RAG1 binding to TCR gene elements is dictated by transcriptional control elements and by transcription itself; these findings provide direct confirmation of the long-held accessibility model.
Abstract
V(D)J recombination assembles antigen receptor genes in a well-defined order during lymphocyte development. This sequential process has long been understood in the context of the accessibility model, which states that V(D)J recombination is regulated by controlling the ability of the recombination machinery to gain access to its chromosomal substrates. Indeed, many features of “open” chromatin correlate with V(D)J recombination, and promoters and enhancers have been strongly implicated in creating a recombinase-accessible configuration in neighboring chromatin. An important prediction of the accessibility model is that cis-elements and transcription control binding of the recombination-activating gene 1 (RAG1) and RAG2 proteins to their DNA targets. However, this prediction has not been tested directly. In this study, we use mutant Tcra and Tcrb alleles to demonstrate that enhancers control RAG1 binding globally at Jα or Dβ/Jβ gene segments, that promoters and transcription direct RAG1 binding locally, and that RAG1 binding can be targeted in the absence of RAG2. These findings reveal important features of the genetic mechanisms that regulate RAG binding and provide a direct confirmation of the accessibility model.
V(D)J recombination assembles the variable portion of antigen receptor loci from component variable (V), diversity (D), and joining (J) gene segments. Each of these gene segments is flanked by a recombination signal sequence (RSS) consisting of relatively well-conserved heptamer and nonamer elements separated by a less well-conserved spacer of 12 or 23 bp. V(D)J recombination is initiated when proteins encoded by the recombination-activating genes, RAG1 and RAG2, probably assisted by the high mobility group protein HMGB1 or HMGB2, bind one RSS and then capture a second RSS to create a synaptic complex. Within this complex, the RAG proteins introduce DNA double strand breaks between the RSSs and the gene segments; the reaction is then completed by the processing and ligation of the broken ends by the classical nonhomologous end joining DNA repair pathway (Swanson, 2004; Cobb et al., 2006). RAG1 plays a major role in RSS binding through its interactions with both the heptamer and nonamer, and subsequently in the catalysis of DNA cleavage (Swanson, 2004). RAG2 is an essential cofactor for DNA cleavage via its interaction with RAG1, enhances RSS binding, and contributes important regulatory functions, such as binding to the N-terminal tail of histone H3 when lysine 4 is trimethylated (H3K4me3; Liu et al., 2007; Matthews et al., 2007).
V(D)J recombination is tightly regulated in both a developmental stage– and a lineage-specific manner (Cobb et al., 2006; Jung et al., 2006; Krangel, 2007). For example, the Tcrb locus undergoes recombination in early CD4−CD8− (double negative, DN) thymocytes, whereas the Tcra locus is assembled at the later CD4+CD8+ (double positive, DP) stage of thymocyte development. Throughout this process, the immunoglobulin loci undergo little or no recombination. Tcrb locus assembly is itself a strictly ordered process, with D-to-J joining occurring before V-to-DJ joining. This precise regulation is achieved despite the use of the same enzymatic machinery for all recombination events and the conserved sequence features shared by all RSSs.
Our understanding of the mechanisms that dictate ordered V(D)J recombination has for many years been guided by the accessibility model (Yancopoulos and Alt, 1985), which proposes that the access of chromatinized RSSs to the V(D)J recombinase is modulated by developmental and stage-specific mechanisms. The model has received support from a wide range of experiments. V(D)J recombination of specific gene segments strongly correlates with features reflecting an open configuration at associated chromatin, including nuclease sensitivity, germline transcription, activating histone modifications, and DNA hypomethylation (Cobb et al., 2006; Jung et al., 2006; Krangel, 2007). Both in vivo (Stanhope-Baker et al., 1996) and biochemical studies (Kwon et al., 1998; Golding et al., 1999) have demonstrated that chromatin represents a significant barrier to the initiation of V(D)J recombination, and numerous findings indicate that promoters, enhancers, transcription factors, and transcription itself play key roles in overcoming this barrier. A central prediction of the accessibility model is therefore that transcriptional control elements and transcription are critical for allowing the recombination machinery to gain access to RSSs. However, this prediction has not been tested directly because methods for measuring RAG binding to DNA in vivo were unavailable.
We recently demonstrated, using chromatin immunoprecipitation (ChIP), that RAG1 and RAG2 bind to a focal region (termed the “recombination center”) containing some or all of the J gene segments within the Ig heavy chain (Igh), Igκ, Tcrb, and Tcra loci (Ji et al., 2010). Importantly, RAG1 and RAG2 were found to be recruited independently of one another into Igκ, Tcrb, and Tcra recombination centers. Although RAG2 binding closely mirrored the distribution of H3K4me3 throughout the entire genome, RAG1 binding was suggested to be strongly dependent on direct recognition of the RSS (Ji et al., 2010). How RAG1 binding is targeted and how this relates to the mechanisms that control accessibility is not known.
