The presence of particular oligosaccharides in mother’s milk influences bacterial colonization of the newborn mouse intestine and susceptibility to dextran sodium sulfate-driven colitis.
Abstract
Milk oligosaccharides contribute to the development of the intestinal environment by acting as decoy receptors for pathogens and as prebiotics, which promote the colonization of commensal bacteria. Here, using α2,3- and α2,6-sialyltransferase-deficient mice, we investigated the role of the sialylated milk oligosaccharides sialyl(α2,3)lactose and sialyl(α2,6)lactose on mucosal immunity. The exposure of newborn mice to milk containing or deficient in sialyllactose had no impact on the development of mucosal leukocyte populations. However, when challenged by dextran sulfate sodium (DSS) in drinking water, adult mice that had been fostered on sialyl(α2,3)lactose-deficient milk were more resistant to colitis compared with mice fostered on normal milk or sialyl(α2,6)lactose-deficient milk. Analysis of intestinal microbiota showed different colonization patterns depending on the presence or absence of sialyl(α2,3)lactose in the milk. Germ-free mice reconstituted with intestinal microbiota isolated from mice fed on sialyl(α2,3)lactose-deficient milk were more resistant to DSS-induced colitis than germ-free mice reconstituted with standard intestinal microbiota. Thus, exposure to sialyllactose during infancy affects bacterial colonization of the intestine, which influences the susceptibility to DSS-induced colitis in adult mice.
Oligosaccharides represent a major fraction of milk constituents. Unique among mammals, human milk contains a tremendous diversity of oligosaccharide structures, which are shaped by extension of lactose through glycosyltransferase enzymes in the mammary gland (Egge, 1993). The most abundant structures are the trisaccharides produced by addition of fucose or sialic acid to lactose. Whereas fucosylated oligosaccharides are missing from most mammalian milks, sialylated oligosaccharides are more widely distributed (Urashima et al., 2001). Because milk oligosaccharides are neither digested nor absorbed in the small intestine (Brand Miller et al., 1995), they have been suggested to contribute to the development of the infant gastrointestinal tract and its colonization by commensal bacteria (Savage, 1977; Frank and Pace, 2008).
Milk oligosaccharides influence the development of the intestinal microbiota by acting as selective nutrients, which support the proliferation of specific bacterial groups (Gibson and Roberfroid, 1995). The prebiotic action of milk oligosaccharides has been demonstrated by comparing the intestinal microbiota of infants fed on oligosaccharide-rich breast milk and infants fed on formula (Harmsen et al., 2000). Furthermore, considering the structural similarity of milk oligosaccharides with cell surface glycans, milk oligosaccharides can function as soluble receptors, thereby preventing the attachment of pathogenic bacteria to intestinal epithelial cells (Newburg, 2009).
Commensal bacteria are mainly found in the large intestine, consisting predominantly of the phyla Firmicutes and Bacteroidetes (Eckburg et al., 2005). Firmicutes themselves are composed of two major clostridial groups, namely the clostridial cluster IV and clostridial cluster XIVa, which comprises Lachnospiraceae. The density and diversity of the intestinal microbiota are highly complex. Culture-independent methods have allowed us to estimate the presence of 500–1,000 different species and >7,000 strains in the human gastrointestinal tract (Ley et al., 2006; Dethlefsen et al., 2007). Commensal bacteria are indispensible for the proper development of the mucosal immune system. In addition to morphological development of immune compartments, bacterial colonization initiates antibody production (Macpherson and Harris, 2004) and the production of antimicrobial proteins such as defensins (Falk et al., 1998). Bacterial cell wall components have also been shown to regulate CD4+ T helper cell activity in the lamina propria (Mazmanian et al., 2008). On the other hand, a defect of the innate immune system has been shown to affect the composition of the intestinal microbiota in mice, and thereby contributes to metabolic imbalance (Vijay-Kumar et al., 2010). In humans, similar relationships have been established between the intestinal microbiota and energy balance, thus leading to the definition of microbiomes typical for disorders such as obesity (Turnbaugh et al., 2006).
Prebiotic oligosaccharides have been said to reduce the susceptibility to allergies in infants (Moro et al., 2006; von Hoffen et al., 2009) and shown to influence the immune response to vaccination in mice (Vos et al., 2007). However, the mechanisms underlying the regulatory action of oligosaccharides remain largely unknown. Considering the structural complexity of human milk oligosaccharides, the mouse with its reduced range of milk oligosaccharides (Prieto et al., 1995) enables addressing the functional impact of specific oligosaccharides on mucosal immunity in vivo. In this study, we have studied the impact of sialylated oligosaccharides using sialyltransferase-knockout mice deficient for these milk oligosaccharides. The fostering of newborn mice by normal or sialyllactose-deficient mothers demonstrated the importance of theses type of milk oligosaccharides on the colonization of intestinal bacteria and on the susceptibility to an experimental colitis model.
RESULTS
The sialyltransferase enzymes St6gal1 and St3gal4 are responsible for the production of sialyllactose in mouse milk
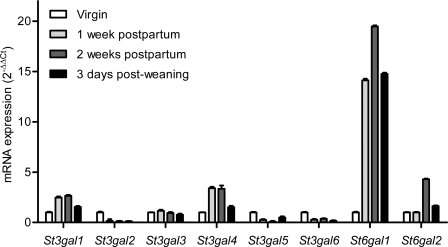
The composition of mouse milk oligosaccharides is limited to sialyllactose with only traces of fucosylated lactose (Kuhn, 1972; Prieto et al., 1995). This low structural complexity makes the mouse a suitable model to investigate the role of these specific milk oligosaccharides on mucosal immunity. Because multiple α2,3 and α2,6 sialyltransferase enzymes (Harduin-Lepers et al., 2001) could be responsible for the production of sialyllactose, we have first examined the expression of these genes in the lactating mammary gland. The messenger RNA (mRNA) levels of the α2,3 sialyltransferase genes St3gal1 to St3gal6 and of the two α2,6 sialyltransferase genes St6gal1 and St6gal2 were determined by real-time PCR. The expression of the St6gal1 gene was induced up to 20-fold during lactation, suggesting this sialyltransferase may account for the biosynthesis of sialyl(α2,6)lactose (6SL; Fig. 1), and thus confirming a previously published observation (Dalziel et al., 2001). Among α2,3 sialyltransferase genes, the expression of St3gal1 and St3gal4 were induced by three- and fourfold during lactation, respectively (Fig. 1). The three St6gal1, St3gal1, and St3gal4 also represented the most abundant sialyltransferase transcripts in lactating mammary gland when mRNA levels were normalized to GAPDH (Fig. S1).
Figure 1.
Sialyltransferase gene expression in mouse mammary gland. The relative levels of mRNA transcripts encoding the six α2,3 sialyltransferases (St3gal1 to St3gal6) and the two α2,6 sialyltransferases (St6gal1 and St6gal2) were measured in virgin mice (mRNA levels set to 1), at 1 wk postpartum, 2 wk postpartum, and 3 d post-weaning. Data represent the mean ± SEM of 3 mice per time point (n = 3). Similar results were obtained in two independent experiments.
