Abstract
At present the three-dimensional structure of the tobacco lectin, further referred to as Nictaba, and its carbohydrate-binding site are unresolved. In this paper, we propose a three-dimensional model for the Nictaba domain based on the homology between Nictaba and the carbohydrate-binding module 22 of Clostridium thermocellum Xyn10B. The suggested model nicely fits with results from circular dichroism experiments, indicating that Nictaba consists mainly of β-sheet. In addition, the previously identified nuclear localization signal is located at the top of the protein as a part of a protruding loop. Judging from this model and sequence alignments with closely related proteins, conserved glutamic acid and tryptophan residues in the Nictaba sequence were selected for mutational analysis. The mutant DNA sequences as well as the original Nictaba sequence have been expressed in Pichia pastoris and the recombinant proteins were purified from the culture medium. Subsequently, the recombinant proteins were characterized and their carbohydrate binding properties analyzed with glycan array technology. It was shown that mutation of glutamic acid residues in the C-terminal half of the protein did not alter the carbohydrate-binding activity of the lectin. In contrast, mutation of tryptophan residues in the N-terminal half of the Nictaba domain resulted in a complete loss of carbohydrate binding activity. These results suggest that tryptophan residues play an important role in the carbohydrate binding site of Nictaba.
Keywords: Nicotiana tabacum, Lectin, Carbohydrate-binding, Structure, Mutant
Introduction
Lectins are defined as proteins that can bind to carbohydrate structures in a reversible and non-catalytic way [1]. So far about 500 lectins from plants have been purified and characterized to some extent. Based on their three-dimensional structure and sequence, all plant lectins can be classified into 12 distinct lectin families each typified by a specific lectin domain [2, 3].
Historically, the first plant lectins discovered were purified from tissues in which they are present in high concentrations. It was shown that most of these lectins locate to the vacuole of the plant cell. Because of their high abundance and the ease of purification with affinity chromatography techniques, these “vacuolar” plant lectins were the favorite proteins for many structural biologists. Hence, a significant amount of structural information on these vacuolar lectins and their interaction with carbohydrate ligands is available [4, 5]. It is now believed that these proteins act as defense and/or storage proteins in plants [2].
In the past decade, several new lectins have been discovered that reside in the nucleocytoplasmic compartment of the plant cell [3]. Typically these lectins are present in very low concentrations and are often inducible by stress factors such as salt, drought or pathogen attack. It is believed that these nucleocytoplasmic lectins fulfill a role in the stress physiology of the plant cell. Although nucleocytoplasmic lectins have been found in at least six lectin families, no information is available yet regarding their three-dimensional structure and the conformation of their glycan binding sites.
The Nicotiana tabacum agglutinin, abbreviated as Nictaba, was first reported in 2002 when it was purified from tobacco leaves treated with jasmonates [6]. Biochemical analysis showed that Nictaba exists as a dimer of two unglycosylated 19 kDa subunits. Molecular cloning of the coding sequence revealed that Nictaba shares more than 40% sequence similarity with a group of carbohydrate-binding proteins known as the Cucurbitaceae phloem lectins. A more extensive database search revealed that the Nictaba sequence is widespread in plants belonging to different taxonomic groups. In some cases the Nictaba sequence is present as a single domain, while in other cases it is part of a larger fusion protein [7]. Therefore, Nictaba can be considered as a representative for the (carbohydrate binding) domain present in the family of so-called Nictaba-related proteins [8].
Using immuno-fluorescence microscopy with a polyclonal antibody directed against Nictaba, it was shown that the localization of this lectin is confined to the nucleus and the cytoplasm of leaf parenchym cells [6]. This nucleocytoplasmic localization was confirmed by confocal microscopy of an EGFP-Nictaba fusion protein. In addition, it was demonstrated that the basic tetrapeptide (102-LysLysLysLys-105) is required and sufficient for transport of Nictaba from the cytoplasm into the nucleus [9]. Hapten inhibition assays using mono- and oligosaccharides revealed that Nictaba exhibits affinity towards β-,4 linked GlcNAc oligomers. With the advent of glycan array technology, it could be demonstrated that Nictaba also shows affinity for high mannose and complex type N-glycans, suggesting that the binding site of Nictaba is most complementary to the core GlcNAc2Man3 structure [9].
The expression level for Nictaba in tobacco leaves is very low, even after treatment of the plant with jasmonates or insect herbivory [10, 11]. Consequently, the purification of Nictaba starting from tobacco leaves is very inefficient. Moreover, Nictaba preparations purified from tobacco leaf material often contain low molecular weight impurities, even after several affinity chromatography purification steps. Since these contaminants—most probably phenolic compounds—hamper the crystallization of the lectin, the three-dimensional structure of Nictaba could not be resolved yet. To overcome some of these problems, Nictaba has been expressed in the methylotrophic yeast Pichia pastoris [12]. Nictaba was purified from the cell pellet and the culture medium of Pichia strain GS115 using a combination of anion exchange chromatography and affinity chromatography on a column with immobilized ovomucoid. It was shown that the purified recombinant Nictaba exhibited similar biochemical properties and carbohydrate binding specificity, compared to native Nictaba from tobacco [12].
Due to the lack of information related to the three-dimensional structure of Nictaba, little is also known about the carbohydrate-binding site of the lectin. In an attempt to unravel which amino acids are important for carbohydrate-binding activity of the protein, a three-dimensional structure model of Nictaba was made based on the homology between Nictaba and the carbohydrate-binding module 22 of Clostridium thermocellum Xyn10B. Judging from this model and sequence alignments with closely related proteins, conserved glutamic acid and tryptophan residues in the Nictaba sequence were selected for mutational analysis. The mutant sequences as well as the original Nictaba sequence were expressed in Pichia pastoris. After purification, carbohydrate binding properties of the mutant proteins were analyzed with glycan array technology and compared to the carbohydrate binding properties of Nictaba.
Materials and methods
Hydrophobic cluster analysis
Multiple amino acid sequence alignments were carried out with CLUSTAL-X [13] using the Risler’s structural matrix for homologous amino acid residues [14]. Hydrophobic Cluster Analysis (HCA) [15] was performed to delineate the structurally conserved strands of β-sheets along the amino acid sequence of Nictaba using the carbohydrate-binding module 22 of Clostridium thermocellum Xyn10B [16] as a model. HCA plots were generated using the HCA server (http://bioserv.rpbs.jussieu.fr).