Here, we demonstrate that promoters, enhancers, and transcription are critical regulators of RAG1 binding to the Tcrb and Tcra loci, thereby validating a central tenet of the accessibility model.
RESULTS AND DISCUSSION
Control of RAG1 binding in the Tcra locus
The 1.6-Mb Tcra locus contains 61 J gene segments distributed throughout a 65-kb region near its 3′ end and ∼100 V gene segments scattered over a large 5′ region of the locus (Fig. 1 A). We recently found that RAG binding to Tcra chromatin occurs in DP but not DN thymocytes and focuses on the most 5′ Jα gene segments (Ji et al., 2010), which are strongly preferred in initial Tcra gene rearrangements (Krangel, 2007). Little or no binding was detected to Vα gene segments, leading us to propose that RAG proteins bind first to Jα segments, forming a “recombination center,” within which the RAG proteins capture a Vα segment for recombination (Ji et al., 2010).
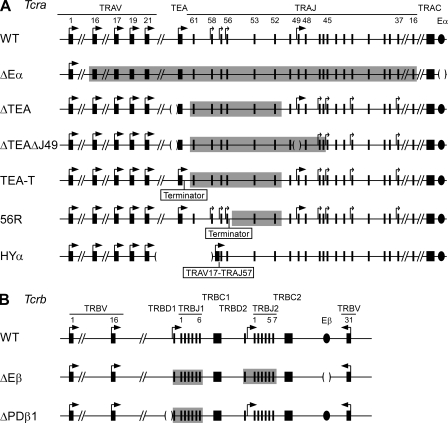
Figure 1.
Analysis of Tcra and Tcrb alleles. (A) Schematic maps of WT and mutant Tcra alleles are provided, with Eα represented as a filled oval and active promoters represented as large or small arrows depending on whether their activity is independent (large arrows) or dependent (small arrows) on the activity of other promoters. Promoters associated with TRAJ58, 57, and 56 are activated by TEA, whereas those associated with TRAJ47, 45, 42, and 37 are inhibited by TEA. Deleted regions are identified by parentheses. Shading identifies regions of mutant alleles that display reduced accessibility as indicated by histone modifications and recombination frequencies. (B) Schematic maps of WT and mutant Tcrb alleles are provided, with Eβ represented as a filled oval and active promoters represented as large arrows. Deleted regions and regions of reduced accessibility are identified as in A.
To investigate how Tcra locus assembly is controlled, we determined the pattern of RAG1 binding to six mutant Tcra alleles in which transcriptional control elements were deleted or repositioned, or in which transcriptional elongation was blocked (Fig. 1 A; shading indicates regions in which recombination is inhibited as a result of the mutation). WT and mutant alleles were analyzed in thymocytes from mice that were deficient in RAG2 and that expressed a rearranged Tcrb transgene. The absence of RAG2 ensured that all Tcra alleles remained in their unrearranged configuration while the Tcrb transgene allowed for the development of DP thymocytes, the cellular subset in which Tcra recombination takes place.
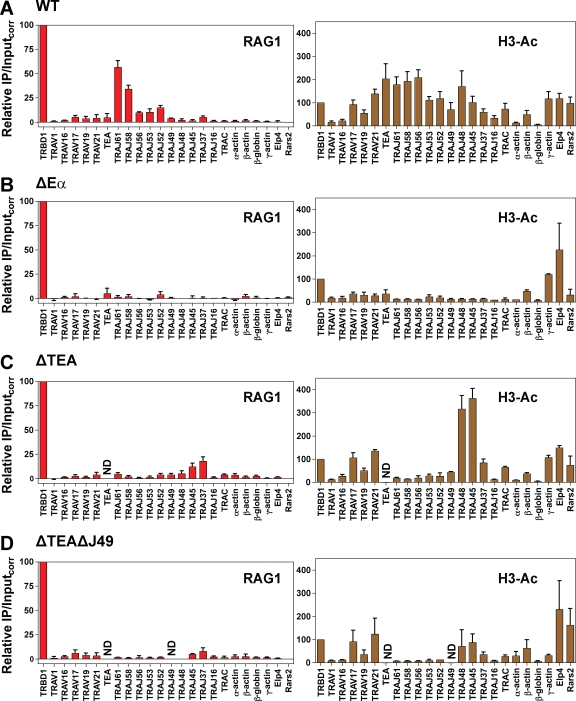
The Tcra enhancer (Eα), which lies 3′ of the Cα constant region, is critical for Tcra locus recombination, germline transcription from the TEA promoter (Sleckman et al., 1997), and histone acetylation across a 500-kb region that spans all of the Jα gene segments and the 3′ portion of the Vα cluster (Hawwari and Krangel, 2005; McMurry and Krangel, 2000). As expected (Ji et al., 2010), the WT Tcra allele showed strong binding of RAG1 at the most 5′ Jα gene segments analyzed (TRAJ61 and TRAJ58) and substantial acetylation of histone H3 across the majority of Vα and Jα gene segments analyzed (Fig. 2 A). In contrast, deletion of Eα (ΔEα allele) resulted in a complete loss of RAG1 binding and a strong reduction of histone H3 acetylation across the locus (Fig. 2 B). Therefore, Eα is required to establish a chromatin state that supports binding of RAG1 to the Tcra locus.
