSGG | SRHMF | TILTQ | UTYWW | |
| QRWGG | LAKMF | YRRWQ | LQIHA | |
| YIWGG | TRSRF | SRRWQ | MYRER | |
| HHYGG | LYUIG | SWRAR | YFRRU | |
| FYYGG | ADSIG | SLRAR* | QVRRL | |
| HVYGG | LRUMG | WQKAR | WWHLR |
M, norleucine; U, (S)-2-aminobutryic acid; X, amino acids whose identity could not be unambiguously determined.
sequences selected for resynthesis and binding analysis.
Binding Affinity of Ubp-M and HDAC6 BUZ Domains to Selected Peptides
To confirm the library screening results, we individually synthesized six representative peptides selected against the two BUZ domains and tested their binding affinities to the BUZ domains by fluorescence polarization (Table 3 peptides 1–6). Peptides IAKGG and NALGG were class I ligands selected against Ubp-M and HDAC6 BUZ domains, respectively. Two class II peptides (LQDGF and FDGFM) were chosen to contain the DGF motif at the C-terminus and an internal position, respectively. Finally, one representative class V peptide was selected for each BUZ domain (PSDHV for Ubp-M and SLRAR for HDAC6). As a control, we also synthesized a peptide corresponding to the C-terminal sequence of ubiquitin (RLRGG). All of the peptides contained a free C-terminus and an N-terminal FITC-β-Ala-β-Ala motif.
Table 3.
Binding Affinity of Peptide Ligands to Ubp-M and HDAC6 BUZ Domains
| Entry No. | Peptide Sequencea | Peptide Source |
KD (μM) |
|
|---|---|---|---|---|
| Ubp-M BUZb | HDAC6 BUZc | |||
| 1 | IAKGG | library | 12 ± 3 | 8.8 ± 1.1 |
| 2 | NALGG | library | 33 ± 7 | 0.79 ± 0.12 |
| 0.23 ± 0.02 (non-GST)d | ||||
| 3 | LQDGF | library | >150 | 4.0 ± 1.3 |
| NA (non-GST) d | ||||
| 4 | FDGFM | library | >150 | 10.8 ± 2.5 |
| 5 | PSDHV | library | >140 | ND |
| 6 | SLRAR | libraryc | ND | NA |
| 7 | RLRGG | ubiquitin | 2.2 ± 0.4 | 0.34 ± 0.02 |
| 8 | RLRGA | NA | NA | |
| 9 | RLRAG | 62 ± 11 | NA | |
| 10 | RLAGG | 16 ± 4 | 0.90 ± 0.10 | |
| 11 | RARGG | 2.0 ± 0.3 | 0.47 ± 0.02 | |
| 12 | ALRGG | 2.3 ± 0.4 | 2.5 ± 0.4 | |
| 13 | STLGG | FBXO11 | 16 ± 4 | 3.7 ± 0.5 |
| 14 | YGFGG | Histone H4 | 13 ± 3 | 6.2 ± 0.6 |
| 15 | RGMGG | PTOV1 | 17 ± 3 | 1.3 ± 0.2 |
| 16 | YUIGG | FAT10 | 12 ± 2 | 1.3 ± 0.1 |
Each peptide was labeled at the N-terminus with FITC via a β-Ala-β-Ala linker and contained a free C-terminus.
Ubp-M BUZ domain contained an N-terminal six-histidine tag.
GST-BUZ fusion protein was used in the binding studies unless otherwise noted.
Isolated BUZ domain (non-GST fusion) was used. NA, no significant binding affinity; ND, not determined; U, (S)-2-aminobutryic acid.