Molecular modeling
Molecular modeling of Nictaba was carried out on a Silicon Graphics O2 R10000 workstation, using the programs InsightII, Homology and Discover3 (Accelrys, San Diego CA, USA). The atomic coordinates of the Clostridium thermocellum Xyn10B carbohydrate-binding module [4] (RCSB Protein Data Bank code 1H6X) were used to build the three-dimensional model of the lectin. Although Nictaba shares low percentages of identity (~14%) and similarity (56%) with the carbohydrate binding domain of C. thermocellum, HCA suggested a very similar structure for both proteins (result not shown). Steric conflicts were corrected during the model building procedure using the rotamer library [17] and the search algorithm of the Homology program [18] to maintain proper side-chain orientation. An energy minimization of the final model was carried out by 200 cycles of steepest descent using the cvff forcefield of Discover. PROCHECK [19] was used to assess the geometric quality of the three-dimensional model. In this respect, about 80% of the residues of Nictaba were correctly assigned to the best allowed regions of the Ramachandran plot, the remaining residues being located in the generously allowed regions of the plot except for five residues (Asp17, Gln20, Val33, Trp41, Leu116), which occur in the non allowed region (result not shown). Electrostatic potentials were calculated and displayed with GRASP using the parse3 parameters [20]. The solvent probe radius used for molecular surfaces was 1.4 Å and a standard 2.0 Å-Stern layer was used to exclude ions from the molecular surface [21]. The inner and outer dielectric constants applied to the protein and the solvent were fixed at 4.0 and 80.0, respectively, and the calculations were performed keeping a salt concentration of 0.145 M. Ribbon diagram and molecular surface of Nictaba were drawn with PyMol (W. L. DeLano (http://pymol.sourceforge.net)).
Bioinformatics analyses
Nictaba domains were identified by performing a pBLAST search against the non-redundant protein sequences database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence analysis was carried out using the ClustalW2 sequence analysis tool (http://www.ebi.ac.uk/).
Expression of Nictaba sequences in Pichia pastoris
Cloning and expression of native and mutant forms of Nictaba was performed using the EasySelect Pichia Expression Kit from Invitrogen (Carlsbad, CA USA). The coding sequence for Nictaba was amplified by PCR from a pUC plasmid (Genbank accession number AF389848, [6]) using primers evd 65 and evd 66. The Nictaba coding sequence was mutated using overlap PCR with primer sequences shown in Table 1. PCR was carried out using the Phusion polymerase with proof reading activity (New England Biolabs, Ipswich, MA USA). All Nictaba sequences were amplified using 5′ and 3′ primers with XbaI and EcoRI overhangs, respectively, to allow cloning in the pPICZαA vector (EasySelect Pichia expression kit, Invitrogen) that was cut with the same restriction enzymes. The ligated DNA was heat shock transformed in Top10F′ cells and transformants were selected on LB agar plates containing 0.25 μg/ml zeocin (Invitrogen). Plasmid DNA was purified using the E.Z.N.A.® Plasmid Mini Kit (Omega Bio-Tek, Norcross, GA USA). Proper insert orientation and sequence of Nictaba and its mutant forms was verified by sequencing using 5′ and 3′ AOX1 specific primers evd 21 (5′ GACTGGTTCCAATTGACAAGC3′) and evd 22 (5′ GCAAATGGCATTCTGACATCC3′) (carried out by LGC Genomics GmbH, Berlin, Germany). The plasmid DNA was linearized with the restriction enzyme SacI (Fermentas, St. Leon-Rot, Germany) and purified before transformation of Pichia. Pichia pastoris strains GS115 (Mut+ His−—Nictaba mutant 1 and recombinant Nictaba) and KM71H (MutS His+Arg+—Nictaba mutant 2, 3) were electroporated with 5 μg of linearized DNA using a GenePulser® (Bio-Rad, Hercules, CA USA) with pulse settings of 25 μF, 1.5 kV and 200Ω, and spread on YPDS plates (1% yeast extract, 2% peptone, 2% dextrose, 2% sorbitol, 2% agar, 100 μg/ml zeocin (Invitrogen)). Selected colonies were grown overnight in 5 ml BMGY medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 6.0, 1.34% YNB, 4×10−5% biotin, 1% glycerol) at 30°C in a shaker incubator at 250 rpm. The next day, cultures were washed with water, resuspended in BMMY medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 6.0, 1.34% YNB, 4×10−5% biotin, 0.5% methanol) and grown for 4 days. Every day, methanol was added to a final concentration of 2%. After 4 days, protein profiles in medium and cell pellet were compared. Proteins were precipitated from the medium with 10% TCA. Protein extraction from the pellet was done by vortexing with glass beads (108 μm diameter, Sigma, St Louis, MO USA) in 20 mM 1,3 diaminopropane buffer. Protein extracts were analyzed by SDS-PAGE and Western blot analysis.
Table 1.