Figure 2.
The effect of enhancer or promoter deletion on RAG1 binding to Tcra. (A–D) Binding of RAG1 (left) or levels of H3 acetylation (H3-Ac, right) at the indicated gene segments or regions were assessed by ChIP in primary thymocytes (almost entirely DP cells) from Rag2−/− Tcrb transgenic mice homozygous for a WT Tcra allele (A), the ΔEα allele (B), the ΔTEA allele (C), or the ΔTEAΔJ49 allele (D). DNA recovery in immunoprecipitates and in input DNA samples was assessed by qPCR and relative immunoprecipitation/inputcorr values were calculated as described in Materials and methods. These values have been corrected for background and are expressed relative to the signal obtained at the TRBD1 (Dβ1) gene segment, which was set arbitrarily to a value of 100. TRBD1 binds RAG1 robustly and exhibits substantial H3 acetylation in Rag2−/− x Tcrb-transgenic thymocytes (Ji et al., 2010 and not depicted). Data are the mean of four (A, RAG1), five (A, H3-Ac), three (C, RAG1), or two (all others) independent experiments involving individual mice, with bars indicating the mean and error bars representing the SEM. ND, not done.
Initial Tcra recombination events are regulated by two germline promoters: TEA, which lies ∼2 kb upstream of TRAJ61 and controls recombination to the most 5′ Jα gene segments (TRAJ61–TRAJ52; Villey et al., 1996; Hawwari et al., 2005), and the Jα49 promoter, which is located within TRAJ49 and directs primary recombination events to the region spanning TRAJ50–TRAJ45 (Hawwari et al., 2005). Deletion of TEA greatly reduced RAG1 binding and H3 acetylation at the 5′ end of the Jα cluster (TRAJ61–TRAJ52; Fig. 2 C), in close agreement with its effect on Tcra recombination (Villey et al., 1996). These data strongly support a role for TEA in the local control of V(D)J recombination through the regulation of RAG binding to RSSs. In the region 3′ of TRAJ52, both RAG1 binding and H3 acetylation were increased on the ΔTEA allele relative to WT (Fig. 2 C), probably because the Jα49 promoter and additional downstream promoters become more active in the absence of TEA (Abarrategui and Krangel, 2007; Hawwari and Krangel, 2007). When both the TEA and Jα49 germline promoters were deleted (ΔTEAΔJ49 allele), RAG1 binding and H3 acetylation were reduced in the region spanning TRAJ48–TRAJ37 (Fig. 2 D) relative to TEA deletion only (Fig. 2 C), which is consistent with a dominant role for the Jα49 promoter in controlling both chromatin structure and RSS accessibility in this region.
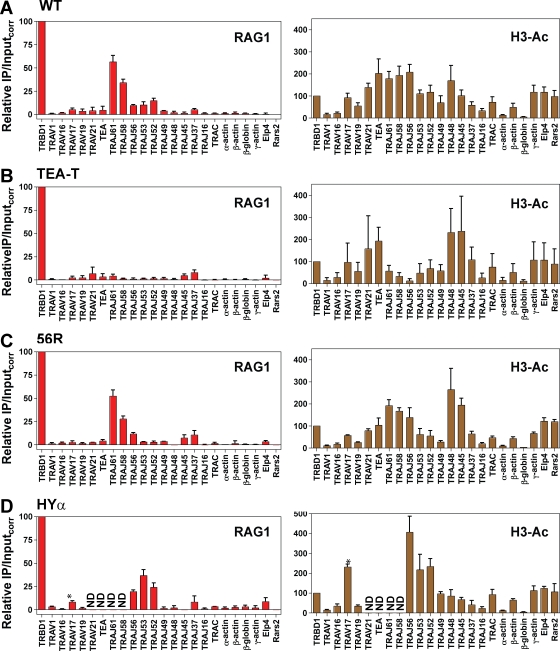
A critical function for transcription elongation in targeting V(D)J recombination has been revealed through the creation of Tcra alleles in which a transcription terminator was inserted immediately downstream of TEA (TEA-T allele) or immediately downstream of TRAJ56 (56R allele; Abarrategui and Krangel, 2006; Abarrategui and Krangel, 2007; Fig. 1 A). The TEA-T allele displays a strong reduction in activating histone marks and recombination in the region spanning TRAJ61–TRAJ52, which is very similar to that caused by complete deletion of TEA (Abarrategui and Krangel, 2007). In contrast, the 56R allele displays defective recombination only in a small region downstream of TRAJ56, including TRAJ53 and TRAJ52 (Abarrategui and Krangel, 2006). When we assessed RAG1 binding to these two alleles, defects closely paralleled those observed for recombination: the TEA-T allele showed greatly diminished RAG1 binding throughout the TRAJ61–TRAJ52 interval (Fig. 3, A and B), whereas the 56R allele displayed robust binding upstream of the terminator (TRAJ61, TRAJ58, and TRAJ56), and weak binding at TRAJ53 and TRAJ52 (Fig. 3 C). These findings strongly argue that transcripts initiating at the TEA promoter facilitate V(D)J recombination by virtue of their elongation through the TRAJ61–TRAJ52 region, thereby rendering RSSs in the transcribed region accessible to RAG1 binding.