As expected, both class I peptides exhibited strong binding to the BUZ domains. Peptide IAKGG, which was selected against Ubp-M, bound to both BUZ domains with similar affinities (KD ~10 μM) (Table 3 and Figure S1 in Supporting Information). Peptide NALGG, on the other hand, bound to its cognate BUZ domain (from HDAC6) with much higher affinity than Ubp-M domain (KD values of 0.79 and 33 μM for HDAC6 and Ubp-M BUZ domains, respectively). This is in excellent agreement with the screening results, which showed that the HDAC6 BUZ domain has a stronger preference for aliphatic hydrophobic residues (Leu, Nle, and Ile) at the -2 position as compared to the Ubp-M domain (Figure 2). As a comparison, the ubiquitin C-terminal peptide had KD values of 2.2 and 0.34 μM to Ubp-M and HDAC6 BUZ domains, respectively. It was previously reported that the Ubp-M BUZ domain binds the full-length ubiquitin with a KD value of 6.5 μM (13). Thus, the binding affinities of the selected peptides (IAKGG and NALGG) to Ubp-M BUZ domain were similar to those of known binding partners of this BUZ domain. To ascertain that the BUZ domain was responsible for the observed binding, we also carried out the binding study with the isolated HDAC6 BUZ domain (without GST tag) and obtained a slightly higher affinity for peptide NALGG (KD = 0.23 μM). This discrepancy is likely caused by GST dimerization (KD = 0.33 μM) (31), because the GST dimer produces higher polarization signal than a monomer and therefore artificially increases the signal at high protein concentrations. Unfortunately, production of GST-free HDAC6 protein was problematic and only small amounts of the protein could be produced at a time. Consequently, most of the binding studies involving the HDAC6 domain were carried out with the GST fusion protein. A KD value of 60 nM was reported for the interaction between the HDAC6 BUZ domain and the full-length ubiquitin (16). To determine whether the class I peptides bind to the same site as the full-length ubiquitin, the fluorescence polarization experiment was carried out in the presence of increasing concentrations of ubiquitin. Ubiquitin addition decreased the anisotropy value in a concentration-dependent manner and completely abolished the binding of peptide FITC-BBRGMGG to the HDAC6 BUZ domain at ≥7 μM ubiquitin (Figure S2). We thus conclude that the selected class I peptides bind to the ubiquitin-binding site of the BUZ domains.
The class II peptides LQDGF and FDGFM bound to GST-HDAC6 BUZ domain with KD values of 4.0 and 10.8 μM, respectively. However, neither peptide showed significant binding to the Ubp-M BUZ domain (KD >150 μM), which contained a six-histidine tag but not the GST tag. When the GST tag was removed by partial proteolysis, the isolated HDAC6 BUZ domain had no detectable binding to the peptides (Figure S3). This suggests that the peptides might be binding to the GST tag. To test this notion, we performed the binding studies with a GST-Abl2 SH2 domain fusion protein and found that the peptide LQDGF bound to the GST-SH2 protein with a KD value of 4.6 μM, essentially the same binding affinity of LQDGF for the GST-BUZ domain. Moreover, the binding of the peptide to the GST-SH2 protein was inhibited by glutathione, with an IC50 of ~200 μM. Therefore, we conclude that the class II peptides were selected from the peptide library because of their ability to bind to the GST portion of the GST-BUZ domain proteins. The class V peptide PSDHV showed very weak binding to the Ubp-M BUZ domain (KD >140 μM), while the class V peptide SLRAR showed no binding towards HDAC6 BUZ (Figure S1).
Determination of Critical Positions for Binding by Alanine Scan
To determine whether all or which of the positions are critical for binding to a BUZ domain, we performed an “alanine scan” of the ubiquitin C-terminal peptide (RLRGG), during which each of the five C-terminal residues of the peptide was replaced by an alanine. For both BUZ domains, the C-terminal dipeptide GG are critical for binding; substitution of Ala for the terminal Gly completely abolished binding, while mutation of the Gly at position -1 either abolished binding (HDAC6) or greatly reduced the binding affinity (~30-fold for Ubp-M) (Table 3). Interestingly, the two BUZ domains display differential sensitivities to Ala substitution at the more N-terminal positions. At the -2 position, while mutation of the Arg to Ala decreased the affinity for Ubp-M BUZ domain by ~8-fold, it had a smaller effect on the HDAC6 BUZ domain (2.6-fold). This is consistent with the library screening results, which showed a clear preference for Arg at this position by Ubp-M but not HDAC6 (Figure 2). The opposite trend was observed at the -4 position, where Arg->Ala mutation reduced the affinity for HDAC6 BUZ domain by 7.4-fold but had minimal effect on the Ubp-M domain, again in agreement with the specificity profile obtained from library screening. Replacement of the -3 Leu with Ala had no effect on Ubp-M BUZ domain and resulted in a minor reduction in affinity for HDAC6 domain (1.5-fold). Thus, we conclude that for Ubp-M and HDAC6 BUZ domains and likely many other BUZ domains, the C-terminal GG motif is the most critical element of specificity determinant. This is consistent with the structure of the ubiquitin C-terminus bound to the BUZ domain of USP5 (isoT), where molecular modeling predicted a steric clash between the Ala-76 side chain of a G76A mutant ubiquitin and tyrosine residues in the binding pocket of USP5 BUZ domain (14). Our results suggest that BUZ domains make additional contacts with the more N-terminal residues, for enhanced affinity and specificity. It appears that different BUZ domains may contact different positions in a peptide (e.g., -2 vs -4 position) as well as recognize different amino acid residues at the same position (e.g., Arg vs Leu at the -2 position).