List of primer sequences used to amplify the original and mutant forms of Nictaba
| Primer | Properties | Sequence (5′-3′) |
|---|---|---|
| Native Nictaba | ||
| Evd 65 | Forward | cag tgg ata gcc gca aga gac ctt tc |
| Evd 66 | Reverse | tta gtt tgg acg aat gtc gaa gcc c |
| Nictaba mutant 1 | ||
| Evd 12 | 5′ EcoRI | ggc gga gaa ttc acc atg caa ggc cag tgg ata gcc gc |
| Evd 303 | Reverse: E128→A128/E135→A135 | att gat tgc cat caa cct cat ttc gac tgc acc atc ctc |
| Evd 302 | Forward: E128→A128;E135→A135 | aga gga tgg tgc agt cga aat gag gtt gat ggc aat caa t |
| Evd 315 | 3′ XbaI | ccc gct ttc tag aca gtt tgg acg aat gtc gaa gcc |
| Nictaba mutant 2/3 | ||
| Evd 376 | 5′ EcoRI | gaa ttc acc atg caa ggc cag tgg ata |
| Evd 377 | Reverse | |
| Mutant 2: W22→L22;W15→L15 | tgt caa gta ctg agg att gtc cac caa tgt | |
| Mutant 3: W15→L15 | tgt caa gta ctg agg att gtc cac cca tgt | |
| Evd 378 | Forward | |
| Mutant 2: W15→L15;W22→L22 | aca ttg gtg gac aat cct cag tac ttg aca | |
| Mutant 3: W15→L15 | aca ttg gtg gac aat cct cag tac tgg aca | |
| Evd 379 | 3′ XbaI | tct aga cag ttt gga cga atg tcg aag |
Large scale culture and purification of recombinant proteins
Transformed yeast colonies were grown overnight in 5 ml BMGY (30°C, 250 rpm). The next day, cultures were transferred to 100 ml BMGY in 250 ml erlenmeyer flasks and allowed to grow until an OD600 of 2–6 was reached. Cells were washed with water, and transferred to 200 ml BMMY in 1 l Erlenmeyer flasks. Methanol was added to a final concentration of 2% every 24 h. After 72 h, the medium was collected by centrifugation and proteins were precipitated overnight with 80% ammonium sulphate. In the case of Nictaba mutant 1, precipitated proteins were resuspended in a small volume of 20 mM diaminopropane and dialysed against the same buffer overnight with several buffer changes. Subsequently, the sample was loaded on a Q Sepharose Fast Flow column (Ø 15 mm, height 4 cm, GE healthcare, Uppsala, Sweden) previously equilibrated with 20 mM diaminopropane. After extensive washing with diaminopropane until OD280<0.3, bound proteins were eluted with 20 mM Tris pH 8.7 containing 0.5 M NaCl. The most concentrated protein fractions from the anion exchange column were pooled and loaded on a gel filtration column (Sephacryl S-100, Ø 2 cm, height 60 cm, GE healthcare). For Nictaba mutants 2 and 3, ammonium sulphate precipitated proteins were resuspended in 20 mM diaminopropane and directly separated on the gel filtration column. Fractions from the gel filtration column were collected and analyzed by SDS-PAGE and Western Blot analysis.
Glycan array screening
The microarrays are printed as described previously [22]. The analyses reported here were performed on Mammalian Printed Array Version 3.0 for the Nictaba mutant 1 and Version 4.0/4.1 for all other proteins (see https://www.functionalglycomics.org/static/consortium/resources/resourcecoreh8.shtml). Recombinant Nictaba (original or mutant forms) purified from Pichia pastoris was labeled using the Alexa Fluor® 488 Protein Labeling Kit from Invitrogen following the manufacturer’s instructions. The labeled proteins were applied to separate microarray slides and incubated under a cover slip for 60 min in a dark, humidified chamber at room temperature. After incubation, the cover slips are gently removed in a solution of Tris-buffered saline containing 0.05% Tween 20 and washed by gently dipping the slides 4 times in successive washes of Tris-buffered saline containing 0.05% Tween 20, Tris-buffered saline, and deionized water. After the last wash, the slides are spun in a slide centrifuge for approximately 15 sec to dry and are immediately scanned in a PerkinElmer ProScanArray MicroArray Scanner using an excitation wavelength of 488 nm and ImaGene software (BioDiscovery Inc., El Segundo, CA USA) to quantify fluorescence. The data are reported as average Relative Fluorescence Units (RFU) of 4 replicate values after removal of the high and low values of the six replicates of each glycan presented on the array.
Amino terminal sequence analysis
Recombinant proteins purified from Pichia pastoris were analyzed by SDS-PAGE and electroblotted on a ProBlot™ polyvinylidene difluoride membrane (Applied Biosystems, Foster City, CA USA). The membrane was stained with a 1:1 mix of Coomassie brilliant blue and methanol. The protein of interest was excised from the blot and the N-terminal sequence determined by Edman degradation performed on a model Procise 491cLC protein sequencer without alkylation of cysteines (Applied Biosystems, Foster City, CA USA).
Analytical methods
The protein content of the samples was estimated using the Coomassie (Bradford) Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL USA), based on the Bradford [23] dye-binding procedure. Extracts from Pichia were analyzed for protein expression by SDS-PAGE using 15% polyacryl-amide gels under reducing conditions as described by Laemmli [24]. Proteins were visualized by staining with Coomassie Brilliant Blue R-250. For Western blot analysis, samples separated by SDS-PAGE were electrotransferred to 0.45 μm polyvinylidene fluoride (PVDF) membranes (Biotrace™ PVDF, PALL, Gelman Laboratory, Ann Arbor, MI USA). After blocking the membrane in Tris-Buffered Saline (TBS: 10 mM Tris, 150 mM NaCl, 0.1% (v/v) Triton X-100, pH 7.6) containing 5% (w/v) BSA, blots were incubated for 1 h with a mouse monoclonal anti-His (C-terminal) antibody (Invitrogen), diluted 1/5000 in TBS. The secondary antibody was a 1/1000 diluted rabbit anti-mouse IgG labeled with horse radish peroxidase (Dako Cytomation, Glostrup, Denmark). Immuno-detection was achieved by a colorimetric assay using 3,3′-diaminobenzidine tetra-hydrochloride (Sigma-Aldrich, St. Louis Missouri, MO USA) as a substrate.
Results
Three-dimensional model for Nictaba
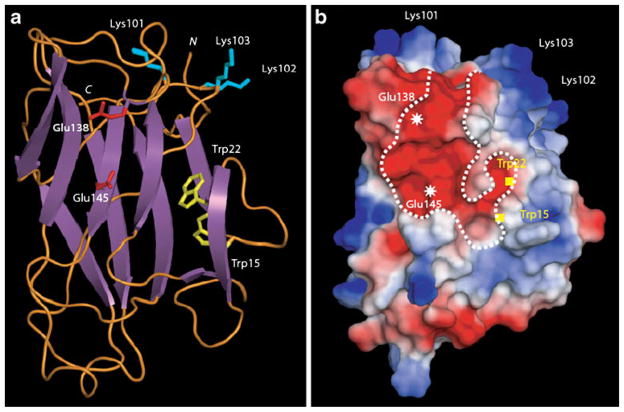
The three-dimensional model built for Nictaba essentially consists of a β-sandwich structure composed of two β-sheets of four and five antiparallel β-strands, respectively, connected by extended loops (Fig. 1a). The previously identified nuclear localization signal [6] (LysLysLysLys) occurs at the top of a well exposed loop which protrudes in the solvent. Mapping of electrostatic potentials on the molecular surface of Nictaba also revealed an extended electronegatively charged groove that most probably corresponds to the glycan binding site of the lectin (Fig. 1b). Two well exposed glutamic acid residues (Glu138 and Glu145) account for the electronegative character of the groove and—according to the model—could be involved in the binding of the GlcNAc2Man3 oligomer. Interestingly, a β-strand forms a sort of barrier, thus separating an electronegatively charged extension from the central groove. This additional electronegative pocket with two tryptophan residues (Trp15 and Trp22) could contribute to the sugar-binding specificity of Nictaba.