Figure 3.
The effect of transcription termination or a rearranged VαJα segment on RAG1 binding to Tcra. (A–D) Binding of RAG1 (left) or levels of H3 acetylation (H3-Ac, right) at the indicated gene segments or regions were assessed by ChIP in primary thymocytes from Rag2−/− Tcrb transgenic mice homozygous for a WT Tcra allele (A), the TEA-T allele (B), the 56R allele (C), or the HYα allele (D). Data in A for the WT allele are reproduced from Fig. 2 A to facilitate comparisons. Data in B–D are the mean of two independent experiments involving individual mice and are presented as in Fig. 2. Asterisk: two copies of TRAV17 are present in the HYα allele (its germline location and the VαJα segment) and both copies are detected by the qPCR assay, which amplifies sequences upstream of the TRAV17 RSS. ND, not done.
Tcra alleles typically undergo multiple V(D)J recombination events that use progressively more 3′ Jα gene segments, with each secondary event deleting the previously formed VαJα segment. The current model to explain the targeting of secondary Tcra recombination events proposes that the promoter of the VαJα segment renders proximal downstream Jα segments accessible for recombination (Hawwari and Krangel, 2007). Evidence for this model derives from a Tcra allele engineered to contain a TRAV17–TRAJ57 junction (HYα allele) in which the earliest subsequent recombination events are focused on the region from TRAV52 to TRAV45 downstream from the VαJα segment (Hawwari and Krangel, 2007). When we examined the HYα allele, we found that RAG1 binding focused strongly on the region immediately downstream of the VαJα segment, from TRAJ56 to TRAJ52 (Fig. 3 D), and was substantially elevated as compared with WT alleles (Fig. 3 A). H3 acetylation was also highest in this interval (Fig. 3 D), as previously reported (Hawwari and Krangel, 2007). We conclude that the presence of a VαJα segment promotes secondary recombination by enhancing the accessibility of immediately downstream RSSs for binding by RAG1.
Control of RAG1 binding in the Tcrb locus
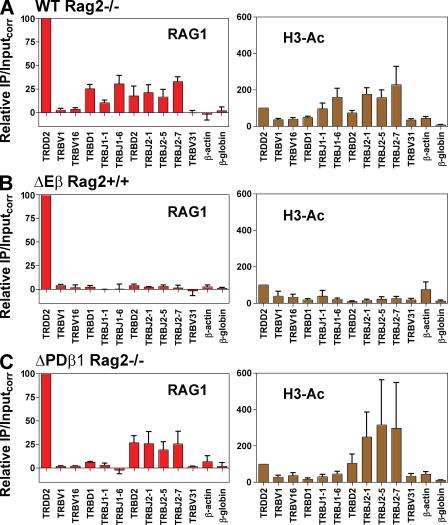
The Tcrb locus contains two Dβ-Jβ clusters in a 10-kb stretch and 31 Vβ gene segments, 30 of which lie in the 380-kb region at the 5′ end of the locus, as well as a single Vβ (TRBV31) that resides at the 3′ end of the locus, downstream of the Tcrb enhancer (Eβ; Fig. 1 B). We previously showed that RAG protein binding focuses on the two Dβ-Jβ clusters and that binding of RAG1 occurs in the presence or absence of RAG2 (Ji et al., 2010). Transcriptional control elements play a critical role in controlling Tcrb assembly. Deletion of Eβ dramatically inhibits recombination of the entire Tcrb locus (Bories et al., 1996; Bouvier et al., 1996) and strongly reduces measures of chromatin accessibility across both Dβ-Jβ clusters (Mathieu et al., 2000). In contrast, deletion of PDβ1, the germline promoter associated with the TRBD1 gene segment, strongly reduces recombination and measures of accessibility at the first Dβ-Jβ cluster, but not the second (Whitehurst et al., 1999, 2000). To determine whether Eβ and PDβ1 regulate V(D)J recombination by controlling RAG protein binding, we performed RAG1 ChIP on WT, ΔEβ, and ΔPDβ1 alleles in DN thymocytes from Rag2−/− mice (WT and ΔPDβ1 alleles) or Rag2+/+ mice (ΔEβ allele). RAG2 deficiency was used to maintain the WT and ΔPDβ1 alleles in germline configuration and arrest development at the DN stage, but was not required for ΔEβ homozygous mice, which have developmental and recombination defects similar to those of Rag2−/− mice (Bories et al., 1996; Bouvier et al., 1996).