Database Search for Potential Interacting Partners of Ubp-M and HDAC6 BUZ Domains
We searched an Expasy proteomics server database (Web site: http://ca.expasy.org/) for human proteins that contain a diglycine motif at the C-terminus. The search was conducted by entering the motif XXXGG> (the sign “>” limits the search to C-terminal GG sequences, X represents any amino acid) into the search site. Only proteins that are known to exist either at the transcript level or at the protein level are included. The search resulted in 73 human proteins with a C-terminal GG motif. On the basis of the sequence specificity determined above, we predict 13 of the proteins as the potential targets of the Ubp-M BUZ domain and 23 proteins as the potential partners of the HDAC6 BUZ domain, one of which is ubiquitin (Table 4).
Table 4.
Potential Ubp-M/HDAC6 BUZ Domain Binding Partners
| C-terminal Sequence | ID number | Proteins |
|---|---|---|
| P98198-2 | Probable phospholipid-transporting ATPase ID, isoform 2 | |
| STLGGb | Q86XK2-4 | Isoform 4 of F-box only protein 11 |
| O60942-4 | Isoform 4 of mRNA-capping enzyme | |
| LERGGa | Q9HC96-6 | Calpain-10, isoform F |
| LEKGGa | Q9NX63 | Coiled-coil-helix-coiled-coil-helix domain-containing protein 3, mitochondrial |
| RVSGGb | O14578-2 | Isoform 2 of Citron Rho-interacting kinase |
| QRLGGb | Q9GZP9 | Derlin-2 |
| KSFGGb | Q6P158-2 | Isoform 2 of Putative ATP-dependent RNA helicase DHX57 |
| LYKGGa | Q8TBM8 | DnaJ homolog subfamily B member 14 |
| LYKGGa | Q8TBM8-2 | Isoform 2 of DnaJ homolog subfamily B member 14 |
| LRRGGa | Q9H819 | DnaJ homolog subfamily C member 18 |
| YGFGGa,b | P62805 | Histone H4 |
| HPKGGa,b | P49641-2 | Isoform Short of Alpha-mannosidase 2x |
| Q9NZB8-3 | Isoform 3 of Molybdenum cofactor biosynthesis protein 1 | |
| ILIGGb | Q9NZB8-5 | Isoform MOCS1A of Molybdenum cofactor biosynthesis protein 1 |
| Q9NZB8-6 | Isoform 2 of Molybdenum cofactor biosynthesis protein 1 | |
| RGCGGb | Q71RS6 | Sodium/potassium/calcium exchanger 5 |
| RWVGGb | Q9H1B4-5 | Isoform E of Nuclear RNA export factor 5 |
| NTLGGb | Q8NGS3 | Olfactory receptor 1J1 |
| YYKGGa | Q9P2J9 | Pyruvate dehydrogenase [acetyl-transferring]]-phosphatase 2, mitochondrial |
| Q86YD1 | Prostate tumor-overexpressed gene 1 protein | |
| RGMGGb | Q86YD1-2 | Isoform 2 of Prostate tumor-overexpressed gene 1 protein |
| Q86YD1-3 | Isoform 3 of Prostate tumor-overexpressed gene 1 protein | |
| GKKGGa | P10155-2 | Isoform Short of 60 kDa SS-A/Ro ribonucleoprotein |
| RLLGGb | Q9BZJ4 | Solute carrier family 25 member 39 |
| Q9BZJ4-2 | Isoform 2 of Solute carrier family 25 member 39 | |
| IIVGGb | P60059 | Protein transport protein Sec61 subunit gamma |
| RGSGGb | Q58EX2-4 | Isoform 4 of Protein sidekick-2 |
| TPRGGa | Q9Y675 | SNRPN upstream reading frame protein |
| YCIGGb | O15205 | FAT10 (Ubiquitin D) |
| RMLGGb | P35544 | Ubiquitin-like protein FUBI |
