Abstract
Plastids are found across the tree of life in a tremendous diversity of life forms. Surprisingly they are not limited to photosynthetic organisms but also found in numerous predators and parasites. An important reason for the pervasiveness of plastids has been their ability to move laterally and to jump from one branch of the tree of life to the next through secondary endosymbiosis. Eukaryotic algae have entered endosymbiotic relationships with other eukaryotes on multiple independent occasions. The descendants of these endosymbiotic events now carry complex plastids, organelles that are bound by three or even four membranes. As in all endosymbiotic organelles most of the symbiont’s genes have been transferred to the host and their protein products have to be imported into the organelle. As four membranes might suggest, this is a complex process. The emerging mechanisms display a series of translocons that mirror the divergent ancestry of the membranes they cross. This review is written from a parasite biologist viewpoint and seeks to provide a brief overview of plastid evolution in particular for readers not already familiar with plant and algal biology and then focuses on recent molecular discoveries using genetically tractable Apicomplexa and diatoms.
Keywords: Apicoplast, chloroplast, Apicomplexa, protein import, targeting
Introduction
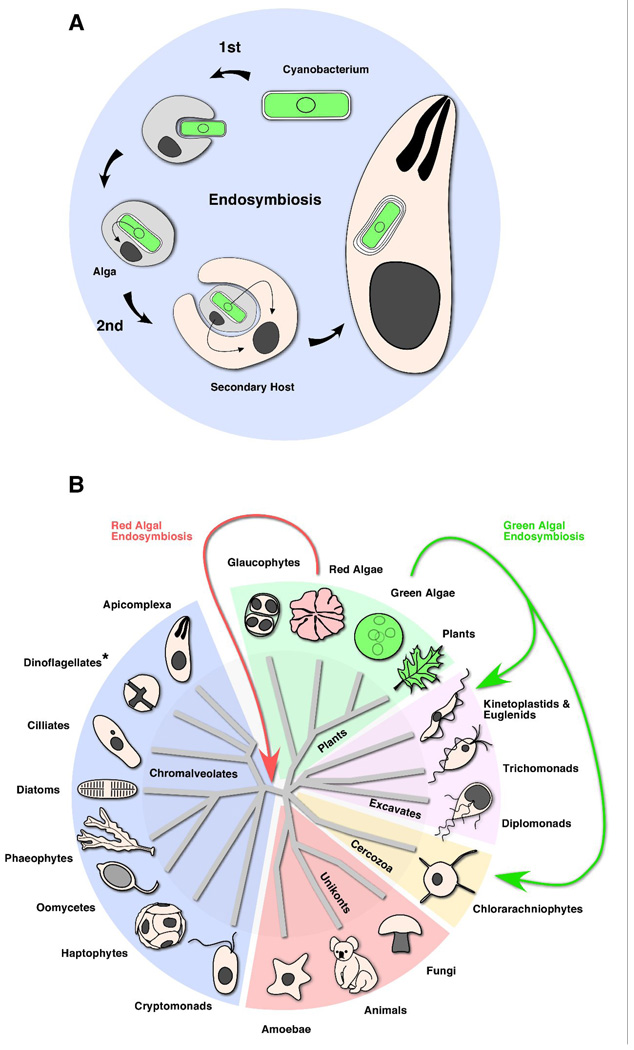
The massive expansion of cyanobacteria and oxygenizing photosynthesis, began to transform the atmosphere of our planet about 2.4 billion years ago. This new atmosphere gave birth to an explosion of complex life forms that took advantage of molecular oxygen and the large amount of energy that can be gained through oxidative phosphorylation. Cyanobacteria also gave rise to the chloroplasts of plants and algae further increasing their numbers and ecological impact (Cavalier-Smith 1982; Gray 1993). There is now broad support for an endosymbiosis model of plastid genesis (Gould et al. 2008). According to this model a cyanobacterium was taken up by an early eukaryote and subsequently domesticated into a dependent organelle (Fig. 1A). It appears that a single endosymbiotic event was responsible for the origin of the three major extant lineages of photosynthetic eukaryotes namely the red and green algae (including their progeny, the green plants) and the glaucophytes. Members of these groups possess plastids surrounded by two membranes and we will refer to these as primary plastids (Adl et al. 2005).
Figure 1.
(A) Diagramatic representation of the complex evolutionary process that gave rise to present day primary and secondary plastids. A cyanobacterium was engulfed by a heterotrophic eukaryote giving rise to the plastids of red and green algae (including land plants) and glaucophytes. In a second endosymbiotic event an alga was taken up by another eukaryote (note that this occurred multiple independent times). The resulting complex plastids are surrounded by four (and sometimes three) membranes. (B) Schematic tree of eukaryotic life highlighting the three major secondary endosymbiotic events that are thought to be responsible for present day complex plastids (*note that we excluded multiple events in dinoflagellates for simplicity). The relationships shown here are based on phylogenetic analyses summarized by Keeling and colleagues (Keeling et al. 2005). The ancestor of present day chromalveolates acquired their plastid through endosymbiotic uptake of a red alga. Diversification and adaptation to different ecological niches led to subsequent loss of photosynthesis (as in Apicomplexa) or loss of entire plastids (as in ciliates or oomycetes). Plastids of euglenids and chlorarachniophytes were acquired by two independent endosymbiotic events involving green algae.