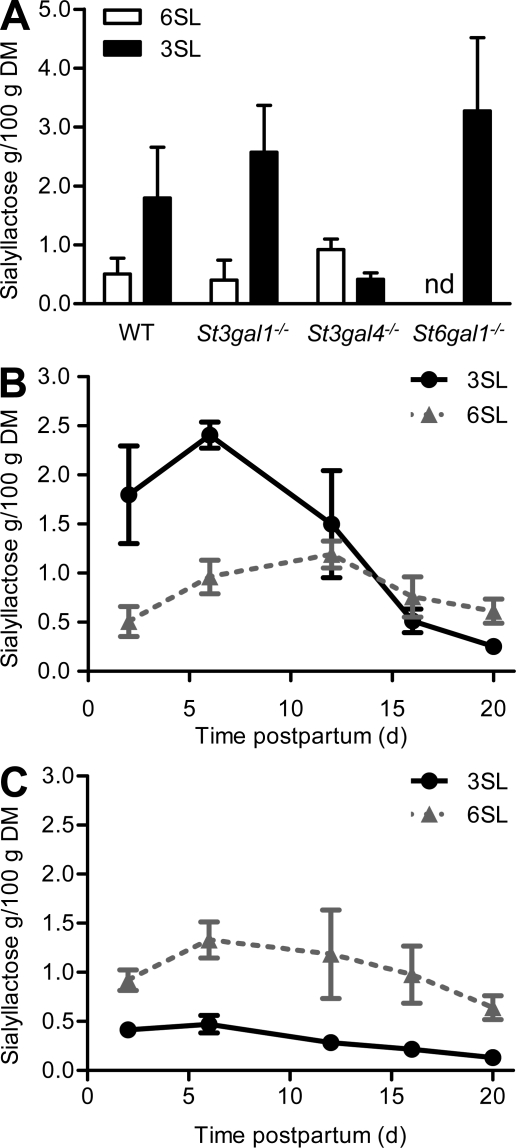
To confirm the involvement of the St3gal1, St3gal4, and St6gal1 sialyltransferases in the production of milk sialyllactose, the oligosaccharide composition of milk isolated from St3gal1, St3gal4, and St6gal1 sialyltransferase-deficient mice was determined by pulsed amperometry-HPAEC. Milk isolated from St3gal1−/− mice showed unaffected or rather increased levels of both sialyl(α2,3)lactose (3SL) and 6SL when compared with the levels measured in WT mice by day 2 of lactation (Fig. 2 A). The analysis of oligosaccharides in milk from St3gal4−/− mice showed a strong decrease of 3SL, indicating that this sialyltransferase accounts for the bulk of 3SL production. The importance of St6gal1 in the production of 6SL was confirmed by the absence of this oligosaccharide in the milk isolated from St6gal1−/− mice (Fig. 2 A). The impact of St3gal4 on 3SL production was investigated in more details by measuring sialyllactose levels across lactation. In WT mice, 3SL levels peaked in the first week of lactation and slowly decreased until day 20, where only minor concentrations were detected. In contrast, 6SL levels showed only a modest increase by mid-lactation (Fig. 2 B). The 3SL peak by the first week of lactation was absent in the milk of St3gal4−/− mice and 3SL levels remained low across lactation, thus demonstrating the importance of St3gal4 for the biosynthesis of 3SL (Fig. 2 C). The identity of 3SL and 6SL and the exact structure of sialic acid in mouse milk were also analyzed after release by neuraminidase treatment. We did find that milk sialyllactose was exclusively composed of N-acetylneuraminic acid because no N-glycolylneuraminic acid could be detected (Fig. S2).
Figure 2.
Sialyllactose concentration in mouse milk during lactation. (A) 3SL and 6SL in milk from WT, St3gal1−/−, St3gal4−/−, and St6gal1−/− mice isolated at 2 d postpartum. nd, not detectable. 3SL and 6SL were measured in WT milk (B) and in St3gal4−/− milk (C) throughout lactation. Amounts are given in grams per 100 g dried milk (DM). Values are given as mean ± SEM of 3 mice from 2 independent experiments (n = 3).
Feeding with milk deficient in 3SL increases the resistance of mice to dextran sulfate sodium–induced colitis
The role of milk 3SL and 6SL in the development of the mucosal immune system was addressed by feeding WT and St3gal4−/− newborn mice with normal or with 3SL-deficient milk by cross-fostering litters with WT and St3gal4−/− mothers. The same cross-fostering approach was applied to study the role of 6SL. Leukocyte populations and IgA secretion were determined in 3-, 6-, and 12-wk-old mice by flow cytometry and enzyme-linked immunosorbent assay, respectively. The T cell–specific markers TCRαβ/γδ, CD4, CD8, and CD8αα/αβ were measured in WT, St3gal4−/−, St6gal1−/−, and correspondingly cross-fostered mice. No differences were noticeable for leukocyte populations and IgA secretion (Fig. S3), indicating that neither the feeding with sialyllactose-deficient milk, nor the disruption of the St3gal4 and St6gal1 genes had any impact on the maturation of intestinal leukocytes.
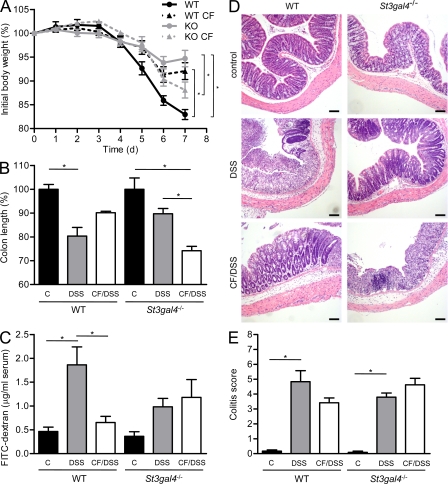
In a second approach, we addressed whether feeding with 3SL- and 6SL-deficient milk affected the response of mice to an intestinal challenge. At 7 wk of age, mice were exposed to dextran sulfate sodium (DSS) in drinking water for 5 d. DSS impairs the integrity of intestinal barrier, thereby inducing an acute colitis (Okayasu et al., 1990). WT mice responded strongly to the treatment, as shown by a loss of body weight of 17% by day 7. In contrast, St3gal4−/− mice were more resistant to DSS, as they lost only 5% of their body weight by day 7 (Fig. 3 A). WT mice that were cross-fostered and fed with 3SL-deficient milk sustained the DSS treatment better than those fed with normal milk, as shown by a reduced loss of body weight of 8%. Correspondingly, St3gal4−/− mice fed with normal milk were more susceptible to DSS-induced colitis than littermates fed with 3SL-deficient milk (Fig. 3 A). DSS-induced colitis in St6gal1−/− mice led to a similar disease as found in WT mice. Accordingly, cross-fostering experiments with normal and 6SL-deficient milk had no impact on the susceptibility to acute colitis (Fig. S4). We therefore focused on the investigation of 3SL in colitis development.
Figure 3.
DSS-induced colitis. (A) 7-wk-old WT mice (black circle, WT), cross-fostered WT mice (black triangle, WT CF), St3gal4−/− mice (gray circle, KO), and cross-fostered St3gal4−/− mice (gray triangle, KO CF) were fed DSS. (A) Body weight (n = 4–8). (B) Colon length. Values are given as percentage of untreated control mice; C, control untreated mice; DSS, DSS-treated mice; CF/DSS, cross-fostered DSS-treated mice (n = 4–8). Similar results were obtained in four independent experiments. (C) Intestinal permeability as measured by FITC-dextran in serum from control mice and colitogenic mice on day 5 of DSS-induced colitis as obtained from 3 independent experiments (n = 4–8). (D) Microscopic analysis of colon tissues. Colons were removed at day 7, fixed, and stained with hematoxylin and eosin. Representative results from 6 mice out of 2 independent experiments are shown. Bars, 100 µm. (E) Histological scoring of colitis by evaluation of epithelial damage and extension of leukocyte infiltration, as previously described (Hausmann et al., 2007). Three mice per group were analyzed in two independent experiments (total 6) and three animals in untreated groups. *, P < 0.05.