As expected (Ji et al., 2010), the WT Tcrb allele exhibited RAG1 binding at both the first and second Dβ-Jβ clusters, but not at the three Vβ gene segments assayed (Fig. 4 A). Deletion of Eβ eliminated RAG1 binding and reduced H3 acetylation across both Dβ-Jβ clusters (Fig. 4 B), whereas deletion of PDβ1 only affected RAG1 binding and H3 acetylation at the first Dβ-Jβ cluster (Fig. 4 C). Hence, in both the Tcra and Tcrb loci, enhancers exert global control of V(D)J recombination, whereas promoters operate in a local manner, and they do so by enabling the recombination machinery access to RSSs. A previous study found that TRBJ1.6 retains substantial nuclease sensitivity on a ΔPDβ1 allele (Oestreich et al., 2006). Our data indicate that this is not sufficient to allow detectable RAG1 binding (Fig. 4 C), and hence that Eβ is not sufficient in the absence of PDβ1 to support RAG1 binding to TRBJ1 gene segments.
Figure 4.
The effect of enhancer or promoter deletion on RAG1 binding to Tcrb. (A–C) Binding of RAG1 (left) or levels of H3 acetylation (H3-Ac, right) at the indicated gene segments or regions were assessed by ChIP in primary thymocytes (almost entirely DN cells) from Rag2−/− mice homozygous for a WT Tcrb allele (A) or the ΔPDβ1 allele (C), or Rag2+/+ mice homozygous for a ΔEβ allele (B). Relative immunoprecipitation/inputcorr values have been normalized to the signal obtained at the TRDD2 gene segment (arbitrarily set to a value of 100), which we have found binds RAG1 and RAG2 strongly in thymocytes (not depicted). Data are the mean of 3 (A) or 2 (B, C) independent experiments involving thymocytes pooled from 5–10 mice and are presented as in Fig. 2.
In the mutant Tcra or Tcrb alleles analyzed, we observed a striking spatial correspondence between the region of the locus that suffers a recombination defect, the region in which RAG1 binding is defective, and the region in which H3 acetylation is reduced. Given the numerous important functions of RAG2, it is remarkable that RAG1 binding in the absence of RAG2 reflects so accurately the recombination defects of the mutant alleles. We infer that transcriptional control elements and transcription elongation directly facilitate RAG–DNA binding, perhaps by disrupting RSS–nucleosome contacts (Du et al., 2008; Kondilis-Mangum et al., 2010) in a manner that is not dependent on RAG2. There are, however, two examples where the correlations are imperfect. First, in the HYα allele, early recombination events are higher at TRAJ49 and TRAJ48 than at TRAJ56–TRAJ50 (Hawwari and Krangel, 2007). In contrast, RAG1 binding (Fig. 3 D) and H3 acetylation (Fig. 3 D; Hawwari and Krangel, 2007) were strongest at TRAJ56, TRAJ53, and TRAJ52. The basis of this discrepancy, which is particularly marked at TRAJ56, is unclear (Hawwari and Krangel, 2007). Second, for all Tcra alleles analyzed, except ΔEα (most notably ΔTEA), RAG1 binding and H3 acetylation were not correlated at TRAJ48 and TRAJ37, with TRAJ48 exhibiting higher H3 acetylation but lower RAG1 binding than TRAJ37 (Fig. 2 and Fig. 3). We hypothesized that this discrepancy might be explained by better binding of RAG1 to the TRAJ37 RSS than to the TRAJ48 RSSs. However, competition gel shift experiments demonstrated that these two RSSs bind equally well to RAG1 in the presence of HMGB1 (which was included to more closely mimic the conditions found in RAG2-deficient cells; Fig. S1). We do not currently have an explanation for the discrepancy between histone acetylation and RAG1 binding at TRAJ48 and TRAJ37.