Endosymbiosis and the Complex Ancestry of Complex Plastids
Primary plastids were massively successful but do not yet represent the end of the journey cyanobacteria have taken through the eukaryotic tree of life. In addition plastids were acquired laterally by secondary endosymbiosis to give rise to “secondary” or “complex” plastids spreading them further into previously non-photosynthetic eukaryotes (Cavalier-Smith 1982). Although the plastid is now the most conspicuous remnant of these events, it is important to bear in mind that as depicted in Figure 1A, an entire eukaryotic alga was the initial endosymbiont in these events. This eukaryotic cell within a second eukaryotic cell was then gradually reduced to the feature most useful to the host, the plastid. This origin is still reflected in the additional membranes found in secondary plastids, and, as we will discuss in more detail below, also in the origin and mechanisms of some of the protein import machinery. Secondary acquisition of plastids appears to have occurred in at least three independent incidents and gave rise to major branches of the eukaryotic tree. The plastids of euglenids and chlorarachniophytes arose by enslavement of two different green algae with chlorophyll a and b. Chlorarachniophytes are a group of unicellular green protists with some purely photosynthetic members (Lotharella globosa), and other species that are both photosynthetic and phagotrophic (Chlorarachnion). Euglenids are nutritionally even more diverse, ranging from photosynthetic species like Euglena spp, to phagotrophic petalomonads and peranemids (specialized on bacterial or eukaryotic prey respectively), to saprotrophs, which show no trace of plastids or have lost photosynthesis but retain a colorless plastid with a reduced genome. Initial studies postulated a common origin for chlorarachniophyptes and euglenids, but this hypothesis was later refuted (Rogers et al. 2007). Phylogenetic analysis of plastid proteins from the chlorachniophyte Bigelowiella natans supported an independent origin of plastids in these two groups (Rogers et al. 2007). In contrast, the plastids found in cryptomonads, haptophytes, stramenopiles, dinoflagellates and Apicomplexa are of red algal origin. The discovery of apicoplasts, plastids in the non-photosynthetic Apicomplexa, has fostered the idea of a common origin, for an at first sight rather incoherent group of organisms. The chromalveolate hypothesis proposes that a single endosymbiotic event followed by subsequent diversification was responsible for acquisition of the present day complex plastids in all these taxa (Cavalier-Smith 1999). The resulting super-phylum Chromalveolata (joining chromists and alveolates) represents as much as half of the thus far described protists and also includes many multicellular groups (see Fig. 1B). In addition to the plastid-containing taxa mentioned above the Chromalveolata also include groups such as the oomycetes or ciliates that may have possessed plastids in the past but have subsequently lost them (Cavalier-Smith and Chao 2006). Overall this represents a highly diverse group that has adapted to a tremendous breadth of ecological niches including autotrophy, predation and parasitism. Additional evidence in support of this hypothesis came from phylogenetic studies on glyceraldehyde 3- phosphate dehydrogenase (Fast et al. 2001; Harper and Keeling 2003; Harper et al. 2005). The common ancestor of different chromalveolate groups, cryptomonads, dinoflagellates and Apicomplexa seem to have replaced their plastid GAPDH gene with a cytosolic form that underwent duplication and acquired plastid targeting signal. It is unlikely that four different groups of organisms independently followed this complex path of locus evolution and it appears more parsimonious to conclude that diversification of chromalveolates was preceded by a single common endosymbiotic event involving a red alga. Further support for the red algal origin of chromalveolate plastids comes from the recent description of a photosynthetic apicomplexan, Chromera velia, a coral symbiont (Moore et al. 2008). This discovery has also produced interesting insights into the common origin of apicomplexan and dinoflagellate plastids (Janouskovec et al. 2010). C. velia plastids have features found in dinoflagellate plastids including a peculiar rubisco that appears to be acquired by horizontal transfer, transcript polyuridylation, and triplet stacking of thylakoids. In contrast, the plastid-encoded psbA gene of C. velia utilises the canonical UGA stop codon to encode tryptophan similar to its counterpart in the apicoplast genome and furthermore the gene order in the ribosomal superoperon is highly conserved. Careful phylogenetic analysis of C. velia plastid genes shows an overall closer affiliation to apicomplexan plastids and confirms a common red algal origin of both apicomplexan and peridinin-containing dinoflagellate plastids (Janouskovec et al. 2010; Moore et al. 2008).
All Plastids Import the Bulk of their Proteome
Plastids are important organelles and home to numerous essential cellular functions. Photosynthesis is an obvious plastid function and its benefits likely drove the initial endosymbiosis. However, plastids are central anabolic hubs that provide cells with additional key metabolites such as heme, aromatic amino acids, fatty acids, isoprenoids, and phospholipids, and this is true for primary as well as secondary plastids. These metabolic functions appear responsible for plastid persistence even after loss of photosynthesis in taxa like the Apicomplexa (Seeber and Soldati-Favre 2010). The metabolic functions of the apicoplast are heavily pursued as potential drug targets in particular for the treatment of malaria. Compatible with their complex functions, plastids have a complex proteome made up of hundreds of proteins. Although all currently known plastids maintain an organellar genome, this genome encodes only a relatively small number of proteins. As an example, the chloroplast proteome of higher plants is estimated to be comprised of about 4000 polypeptides (Leister 2003), a number far in excess of the 130 gene products encoded by the organellar genome (Wakasugi et al. 2001). Much of the symbiont’s genetic information has been transferred to the host thus providing central control over function and inheritance (Kleine et al. 2009; Martin et al. 1993; Martin and Herrmann 1998). This applies to primary and secondary endosymbiosis. In the case of secondary plastids, genes were largely transferred from the nucleus of the algal symbiont to the nucleus of the host, however additional transfer may also have occurred from the symbiont’s plastid genome to the host nucleus (McFadden 1999). The endosymbiont nucleus underwent severe reduction through this process and in most cases (as in the apicomplexan plastid) was lost entirely. Fascinatingly, in cryptophytes and chlorarachinophytes a small remnant of the nucleus, the nucleomorph, resides between the second and third membrane of the organelle (Douglas et al. 2001; Gilson and McFadden 1996) (see Fig. 2 B and E for a schematic representation). The nucleomorph genomes of both groups are very compact and organized into three small linear chromosomes with telomeres at their ends (Gilson and McFadden 1995, 2002). An obvious consequence of gene transfer is that the bulk of the proteome now has to be post-translationally imported. The relative complexity of establishing protein import mechanisms to accomplish this might have limited the number of endosymbiotic events throughout evolution (Cavalier-Smith 1999).