The severity of colitis was also registered by measuring colon length, epithelial permeability, and histology. At day 7, colon length was shortened by 20–25% in WT mice, whereas it was shortened by 10% in St3gal4−/− mice (Fig. 3 B). Cross-fostered mice showed a degree of colon shortening that matched the loss of body weight. WT mice fed with 3SL-deficient milk showed only a 10% colon shortening, and St3gal4−/− mice fed with normal milk showed a more pronounced colon shortening (Fig. 3 B). A similar picture was obtained by examining epithelial permeability in the intestine. DSS treatment increased epithelial leakiness in WT mice, but not in St3gal4−/− mice. The finding was reversed when looking at cross-fostered WT and St3gal4−/− mice fed with 3SL-deficient and normal milk, respectively (Fig. 3 C). Histological examination of DSS-treated mice revealed severe inflammation with massive infiltration of leukocytes into the mucosa and loss of entire crypts and surface epithelium, particularly in the middle to distal colon at day 7, for WT and cross-fostered St3gal4−/− mice (Fig. 3 D). St3gal4−/− mice were less affected by DSS treatment as focal inflammation was less pronounced. The extent of tissue damage and leukocyte infiltration in WT and St3gal4−/− mice correlated with the exposure to milk 3SL during feeding. The impact of cross fostering on the colitis of WT and St3gal4−/− mice was confirmed by grading the extent of epithelial damage and leukocyte infiltration in the medial and distal colon (Hausmann et al., 2007). The colitis score of WT mice fed with 3SL-deficient milk showed a nonstatistically significant lower trend than that of WT mice fed with normal milk. Correspondingly, the score of St3gal4−/− mice fed with normal milk tended to be higher, yet nonstatistically significantly, than that of St3gal4−/− mice fed with 3SL-deficient milk (Fig. 3 E).
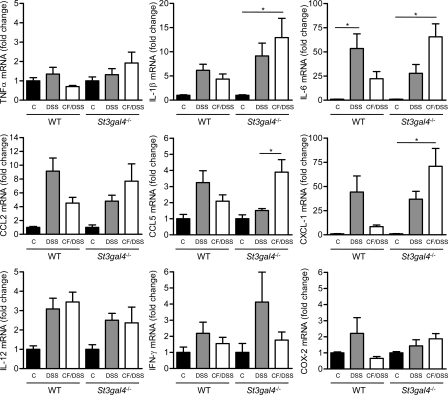
To better characterize the inflammatory response, we first measured the expression of chemokine and inflammatory marker genes in colon tissue on day 7 of DSS treatment. Whereas the levels of TNF and IL-1β expression were only partially altered by milk feeding history, IL-6 expression was induced 59-fold in St3gal4−/− mice, which had been fed with normal milk, when compared with an ∼25-fold increase observed in mice exposed to 3SL-deficient milk (Fig. 4). WT mice exposed to normal milk during lactation contained elevated levels of IL-6 that was again reduced in mice previously exposed to 3SL-deficient milk. The expression of the chemokines CCL2, CCL5, and CXCL-1 was elevated in mice with severe inflammation and correlated with the exposure to normal milk. Similarly, the expression of other inflammatory markers, such as IL-12, IFN-γ, and COX-2 was also increased in DSS-treated mice, yet to lower levels (Fig. 4).
Figure 4.
Expression of mRNA encoding inflammation markers and chemokines in colons of mice at day 7 of DSS treatment. 7-wk-old WT mice, cross-fostered WT mice, St3gal4−/− mice and cross-fostered St3gal4−/− mice were fed DSS. C, control untreated mice, DSS, DSS-treated mice, CF/DSS, cross-fostered DSS-treated mice. mRNA levels were measured using the 2−ΔΔct method and normalized to the levels of the GAPDH mRNA. Values are shown relative to the mRNA levels of untreated control mice. Data are represented by mean ± SEM from 4 samples out of 2 independent experiments (n = 8). *, P < 0.05.
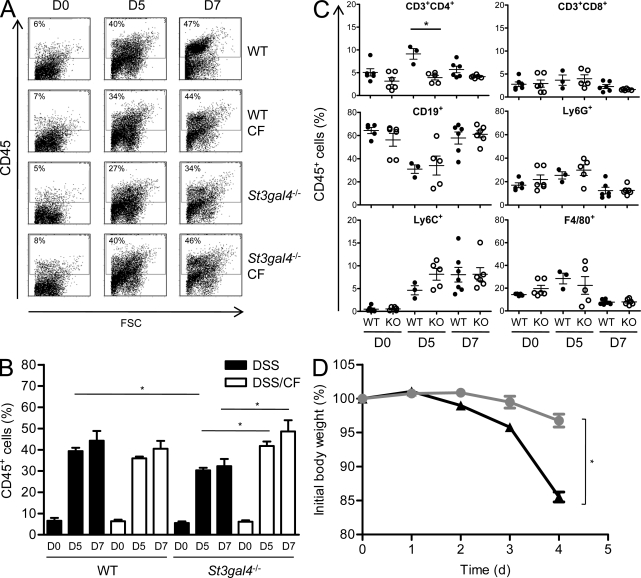
The extent and types of cells infiltrating the inflamed colons were analyzed by flow cytometry in WT, St3gal4−/− mice, and correspondingly cross-fostered mice. By day 5, 27–40% of cells recovered from the lamina propria were CD45+ (Fig. 5 A). By day 7, an additional 5–10% CD45+ cells were detected. Leukocyte infiltration was highest in mice with normal milk feeding history, but infiltration was only slightly decreased in mice that had been fed with 3SL-deficient milk (Fig. 5 B). The CD45+ cells were identified as B cells (68%, CD19+) and granulocytes (20%, Ly6G+), with a minor population of T cells (7%, CD3+), inflammatory monocytes (6%, Ly6C+ high), and macrophages (7%, F4/80+; Fig. 5 C and Fig. S5). The number of Th1 and Th17 cells, as assessed by respective IFN-γ and IL-17 detection in CD4+ T cells, was equal in WT mice by day 7 of DSS treatment, whereas a threefold decrease of Th1 against Th17 cells was found in St3gal4−/− mice (Fig. S5). The main increase in infiltrating leukocytes was caused by Ly6C+ inflammatory monocytes, as previously observed in IBD patients (Flanagan et al., 2008). To determine whether the genotype of the infiltrating leukocytes affected the extent of DSS-induced colitis, we subjected irradiated WT mice transplanted with St3gal4−/− bone marrow and irradiated St3gal4−/− mice transplanted with WT bone marrow to DSS treatment. After transplantation, mice were given 7 wk to recover from mucosal injury sustained during irradiation before being subjected to DSS treatment. WT mice bearing immune cells from St3gal4−/− mice rapidly developed colitis and lost 15% of body weight, whereas St3gal4−/− mice bearing WT immune cells did only lose 3% of weight loss at the same time point (Fig. 5 D). The fact that St3gal4−/− mice remained resistant to DSS treatment regardless of the genotype of infiltrating leukocytes, i.e., WT or St3gal4−/−, established that the resistance to DSS was inherent to the intestinal tissue and not affected by the deletion of St3gal4 in blood leukocytes.
Figure 5.
Hematopoietic cell infiltration and bone marrow chimera of WT and St3gal4−/− mice. Colonic cells were isolated and stained with anti-CD45 APC-Cy7 and analyzed by FACS. Mice were analyzed on day 0 (D0), 5 (D5), and 7 (D7) of DSS treatment. (A) Representative dot plots of WT, cross-fostered WT mice (WT CF), St3gal4−/−, and cross-fostered St3gal4−/− mice (St3gal4−/− CF) from three independent experiments are shown. The percentage of gated cells is indicated at the top left corner of each plot. (B) Quantitation of CD45+ cells in colon infiltrates from 3 independent experiments (n = 4–8). *, P < 0.05. (C) Leukocyte subsets in colons on day 0 (D0), 5 (D5), and 7 (D7) of DSS treatment as obtained from 2 independent experiments (n = 3–6). *, P < 0.05, KO: St3gal4−/−. (D) Body weight of WT mice transplanted with St3gal4−/− bone marrow cells (black triangles; n = 10) and St3gal4−/− mice transplanted WT bone marrow cells (gray circles; n = 11) during DSS treatment. Data represent the mean ± SEM of 10–11 mice from 2 independent experiments. *, P < 0.05.