Fig. 1.
Molecular modeling of Nictaba sequence. a Ribbon diagram showing the β-sandwich organization of Nictaba. Exposed Lys residues participating to the nuclear localization signal are represented in sticks colored cyan. Glu residues occurring at the center of the putative chitin-binding groove are represented in sticks colored red. Trp residues possibly participating to the putative carbohydrate-binding pocket are represented in sticks colored yellow. N and C correspond to the N- and C-terminal ends of the polypeptide chain, respectively. b Mapping of electrostatic potentials on the molecular surface of the modeled Nictaba. The putative carbohydrate-binding groove is delineated by a dotted white line and the location of important residues is indicated. Electronegatively and electropositively charged areas are colored red and blue, respectively; neutral regions are shown in white
Trp15/22 and Glu138/145 are conserved amino acids in the Nictaba domain
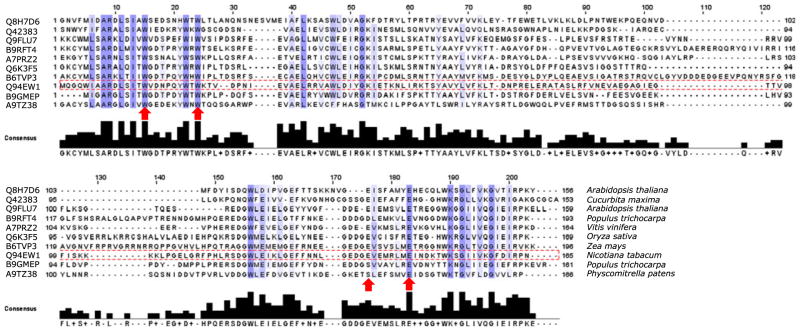
Sequence alignment of the Nictaba domain present in the tobacco leaf lectin and several Nictaba-related proteins from different plant species revealed that Trp15, Trp22 and Glu145 in the Nictaba sequence are strongly conserved in all sequences analyzed (Fig. 2). Although Glu138 is less conserved, the electronegative character of the presumed carbohydrate-binding site is always preserved by the presence of nearby glutamic acid residues.
Fig. 2.
Multiple sequence alignment of the amino acid sequences of the Nictaba domain from different plant species. Amino acids that were selected for mutation are indicated by the arrows. The Nictaba sequence is boxed. Accession numbers and species names are indicated in front of the sequence and after the sequence, respectively. Proteins that were aligned are, from top to bottom: hypothetical protein containing a TIR domain from Arabidopsis thaliana, dimeric phloem specific lectin PP2 from Cucurbita maxima, Nictaba from Nicotiana tabacum, F-box family protein from Populus trichocarpa, Putative F-box protein PP2-B12 from Arabidopsis thaliana, F-box family protein-like from Oryza sativa, F-box containing protein from Vitis vinifera, putative uncharacterized protein containing a F-Box domain from Populus trichocarpa, Phloem-specific lectin from Zea mays and predicted protein containing an F-box domain from Physcomitrella patens
Purification of recombinant proteins from Pichia pastoris
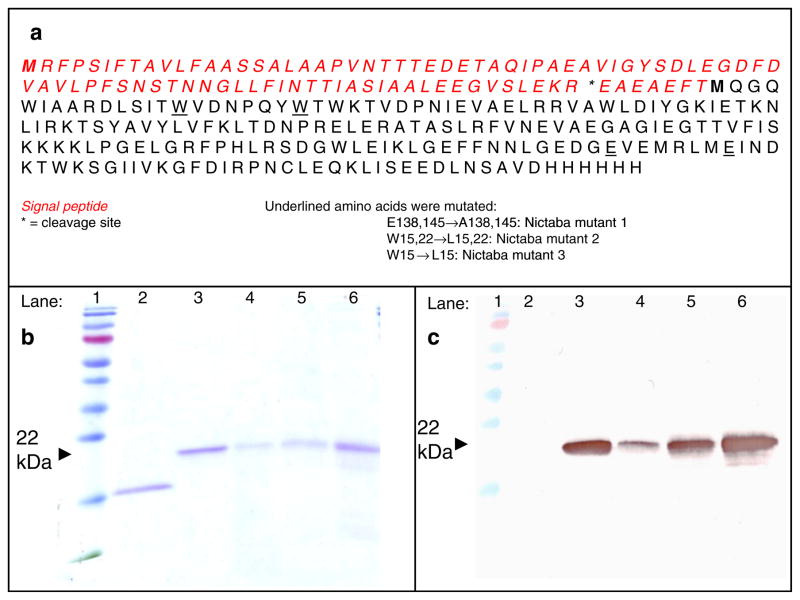
To validate the proposed three-dimensional model, a mutational analysis of Nictaba was performed. Mutagenesis of the Nictaba coding sequence was done by overlap PCR. Afterwards, the mutated sequences and the original Nictaba sequence were cloned into a Pichia pastoris pPICZαA expression vector. In the sequence of Nictaba mutant 1, Glu138 and Glu145 were mutated into Ala. In the Nictaba mutant 2 Trp15 and Trp22 were changed into Leu, whereas in mutant 3 only Trp15 was modified (Fig. 3a). The resulting constructs were electroporated in Pichia pastoris strains GS115 (Nictaba mutant 1 and recombinant Nictaba) or KM71H (Nictaba mutants 2 and 3). Transformed colonies were grown in 1 l cultures and the recombinant proteins purified from the culture medium.