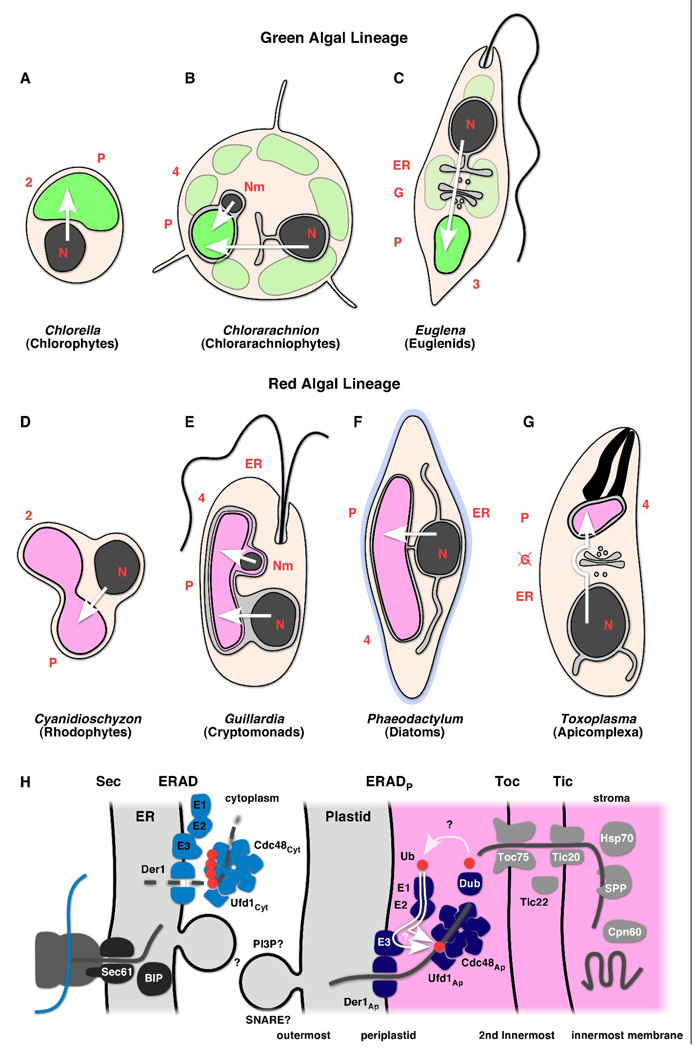
Figure 2.
(A) Schematic depiction of cellular morphology and the trafficking routes to the plastid for a selection of plastid containing organisms. Arrows indicate the path taken by nuclear/nucleomorph encoded proteins to reach their final destination in the plastid. (A, D) The green algae Chlorella and the red alga Cyanidioschyzon: nuclear encoded plastid proteins are synthesized in the cytosol and transported across the two plastid membrane with the help of Tic-Toc translocons. Engulfment of a red or green plastid by a heterotrophic eukaryote gave rise to organisms with multiple membrane bound secondary plastids. Secondary symbiosis gave rise to two main green lineages, chlorarachniophytes (B) euglenids (C). The main secondary red lineage has a common origin and includes cryptomonads (E), diatoms (F) and Apicomplexa (G). A remnant of the algal nucleus, the nucleomorph (Nm) resides between the second and third outermost compartments in chlorarachniophytes and cryptomonads (B , E). Note that in cryptomonads and diatoms the plastid resides within the endoplasmic reticulum (E, F). N, nucleus, G, Golgi, P, plastid, ER, endoplasmic reticulum. H. Schematic outline of a molecular model of the plastid protein import machinery based on results from Apicomplexa and diatoms. Note that not all elements have been experimentally validated (several such points are highlighted by a question mark). The pathway taken by apicomplexan proteins from the ER to the apicoplast remains highly speculative. Cargo proteins are shown as grey lines, proteins destined for degradation as dashed lines. Please refer to Table 1 for further detailed reference on specific proteins.
Our understanding of the molecular detail of protein import mechanisms in different complex plastids has benefited tremendously from the quickly growing number of complete genome sequences (nuclear, nucleomorph and plastid) and the successful establishment of genetic transfection models in apicomplexan parasites (mostly Toxoplasma and Plasmodium), diatoms (Phaeodactylum and Thalassiosira), and most recently in chlorarachniophytes (Lotharella amoebiformis). In this review we will try to capture the complex journey of nuclear- encoded plastid proteins, the similarities and differences in the pathways between different taxa and between plastids bounded by four versus three membranes.
N-terminal Leaders Guide Trafficking to the Plastid
Most plastid proteins initially possess sequences at their N-termini that guide targeting to the organelle, and these are commonly referred to as transit peptides. Plant chloroplast transit peptides are characterized by an overall positive charge and are enriched for the hydroxylated amino acids, serine and threonine. These transit peptides have been shown to be necessary and sufficient for targeting GFP and other reporters to the chloroplast. Secondary plastid proteins of heterokonts, cryptomonads, Apicomplexa, chlorarachinophytes, dinoflagellates and euglenids all seem to require additional targeting information that is also encoded at the N-terminus. The targeting information for complex plastids consists minimally of a bipartite signal made up of at least two sequential elements, the signal peptide and the transit peptide. The first part is the signal peptide that allows co-translational insertion of preproteins into the ER (Bhaya and Grossman 1991). This is thought to occur using the Sec61 translocon, a mechanism described for a large variety of secretory proteins (Rapoport 2007). Numerous studies have shown that this signal peptide once isolated confers secretion to a reporter (DeRocher et al. 2000; Waller et al. 2000). Furthermore, in the bipartite context, the signal peptide can be replaced with a canonical signal peptide from a secretory protein without loss of plastid targeting (Tonkin et al. 2006b). The cleavage of the signal peptide, which is typical for this process would then reveal the second part, the transit peptide, which appears to be responsible for the remainder of the journey of plastid proteins with the stromal compartment as final destination. Several groups have now confirmed that transit peptides from secondary plastids (euglenids, Apicomplexa, cryptomonads and heterokonts) when fused to a reporter such as GFP can mediate protein import into isolated primary plastids of higher plants in vitro suggesting that they share structural and functional characteristics with plant transit peptides (Ralph et al. 2004b). For example a truncated form of plastid GAPDH from the cryptophyte Guillardia theta that lacks a signal peptide is translocated across the two membranes of the pea chloroplast (Wastl and Maier 2000). The processing of signal peptides is thought to be fast and likely occurs before the protein is synthesized to full length; transit peptide processing is slower. Using pulse chase labeling with radioactive amino acids and immunoprecipitation of an apicoplast-targeted acyl carrier protein-GFP fusion protein in P. falciparum, van Dooren et al. estimated that plastid proteins take an average of 90 minutes to reach the stroma (van Dooren et al. 2002).