Feeding with 3SL-deficient milk modifies the bacterial colonization of the mouse intestine
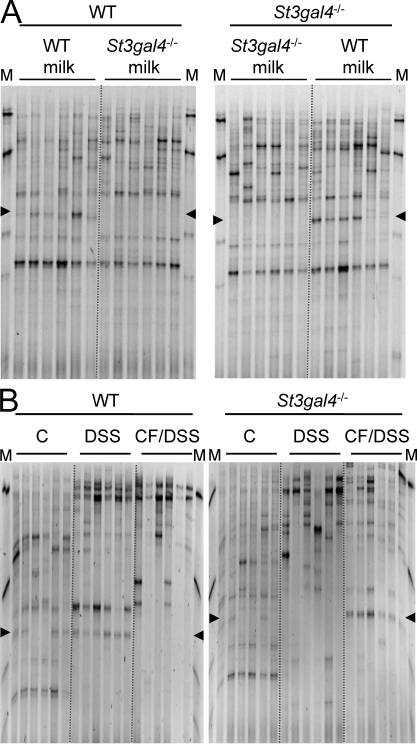
We did show so far that the exposition to milk in the first 3 wk of life had an impact on the susceptibility to DSS-induced colitis tested in adult mice. Considering the known effect of milk oligosaccharides as prebiotics (Bode, 2009), we addressed whether the presence or absence of 3SL affected the composition of the intestinal microbiota and thereby the outcome of DSS-induced colitis. To address the first point, we fingerprinted the intestinal microbiota of WT, St3gal4−/−, and cross-fostered mice by temporal temperature gradient gel electrophoresis (TTGE) and by real-time PCR. TTGE fingerprints from each 6 mice of each group were analyzed at 3, 6 (Fig. 6 A), and 12 wk of age. Despite internal differences within groups, cluster analysis indicated a higher degree of similarity within than between these groups. A specific band, identified by sequencing as representing a Ruminococcaceae species (Fig. S6), was only detectable by TTGE in the microbiota of mice exposed to 3SL, namely in WT mice and in St3gal4−/− mice fed with WT milk (Fig. 6 A). The sequence obtained from the TTGE band indicated that the Ruminococcaceae species was very close to Ruminococcaceae from the clostridial cluster IV, yet different from the species known to date. By comparison, the presence or absence of 6SL as tested with St6gal1−/− mice did not affect the occurrence of the Ruminococcaceae in the intestinal microbiota (Fig. S7), indicating that these bacteria require 3SL for gut colonization.
Figure 6.
Microbiota analysis in the mouse gastrointestinal tract. Temporal temperature gradient gel electrophoresis profiles of 16S rDNA gene amplification products from cecum of 6 wk-old WT and St3gal4−/− mice fed with either WT or St3gal4−/− milk (A) and after DSS-induced colitis (B). The data represent the results of one out of two independent experiments with six animals per group. The marker (M) shows amplification products from the species Lactobacillus plantarum, Lactococcus lactis, Corynebacterium variabile, Brevibacterium linens, and Arthrobacter protophormiae from top to bottom. Arrowheads mark the position corresponding to the Ruminococcaceae bands.
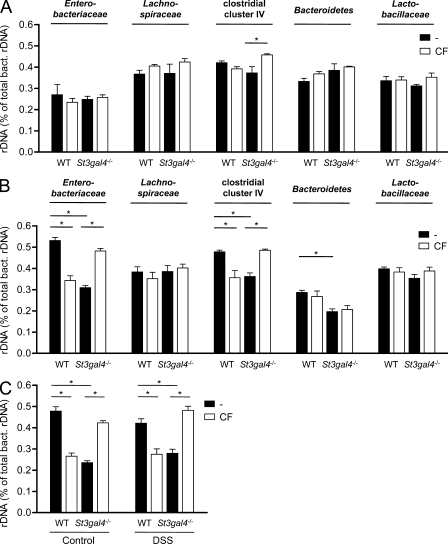
The diversity of the intestinal microbiota was also determined during DSS-induced colitis. A lower amount of bands on the gels were detected in DSS-treated mice compared with healthy animals, indicating a decrease in microbial diversity (Fig. 6 B). The band distribution on the gel was also shifted in mice treated with DSS, showing a change in microbial composition. Interestingly, the Ruminococcaceae-related band remained unchanged during colitis. The intestinal microbiota DNA samples investigated by TTGE (Fig. 6, A and B) were also tested by real-time PCR analysis to determine the distribution of the five phylogenic groups (Enterobacteriaceae, Lachnospiraceae, clostridial cluster IV, Bacteroidetes, and Lactobacillaceae) in WT, St3gal4−/−, and cross-fostered mice. This analysis confirmed that 3SL deficiency in the milk affected the colonization of clostridial cluster IV bacteria (Fig. 7 A). However, during DSS-induced colitis, the previous exposure to 3SL in milk influenced the relative composition of Enterobacteriaceae and the clostridial cluster IV to total bacteria. Enterobacteriaceae and the clostridial cluster IV were more abundant in WT and cross-fostered St3gal4−/− mice after DSS treatment (Fig. 7 B).
Figure 7.
Quantitative analysis of gastrointestinal microbiota. The microbiota composition of WT, St3gal4−/−, and respective cross-fostered (CF) mice was determined by real-time PCR in native microbiota (A) and microbiota on day 7 of DSS treatment (B). (C) Real-time PCR of Ruminococcaceae species in WT, St3gal4−/−, and CF mice in control conditions and after 7 d of DSS-induced colitis. Values are shown as relative amount to total bacteria 16S rDNA measured by the 2−ΔΔCt method. Two independent experiments were performed and data show means ± SEM of 4–6 animals (n = 4–6). *, P < 0.05.
Considering the differential detection of a Ruminococcaceae species by TTGE analysis and the fact that several Ruminococcaceae are part of the clostridial cluster IV (Van Dyke and McCarthy, 2002), we have measured the abundance of these bacteria using PCR primers that specifically target 16S recombinant DNA (rDNA) from Ruminococcus genera (Fig. S8). In agreement with the TTGE data, the Ruminococcaceae species was more abundant in mice that had been exposed to 3SL containing milk, i.e., in WT and cross-fostered St3gal4−/− mice (Fig. 7 C). The treatment with DSS did not alter the levels of Ruminococcaceae, thereby confirming the TTGE findings (Fig. 6 B).
Microbiota isolated from mice fed with 3SL-deficient milk increase the resistance of reconstituted germ-free mice to DSS-induced colitis
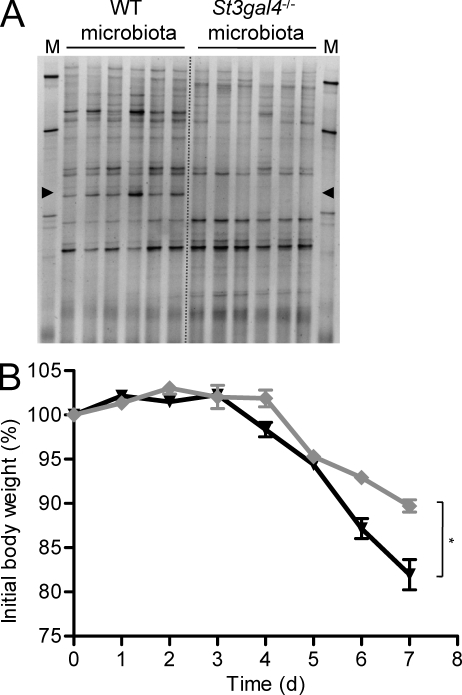
To demonstrate that the effect of 3SL-deficiency toward DSS treatment was mediated by selective bacterial colonization of the intestine, we have reconstituted germ-free mice with intestinal microbiota isolated from the cecum of WT and St3gal4−/− mice. Colonization success was confirmed by TTGE analysis of fecal pellets 2 wk after reconstitution. The microbiota from WT mice included the Ruminococcus-related specific band, whereas the microbiota from St3gal4−/− mice lacked this major band (Fig. 8 A). The mice with reconstituted microbiota were then subjected to DSS treatment by 12 wk of age. The mice with the WT-derived microbiota showed a progressive loss of body weight similar to WT mice, reaching 82% of their initial body weight by day 7 (Fig. 8 B). In contrast, the mice with the St3gal4−/−-derived microbiota showed a moderate weight loss to 90% of initial body weight, thus demonstrating that the composition of the intestinal microbiota had a direct influence on the susceptibility to DSS-induced colitis.