Although it was not possible to assess RAG2 binding in our experiments, we expect that the pattern of RAG2 binding would closely resemble that of RAG1 in these mutant Tcra and Tcrb alleles, for two reasons. First, we have not previously observed a substantial difference between RAG1 and RAG2 binding patterns in antigen receptor loci (Ji et al., 2010). And second, for the mutant Tcra alleles for which it has been determined (ΔTEA, TEA-T, and 56R), H3K4me3 patterns (which should accurately predict RAG2 binding) are similar to those we observe for RAG1 (Abarrategui and Krangel, 2006, 2007), and clearly depend on transcription. Because the ΔEα and ΔEβ alleles are transcriptionally silent (Bories et al., 1996; Bouvier et al., 1996; Sleckman et al., 1997), they almost certainly lack substantial levels of both H3K4me3 and RAG2 binding, as we have shown is the case for RAG1 binding (Fig. 2 B and Fig. 4 B). The absence of RAG2 was unlikely to compromise RAG1 analysis because RAG1 binding to Tcra and Tcrb was very similar in the presence or absence of RAG2 (Ji et al., 2010).
The accessibility model grew out of observations that transcription of germline gene segments correlated developmentally with their recombination (Yancopoulos and Alt, 1985). Subsequently, the model has been strengthened by numerous findings that link V(D)J recombination to transcriptional control elements, transcription factors, transcription elongation, activating histone modifications, nuclease hypersensitivity, DNA hypomethylation, chromatin structure and chromatin remodeling enzymes (Cobb et al., 2006; Jung et al., 2006; Krangel, 2007). At the core of the model is the idea that all of these processes operate together to achieve a single goal: to allow a common recombination machinery (RAG1/RAG2) access to the appropriate DNA substrates (RSSs) so that binding can take place. Our experiments provide the first direct test of this idea and demonstrate that enhancers, promoters, and transcription elongation indeed control the binding of RAG1 to RSSs—and hence are critical for the formation of recombination centers, within which V(D)J recombination has been proposed to take place (Ji et al., 2010). Although regulated accessibility of RSS substrates is not the only means by which V(D)J recombination is controlled (e.g., higher order chromatin architecture plays a significant role; Jhunjhunwala et al., 2009), our findings emphasize the fundamental importance of the accessibility model in understanding the biology of V(D)J recombination.
MATERIALS AND METHODS
Mice and alleles.
The ΔEα allele (Sleckman et al., 1997), ΔTEA allele, ΔTEAΔJ49 allele (Hawwari et al., 2005), TEA-T allele (Abarrategui and Krangel, 2007), 56R allele (Abarrategui and Krangel, 2006), and HYα allele (Buch et al., 2002) were bred to homozygosity on the Rag2−/− Tcrb transgene background as described previously (Hawwari et al., 2005). The ΔPDβ1 allele (Whitehurst et al., 1999) was bred to homozygosity on the Rag2−/− background and the ΔEβ allele (Bouvier et al., 1996) was bred to homozygosity on the C57BL/6 background (Oestreich et al., 2006). All animal procedures were approved by the Institutional Animal Care and Use Committee of Duke University Medical Center and Washington University School of Medicine.
ChIP.
The antibodies and procedures used for the ChIP assay have been described in detail previously (Ji et al., 2010). In brief, total thymocytes were harvested, cross-linked with 1% HCHO, and after quenching with 0.125 M glycine, cells were washed and frozen as cell pellets. Cell pellets were resuspended in RIPA buffer (10 mM, Tris pH 7.4, 1 mM EDTA, 1% Triton X-100, and 0.1% sodium deoxycholate, 0.1% SDS) containing 0.8 M NaCl and sonicated to achieve a DNA length of approximately 300–500 bp. The resulting chromatin was incubated with anti-RAG1 polyclonal antibody (Ji et al., 2010), anti-acetylated H3 antibody (recognizing H3 acetylated on K9 or K14; Millipore), or normal rabbit IgG (Millipore), and immune complexes were isolated with Protein A agarose beads (Millipore). Input and immunoprecipitated DNAs were quantitated by duplicate Taqman qPCR, and after correction for the background signal obtained with normal rabbit IgG, the immunoprecipitation/inputcorr values were calculated as described previously (Ji et al., 2010). These values were then divided by those obtained for the TRBD1 gene segment (Fig. 2 and Fig. 3) or the TRDD2 gene segment (Fig. 4), and multiplied by 100 to yield the plotted values. Most PCR primer and Taqman probe sequences have been described previously (Ji et al., 2010). For TRDD2, the following oligonucleotides were used: forward primer, 5′-GGGATACGAGCACAGTGTTG-3′; reverse primer, 5′-GGGCTGTGTTTACCTTCCAT-3′; and probe, 5′-TCTCCCAGGCCTCCTGCCTG-3′.
Gel shift experiments.