Although the transit peptide is not clearly defined at the primary sequence level, some common features have emerged. The transit peptides of apicoplast proteins in T. gondii and P. falciparum are characterized by an overall abundance of positively charged residues. Analysis of transit peptide variants generated by mutagenesis or de novo design suggest that a net positive charge is necessary for import but that the exact position of positively charged residue can be flexible (Foth et al. 2003; Tonkin et al. 2006a). Complex plastids with their multiple membranes have numerous destinations that require differential targeting. An important example is the space between the 2nd and 3rd membrane, the periplastid compartment or PPC. In cryptomonads and chlorarachniophytes this is home to the reduced algal nucleus and thus likely represents the remnant of the algal cytoplasm. The nucleomorph genome encodes many of the housekeeping proteins required to maintain this nucleus (in addition to a smaller subset that targets to the stroma of the plastid). However, a significant number of proteins essential for DNA replication and protein synthesis in this space are encoded by the host nucleus. Proteins targeted to the stroma in cryptophytes are distinguished from those targeted to the periplastid compartment by the presence of an aromatic amino acid at the +1 position of the transit peptide (Gould et al. 2006; Kilian and Kroth 2005). A similar pattern was subsequently found in plastid proteins from organisms that have lost the nucleomorph, like diatoms (Kilian and Kroth 2005), haptophytes (Patron et al. 2006), dinoflagellates (Patron et al. 2005) and P. falciparum (Ralph et al. 2004a). Recently Ishida and colleagues found that in the chlorarachniophyte L. amoebiformis, the presence of negative charges in the transit peptide of an elongation factor-like protein was important for accurate targeting to the PPC (Hirakawa et al. 2010).
Protein targeting to the three membrane bound plastid of Euglena occurs via the secretory pathway analogous to other complex plastids, however in some cases this requires a tripartite presequence (Durnford and Gray 2006; Patron et al. 2005). In addition to the signal and transit peptide sequences found in other complex plastids which ensure transport into the ER and across inner membranes of the plastid, Euglena plastid proteins feature a third domain (also present in some dinoflagellates plastid proteins). This domain acts as a hydrophobic stop transfer signal and allows precursor proteins to be transported to the plastid as integral membrane proteins (Sulli et al. 1999; van Dooren et al. 2001). Experiments using the Euglena light harvesting chlorophyll a/b-binding protein of photosystem II and a canine microsome membrane system have validated this model (Kishore et al. 1993). The first segment (a signal peptide) is required for co-translation insertion and is cleaved. The transit peptide is followed by the second hydrophobic region that prevents full translocation. Thus plastid directed proteins remain associated with the ER (and Golgi) membranes, instead of being soluble in the lumen of the endomembrane system. Note that the bulk of the proteins remains exposed to the cytoplasm under this model, a prediction that is supported by protease digestion experiments (Sulli et al. 1999). Dinoflagellate plastid-directed proteins feature leader sequences that are functionally similar to those found in Euglena suggesting that the second hydrophobic region might be linked to the three membrane topology of euglenid and dinoflagellate plastids (Nassoury et al. 2003; Patron et al. 2005). It appears that the transit peptide is required and sufficient to cross the remaining inner membranes in all complex plastids. Once inside the plastid stroma, a protease related to plant stromal processing peptidase is thought to cleave the transit peptide to release the mature protein. A SPP homolog has been identified in P. falciparum and is speculated to reside in the apicoplast lumen (van Dooren et al. 2002).
While bipartite leader sequences have been identified and validated in many proteins in different organisms they are not found on all plastid proteins. Several apicomplexan proteins, in particular membrane and intermembrane proteins have been reported that lack canonical plastid signaling sequences. Examples of such proteins include FtsH1, a zinc finger metalloprotease, that undergoes both N and C-terminal processing, (Karnataki et al. 2007b, 2009) ATRX1, a thioredoxin like protein that is targeted to the outermost apicoplast membrane (DeRocher et al. 2008) and apicoplast phosphate translocators that show differential targeting in T. gondii and P. falciparum (Karnataki et al. 2007a; Mullin et al. 2006). It is not clear how these proteins traverse the plastid membranes at this time but one of the transmembrane domains of these proteins may act as cryptic signal anchor. It is important to consider here that there are many distinct destinations within secondary plastids (4 membranes and 4 luminal or intermembrane compartments) that proteins need to be targeted to. The differences in the structure and topology of transit peptides might be an important clue for differential targeting mechanisms.
Crossing Multiple Membranes Using Specific and Distinct Translocons
A variety of mechanistic models have been developed to explain how nuclear-encoded proteins cross the many membranes that define complex plastids. Over time these models invoked internal vesicular trafficking and membrane fission and fusion (Gibbs 1981; Kilian and Kroth 2005), transport by an unspecific pore, and more recently a series of protein translocons of varied origin (Bodyl 2004; Cavalier-Smith 1999). Our view of the precise nature and sequence of these translocons has evolved rapidly over recent years as more and more molecular and functional data became available. An important observation from this work is that in general membranes are crossed with the help of translocons derived from the organism that “donated” the membrane to the complex organelle. In other words evolution matters, and mechanisms set up at the beginning are very stable and remain in place over hundreds of millions of years despite dramatic cellular reduction and reorganization. To fully appreciate this it might be helpful to the reader less familiar with the diverse set of organisms that harbor secondary plastids to quickly reconsider the origin of the various membranes involved. During secondary endosymbiosis an alga was endocytosed into a vacuole most likely derived from the host endomembrane system (Fig. 1A; Archibald and Keeling 2002; Delwiche 1999; Gould et al. 2008; van Dooren et al. 2001). This is believed to now represent the outermost membrane. Following this line of thought the next membrane (the periplastid membrane) would be a derivative of the plasma membrane of the alga. The two innermost membranes are likely equivalent to the two chloroplast envelope membranes. Note that as in the original chloroplast these two membrane show tight physical apposition (Tomova et al. 2006). Chlorarachinophytes, heterokontophytes, haptophytes, cryptophytes and Apicomplexa have four membrane bound plastids. Phototrophic euglenophytes and peridinin containing dinoflagellates posses plastids that are bound by three membranes (Cavalier-Smith 1999). The third membrane of euglenid plastids has been argued to be either a remnant of the algal plasma membrane, or the host food vacuole membrane (Whatley et al. 1979). It is not inconceivable that the difference in numbers of membranes may be linked to a particular form of predatory “vampirism” found in members of these taxa called myzocytosis. In this process the membrane of a prey organism is punctured and the cellular contents are sucked into a phagosome leaving the empty membrane behind (Lukes et al. 2009).