Fig. 3.
Primary sequence of recombinant Nictaba, preceded by an N-terminal signal peptide necessary for secretion and a C-terminal tag containing of a c-myc epitope and a (His)6 tag (a). Proteins were analyzed by SDS-PAGE (b) and Western blot analysis with a monoclonal anti-His antibody (c). Samples are loaded as follows: lane 1: molecular weight marker, lane 2: native Nictaba from tobacco; lane 3: recombinant Nictaba; lane 4: Nictaba mutant 1; lane 5: Nictaba mutant 2; lane 6: Nictaba mutant 3. Approximately 0.5 μg of each protein was loaded on the gel
Biochemical characterization of recombinant proteins
SDS-PAGE of recombinant proteins purified from Pichia pastoris revealed polypeptides with a molecular mass of approximately 22 kDa (Fig. 3a). The size of these polypeptide bands is in good agreement with the molecular mass calculated from the primary sequence taking into account that the recombinant protein contains an additional c-myc epitope and 6×His tag compared to the native Nictaba from tobacco. Further analysis of the purified proteins by Western blot, using a monoclonal antibody directed against the C-terminal polyHis tag (Fig. 3b) and N-terminal sequencing confirmed that the purified proteins correspond to the different Nictaba forms. Edman degradation also showed that the signal peptide of Nictaba mutant 1 was correctly processed at the predicted cleavage site (Fig. 3a). Apart from this form, Nictaba mutant 2 and 3 also exist as slightly smaller proteins, two or four amino acids shorter than the predicted protein (starting at EAEFTMQG or EFTMQGQW). Dynamic light scattering measurements revealed that part of the purified recombinant protein for Nictaba mutants 2 and 3 forms aggregates, making further secondary structure analysis impossible.
Carbohydrate-binding specificity of recombinant proteins expressed in Pichia
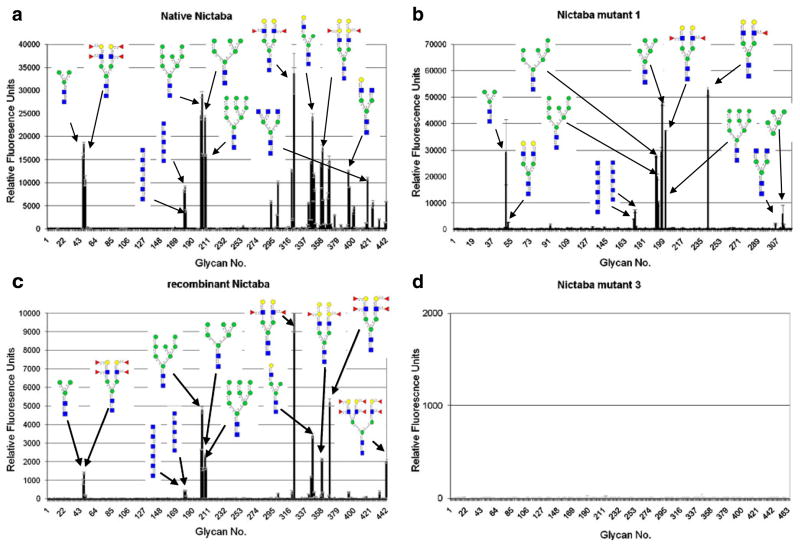
The carbohydrate-binding properties of the recombinant Nictaba proteins (both the original and the mutant forms) were investigated by screening the labeled proteins on a glycan array, and comparison to the sugar-binding specificity of the native Nictaba from tobacco. Analyses of the native tobacco lectin and the recombinant Nictaba expressed in Pichia revealed high affinity for complex and high mannose N-type glycans and to a lesser extent for GlcNAc oligomers (Fig. 4a and c). It can thus be concluded that the recombinant Nictaba protein expressed in Pichia preserves the carbohydrate binding properties of the native tobacco protein. Mutation of Glu138 and Glu145 in the Nictaba sequence (Nictaba mutant 1) did not significantly change the apparent specificity of the protein for the above-mentioned carbohydrate structures (Fig. 4b). However, mutation of Trp15 and Trp22 (Nictaba mutant 2) drastically altered the carbohydrate-binding properties of the protein and almost completely abolished binding to carbohydrate structures. Similarly, when only Trp22 was changed into a leucine residue (Nictaba mutant 3) no glycan binding could be observed (Fig. 4d). Evidently, the combination of both Glu and both Trp residues in one mutant protein also completely abolished the interaction of this mutant protein with all glycans on the array (data not shown).
Fig. 4.
Comparative analysis of binding of native Nictaba and recombinant Nictaba proteins purified from Pichia on the glycan array. Different panels show interaction of a, native Nictaba (150 μg/ml, v4.0): b, Nictaba mutant 1 (100 μg/ml, v3.0); c, recombinant Nictaba (210 μg/ml, v4.0): and d, Nictaba mutant 3 (200 μg/ml v4.1). The complete primary data set for each protein is available on the website of the Consortium for Functional Glycomics (www.functionalglycomics.org)
A comparative analysis of the glycan array results for native and recombinant Nictaba as well as for Nictaba mutants 1 and 3 was performed. Therefore, all glycan array data were compared by first doing a ranking calculation that “normalizes the values” of different data sets, and assigning a percentile ranking to each RFU. Only the results for the top 25 glycans are shown in Table 2. A detailed analysis of these data demonstrates that the glycan structures with the highest rank for native Nictaba, recombinant Nictaba and Nictaba mutant 1 are very similar and all belong to the high mannose or more complex type N-glycans, whereas the interaction with GlcNAc or GlcNAc oligomers is much weaker.
Table 2.