Figure 8.
Colitis in reconstituted germ-free mice. (A) Temporal temperature gradient gel electrophoresis profiles of 16S rDNA amplification products from feces of mice colonized with WT or St3gal4−/− microbiota at 2 wk after colonization (n = 6). The marker (M) shows amplification products from the species Lactobacillus plantarum, Lactococcus lactis, Corynebacterium variabile, Brevibacterium linens, and Arthrobacter protophormiae from top to bottom. Arrowheads mark the position corresponding to the Ruminococcaceae bands. (B) Body weight change in DSS-induced colitis of reconstituted germ-free mice colonized with WT (black triangles) or St3gal4−/− (gray diamonds) microbiota (n = 6). The difference in body weights measured at day 7 was significant (P < 0.05). The data show the results from 1 experiment performed with 6 animals per group (n = 6).
DISCUSSION
This study demonstrated the impact of the milk sialyllactose 3SL on the colonization of the mouse intestinal microbiota, and thereby on the susceptibility to DSS-induced colitis. The application of the cross-fostering setup between WT and St3gal4−/− mice demonstrated for the first time that a single oligosaccharide structure influences microbial composition in vivo. The different abundance of specific bacterial groups like Ruminococcaceae in mice fed with normal or 3SL-deficient milk correlated with the susceptibility to DSS-induced colitis. The first question arising when considering the model investigated is whether the oligosaccharide 3SL exerts regulatory functions on the mucosal immune system, thereby influencing the immune response to DSS exposure. Our survey of leukocyte populations did not reveal any differences between WT, St3gal4−/− and cross-fostered mice, thus speaking against such an immunoregulatory effect of 3SL. Furthermore, the bone marrow transplantation between WT and St3gal4−/− mice showed that the susceptibility to DSS did not correlate with the genotype of leukocytes. Finally, the differential susceptibility of reconstituted germ-free mice to DSS demonstrated that the effect of 3SL-deficient milk was mediated by microbiota and not by the mucosal immune system.
The exposure to 3SL during lactation could influence the colonization of intestinal bacteria by affecting their adhesion to the intestinal epithelium or by serving as nutrients for specific groups of bacteria. Milk 3SL could impair the attachment of bacteria binding to sialylated surfaces (Sakarya et al., 2003) or induce phase variation, thereby decreasing type 1 fimbriae expression on some bacteria (Sohanpal et al., 2004). However, we could not address such an effect of 3SL on the colonization of Ruminococcaceae because we have not succeeded at isolating this bacterial group in culture. Alternatively, 3SL could be used as a carbon and nitrogen source, which would facilitate the proliferation of bacteria capable of metabolizing sialic acid, as shown for the intestinal colonization of E. coli and V. cholerae (Chang et al., 2004; Almagro-Moreno and Boyd, 2009b). The Nan cluster of genes required for the catabolism of sialic acid is found in several pathogenic and commensal bacteria including, Ruminococcus gnavus (Almagro-Moreno and Boyd, 2009a). Thus, the selective colonization of Ruminococcaceae in the presence of 3SL might be mediated through their ability to use 3SL for energy gain.
The Gram-positive Ruminococcaceae are obligate anaerobes that are commonly found in the colon of mammals such as mice and humans (Collins et al., 1994). Ruminococcaceae are known to ferment polysaccharides like cellulose and starch (Herbeck and Bryant, 1974; Wang et al., 1997; Leitch et al., 2007). The correlation between the abundance of the Ruminococcus-related species and the susceptibility to DSS-induced colitis suggests that this bacterium may have a proinflammatory action. The fact that Ruminococcaceae have been found enriched in patients with inflammatory bowel disease (Prindiville et al., 2004; Martinez-Medina et al., 2006; Andoh et al., 2007) supports their potential role as proinflammatory bacteria. Moreover, the intestinal colonization of Ruminococcaceae may impair the settlement of other bacterial groups, which are known to attenuate the extent of the inflammatory response (Nanda Kumar et al., 2008; Im et al., 2009)
At the present stage, it is not possible to demonstrate a direct relationship between the intestinal abundance of Ruminococcaceae and the severity of DSS-induced colitis. The isolation and culture of Ruminococcaceae in vitro would allow the selective reconstitution of germ-free mice with these bacteria and subsequently to address the susceptibility of the mice to DSS treatment. We are currently trying to enrich for Ruminococcaceae on 3SL-containing media to assess their exact role in the development of colitis.
Our study has demonstrated that the exposure to a single milk oligosaccharide structure can significantly influence intestinal bacterial colonization and thereby affect the susceptibility of the host to DSS-induced colitis. The fact that mice fed with 3SL-deficient milk were more resistant to DSS treatment is somehow paradoxical, considering that this oligosaccharide is evolutionary conserved in most mammals. Although 3SL has a proinflammatory effect in the DSS model, it can be assumed that 3SL may mediate protective actions, e.g., by preventing the adhesion of pathogenic viruses and bacteria during infancy. Further, the 3SL-dependent microbiota might confer an evolutionary advantage by promoting an inflammatory defense reaction upon an infection challenge. The fact that 3SL levels in milk are elevated in the first days postpartum and strongly decrease until weaning may indicate the need for a balanced availability of the oligosaccharide in the developing gastrointestinal tract. The study of additional immunological challenges and infectious models in sialyltransferase−/− mice will further clarify the biological importance of sialylated milk oligosaccharides in the physiology of the gastrointestinal tract.
MATERIALS AND METHODS
Mouse models.
Sialyltransferase St6gal1−/− (Hennet et al., 1998), St3gal1−/− (Priatel et al., 2000), and St3gal4−/− (Ellies et al., 2002) were provided by J. Marth (University of California, Santa Barbara, Santa Barbara, CA). All mice were in the C57BL/6 background. Sialyltransferase-deficient and WT control mice were housed in light-cycled and climate-controlled rooms. All experiments were performed in compliance with the Swiss Animal Protection Ordinance and approved by the Veterinary Office of the Canton of Zürich, Switzerland. Synchronized matings were set up for sialyltransferase and WT control mice to allow the exchange of newborn mice for cross-fostering experiments. To this end, both the mothers and the litter of the other genotype were transferred to new cages, including parts of the mother’s nests.
Sialyltransferase gene expression in mammary glands.
Commercial RNA (Axxora Ltd.) isolated from mammary glands of virgin mice and mice after 1 and 2 wk postpartum and 3 d after weaning was used to monitor sialyltransferase expression profiles by real-time PCR (ABI Prism 7000; Applied Biosystems) using a SybrGreen protocol. In brief, RNA samples were treated with DNase (DNA-Free; Ambion) according to the manufacturer’s instructions. RT reactions were performed with 2 µg of total RNA and random hexamer primers using the Thermoscript RT-PCR System (Invitrogen) according to manufacturer’s instructions. Real-time PCR reactions were performed using the SYBRGreen PCR Master Mix (Applied Biosystems) of 24 µl and 1 µl of test cDNA per reaction. The primers used for amplification of mouse sialyltransferase genes are listed in Table S1. After an initial denaturation step of 10 min at 95°C, 40 cycles were performed at 95°C for 15 s, 60°C for 1 min, and 72°C for 1 min. Gene expression was normalized to GAPDH expression and calculations were done according to the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Mouse milking.
Lactating mothers were separated from their suckling newborns for 4 h. Milk ejection was stimulated by intraperitoneal injection of 0.5 IU of oxytocin (Sigma-Aldrich). Mice were anesthetized by ketamine (0.65 ml/kg) and xylazine (0.5 ml/kg). Milk was collected by aspiration (Haberman, 1974; Nagasawa, 1979), and then frozen and lyophilized. Dry matter was determined by weighing, and samples were resuspended in water to yield a stock solution of 100 mg/ml.