| RLRGGa,b | P62988 | Ubiquitin |
Potential binding partners of Ubp-M BUZ domain;
potential binding partners of HDAC6 BUZ
In Vitro Interaction between Ubp-M and HDAC6 BUZ Domains and Protein C-Termini
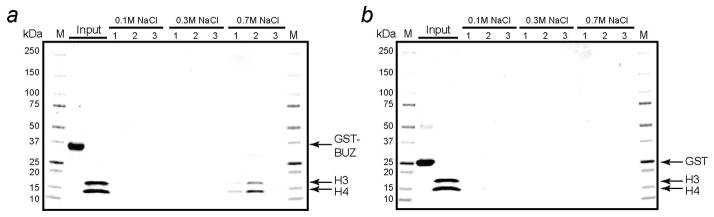
We next synthesized four peptides corresponding to the C-terminal sequences of four of the predicted BUZ-binding partners, F-box only protein 11 (FBXO11, STLGG), histone H4 (YGFGG), prostate tumor-overexpressed gene 1 protein (PTOV1, RGMGG), and FAT10 (or ubiquitin D, YCIGG), respectively (Table 3). FBXO11 contains an F-box motif, which potentially links it to the protein ubiquitination pathway (32). Histone H4 was chosen because it has already been established that Ubp-M interacts with another histone protein, histone H2A (33). PTOV1 is a protein overexpressed in prostate cancer (34). It has previously been reported that HDAC6 interacts with FAT10 and both the catalytic and BUZ domains of HDAC6 contribute to the binding interaction (35). It was not clear, however, whether the BUZ domain recognizes the C-terminal sequence of FAT10. All four peptides bound to the HDAC6 BUZ domain with KD values in the low μM range (1.3–6.2 μM) (Figure S4). We therefore propose that binding to the C-terminus of FAT10 by the BUZ domain is a key mechanism for the observed HDAC6-FAT10 interaction. The four peptides also bound to the Ubp-M BUZ domain, although the binding affinities were somewhat lower as compared to the HDAC6 domain (KD = 12–17 μM). To test whether the Ubp-M BUZ domain is capable of binding to intact proteins (other than ubiquitin), we performed in vitro GST pull-down assays using recombinant Xenopus laevis histone proteins H3 and H4 heterotetramer [(H3 + H4)2] and GST-Ubp-M BUZ domain. As shown in Figure 3, (H3 + H4)2 bound to GST-BUZ domain but not to GST alone. The bound histone protein required 0.7 M NaCl to elute, indicating that the interaction was relatively tight. Taken together, we conclude that the BUZ domains are sequence-specific protein-binding modules that recognize the free C-termini of cellular proteins, with each interacting with a specific subset of partner proteins (including ubiquitin).
Figure 3.
GST pull-down assay showing the interaction between Ubp-M BUZ domain and histone (H3 + H4)2 tetramer. GST-BUZ (or GST) protein and (H3+H4)2 tetramer were sequentially loaded onto the column as input. The column was washed with 10 mL of PBS containing 0.1 M NaCl and then eluted with 3 × 200 μL of PBS containing 0.1 M NaCl (fractions 1–3), 3 × 200 μL of PBS containing 0.3 M NaCl (fractions 1–3), and 3 × 200 μL of PBS containing 0.7 M NaCl (fractions 1–3). Each fraction was analyzed by SDS-PAGE and stained by Coomassie blue. (a) Elution profile of the column loaded with GST-BUZ protein; (b) elution profile of the column loaded with GST alone. M, molecular weight markers.