The Search for Tic and Toc in Secondary Plastids
As detailed above, the two innermost membranes of all secondary plastids are considered to be equivalent to membranes of the algal chloroplast. Plant chloroplast protein import has been studied in considerable detail and is mediated by two macromolecular protein complexes that translocate proteins across the two membranes. These are: the translocon of the outer chloroplast membrane (Toc) and the translocon of the inner chloroplast membrane (Tic) (Soll and Schleiff 2004). Numerous proteins have been identified in these complexes; we focus here on a smaller subset that has also been found in secondary plastids. Critical components of the Toc complex in plants are the receptor proteins Toc34 and Toc159 that recognize transit peptide bearing proteins and direct them to the β barrel pore in the outer membrane Toc75 (Schleiff et al. 2003). Only homologs of Toc75 and Toc34 have been identified so far in the genome of the red alga Cyanidioschyzon merolae (McFadden and van Dooren 2004). Once through the outer membrane, proteins interact with and subsequently pass through the Tic complex. Tic 22 is located in the space between the two membranes and interacts with both Tic and Toc components and may function to hand over proteins from one to the other complex (Becker et al. 2004; Kouranov et al. 1998). Tic20 is an integral membrane protein and together with Tic 110 has been thought to function as part of the protein pore that facilitates protein transport across the inner membrane. Note that some researchers think of Tic20 as the actual pore while others favor Tic110 (Becker et al. 2005). Inside the chloroplast lumen, the AAA-ATPase chaperon ClpC interacts with Tic40 and Tic110 and appears to provide the mechanical force needed for pulling proteins across the inner membrane (Kovacheva et al. 2005). On arrival in the chloroplast stroma the transit peptide is removed by the stromal processing peptidase (Richter and Lamppa 1998) thus releasing the mature protein to be folded properly by the stromal chaperones (Li and Chiu 2010). The transit peptide is further degraded by the presequence peptidase (Stahl et al. 2005).
Genome sequencing efforts for numerous organisms with secondary plastids has opened the hunt for components of protein translocons using bioinformatic approaches. Comparative genome-wide searches have since identified several members of the Tic complex in secondary plastid containing organisms (see Table 1 for a list of Tic and Toc components). The nucleomorph genome of the cryptophyte G. theta encodes homologs of Tic110, Tic22 and a chaperone-binding Tic complex-associated protein called IAP100 (McFadden and van Dooren 2004), while a Tic20 gene is present on the B. natans nucleomorph (Gilson et al. 2006). Importantly, all of these components are also found in the genome of the red alga C. merolae (McFadden and van Dooren 2004). Homologs of Tic20 and Tic22 have also been identified in a variety of Apicomplexa with the marked exception of the plastid-less genus Cryptosporidium (van Dooren et al. 2008). Toxoplasma Tic20 (TgTic20) possesses a canonical N-terminal bipartite signal sequence that is cleaved and has four transmembrane domains. Adapting split-GFP technology van Dooren and colleagues further demonstrated that TgTic20 is an integral protein of the innermost membrane with both the N- and C-terminus of the protein facing the inside of apicoplast stroma (van Dooren et al. 2008). The split-GFP assay uses two fragments of GFP that are inserted into candidate genes to produce translational fusions (Cabantous and Waldo 2006). If the resulting proteins co-localize to a particular compartment they assemble into a fluorescent reporter. This system has proven to be very useful to dissect the complex compartmentalization of secondary plastids in Apicomplexa and more recently in diatoms. The human parasite Toxoplasma has become a somewhat surprising yet quite powerful experimental model organism to study plastid biology. One of the most helpful tools is the ability to construct conditional null mutants. Such genetic analysis provides a rigorous test for the importance and function of presumptive translocon components. This is particularly important for components that only share modest sequence conservation across taxa or that are specific to secondary plastids. Genetic ablation of Toxoplasma TgTic20 leads to parasite death (van Dooren et al. 2008). While the Toxoplasma plastid cannot be isolated in significant quantity at this point several assays have been recently developed to measure protein import following posttranslational modification of reporter proteins. These analyses demonstrate that the TgTic20 mutant shows profound loss of protein import followed by organelle loss and cell death (van Dooren et al. 2008). Similarly a second putative member of the Tic complex, Tic22, has been identified in P. falciparum and T. gondii apicoplast (Kalanon et al. 2009). Inducible knock down of TgTic22 results in loss of import and parasite viability similar to TgTic20 ablation suggesting an important role in the apicoplast protein import apparatus (Giel van Dooren, Swati Agrawal and Boris Striepen unpublished). It remains to be tested whether Apicomplexa have lost other Tic components, which may suggest fundamental differences in protein import mechanisms between the chromists and Apicomplexa, or if these components have simply diverged sufficiently to preclude identification by primary sequence comparison.
Table 1.
Proteins associated with membrane translocons in secondary plastids. Note that not all proteins have been experimentally validated. Numbers refer to PlasmoDB, ToxoDB, NCBI (G. theta and where indicated), or PhatrDB protein identifiers respectively.
| P. falciparum | T. gondii | G. theta | P. tricornutum | |
|---|---|---|---|---|
| Innermost membrane | ||||
| Tic20 | PF11_0459 | EU427053 (NCBI)1 | ||
| Tic22 | PFE1460w7 | TGME49_086050 | ||
| Second innermost membrane | ||||
| Toc75 |
ADG291232 (NCBI) |
TGME49_0723902 | 12881482 | |
| Periplastid membrane | ||||
| Der1-1 | PF14_04983,7 | TGME49_0819404 | AAK398105 | 316976 |
| Der1-2 | PFC0590c13 | TGME49_0374604 | 359656 | |
| Cdc48 | PF07_00473 | TGME49_1216404 | AAK397735 | 509786 |
| Ufd-1 | TGME49_857004 | AAF240065 | 493195 | |
| E1 | PF13_0182 PF13_03443 |
TGME49_114890 | 544605 | |
| E2 | Mal13P1.2273 | TGME49_095990 | 564315 | |
| E3 | CAC270645 | 480346 | ||
| DeUb | ADK55599 (NCBI) 6 | |||
| Ub | PF08_00673 | |||
In contrast to the Tic components, homologs of the Toc complex have been surprisingly hard to find in complex plastids. This observation could be attributed to the loss of Toc components in which case a different translocon might have replaced the Toc complex (a second Der1 translocon has been speculated to serve in this function (Tonkin et al. 2008)). Alternatively it is possible that the Toc protein homologs in secondary plastids are too divergent to be identified. Nonetheless, Bullmann and colleagues recently reported the presence of an omp85 like protein in the second innermost membrane of the complex plastid of the diatom Phaeodactylum tricornutum (Bullmann et al. 2010). Physiochemical studies using recombinant protein reconstituted into membranes show that ptOmp85 shares electrophysiological characteristics with its cyanobacterial counterparts and the Toc75 protein from land plants. Taken together the size and gating behavior of the pore and the presence of primary and secondary sequence features suggest that this protein is a strong candidate for a Toc component in complex plastids. There are similar beta barrel proteins in Apicomplexa, their localization and function is currently unknown. A Toc75 homolog is also encoded on the nucleomorph genome of the chlorarachniophyte B. natans (Gilson et al. 2006). Lastly, a putative Toc34 homolog has also been described in P. falciparum, but its function and role in import has not been tested (Waller and McFadden 2005). The overall working model that emerges is that the two innermost membranes of complex plastids share the import machinery of primary plastids in broad strokes, but that the system has experienced considerable diversification and adaptation in different taxa.