Milk oligosaccharide analysis.
For each sample, an equivalent of 16.5 µg dry matter was separated on a CarboPac PA200 analytical column (Dionex) with an amino trap guard (Dionex) using a high performance anion exchange chromatography system ICS3000 (Dionex) equipped with a pulsed amperometry detector. The column compartment was set to 25°C and the flow speed to 0.38 ml/min. The running conditions were as follows: isocratic 30 mM NaOH (Avantor) for 10 min followed by a linear gradient to 100 mM NaOH for 10 min followed by isocratic 100 mM NaOH for 10 min and a linear gradient from 0 to 100 mM Na-acetate (Merck) for an additional 35 min. Each run was preceded by a washing and equilibration step: isocratic 500 mM Na-acetate for 5 min, followed by isocratic 300 mM NaOH for 10 min, followed by isocratic 30 mM NaOH for 10 min. The retention times for 6SL and 3SL were 38.3 min and 38.7 min, respectively. Peak identification was done based on retention time comparison with authentic external sialyllactose standards (Dextra Laboratories) and disappearance of sialyllactose peaks upon neuraminidase treatment with simultaneous appearance of N-acetylneuraminic acid. For quantification, a standard curve with 50, 100, and 250 ng authentic 6SL and 3Sl standards was established before and after injection of five milk samples.
IgA analysis.
Fresh feces pellets were homogenized in PBS containing 0.1 mg/ml trypsin inhibitor (Sigma-Aldrich) at 4°C and cleared by centrifugation at 10,000 g for 10 min. Supernatants were serially diluted in PBS containing 0.05% Tween (Fluka)/0.1% BSA (Fluka) and 50 µl were applied per well. IgA was determined by sandwich-ELISA using anti–mouse IgA antibody (BD) as coating antibody and biotinylated rat anti–mouse IgA antibody (BD) as detection antibody. The antibody complex was detected using streptavidin-HRP (Sigma-Aldrich) and tetramethylbenzidine substrate solution (BD) at 440 nm in an ELISA plate photometer (Tecan). A purified mouse IgA standard (BD) was used for quantification.
DSS-induced colitis.
7-wk-old sex-matched mice were treated with 3.75% (wt/vol) DSS (molecular mass = 36–50 kD; MP Biomedicals) in drinking water for 5 d, followed by a supply of normal water until sacrifice of the animals (Okayasu et al., 1990). The lowest possible DSS dosage was chosen to achieve acute inflammation within 7 d. Body weight and physical activity were monitored daily. Animal pain was kept to a minimum by following the Swiss Animal Protection Ordinance and euthanizing animals reaching <85% of initial body weight.
Transepithelial permeability assay.
Mice were gavaged with 60 mg/100 g body weight of FITC-dextran (MW 3,000–5,000; Sigma-Aldrich; Napolitano et al., 1996). Mice were sacrificed and blood was isolated by cardiac puncture. Serum fluorescence (485/535 nm) was measured immediately using a Genios Multi-Detection Microplate Reader (Tecan, Switzerland). Concentrations were calculated from standard curves using serial dilutions of FITC-dextran in serum.
Histology.
The distal third of the colon was mechanically cleaned, cut longitudinally, and fixed in 4% paraformaldehyde, and then embedded in paraffin. Tissue samples were cut in serial 3-µm sections, which were stained with hematoxylin–eosin. Histological examination was performed in a blinded fashion. Histological scoring of 3 sections obtained from 3 sites at 100 µm distance was graded on a scale of 0 (normal morphology, no infiltrate) to 8 (loss of crypts in large areas, infiltration of the lamina submucosa) as described previously (Hausmann et al., 2007).
Cytokine gene expression.
RNA from frozen colon tissue was isolated using the RNeasy Protect Mini kit (QIAGEN) according to the manufacturer’s instructions. RT was performed with 2 µg total RNA using oligo(dT) primers and an Omniscript RT kit (QIAGEN). Real-time PCR was performed using SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) with specific primers for IL-1β, IL-6, TNF, IL-12, IFN-γ, and GAPDH (QuantiTect Primer; QIAGEN) and CCL2, CCL5, CXCL-1, and COX-2 (Table S1) in a Mx3000P thermocycler (Stratagene). Cycling conditions were 40 cycles at 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s after initial denaturation at 95°C for 10 min. Gene expression was normalized to GAPDH expression using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Lamina propria leukocyte isolation.
Colon lamina propria leukocytes were isolated as described previously (Lefrançois and Lycke, 2001). In brief, to detach intraepithelial lymphocytes, chopped colon segments were incubated two times for 30 min at 37°C under constant stirring condition in 50 ml of Ca2+- and Mg2+-free Hanks’ balanced salt solution containing 10 mM Hepes, 2% horse serum, 2 mM DTT, and 0.5 mM EDTA. Leukocytes were released by additional incubation with 0.5 mg/ml of collagenase type IV (Sigma-Aldrich) and 30 µg/ml of DNase I (Sigma-Aldrich) for 2 × 45 min at 37°C, and cells were filtered through a 40-µm nylon mesh cell strainer.
Flow cytometry.
Cells were stained on ice for 30 min with the following: anti-CD45 APC-Cy7, anti-CD19 APC, anti-CD3 FITC, anti-Gr.1 PE, anti-TCRγδ PE, anti-TCRβ APC, anti-CD4 PE, anti-CD8β PE, anti-CD8α PE-Cy5, anti-CD11c APC, anti-Ly-6G PerCP, anti-Ly-6C APC, anti-F4/80 Alexa Fluor 488, anti-CD11b PE, anti-CD4 PerCP-Cy5, anti–IL-17A PE, anti–IFN-γ FITC, and anti–IL-17 PE antibodies (BD). Cells were analyzed with a FACSCanto II flow cytometer (BD).
Bone marrow transfer.
10-wk-old male WT and St3gal4−/− recipient mice were lethally exposed to 9.33 Gy radiations (3.11 Gy/min). Femur and tibia from WT and St3gal4−/− donor mice were removed and flushed with RPMI 10% FCS (Invitrogen) to harvest BM cells. Irradiated recipient mice were reconstituted with 2 × 106 BM cells by intravenous injection (Spangrude, 2008). Mice were treated with antibiotics (Borgal 24% ad us. vet.; Veterinaria AG) for 3 wk.
Temporal temperature gradient gel electrophoresis.
DNA was isolated from freshly isolated cecal content using the QIAamp DNA Stool Mini kit (QIAGEN). Bacterial 16S rDNA was amplified using the universal primers HDA1-GC 5′-CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGGACTCCTACGGGAGGCAGCAGT-3′ and HDA-2 5′-GTATTACCGCGGCTGCTGGCAC-3′ according to Ogier (Ogier et al., 2002). The PCR conditions were 30 cycles at 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min, flanked by an initial denaturation at 94°C for 4 min and a final elongation at 72°C for 4 min. PCR products were loaded on 8.5% polyacrylamide gels containing 8 M urea and separated by TTGE using the D-Code universal mutation detection system (Bio-Rad Laboratories). Electrophoresis was performed in 60 mM Tris-acetate, 30 mM acetic acid, and 1.5 mM EDTA first at 20 V for 15 min, followed by a constant voltage at 80 V for 18 h with a temperature increase of 0.2°C/h from 66 to 70°C. TTGE profiles were analyzed by using GelCompar II software (Applied Maths).
Bacterial typization.