Discussion
In this work, we have systematically profiled the sequence specificities of Ubp-M and HDAC6 BUZ domains. Several conclusions have been borne out of our studies. First, the BUZ domain is a sequence-specific protein-binding module that recognizes the free C-termini of proteins. Second, different BUZ domains apparently have distinct specificity profiles, with each domain recognizing a different subset of peptide sequences, although overlapping specificities are possible. For the two BUZ domains examined in this work, a Gly-Gly motif at the extreme C-terminus is required for binding. This specificity is unique among all of the protein-binding modules that have been biochemically and/or structurally characterized and is endowed by having a deep, narrow pocket on the BUZ domain surface that is large enough only to fit the small Gly residues (14). At positions N-terminal to the Gly-Gly motif, the two BUZ domains exhibit different specificities. For example, while the Ubp-M BUZ domain strongly prefers a basic residue at the -2 position but has broad specificity at the -3 and -4 positions, the BUZ domain of HDAC6 prefers a basic residue at the -4 position and tolerates a variety of amino acids at the -2 and -3 positions. The ability of the BUZ domains to bind with high affinity to peptide sequences that are different from the ubiquitin C-terminus suggests that they may bind to other cellular proteins in vivo, in addition to ubiquitin. A database search identified 12 and 22 other human proteins as potential binding partners of Ubp-M and HDAC6 BUZ domains, respectively (Table 4). Peptides corresponding to the C-terminal sequences of four of these potential targets that were selected for further testing all bound to the two BUZ domains with low μM KD values. Furthermore, one of the four proteins (FAT10) has previously been reported to bind to HDAC6 via its BUZ domain (35), while our current work has demonstrated that histone H4 also binds to GST-Ubp-M BUZ domain in an in vitro pull-down assay (Figure 3). We propose that many of the other predicted interactions in Table 4 may be physiologically relevant. In addition, proteolytic cleavage may generate protein fragments that bear C-terminal Gly-Gly motifs, which may bind to the BUZ domains. In this regard, several BUZ domains including those of Ubp8p, USP20, USP22, and USP33 have either been shown or are expected to be incapable of binding to ubiquitin (6, 15, 36). We suggest that they may function by binding to non-ubiquitin proteins. Finally, the BUZ domain of HDAC6 generally has higher affinity to peptide ligands than the Ubp-M domain (Table 3). It is likely that BUZ domains interact with their physiological targets with a wide range of affinities to perform proper functions.
Screening of the inverted peptide library also resulted in a large number of other peptide sequences (classes II to V in Tables 1 and 2). We have shown that the class II peptides with the consensus motif of DG(F/Y) bind to the active site of GST with KD values of ~4 μM. To the best of our knowledge, this represents a novel class of high-affinity peptide ligands for GST (37). Since GST fusion proteins are widely used in numerous applications, the new peptide ligands may provide useful tools for research and development purposes. For example, since DG(F/Y) peptides do not contain free thiol or amine functionalities (unlike free glutathione), DG(F/Y) peptides could be used to elute GST fusion proteins from glutathione columns and the eluted protein could be directly labeled with amine or thiol reactive reagents without the need for dialysis/size exclusion chromatography. They may also be used to immobilize GST fusion proteins onto solid surfaces to generate protein chips of uniform orientation (38). The class III and IV ligands have previously been shown to be caused by either binding directly to SA-AP (used in library screening) or nonspecific binding of unknown origin. The origin of the class V peptides is currently unknown. Although the two peptides we have tested failed to bind to the BUZ domains, we cannot rule out the possibility that some of the peptides may be bona fide ligands of the BUZ domains. It should be pointed out that among the numerous protein domains that have been subjected to peptide library screening in this laboratory, the two BUZ domains were unusual in that screening led to so many non-cognate peptides. The underlying reason for their appearance is yet unclear. However, our ability to generate individual peptide sequences and therefore the ability to classify the selected peptides into different families speaks highly for the advantage of the OBOC method over other library methods (e.g., oriented peptide libraries or SPOT libraries) (39–41).
In conclusion, we have demonstrated the BUZ domains as a family of sequence-specific protein-binding domains and determined the sequence specificity profiles for two BUZ domains. Our results suggest that the BUZ domains may bind to cellular proteins other than ubiquitin and the specificity data should be useful for the identification of these protein targets.