Crossing the Algal Plasma Membrane by Retooling an ER Protein Extraction Machinery
The second outermost membrane (in plastids with four membranes) or periplastid membrane is thought to be a remnant of the algal plasma membrane. The establishment of a protein import apparatus across this membrane can be considered a key event of secondary endosymbiosis as it directly links host and endosymbiont opening the door for communication and gene transfer. Several hypotheses have been put forward in the past to explain traffic across this border, but the lack of experimental evidence made it hard to distinguish between these different models. Recently, substantial insights into the molecular determinants that might mediate import across this membrane have emerged. A key observation came from sequencing the nucleomorph genome of the cryptophyte G. theta. The highly reduced nucleomorph genome surprisingly retains the genes for key elements of the endoplasmic reticulum-associated degradation system (Sommer et al. 2007) (see Table 1 for list of plastid specific ERAD and ubiquitination components in cryptomonads, diatoms and Apicomplexa). The ERAD pathway is generally responsible for ER homeostasis by retro-translocating misfolded secretory proteins from the ER followed by degradation by the proteasome in the cytosol. Elements of the ERAD machinery found in the cryptomonad nucleomorph genome are Der-1 (a candidate for the translocation channel), the AAA-ATPase Cdc48 and its cofactor Ufd-1. Note that there is no indication of Sec61, an alternative candidate for the ERAD pore. Der-1 is a small four trans-membrane domain containing protein, shown to be essential for retrotranslocation of misfolded luminal proteins from the ER in yeast and human cells (Ye et al. 2004). Translocating proteins are marked for degradation by conjugation of ubiquitin resulting in polyubiquitin chains. This appears to occur during translocation on the cytoplasmic face of the membrane. Cdc48 subsequently extracts these substrate proteins from the pore with the help of its cofactors, Ufd-1–Npl4 complex (Ye et al. 2004). Cdc48 has additional functions in protein extraction and degradation and many of these functions are linked to ubiquitination. Sommer and colleagues argued that protein degradation might not be the primary function of these proteins in secondary plastids and proposed a new role for this ERAD derived translocon (Sommer et al. 2007). They speculated that components of this ERAD complex might be involved in transport across the periplastid membrane in complex plastids. The hypothesis was subsequently tested in several organisms including cryptomonads, diatoms, and Apicomplexa (Agrawal et al. 2009; Kalanon et al. 2009; Sommer et al. 2007; Spork et al. 2009). As an example we demonstrated that the T. gondii nuclear genome encodes multiple paralogs of key components of ERAD including the pore candidate Der1, Cdc48 and Ufd-1. Immunofluorescence analysis of epitope tagged fusion products of these genes demonstrated the presence of two putative complexes. One set of components localizes to the ER and the cytoplasm; consistent with the well-described role of ERAD in degradation of misfolded ER proteins. A second set of ERAD components however localized to the apicoplast. EM analysis further confirmed peripheral localization of these components in the apicoplast. Using split GFP assays in the diatom P. tricornutum Hempel and colleagues confirmed that Der-1 is associated with the third membrane as proposed in the initial hypothesis (Hempel et al. 2009; Spork et al. 2009). To further investigate the role of these plastid localized proteins, we generated a conditional knock out in the apicoplast specific Der-1 paralog in Toxoplasma. Genetic ablation of the protein leads to a rapid loss of protein import as measured by pulse chase assay of post-translational modification associated with plastid targeting (Agrawal et al. 2009). Loss of protein import results in apicoplast biogenesis defects and ultimately parasite death. Phylogenetic analysis of the two Cdc48 paralogues found in organisms with complex plastids showed a divergent evolutionary origin of these two proteins. The plastid specific Cdc48 paralogs of Apicomplexa and diatoms show strong affinity with their red algal counterparts (Agrawal et al. 2009) while the cytoplasmic proteins of Apicomplexa are most closely related to Cdc48 proteins of their sister phyla of ciliates and dinoflagellates. This phylogeny, along with the presence of ERAD genes on the nucleomorph genome, strongly suggests that the plastid system evolved through the adaptation of the endosymbiont’s ERAD machinery. Interestingly, such an adaptation might have been relatively straightforward. Duplication of Der1 and relocation to the plasma membrane of the endosymbiont likely represented the first step. Topologically the ERAD system still imported into same compartment, the cytoplasm of the endosymbiont, and the numerous factors required to pull, fold and modify cargo proteins were already in place.
As mentioned earlier, classical ERAD is coupled with ubiquitination to mark proteins for subsequent degradation by the proteasome. Ubiquitination is a highly regulated process that in most systems requires the activity of three key proteins, namely the ubiquitin activating (E1), conjugating (E2) and ligase (E3) enzymes to transfer ubiquitin onto target proteins. The covalent addition of ubiquitin to a substrate starts with the activation of ubiquitin using ATP to form an ubiquitin-adenylate intermediate followed by transfer to the active site cysteine residue of E1 enzyme and the release of AMP. Ubiquitin is then transferred to the next enzyme of the pathway, E2 by a trans-thioesterification reaction. In the final step the C-terminal glycine of ubiquitin is ligated through an isopeptide bond to a lysine residue of the target protein. This step is mediated by an E3 enzyme and this step confers specificity for the final substrate (Haas and Siepmann 1997; Siepmann et al. 2003). Through this process single as well as multiple ubiquitin molecules can be added to a substrate protein. Polyubiquitin chains form by addition of ubiquitin on the lysine residues of a previously attached ubiquitin. Although the earliest reported functions of ubiquitin focused on its role in proteasomal degradation (Elsasser and Finley 2005; Miller and Gordon 2005), we now know that ubiquitin is important for many other physiological processes, including endocytosis, vesicular trafficking (Hicke 2001; Raiborg et al. 2003; Staub and Rotin 2006), cell-cycle control, stress response, DNA repair, signalling, transcription and gene silencing (Di Fiore et al. 2003; Haglund and Dikic 2005; Huang and D'Andrea 2006). Ubiquitin conjugation has been reported to be critical for both the ERAD translocation step and the subsequent degradation in the proteasome. There is accumulating evidence that cryptomonads, Apicomplexa and diatoms, all possess plastid-specific ubiquitination factors (Hempel et al. 2010; Spork et al. 2009; Swati Agrawal, Giel van Dooren, and Boris Striepen unpublished). Biochemical studies using recombinant proteins further suggest that these enzymes have the ability to activate and transfer ubiquitin. However, some questions remain. Most importantly, is ubiquitination required for plastid import? The model would suggest that plastid-targeted preproteins are ubiquitinated at some point of their journey. Unfortunately this has not been robustly demonstrated so far. However, there may be technical reasons for this, e.g. modification is only transient and therefore difficult to detect. It may be interesting to note in this context that plastid specific deubiquitinases have been reported in the P. tricornitum (Hempel et al. 2010). Ubiquitylation of plastid cargo may serve multiple functions. The first function that comes to mind is as an additional signal for movement of preproteins across the membranes, and this may be a requirement for the activity of Cdc48 in this process. Alternatively, ubiquitination may provide the signal necessary for retention of proteins in intermediary compartments. Ubiquitin may also serve as a specific signal for only a subset of plastid proteins. Ubiquitin has emerged at the center of numerous sorting and transport processes (Mukhopadhyay and Riezman 2007), and further work is needed to fully define its specific role(s) in the biology of complex plastids.