TTGE bands were excised from the gels and DNA was diffused over night at 4°C in 100 µl H2O. Standard HDA primers were used to reamplify the PCR product from 3 µl of eluate. PCR products were purified using QIAquick PCR purification kit (QIAGEN) and sequenced (Synergene Biotech). The proportion of bacterial phyla in intestinal samples was determined by real-time PCR using the SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) as described for cytokine gene expression and primer pairs specific for the lineages Bacteroidetes, Enterobacteriaceae, Lachnospiraceae, clostridial cluster IV, and Lactobacillaceae (Table S1). The forward primer specific for the Ruminococcaceae species was designed by choosing a stretch of 16S rDNA gene sequence that was distinctive from the corresponding sequences of other Ruminococcaceae of the clostridial cluster IV (Fig. S8). Cycling conditions were 40 cycles at 95°C for 15 s, 66°C for 20 s, and 72°C for 20 s after an initial denaturation at 95°C for 10 min. Quantification values were calculated by the 2−ΔΔCt method relative to total bacteria 16S rDNA amplification.
Germ-free colonization.
Cecal contents (100 mg) of WT and St3gal4−/− donor mice were collected under anaerobic conditions and diluted in 10 ml of anaerobic mineral solution containing 5 g/liter NaCl, 2 g/liter glucose, and 0.3 g/liter cysteine-HCl (de Sablet et al., 2009). 3–4-wk-old C57BL/6 germ-free males (Institute of Laboratory Animal Science, University of Zürich, Switzerland) were colonized with 200 µl of 1:100 diluted cecal microbiota by gavage and kept in isolators for 4 wk.
Statistics.
Results were expressed as mean ± SEM. Difference between groups was analyzed using one-way analysis of variance with Bonferroni’s Multiple Comparison Post-test. Significance was accepted for P < 0.05.
Online supplemental material.
Fig. S1 shows the mRNA levels of sialyltransferases normalized to GAPDH expression in mammary gland tissue. Fig. S2 outlines the determination of sialic acid in mouse milk. Fig. S3 shows unchanged T cell development and IgA production in the intestine of St3gal4−/− and St6gal1−/− mice. Fig. S4 shows the normal susceptibility of St6gal1−/− mice to DSS-induced colitis. Fig. S5 shows the typing of leukocyte subsets in the colon of DSS-treated mice. Fig. S6 outlines the identification of a Ruminococcaceae species from TTGE analysis. Fig. S7 shows the intestinal microbiota analysis in St6gal1−/− mice. Fig. S8 explains the definition of a PCR primer specific for the new Ruminococcaceae species. Table S1 lists the primers used in the present study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20101098/DC1.
Acknowledgments
The authors kindly thank Monique Julita and John Newell for excellent technical assistance with milk oligosaccharide and mammary gland gene expression analysis as well as Charlotte Burger and Isabelle Frey for assistance with histology.
This work was supported by the Zürich Center for Integrative Human Physiology and by the Swiss National Foundation grant 31003A-116039 to T. Hennet.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- 3SL
- sialyl(α2,3)lactose
- 6SL
- sialyl(α2,6)lactose
- DSS
- dextran sulfate sodium
- mRNA
- messenger RNA
- rDNA
- ribosomal DNA
- TTGE
- temporal temperature gradient gel electrophoresis
- WT
- wild-type
References
- Almagro-Moreno S., Boyd E.F. 2009a. Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol. Biol. 9:118 10.1186/1471-2148-9-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro-Moreno S., Boyd E.F. 2009b. Sialic acid catabolism confers a competitive advantage to pathogenic vibrio cholerae in the mouse intestine. Infect. Immun. 77:3807–3816 10.1128/IAI.00279-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andoh A., Sakata S., Koizumi Y., Mitsuyama K., Fujiyama Y., Benno Y. 2007. Terminal restriction fragment length polymorphism analysis of the diversity of fecal microbiota in patients with ulcerative colitis. Inflamm. Bowel Dis. 13:955–962 10.1002/ibd.20151 [DOI] [PubMed] [Google Scholar]
- Bode L. 2009. Human milk oligosaccharides: prebiotics and beyond. Nutr. Rev. 67:S183–S191 10.1111/j.1753-4887.2009.00239.x [DOI] [PubMed] [Google Scholar]
- Brand Miller J., McVeagh P., McNeil Y., Gillard B. 1995. Human milk oligosaccharides are not digested and absorbed in the small intestine of young infants. Proc. Nutr. Soc. Aus. 19:44 [Google Scholar]
- Chang D.-E., Smalley D.J., Tucker D.L., Leatham M.P., Norris W.E., Stevenson S.J., Anderson A.B., Grissom J.E., Laux D.C., Cohen P.S., Conway T. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA. 101:7427–7432 10.1073/pnas.0307888101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M.D., Lawson P.A., Willems A., Cordoba J.J., Fernandez-Garayzabal J., Garcia P., Cai J., Hippe H., Farrow J.A.E. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812–826 10.1099/00207713-44-4-812 [DOI] [PubMed] [Google Scholar]
- Dalziel M., Huang R.Y., Dall’Olio F., Morris J.R., Taylor-Papadimitriou J., Lau J.T.Y. 2001. Mouse ST6Gal sialyltransferase gene expression during mammary gland lactation. Glycobiology. 11:407–412 10.1093/glycob/11.5.407 [DOI] [PubMed] [Google Scholar]
- de Sablet T., Chassard C., Bernalier-Donadille A., Vareille M., Gobert A.P., Martin C. 2009. Human microbiota-secreted factors inhibit shiga toxin synthesis by enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 77:783–790 10.1128/IAI.01048-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L., McFall-Ngai M., Relman D.A. 2007. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 449:811–818 10.1038/nature06245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M., Gill S.R., Nelson K.E., Relman D.A. 2005. Diversity of the human intestinal microbial flora. Science. 308:1635–1638 10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egge H. 1993. The diversity of oligosaccharides in human milk. New Perspectives in Infant Nutrition. Renner B., Sawatzki G., 12–26 [Google Scholar]
- Ellies L.G., Ditto D., Levy G.G., Wahrenbrock M., Ginsburg D., Varki A., Le D.T., Marth J.D. 2002. Sialyltransferase ST3Gal-IV operates as a dominant modifier of hemostasis by concealing asialoglycoprotein receptor ligands. Proc. Natl. Acad. Sci. USA. 99:10042–10047 10.1073/pnas.142005099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk P.G., Hooper L.V., Midtvedt T., Gordon J.I. 1998. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol. Mol. Biol. Rev. 62:1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan K., Modrusan Z., Cornelius J., Chavali A., Kasman I., Komuves L., Mo L., Diehl L. 2008. Intestinal epithelial cell up-regulation of LY6 molecules during colitis results in enhanced chemokine secretion. J. Immunol. 180:3874–3881 [DOI] [PubMed] [Google Scholar]
- Frank D.N., Pace N.R. 2008. Gastrointestinal microbiology enters the metagenomics era. Curr. Opin. Gastroenterol. 24:4–10 10.1097/MOG.0b013e3282f2b0e8 [DOI] [PubMed] [Google Scholar]
- Gibson G.R., Roberfroid M.B. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401–1412 [DOI] [PubMed] [Google Scholar]
- Haberman B. 1974. Mechanical milk collection from mice for Bittner virus isolation. Lab. Anim. Sci. 6:935–937 [PubMed] [Google Scholar]
- Harduin-Lepers A., Vallejo-Ruiz V., Krzewinski-Recchi M.A., Samyn-Petit B., Julien S., Delannoy P. 2001. The human sialyltransferase family. BMC Evol. Biol. 83:727–737 [DOI] [PubMed] [Google Scholar]
- Harmsen H.J.M., Wildeboer-Veloo A.C.M., Raangs G.C., Wagendorp A.A., Klijn N., Bindels J.G., Welling G.W. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30:61–67 10.1097/00005176-200001000-00019 [DOI] [PubMed] [Google Scholar]