Comparative analysis of glycan array results for native and recombinant Nictaba as well as Nictaba mutants 1 and 3
| Native Nictaba |
Recombinant Nictaba |
Nictaba mutant 1 v3.0 |
Nictaba mutant 3 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Chart number v4.0 | Structure | RFU | Ranka | RFU | Ranka | RFU | Ranka | RFU | Ranka |
| NA | Galβ1-4GlcNAcβ1-2Manα1-3(Fucα1-3(Galβ1-4)GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp20 | NAb | NA | NA | NA | 52664 | 100 | NA | NA |
| 202 | Manα1-2Manα1-6(Manα1-3)Manα1-6[Manα1-2Manα1-2Manα1-3]Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 32124 | 100 | 4775 | 46 | 19894 | 38 | 1 | 0 |
| 201 | Manα1-6[Manα1-2Manα1-3]Manα1-6[Manα1-2Manα1-3] Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 30941 | 96 | 2651 | 26 | 27651 | 53 | 6 | 0 |
| 368 | Galα1-3Galβ1-4(Fucα1-3)GlcNAcβ1-2Manα1-3(Galα1-3Galβ1-4(Fucα1-3)GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp20 | 26138 | 81 | 5182 | 50 | NA | NA | 1 | 0 |
| 322 | Fucα1-3(Galβ1-4)GlcNAcβ1-2Manα1-3(Fucα1-3(Galβ1-4) GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp20 | 25973 | 81 | 10296 | 100 | 36925 | 70 | −2 | 0 |
| 346 | Galβ1-4GlcNAcβ1-2Manα1-3(Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 25012 | 78 | 3395 | 33 | NA | NA | 3 | 0 |
| 206 | Manα1-6(Manα1-3)Manα1-6[Manα1-2Manα1-3] Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 23426 | 73 | 2321 | 23 | 29479 | 56 | 1 | 0 |
| 48 | Manα1-3(Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp13 | 19194 | 60 | 1394 | 14 | 29183 | 55 | 12 | 0 |
| 207 | Manα1-6(Manα1-3)Manα1-6(Manα1-3)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 18865 | 59 | 1616 | 16 | 47537 | 90 | 7 | 0 |
| 47 | Manα1-3(Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 18598 | 58 | 913 | 9 | NA | NA | 1 | 0 |
| 319 | Galβ1-3GlcNAcβ1-2Manα1-3(Galβ1-3GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp19 | 16215 | 50 | 360 | 3 | 124 | 0 | 2 | 0 |
| 301 | GlcNAcβ1-2Manα1-3(GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 15637 | 49 | 273 | 3 | 2015 | 4 | 2 | 0 |
| 359 | Galα1-3Galβ1-4GlcNAcβ1-2Manα1-3(Galα1-3Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp20 | 15277 | 48 | 334 | 3 | NA | NA | 6 | 0 |
| 344 | Galβ1-4GlcNAcβ1-2Manα1-3Manβ1-4GlcNAcβ1-4GlcNAc-Sp12 | 14927 | 46 | 1188 | 12 | NA | NA | 3 | 0 |
| 203 | Manα1-2Manα1-2Manα1-3(Manα1-2Manα1-3(Manα1-2Manα1-6)Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 14796 | 46 | 1605 | 16 | 9830 | 19 | 2 | 0 |
| 357 | Fucα1-2Galβ1-4GlcNAcβ1-2Manα1-3(Fucα1-2Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp20 | 14054 | 44 | 169 | 2 | NA | NA | 4 | 0 |
| 367 | Galα1-3(Fucα1-2)Galβ1-4GlcNAcβ1-2Manα1-3(Galα1-3(Fucα1-2)Galβ1-4GlcNAcβ1-2Manα1-6) Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp20 | 13121 | 41 | 168 | 2 | NA | NA | 7 | 0 |
| 348 | Galβ1-4GlcNAcβ1-2Manα1-3(Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4(Fucα1-6)GlcNAcβ-Sp22 | 12978 | 40 | 352 | 3 | NA | NA | 6 | 0 |
| 180 | (GlcNAcβ1-4)5β-Sp8 | 12695 | 40 | 446 | 4 | 6910 | 13 | 2 | 0 |
| 349 | Galβ1-3GlcNAcβ1-2Manα1-3(Galβ1-3GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4(Fucα1-6)GlcNAcβ-Sp22 | 12408 | 39 | 115 | 1 | NA | NA | 4 | 0 |
| 393 | Galβ1-4GlcNAcβ1-2Manα1-3(GlcNAcβ1-2Manα1-6) Manβ1-4GlcNAcβ1-4GlcNAc-Sp12 | 11677 | 36 | 331 | 3 | NA | NA | −1 | 0 |
| 179 | (GlcNAcβ1-4)6β-Sp8 | 11325 | 35 | 439 | 4 | 3909 | 7 | 5 | 0 |
| 50 | Galβ1-4GlcNAcβ1-2Manα1-3(Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 10836 | 34 | 163 | 2 | 2585 | 5 | 4 | 0 |
| 442 | Fucα1-2Galβ1-4(Fucα1-3)GlcNAcβ1-2(Fucα1-2Galβ1-4(Fucα1-3)GlcNAcβ1-4)Manα1-3(Fucα1-2Galβ1-4 (Fucα1-3)GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-N | 10726 | 33 | 2006 | 19 | NA | NA | 1 | 0 |
| 358 | Fucα1-2Galβ1-4(Fucα1-3)GlcNAcβ1-2Manα1-3(Fucα1-2Galβ1-4(Fucα1-3)GlcNAcβ1-2Manα1-6) Manβ1-4GlcNAcβ1-4GlcNAβ-Sp20 | 10151 | 32 | 2110 | 20 | NA | NA | 1 | 0 |
Percentile ranking: The glycan with the highest RFU is assigned a value of 100
NA means that the glycan was not available in that assay; i.e., present on version 4 but not version 3, or present on version 3 and not on version 4
Discussion
The tobacco lectin or Nictaba can be regarded as a prototype for a new group of nucleocytoplasmic proteins. Hitherto, the function of the Nictaba domain is unknown and subject to speculation. It is known that Nictaba shares significant sequence similarity with the Cucurbitaceae phloem lectins (also referred to as PP2 proteins) that are highly conserved within Cucurbita species. In these plants, PP2 is one of the most abundant proteins present in the phloem sap. In contrast to Nictaba, PP2 proteins are expressed in the vascular tissue and contain an additional N-terminal peptide as well as a C-terminal domain that is thought to crosslink with the structural P-protein (PP1) through disulfide bridges. The PP2 protein was shown to be secreted into the assimilate stream where it can travel over long distances through the plant and exert its carbohydrate and RNA-binding activities [25]. Extensive database searches with the Nictaba domain signature revealed the existence of Nictaba or PP2-related proteins in a variety of genera belonging to the angiosperms and gymnosperms, of which the latter are known to be devoid of the structural P-protein. In Arabidopsis, many Nictaba-like proteins have been identified that have acquired an additional modular N-terminal domain such as an F-box- or TIR-domain. It can therefore be concluded that the Nictaba-domain is widespread in the plant kingdom [8]. It remains to be shown if all these proteins possess carbohydrate-binding activity.