Supplementary Material
Abbreviations
- BCIP
5-bromo-4-chloro-3-indolyl phosphate
- BUZ
binder of ubiquitin zinc finger
- DIC
diisopropylcarbodiimide
- FITC
fluorescein isothiocyanate
- Fmoc-OSU
N-(9-fluorenylmethoxycarbonyloxy) succinimide
- HBTU
O-Benzotriazole-N,N,N′,N′-tetramethyl-uronium-hexafluoro-phosphate
- HDAC6
histone deacetylase 6
- HMPA
4-hydroxymethylphenoxyacetic acid
- HOBt
N-hydroxybenzotriazole
- MALDI-TOF
matrix assisted laser desorption ionization-time of flight
- NMM
N-methylmorpholine
- OBOC
one-bead-one-compound
- PED-MS
partial Edman degradation-mass spectrometry
- PITC
phenyl isothiocyanate
- PyBOP
Benzotriazole-1-yl-oxy-tris-pyrrolidino-phosphonium hexafluorophosphate
- SA-AP
streptavidin-alkaline phosphatase
- TFA
trifluoroacetic acid
- Ubp-M
mutant ubiquitin-processing protease
- USP
ubiquitin-specific processing protease
Footnotes
This work was supported by the National Institutes of Health (GM062820 to D.P. and GM079376 to P.Z.). Ryan Hard was supported by a fellowship from the NIH Chemistry-Biology Interface Training Program (T32 GM08512).
Supporting Information Available. Additional experimental details and fluorescence anisotropy data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ciechanover A. The Ubiquitin-Proteasome Pathway: On Protein Death and Cell Life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wickliffe K, Williamson A, Jin L, Rape M. The Multiple Layers of Ubiquitin Dependent Cell Cycle Control. Chem Rev. 2009;109:1537–1548. doi: 10.1021/cr800414e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaiser P, Flick K, Wittenberg C, Reed S. Regulation of Transcription by Ubiquitination without Proteolysis: Cdc34/SCFMet30-Mediated Inactivation of the Transcription Factor Met4. Cell. 2000;102:303–314. doi: 10.1016/s0092-8674(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 4.Conaway R. Emerging Roles of Ubiquitin in Transcription Regulation. Science. 2002;296:1254–1258. doi: 10.1126/science.1067466. [DOI] [PubMed] [Google Scholar]
- 5.Reyes-Turcu F, Ventii K, Wilkinson K. Regulation and Cellular Roles of Ubiquitin-Specific Deubiquitinating Enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnet J, Romier C, Tora L, Devys D. Zinc-finger UBPs: Regulators of Deubiquitylation. Trends Biochem Sci. 2008;33:369–375. doi: 10.1016/j.tibs.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Bilodeau P, Urbanowski J, Winistorfer S, Piper R. The Vps27p-Hse1p Complex Binds Ubiquitin and Mediates Endosomal Protein Sorting. Nature Cell Biology. 2002;4:534–539. doi: 10.1038/ncb815. [DOI] [PubMed] [Google Scholar]
- 8.Kawaguchi Y, Kovacs J, McLaurin A, Vance J, Ito A, Yao T. The Deacetylase HDAC6 Regulates Aggresome Formation and Cell Viability in Response to Misfolded Protein Stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann K, Bucher P. The UBA Domain: A Sequence Motif Present in Multiple Enzyme Classes of the Ubiquitination Pathway. Trends Biochem Sci. 1996;21:172–173. [PubMed] [Google Scholar]
- 10.Hurley J, Lee S, Prag G. Ubiquitin Binding Domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beal RE, Toscano-Cantaffa D, Young P, Rechsteiner M, Pickart CM. The Hydrophobic Effect Contributes to Polyubiquitin Chain Recognition. Biochemistry. 1998;37:2925–2934. doi: 10.1021/bi972514p. [DOI] [PubMed] [Google Scholar]
- 12.Mueller TD, Feigon J. Solution Structures of UBA Domains Reveal a Conserved Hydrophobic Surface for Protein-Protein Interactions. J Mol Biol. 2002;319:1243–1255. doi: 10.1016/S0022-2836(02)00302-9. [DOI] [PubMed] [Google Scholar]