- Hausmann M., Obermeier F., Paper D.H., Balan K., Dunger N., Menzel K., Falk W., Schoelmerich J., Herfarth H., Rogler G. 2007. In vivo treatment with the herbal phenylethanoid acteoside ameliorates intestinal inflammation in dextran sulphate sodium-induced colitis. Clin. Exp. Immunol. 148:373–381 10.1111/j.1365-2249.2007.03350.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennet T., Chui D., Paulson J.C., Marth J.D. 1998. Immune regulation by the ST6Gal sialyltransferase. Proc. Natl. Acad. Sci. USA. 95:4504–4509 10.1073/pnas.95.8.4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbeck J.L., Bryant M.P. 1974. Nutritional features of the intestinal anaerobe Ruminococcus bromii. Appl. Microbiol. 28:1018–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im E., Choi Y.J., Pothoulakis C., Rhee S.H. 2009. Bacillus polyfermenticus ameliorates colonic inflammation by promoting cytoprotective effects in colitic mice. J. Nutr. 139:1848–1854 10.3945/jn.109.108613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn N.J. 1972. The lactose and neuraminlactose content of rat milk and mammary tissue. Biochem. J. 130:177–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrançois L., Lycke N. 2001. Isolation of mouse small intestinal intraepithelial lymphocytes, Peyer’s patch, and lamina propria cells. Curr Protoc Immunol. Chapter3:Unit 3.19. [DOI] [PubMed] [Google Scholar]
- Leitch E.C.M., Walker A.W., Duncan S.H., Holtrop G., Flint H.J. 2007. Selective colonization of insoluble substrates by human faecal bacteria. Environ. Microbiol. 9:667–679 10.1111/j.1462-2920.2006.01186.x [DOI] [PubMed] [Google Scholar]
- Ley R.E., Peterson D.A., Gordon J.I. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 124:837–848 10.1016/j.cell.2006.02.017 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Macpherson A.J., Harris N.L. 2004. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 4:478–485 10.1038/nri1373 [DOI] [PubMed] [Google Scholar]
- Martinez-Medina M., Aldeguer X., Gonzalez-Huix F., Acero D., Garcia-Gil L.J. 2006. Abnormal microbiota composition in the ileocolonic mucosa of Crohn’s disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm. Bowel Dis. 12:1136–1145 10.1097/01.mib.0000235828.09305.0c [DOI] [PubMed] [Google Scholar]
- Mazmanian S.K., Round J.L., Kasper D.L. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 453:620–625 10.1038/nature07008 [DOI] [PubMed] [Google Scholar]
- Moro G., Arslanoglu S., Stahl B., Jelinek J., Wahn U., Boehm G. 2006. A mixture of prebiotic oligosaccharides reduces the incidence of atopic dermatitis during the first six months of age. Arch. Dis. Child. 91:814–819 10.1136/adc.2006.098251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa H. 1979. A device for milk collection from mice. Lab. Anim. Sci. 5:633–635 [PubMed] [Google Scholar]
- Nanda Kumar N.S., Balamurugan R., Jayakanthan K., Pulimood A., Pugazhendhi S., Ramakrishna B.S. 2008. Probiotic administration alters the gut flora and attenuates colitis in mice administered dextran sodium sulfate. J. Gastroenterol. Hepatol. 23:1834–1839 10.1111/j.1440-1746.2008.05723.x [DOI] [PubMed] [Google Scholar]
- Napolitano L.M., Koruda M.J., Meyer A.A., Baker C.C. 1996. The impact of femur fracture with associated soft tissue injury on immune function and intestinal permeability. Shock. 5:202–207 10.1097/00024382-199603000-00006 [DOI] [PubMed] [Google Scholar]
- Newburg D.S. 2009. Neonatal protection by an innate immune system of human milk consisting of oligosaccharides and glycans. J. Anim. Sci. 87:26–34 10.2527/jas.2008-1347 [DOI] [PubMed] [Google Scholar]
- Ogier J.-C., Son O., Gruss A., Tailliez P., Delacroix-Buchet A. 2002. Identification of the bacterial microflora in dairy products by temporal temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 68:3691–3701 10.1128/AEM.68.8.3691-3701.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayasu I., Hatakeyama S., Yamada M., Ohkusa T., Inagaki Y., Nakaya R. 1990. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 98:694–702 [DOI] [PubMed] [Google Scholar]
- Priatel J.J., Chui D., Hiraoka N., Simmons C.J., Richardson K.B., Page D.M., Fukuda M., Varki N.M., Marth J.D. 2000. The ST3Gal-I sialyltransferase controls CD8+ T lymphocyte homeostasis by modulating O-glycan biosynthesis. Immunity. 12:273–283 10.1016/S1074-7613(00)80180-6 [DOI] [PubMed] [Google Scholar]
- Prieto P.A., Mukerji P., Kelder B., Erney R., Gonzalez D., Yun J.S., Smith D.F., Moremen K.W., Nardelli C., Pierce M., et al. 1995. Remodeling of mouse milk glycoconjugates by transgenic expression of a human glycosyltransferase. J. Biol. Chem. 270:29515–29519 10.1074/jbc.270.49.29515 [DOI] [PubMed] [Google Scholar]
- Prindiville T., Cantrell M., Wilson K.H. 2004. Ribosomal DNA sequence analysis of mucosa-associated bacteria in Crohn’s disease. Inflamm. Bowel Dis. 10:824–833 10.1097/00054725-200411000-00017 [DOI] [PubMed] [Google Scholar]
- Sakarya S., Ertem G.T., Oncu S., Kocak I., Erol N., Oncu S. 2003. Escherichia coli bind to urinary bladder epithelium through nonspecific sialic acid mediated adherence. FEMS Immunol. Med. Microbiol. 39:45–50 10.1016/S0928-8244(03)00185-8 [DOI] [PubMed] [Google Scholar]
- Savage D.C. 1977. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31:107–133 10.1146/annurev.mi.31.100177.000543 [DOI] [PubMed] [Google Scholar]
- Sohanpal B.K., El-Labany S., Lahooti M., Plumbridge J.A., Blomfield I.C. 2004. Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA. 101:16322–16327 10.1073/pnas.0405821101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangrude G.J. 2008. Assessment of lymphocyte development in radiation bone marrow chimeras. Curr. Protoc. Immunol. Chapter 4:Unit 4.6. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 444:1027–1031 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- Urashima T., Saito T., Nakamura T., Messer M. 2001. Oligosaccharides of milk and colostrum in non-human mammals. Glycoconj. J. 18:357–371 10.1023/A:1014881913541 [DOI] [PubMed] [Google Scholar]
- Van Dyke M.I., McCarthy A.J. 2002. Molecular biological detection and characterization of Clostridium populations in municipal landfill sites. Appl. Environ. Microbiol. 68:2049–2053 10.1128/AEM.68.4.2049-2053.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoffen E., Ruiter B., Faber J., M’Rabet L., Knol E.F., Stahl B., Arslanoglu S., Moro G., Boehm G., Garssen J. 2009. A specific mixture of short-chain galacto-oligosaccharides and long-chain fructo-oligosaccharides induces a beneficial immunoglobulin profile in infants at high risk for allergy. Allergy. 64:484–487 10.1111/j.1398-9995.2008.01765.x [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar M., Aitken J.D., Carvalho F.A., Cullender T.C., Mwangi S., Srinivasan S., Sitaraman S.V., Knight R., Ley R.E., Gewirtz A.T. 2010. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 328:228–231 10.1126/science.1179721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos A.P., Haarman M., van Ginkel J.-W.H., Knol J., Garssen J., Stahl B., Boehm G., M’Rabet L. 2007. Dietary supplementation of neutral and acidic oligosaccharides enhances Th1-dependent vaccination responses in mice. Pediatr. Allergy Immunol. 18:304–312 10.1111/j.1399-3038.2007.00515.x [DOI] [PubMed] [Google Scholar]
- Wang R.F., Cao W.W., Cerniglia C.E. 1997. PCR detection of Ruminococcus spp. in human and animal faecal samples. Mol. Cell. Probes. 11:259–265 10.1006/mcpr.1997.0111 [DOI] [PubMed] [Google Scholar]