Competition gel shift experiments were performed as described previously (Rodgers et al., 1999), with the exception that 185 nM HMGB1 protein was included in the analysis. The double strand DNA oligonucleotides used were (top strand sequence): [32P]-labeled consensus 12RSS, 5′-GATCTGGCCTGTCTTACACAGTGATACAGACCTTAACAAAAACCTGCACTCGAGCGGAG-3′; competitor consensus 12RSS, 5′-GATCTGGCCTGTCTTACACAGTGATACAGACCTTAACAAAAACCTGCACTC-3′; TRAJ48 RSS, 5′-TCATTTCCATAGTTGGCACAGTGTGCCAAGCCATTACAAAATCCACCGTGCCAGCTCTG-3′; TRAJ37 RSS, 5′-CCGGTATTGCCTGTTACACCCCAATGCTGCACTTTACAAAAACTGTCAAGAGGGCTTAT-3′; nonspecific competitor, 5′-GATCTGTGTCTTGGTTAGGTTATGAGATCTAG-GAGCATGGCGAGTGCACTCGAGCGGAG-3′.
Online supplemental material.
Fig. S1 shows the quantitation of competitive gel shift experiments that measure the relative binding affinities of the TRAJ48 and TRAJ37 RSSs for RAG1 in the presence of HMGB1 protein. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20101136/DC1.
Acknowledgments
The authors thank Steven Pierce, Jennifer Cayer, and Zanchun Huang for help in mouse breeding and preparation of cell pellets; Karla Rodgers and Mihai Ciubotaru for providing advice and reagents helpful in establishing the RAG1 gel shift assay; and Alexander Little for statistical analysis of gel shift data.
This work was supported in part by Public Health Service grant AI32524 to D.G. Schatz, grants AI079732 and AI081224 to E.M. Oltz, GM41052 to M.S. Krangel, National Institutes of Health (NIH) training grant T32GM07223, and NIH MSTP training grant 2T32GM07205. D.G. Schatz is an investigator of the Howard Hughes Medical Institute.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- DN
- double negative
- DP
- double positive
- Eα
- Tcra enhancer
- Eβ
- Tcrb enhancer
- HMGB
- high mobility group B
- PDβ1
- TRBD1 germline promoter
- RAG
- recombination-activating gene
- RSS
- recombination signal sequence
- TEA
- T early α
References
- Abarrategui I., Krangel M.S. 2006. Regulation of T cell receptor-α gene recombination by transcription. Nat. Immunol. 7:1109–1115 10.1038/ni1379 [DOI] [PubMed] [Google Scholar]
- Abarrategui I., Krangel M.S. 2007. Noncoding transcription controls downstream promoters to regulate T-cell receptor α recombination. EMBO J. 26:4380–4390 10.1038/sj.emboj.7601866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bories J.C., Demengeot J., Davidson L., Alt F.W. 1996. Gene-targeted deletion and replacement mutations of the T-cell receptor β-chain enhancer: the role of enhancer elements in controlling V(D)J recombination accessibility. Proc. Natl. Acad. Sci. USA. 93:7871–7876 10.1073/pnas.93.15.7871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier G., Watrin F., Naspetti M., Verthuy C., Naquet P., Ferrier P. 1996. Deletion of the mouse T-cell receptor β gene enhancer blocks alphabeta T-cell development. Proc. Natl. Acad. Sci. USA. 93:7877–7881 10.1073/pnas.93.15.7877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch T., Rieux-Laucat F., Förster I., Rajewsky K. 2002. Failure of HY-specific thymocytes to escape negative selection by receptor editing. Immunity. 16:707–718 10.1016/S1074-7613(02)00312-6 [DOI] [PubMed] [Google Scholar]
- Cobb R.M., Oestreich K.J., Osipovich O.A., Oltz E.M. 2006. Accessibility control of V(D)J recombination. Adv. Immunol. 91:45–109 10.1016/S0065-2776(06)91002-5 [DOI] [PubMed] [Google Scholar]
- Du H., Ishii H., Pazin M.J., Sen R. 2008. Activation of 12/23-RSS-dependent RAG cleavage by hSWI/SNF complex in the absence of transcription. Mol. Cell. 31:641–649 10.1016/j.molcel.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding A., Chandler S., Ballestar E., Wolffe A.P., Schlissel M.S. 1999. Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. EMBO J. 18:3712–3723 10.1093/emboj/18.13.3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawwari A., Krangel M.S. 2005. Regulation of TCR δ and α repertoires by local and long-distance control of variable gene segment chromatin structure. J. Exp. Med. 202:467–472 10.1084/jem.20050680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawwari A., Krangel M.S. 2007. Role for rearranged variable gene segments in directing secondary T cell receptor α recombination. Proc. Natl. Acad. Sci. USA. 104:903–907 10.1073/pnas.0608248104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawwari A., Bock C., Krangel M.S. 2005. Regulation of T cell receptor alpha gene assembly by a complex hierarchy of germline Jalpha promoters. Nat. Immunol. 