Using hapten inhibition assays and glycan array analyses it was demonstrated that Nictaba exhibits specificity towards β(1–4)GlcNAc oligomers as well as complex and high mannose N-linked glycans. Because of this dual specificity, it was hypothesized that Nictaba specifically recognizes the GlcNAc2Man3 core of N-glycan structures. Despite numerous efforts, the three-dimensional structure of Nictaba and its carbohydrate-binding site could not yet be elucidated. Here we present a three-dimensional model of the Nictaba structure based on the structural homology with the carbohydrate-binding module 22 of Clostridium thermocellum Xyn10B [16]. According to this model, Nictaba consists of a β-sandwich composed of two β-sheets. Similar to many plant lectins the Nictaba model predicts a structure that consists mainly of β-sheet. These results are in agreement with circular dichroism analyses which revealed that Nictaba consists of 45% β-sheet, 55% β-coil, but no α-helix [6].
Nictaba contains a typical nuclear localization signal made up of four basic lysine residues. According to the model, this nuclear localization signal is located at the top of the protein as part of a protruding loop. It is therefore nicely exposed and can be readily recognized by an import protein that can facilitate transport of the lectin into the nucleus through the nuclear pore protein complex. The Nictaba model also shows the presence of a large electro-negatively charged pocket. A β-strand separates the large central groove from a smaller electronegatively charged extension. The electronegative character of the central pocket is mainly due to the presence of several glutamic acid residues, of which Glu138 and Glu145 stand out most prominently. From the second, smaller pocket, two tryptophan residues (Trp15, Trp22) protrude. As commonly observed in many other carbohydrate-binding sites of plant lectins, the conserved tryptophan residues would complete the interaction with the sugar by an aromatic stacking with the pyranose ring of the H-bound sugar. This hypothesis is corroborated by research on other Nictaba-like proteins from the Cucurbitales. Studies of the carbohydrate binding specificity of the Nictaba-like Luffa acutangula lectin strongly suggest a role for a tryptophan residue in the carbohydrate binding site, since fluorescence in the near UV-CD spectrum changes upon interaction with its ligand [26, 27]. In addition, thermodynamic studies and fluorescence analysis of the Coccinia indica agglutinin with a labeled chito-oligomer of variable length also suggest involvement of a tryptophan residue in the binding site [28]. These results are enforced by chemical modification experiments. However, these results are in sharp contrast with recent findings on the Cucurbita maxima phloem exudate lectin. For this lectin it was shown that tryptophan residues are only partially exposed to the aqueous environment and are probably not involved in ligand interaction, because binding of GlcNAc oligomers—which are specifically recognized by the lectin—did not significantly alter the quenching pattern [29].
To validate our proposed three-dimensional model, the Nictaba coding sequence was mutated and recombinant proteins were expressed in and purified from Pichia pastoris. Residues Glu138 and Glu148 have been mutated to alanine residues whereas Trp15 and Trp22 have been changed into leucine. The carbohydrate binding properties of these mutant proteins were investigated using the glycan array technology. It was shown that mutation of both designated glutamic acid residues (Nictaba mutant 1) did not influence the carbohydrate binding specificity. Possibly, this could be explained by the presence of other glutamic acid residues nearby, thus preserving the electronegative character of the binding site. In contrast, mutation of the two tryptophan residues (Nictaba mutant 2) located in a smaller extension of the proposed binding pocket, led to a complete loss of binding of the protein to any of the carbohydrate structures present on the array. Similarly, when only Trp15 was mutated to leucine (Nictaba mutant 3), again no carbohydrate binding was observed, suggesting a complete loss of lectin activity. These results strongly endorse the three-dimensional structure of Nictaba as proposed in the model. We suggest that carbohydrate binding activity of the Nictaba domain is mediated through the large electronegatively charged groove and hypothesize that hydrophobic interactions between the indole group of Trp15 and the pyranose groups of the glycan molecule stabilize the interactions in the lectin binding site.
Acknowledgments
This work was funded primarily by the Fund for Scientific Research-Flanders (FWO grant G.0022.08) and the Research Council of Ghent University (projects BOF2005/GOA/008 and BOF2007/GOA/0017). The authors want to thank The Consortium for Functional Glycomics funded by the NIGMS GM62116 for the glycan array analysis.
Contributor Information
Dieter Schouppe, Department of Molecular Biotechnology, Laboratory of Biochemistry and Glycobiology, Ghent University, Coupure Links 653, 9000 Ghent, Belgium.
Pierre Rougé, Surfaces Cellulaires et Signalisation chez les Végétaux, UMR-CNRS 5546, Pôle de Biotechnologie Végétale, Toulouse, France.
Yi Lasanajak, Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322, USA.
Annick Barre, Surfaces Cellulaires et Signalisation chez les Végétaux, UMR-CNRS 5546, Pôle de Biotechnologie Végétale, Toulouse, France.
David F. Smith, Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322, USA
Paul Proost, Laboratory of Molecular Immunology, Rega Institute for Medical Research, University of Leuven, Minderbroedersstraat 10, 3000 Leuven, Belgium.