From ER to the Outermost Plastid Membrane
Protein targeting in three membrane bound complex plastids like euglenophytes and dinoflagellates appears to proceed from the ER to Golgi and finally to the plastid (Inagaki et al. 2000; Nassoury et al. 2003; Sulli et al. 1999). This model is supported by strong biochemical evidence using homologous and heterologous cell-free protein trafficking systems in Euglena. Slavikova and colleagues further suggested that in euglenophytes vesicles bud off from the Golgi and fuse with the outermost plastid membrane in a fashion independent of SNARE proteins (Slavikova et al. 2005). Although plastids of these two groups have divergent phylogenetic origins (Euglenophytes have a plastid of green algal lineage whereas the dinoflagellates plastids have a red algal origin) they appear to have evolved similar transport mechanisms (Yoon et al. 2002). The plastid takes its place as a “vacuole type” organelle and proteins are sorted via the Golgi apparatus following typical vesicular steps. These initial steps may be less conventional in plastids with four membranes. The outermost membrane in cryptophytes, haptophytes and heterokontophytes is decorated with 80S ribosomes and is thought to have evolved by fusion of the membrane of the phagosome surrounding the symbiont with the host ER (Cavalier-Smith 2002). It may be worthwhile here to also consider recent studies that suggest a more direct functional link between the ER in certain types of endocytosis dependent on synagmotagmin VII (Idone et al. 2008; Lewis and Lazarowitz 2010). Under such a scenario an endosymbiont might reside within the ER of the host cell from the start. Nucleus-encoded plastid proteins in these organisms are imported co-translationally into the ER lumen (Bhaya and Grossman 1991). Once in the ER, proteins have crossed the outermost membrane of the complex plastid and the transit peptide could then interact with the ERAD system of the periplastid membrane. In vitro experiments in canine microsomes demonstrated that signal peptide is required for microsomal import, which mimics protein import into the ER lumen. Subsequent in vivo experiments using reporter constructs that fuse signal peptides of plastid proteins from heterokontophytes, cryptophytes and Apicomplexa, showed targeting to the ER lumen or the secretory system (Gould et al. 2006; Kilian and Kroth 2005). How do proteins reach the plastid in organisms where the plastid does not reside within the ER? There is no solid evidence for permanent connections between the ER and the apicoplast but some electron microscopy studies suggested that the two organelles come into close contact, which may reflect functional interaction (Tomova et al. 2006, 2009). One might caution that connections may nonetheless be present, but too transient or too labile to be detected by electron microscopy. Brefeldin A is a fungal metabolite known to result in the redistribution of the Golgi apparatus thus blocking Golgi mediated transport steps. Interestingly in apicomplexan parasites like T. gondii and P. falciparum Brefeldin A does not perturb the trafficking of GFP reporter constructs destined to the apicoplast (DeRocher et al. 2005). Nonetheless, vesicles carrying apicoplast proteins have been reported by several independent groups using light and electron microscopy, in particular in apicoplast import mutants or mutants that have apicoplast biogenesis defects (DeRocher et al. 2008; Karnataki et al. 2007a; van Dooren et al. 2008, 2009). Overall these observations have been interpreted as evidence for a vesicular step in trafficking from the ER to the plastid that sidesteps the Golgi and is thus resistant to Brefeldin A. Such a mechanism would require a system to sort plastid-destined proteins into vesicles budding from the ER, and further imply a likely SNARE based system to ensure correct fusion of these vesicles with the plastid. There may be alternatives to this model. A new study in P. falciparum has detected phosphatidylinositol 3-monophosphate on the membrane of the apicoplast, the food vacuole and a subset of vesicles (Tawk et al. 2010). Additional experiments showed that interference with PI3P in T. gondii either by drug treatment or overexpression of PI3P binding proteins leads to severe plastid biogenesis defects (Maryse Lebrun, personal communication). More work is needed to understand the nature of these defects but they might suggest that one should consider the mechanisms active in the endosomal pathway and not solely focus on models that assume direct forward targeting from the ER.
Conclusion
The understanding of protein import into complex plastids has made several important leaps in recent years. The ability to perform genetic experiments in an ever-growing number of systems has produced quite detailed molecular models involving numerous protein factors. The list of new candidates and new candidate mechanisms is constantly fueled by the ongoing genome mining effort. Although the process is not fully understood in any of the models collectively, a working model of subsequent translocons is gaining more and more support. Although individual translocons appear well supported, where and when they act exactly and how they are coordinated is less clear. The tools available now should be suitable to tackle open questions like the precise role of ubiquitination or phosphatidylinositol phosphorylation in plastid protein import. Overall the mechanisms at work appear remarkably conserved, in some cases there are similar mechanisms even when the organelles emerged from distinct endosymbioses. Interestingly, how proteins take the first step, from ER to the plastid’s outermost compartment, shows the most diversity. For some systems like the apicoplast this also remains the least understood part of the journey. There is a fundamental distinction between three and four membrane systems. Three membrane systems follow the conventional secretory route of ER to Golgi to final target, and four membrane systems bypass the Golgi. Shedding light on the initial routing of plastid proteins and the different choices made by different organisms in this step may hold significant insights into the evolution of plastids and the eukaryotic cell in general.