6:481–489 10.1038/ni1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhunjhunwala S., van Zelm M.C., Peak M.M., Murre C. 2009. Chromatin architecture and the generation of antigen receptor diversity. Cell. 138:435–448 10.1016/j.cell.2009.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Resch W., Corbett E., Yamane A., Casellas R., Schatz D.G. 2010. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 141:419–431 10.1016/j.cell.2010.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D., Giallourakis C., Mostoslavsky R., Alt F.W. 2006. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 24:541–570 10.1146/annurev.immunol.23.021704.115830 [DOI] [PubMed] [Google Scholar]
- Kondilis-Mangum H.D., Cobb R.M., Osipovich O., Srivatsan S., Oltz E.M., Krangel M.S. 2010. Transcription-dependent mobilization of nucleosomes at accessible TCR gene segments in vivo. J. Immunol. 184:6970–6977 10.4049/jimmunol.0903923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krangel M.S. 2007. T cell development: better living through chromatin. Nat. Immunol. 8:687–694 10.1038/ni1484 [DOI] [PubMed] [Google Scholar]
- Kwon J., Imbalzano A.N., Matthews A., Oettinger M.A. 1998. Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Mol. Cell. 2:829–839 10.1016/S1097-2765(00)80297-X [DOI] [PubMed] [Google Scholar]
- Liu Y., Subrahmanyam R., Chakraborty T., Sen R., Desiderio S. 2007. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 27:561–571 10.1016/j.immuni.2007.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu N., Hempel W.M., Spicuglia S., Verthuy C., Ferrier P. 2000. Chromatin remodeling by the T cell receptor (TCR)-β gene enhancer during early T cell development: implications for the control of TCR-β locus recombination. J. Exp. Med. 192:625–636 10.1084/jem.192.5.625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews A.G., Kuo A.J., Ramón-Maiques S., Han S., Champagne K.S., Ivanov D., Gallardo M., Carney D., Cheung P., Ciccone D.N., et al. 2007. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 450:1106–1110 10.1038/nature06431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry M.T., Krangel M.S. 2000. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 287:495–498 10.1126/science.287.5452.495 [DOI] [PubMed] [Google Scholar]
- Oestreich K.J., Cobb R.M., Pierce S., Chen J.Z., Ferrier P., Oltz E.M. 2006. Regulation of TCRbeta gene assembly by a promoter/enhancer holocomplex. Immunity. 24:381–391 10.1016/j.immuni.2006.02.009 [DOI] [PubMed] [Google Scholar]
- Rodgers K.K., Villey I.J., Ptaszek L., Corbett E., Schatz D.G., Coleman J.E. 1999. A dimer of the lymphoid protein RAG1 recognizes the recombination signal sequence and the complex stably incorporates the high mobility group protein HMG2. Nucleic Acids Res. 27:2938–2946 10.1093/nar/27.14.2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleckman B.P., Bardon C.G., Ferrini R., Davidson L., Alt F.W. 1997. Function of the TCR α enhancer in alphabeta and gammadelta T cells. Immunity. 7:505–515 10.1016/S1074-7613(00)80372-6 [DOI] [PubMed] [Google Scholar]
- Stanhope-Baker P., Hudson K.M., Shaffer A.L., Constantinescu A., Schlissel M.S. 1996. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 85:887–897 10.1016/S0092-8674(00)81272-6 [DOI] [PubMed] [Google Scholar]
- Swanson P.C. 2004. The bounty of RAGs: recombination signal complexes and reaction outcomes. Immunol. Rev. 200:90–114 10.1111/j.0105-2896.2004.00159.x [DOI] [PubMed] [Google Scholar]
- Villey I., Caillol D., Selz F., Ferrier P., de Villartay J.P. 1996. Defect in rearrangement of the most 5′ TCR-J α following targeted deletion of T early α (TEA): implications for TCR α locus accessibility. Immunity. 5:331–342 10.1016/S1074-7613(00)80259-9 [DOI] [PubMed] [Google Scholar]
- Whitehurst C.E., Chattopadhyay S., Chen J. 1999. Control of V(D)J recombinational accessibility of the D β 1 gene segment at the TCR β locus by a germline promoter. Immunity. 10:313–322 10.1016/S1074-7613(00)80031-X [DOI] [PubMed] [Google Scholar]
- Whitehurst C.E., Schlissel M.S., Chen J. 2000. Deletion of germline promoter PD β 1 from the TCR β locus causes hypermethylation that impairs D β 1 recombination by multiple mechanisms. Immunity. 13:703–714 10.1016/S1074-7613(00)00069-8 [DOI] [PubMed] [Google Scholar]
- Yancopoulos G.D., Alt F.W. 1985. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 40:271–281 10.1016/0092-8674(85)90141-2 [DOI] [PubMed] [Google Scholar]