Els J. M. Van Damme, Email: ElsJM.VanDamme@UGent.be, Department of Molecular Biotechnology, Laboratory of Biochemistry and Glycobiology, Ghent University, Coupure Links 653, 9000 Ghent, Belgium
References
- 1.Peumans WJ, Van Damme EJM. Lectins as plant defense proteins. Plant Physiol. 1995;109:347–352. doi: 10.1104/pp.109.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Damme EJM, Lannoo N, Peumans WJ. Plant lectins. Adv Bot Res. 2009;48:107–209. [Google Scholar]
- 3.Lannoo N, Van Damme EJM. Nucleocytoplasmic plant lectins. Biochem Biophys Acta. 2009;1800:190–201. doi: 10.1016/j.bbagen.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 4.Rini JM. Lectin structure. Annu Rev Biophys Biomol Struct. 1995;24:551–577. doi: 10.1146/annurev.bb.24.060195.003003. [DOI] [PubMed] [Google Scholar]
- 5.Sinha S, Gupta G, Vijayan M, Surolia A. Subunit assembly of plant lectins. Curr Opin Struct Biol. 2007;17:498–505. doi: 10.1016/j.sbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Peumans WJ, Hause B, Bras J, Kumar M, Proost P, Barre A, Rougé P, Van Damme EJM. Jasmonate methyl ester induces the synthesis of a cytoplasmic/nuclear chitooligosaccharide-binding lectin in tobacco leaves. FASEB J. 2002;16:905–907. doi: 10.1096/fj.01-0598fje. [DOI] [PubMed] [Google Scholar]
- 7.Lannoo N, Peumans WJ, Van Damme EJM. Do F-box proteins with a C-terminal domain homologous with the tobacco lectin play a role in protein degradation in plants? Biochem Soc Trans. 2008;36:843–847. doi: 10.1042/BST0360843. [DOI] [PubMed] [Google Scholar]
- 8.Dinant S, Clark AM, Zhu YM, Vilaine F, Palauqui JC, Kusiak C, Thompson GA. Diversity of the superfamily of phloem lectins (Phloem protein 2) in angiosperms. Plant Physiol. 2003;131:114–128. doi: 10.1104/pp.013086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lannoo N, Peumans WJ, Van Pamel E, Alvarez R, Xiong TC, Hause G, Mazars C, Van Damme EJM. Localization and in vitro binding studies suggest that the cytoplasmic/nuclear tobacco lectin can interact in situ with high mannose and complex N-glycans. FEBS Lett. 2006;580:6329–6337. doi: 10.1016/j.febslet.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 10.Lannoo N, Vandenborre G, Miersch O, Smagghe G, Wasternack C, Peumans WJ, Van Damme EJM. The jasmonate-induced expression of the Nicotiana tabacum leaf lectin. Plant Cell Physiol. 2007;48:1207–1218. doi: 10.1093/pcp/pcm090. [DOI] [PubMed] [Google Scholar]
- 11.Vandenborre G, Miersch O, Hause B, Smagghe G, Wasternack C, Van Damme EJM. Spodoptera littoralis induced lectin expression in tobacco. Plant Cell Physiol. 2009;50:1142–1155. doi: 10.1093/pcp/pcp065. [DOI] [PubMed] [Google Scholar]
- 12.Lannoo N, Vervecken W, Proost P, Rougé P, Van Damme EJM. Expression of the nucleocytoplasmic tobacco lectin in the yeast Pichia pastoris. Protein Expr Purif. 2007;53:275–282. doi: 10.1016/j.pep.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Risler JL, Delorme MO, Delacroix H, Henaut A. Aminoacid substitutions in structurally related proteins—a pattern-recognition approach—determination of a new and efficient scoring matrix. J Mol Biol. 1988;204:1019–1029. doi: 10.1016/0022-2836(88)90058-7. [DOI] [PubMed] [Google Scholar]
- 15.Gaboriaud C, Bissery V, Benchetrit T, Mornon JP. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987;224:149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- 16.Xie HF, Gilbert HJ, Charnock SJ, Davies GJ, Williamson MP, Simpson PJ, Raghothama S, Fontes CMGA, Dias FMV, Ferreira LMA, Bolam DN. Clostridium thermocellum Xyn10B carbohydrate-binding module 22-2: the role of conserved amino acids in ligand binding. Biochemistry. 2001;40:9167–9176. doi: 10.1021/bi0106742. [DOI] [PubMed] [Google Scholar]
- 17.Ponder JW, Richards FM. Tertiary templates for proteins—use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987;193:775–779. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 18.Mas MT, Smith KC, Yarmush DL, Aisaka K, Fine RM. Modeling the anti-CEA antibody combining site by homology and conformational search. Protein Struct Funct Genet. 1992;14:483–498. doi: 10.1002/prot.340140409. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck—a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 20.Nicholls A, Sharp KA, Honig B. Protein folding and association—insights from the interfacial and thermodynamic properties of hydrocarbons. Protein Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 21.Gilson MK, Honig BH. Calculation of electrostatic potential in an enzyme active site. Nature. 1987;330:84–86. doi: 10.1038/330084a0. [DOI] [PubMed] [Google Scholar]
- 22.Blixt O, Head S, Mondala T, Scanlan C, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, van Die I, Burton D, Wilson IA, Cummings R, Huflejt ME, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bradford MM. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli UK. Cleavage of structural proteins during assembly of head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Gomez G, Torres H, Pallas V. Identification of translocatable RNA-binding phloem proteins from melon, potential components of the long distance RNA transport system. Plant J. 2005;41:107–116. doi: 10.1111/j.1365-313X.2004.02278.x. [DOI] [PubMed] [Google Scholar]
- 26.Anantharam V, Patanjali SR, Surolia A. A chitotetrose specific lectin from Luffa acutangula—physicochemical properties and the assignment of orientation of sugars in the lectin binding site. J Biosci. 1985;8:403–411. [Google Scholar]
- 27.Anantharam V, Patanjali SR, Swamy MJ, Sanadi AR, Goldstein IJ, Surolia A. Isolation, macromolecular properties, and combining site of a chitooligosaccharide-specific lectin from the exudate of ridge gourd (Luffa acutangula) J Biol Chem. 1986;261:4621–4627. [PubMed] [Google Scholar]
- 28.Sanadi AR, Ananthram V, Surolia A. Elucidation of the combining site of Coccinia indica agglutinin (CIA) by thermodynamic analyses of its ligand binding. Pure Appl Chem. 1998;70:677–686. [Google Scholar]
- 29.Narahari A, Swamy MJ. Tryptophan exposure and accessibility in the chitooligosaccharide-specific phloem exudate lectin from pumpkin (Cucurbita maxima). A fluorescence study. J Photochem Photobiol B. 2009;97:40–47. doi: 10.1016/j.jphotobiol.2009.07.008. [DOI] [PubMed] [Google Scholar]