- 13.Pai M, Tzeng S, Kovacs J, Keaton M, Li S, Yao T, Zhou P. Solution Structure of the Ubp-M BUZ Domain, a Highly Specific Protein Module that Recognizes the C-terminal Tail of Free Ubiquitin. J Mol Biol. 2007;370:290–302. doi: 10.1016/j.jmb.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The Ubiquitin Binding Domain Znf UBP Recognizes the C-terminal Diglycine Motif of Unanchored Ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 15.Allen MD, Bycroft M. The Solution Structure of the Znf UBP Domain of USP33/VDU1. Protein Sci. 2007;16:2072–2075. doi: 10.1110/ps.072967807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyault C, Gilquin B, Zhang Y, Rybin V, Garman E, Meyer-Klaucke W, Matthias P, Müller C, Khochbin S. HDAC6-p97/VCP Controlled Polyubiquitin Chain Turnover. EMBO J. 2006;25:3357–3366. doi: 10.1038/sj.emboj.7601210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matheny SA, Chen C, Kortum R, Razidlo G, Lewis R, White M. Ras Regulates Assembly of Mitogenic Signaling Complexes Through the Effector Protein IMP. Nature. 2004;427:256–260. doi: 10.1038/nature02237. [DOI] [PubMed] [Google Scholar]
- 18.Hook SS, Orian A, Cowley S, Eisenman R. Histone Deacetylase 6 Binds Polyubiquitin Through its Zinc Finger (PAZ) Domain and Co-Purifies with Deubiquitinating Enzymes. Proc Natl Acad Sci U S A. 2002;99:13425–13430. doi: 10.1073/pnas.172511699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seigneurin-Berny D, Verdel A, Curtet S, Lemercier C, Garin J, Rousseaux S, Kochbin S. Identification of Components of the Murine Histone Deacetylase 6 Complex: Link Between Acetylation and Ubiquitination Signaling Pathways. Mol Cell Biol. 2001;21:8035–8044. doi: 10.1128/MCB.21.23.8035-8044.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dang LC, Stein R, Merli F. Kinetic and Mechanistic Studies on the Hydrolysis of Ubiquitin C-terminal 7-amido-4-methylcoumarin by Deubiquitinating Enzymes. Biochemistry. 1998;37:1868–1879. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- 21.Stein RL, Chen Z, Melandri F. Kinetic Studies of Isopeptidase T: Modulation of Peptidase Activity by Ubiquitin. Biochemistry. 1995;34:12616–12623. doi: 10.1021/bi00039a017. [DOI] [PubMed] [Google Scholar]
- 22.Pei D, Wavreille AS. Reverse Interactomics: Decoding protein-protein interactions with combinatorial chemistry. Mol BioSyst. 2007;3:536–541. doi: 10.1039/b706041f. [DOI] [PubMed] [Google Scholar]
- 23.Joo SH, Pei D. Synthesis and Screening of Support-Bound Combinatorial Peptide Libraries with Free C-Termini: Determination of the Sequence Specificity of PDZ Domains. Biochemistry. 2008;47:3061–3072. doi: 10.1021/bi7023628. [DOI] [PubMed] [Google Scholar]
- 24.Sweeney MC, Pei D. An improved method for rapid sequencing of support-bound peptides by partial Edman degradation and mass spectrometry. J Comb Chem. 2003;5:218–222. doi: 10.1021/cc020113+. [DOI] [PubMed] [Google Scholar]
- 25.Thakkar A, Wavreille AS, Pei D. Traceless capping agent for peptide sequencing by partial Edman degradation and mass spectrometry. Anal Chem. 2006;78:5935–5939. doi: 10.1021/ac0607414. [DOI] [PubMed] [Google Scholar]
- 26.Luger K, Rechsteiner T, Richmond TJ. Expression and purification of recombinant histones, and nucleosome reconstitution. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 27.Wavreille AS, Garaud M, Zhang Y, Pei D. Defining SH2 domain and PTP specificity by screening combinatorial peptide libraries. Methods. 2007;42:207–219. doi: 10.1016/j.ymeth.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sweeney MC, Wavreille AS, Park J, Butchar JP, Tridandapani S, Pei D. Decoding protein-protein interactions through combinatorial chemistry: Sequence Specificity of SHP-1, SHP-2, and SHIP SH2 Domains. Biochemistry. 2005;44:14932–14947. doi: 10.1021/bi051408h. [DOI] [PubMed] [Google Scholar]
- 29.Garaud M, Pei D. Substrate profiling of protein tyrosine phosphatase PTP1B by screening a combinatorial library. J Am Chem Soc. 2007;129:5366–5367. doi: 10.1021/ja071275i. [DOI] [PubMed] [Google Scholar]
- 30.Lam KS, Lebl M. Streptavidin and avidin recognize peptide ligands with different motifs. Immunomethods. 1992;1:11–15. [Google Scholar]
- 31.Barylko B, Binns D, Lin K-M, Atkinson M, Jameson D, Yin H, Albanesi J. Synergistic Activation of Dynamin GTPase by Grb2 and Phosphoinositides. J Biol Chem. 1998;273:3791–3797. doi: 10.1074/jbc.273.6.3791. [DOI] [PubMed] [Google Scholar]
- 32.Craig KL, Tyers M. The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog Biophys Mol Biol. 1999;72:299–328. doi: 10.1016/s0079-6107(99)00010-3. [DOI] [PubMed] [Google Scholar]
- 33.Mimnaugh EG, Kayastha G, McGovern NB, Hwang SG, Marcu MG, Trepel J, Cai S, Marchesi V, Neckers L. Caspase-dependent deubiquitination of monoubiquitinated nucleosomal histone H2A induced by diverse apoptogenic stimuli. Cell Death Differ. 2001;8:1182–1196. doi: 10.1038/sj.cdd.4400924. [DOI] [PubMed] [Google Scholar]
- 34.Benedit P, Paciucci R, Thompson T, Valeri M, Nadal M, Càceres C, Torres I, Estivill X, Lozano J, Morote J, Reventós J. PTOV1, a novel protein overexpressed in prostate cancer containing a new class of protein homology blocks. Oncogene. 2001;20:1455–1464. doi: 10.1038/sj.onc.1204233. [DOI] [PubMed] [Google Scholar]
- 35.Kalveram B, Schmidtke G, Groettrup M. The ubiquitin-like modifier FAT10 interacts with HDAC6 and localizes to aggresomes under proteasome inhibition. J Cell Sci. 2008;121:4079–4088. doi: 10.1242/jcs.035006. [DOI] [PubMed] [Google Scholar]
- 36.Ingvarsdottir K, Krogan N, Emre N, Wyce A, Thompson N, Emili A, Hughes T, Greenblatt J, Berger S. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisae SAGA complex. Mol Cell Biol. 2005;25:1162–1172. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kay B, Adey N, Sparks A. Reagents binding vinculin, dynein, and glutathione S-transferase from peptide libraries. CODEN: PIXXD2 WO9520601 A1 19950803 CAN 123:250695 AN 1995:826768 CAPLUS PCT Int Appl. 1995:110.
- 38.Propheter DC, Hsu KL, Mahal LK. Fabrication of an oriented lectin microarray. ChemBioChem. 2010;11:1–5. doi: 10.1002/cbic.201000106. [DOI] [PubMed] [Google Scholar]
- 39.Songyang Z, Fanning A, Fu C, Xu J, Marfatia S, Chishti A, Crompton A, Chan A, Anderson J, Cantley L. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 40.Hoffmuller U, Russwurm M, Kleinjung F, Ashurst J, Oschkinat H, Volker-Engert R, Koesling D, Schneider-Mergener J. Interaction of a PDZ protein domain with a synthetic library of all human protein C-termini. Angew Chem, Int Ed. 1999;38:2000–2004. doi: 10.1002/(SICI)1521-3773(19990712)38:13/14<2000::AID-ANIE2000>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 41.Boisguerin P, Leben R, Ay B, Radziwill G, Moelling K, Dong L, Volkmer-Engert R. An improved method for the synthesis of cellulose membrane-bound peptides with free C-termini is useful for PDZ domain binding studies. Chem Biol. 2004;11:449–459. doi: 10.1016/j.chembiol.2004.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.