Acknowledgements
The authors thank their many collaborators and in particular Giel van Dooren for many discussions and inspiration. Our work on apicoplast biology is currently funded by a predoctoral fellowship from the American Heart Association to S.A. and grants from the National Institutes of Health to B.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup O, Mozley-Standridge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor MF. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- Agrawal S, van Dooren GG, Beatty WL, Striepen B. Genetic evidence that an endosymbiont-derived endoplasmic reticulum-associated protein degradation (ERAD) system functions in import of apicoplast proteins. J Biol Chem. 2009;284:33683–33691. doi: 10.1074/jbc.M109.044024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JM, Keeling PJ. Recycled plastids: a 'green movement' in eukaryotic evolution. Trends Genet. 2002;18:577–584. doi: 10.1016/s0168-9525(02)02777-4. [DOI] [PubMed] [Google Scholar]
- Becker T, Hritz J, Vogel M, Caliebe A, Bukau B, Soll J, Schleiff E. Toc12, a novel subunit of the intermembrane space preprotein translocon of chloroplasts. Mol Biol Cell. 2004;15:5130–5144. doi: 10.1091/mbc.E04-05-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Qbadou S, Jelic M, Schleiff E. Let’s talk about.chloroplast import. Plant Biol. 2005;7:1–14. doi: 10.1055/s-2004-830447. [DOI] [PubMed] [Google Scholar]
- Bhaya D, Grossman A. Targeting proteins to diatom plastids involves transport through an endoplasmic reticulum. Mol Gen Genet. 1991;229:400–404. doi: 10.1007/BF00267462. [DOI] [PubMed] [Google Scholar]
- Bodyl A. Evolutionary origin of a preprotein translocase in the periplastid membrane of complex plastids: a hypothesis. Plant Biol. 2004;6:513–518. doi: 10.1055/s-2004-821092. [DOI] [PubMed] [Google Scholar]
- Bullmann L, Haarmann R, Mirus O, Bredemeier R, Hempel F, Maier UG, Schleiff E. Filling the gap, evolutionarily conserved Omp85 in plastids of chromalveolates. J Biol Chem. 2010;285:6848–6856. doi: 10.1074/jbc.M109.074807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabantous S, Waldo GS. In vivo and in vitro protein solubility assays using split GFP. Nat Methods. 2006;3:845–854. doi: 10.1038/nmeth932. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The evolutionary origin and phylogeny of eukaryote flagella. Symp Soc Exp Biol. 1982;35:465–493. [PubMed] [Google Scholar]
- Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J Eukaryot Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Chloroplast evolution: secondary symbiogenesis and multiple losses. Curr Biol. 2002;12:R62–R64. doi: 10.1016/s0960-9822(01)00675-3. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE. Phylogeny and megasystematics of phagotrophic heterokonts (kingdom Chromista) J Mol Evol. 2006;62:388–420. doi: 10.1007/s00239-004-0353-8. [DOI] [PubMed] [Google Scholar]
- Delwiche CF. Tracing the thread of plastid diversity through the tapestry of life. Am Nat. 1999;154:S164–S177. doi: 10.1086/303291. [DOI] [PubMed] [Google Scholar]
- DeRocher A, Hagen CB, Froehlich JE, Feagin JE, Parsons M. Analysis of targeting sequences demonstrates that trafficking to the Toxoplasma gondii plastid branches off the secretory system. J Cell Sci. 2000;113:3969–3977. doi: 10.1242/jcs.113.22.3969. [DOI] [PubMed] [Google Scholar]
- DeRocher A, Gilbert B, Feagin JE, Parsons M. Dissection of brefeldin A-sensitive and -insensitive steps in apicoplast protein targeting. J Cell Sci. 2005;118:565–574. doi: 10.1242/jcs.01627. [DOI] [PubMed] [Google Scholar]
- DeRocher AE, Coppens I, Karnataki A, Gilbert LA, Rome ME, Feagin JE, Bradley PJ, Parsons M. A thioredoxin family protein of the apicoplast periphery identifies abundant candidate transport vesicles in Toxoplasma gondii. Eukaryot Cell. 2008;7:1518–1529. doi: 10.1128/EC.00081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore PP, Polo S, Hofmann K. When ubiquitin meets ubiquitin receptors: a signalling connection. Nat Rev Mol Cell Biol. 2003;4:491–497. doi: 10.1038/nrm1124. [DOI] [PubMed] [Google Scholar]
- Douglas S, Zauner S, Fraunholz M, Beaton M, Penny S, Deng LT, Wu X, Reith M, Cavalier-Smith T, Maier UG. The highly reduced genome of an enslaved algal nucleus. Nature. 2001;410:1091–1096. doi: 10.1038/35074092. [DOI] [PubMed] [Google Scholar]
- Durnford DG, Gray MW. Analysis of Euglena gracilis plastid-targeted proteins reveals different classes of transit sequences. Eukaryot Cell. 2006;5:2079–2091. doi: 10.1128/EC.00222-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- Fast NM, Kissinger JC, Roos DS, Keeling PJ. Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol Biol Evol. 2001;18:418–426. doi: 10.1093/oxfordjournals.molbev.a003818. [DOI] [PubMed] [Google Scholar]
- Foth BJ, Ralph SA, Tonkin CJ, Struck NS, Fraunholz M, Roos DS, Cowman AF, McFadden GI. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science. 2003;299:705–708. doi: 10.1126/science.1078599. [DOI] [PubMed] [Google Scholar]
- Gibbs SP. The chloroplasts of some algal groups may have evolved from endosymbiotic eukaryotic algae. Ann N Y Acad Sci. 1981;361:193–208. doi: 10.1111/j.1749-6632.1981.tb46519.x. [DOI] [PubMed] [Google Scholar]
- Gilson P, McFadden GI. The chlorarachniophyte: a cell with two different nuclei and two different telomeres. Chromosoma. 1995;103:635–641. doi: 10.1007/BF00357690. [DOI] [PubMed] [Google Scholar]
- Gilson PR, McFadden GI. The miniaturized nuclear genome of eukaryotic endosymbiont contains genes that overlap, genes that are cotranscribed, and the smallest known spliceosomal introns. Proc Natl Acad Sci USA. 1996;93:7737–7742. doi: 10.1073/pnas.93.15.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson PR, McFadden GI. Jam packed genomes--a preliminary, comparative analysis of nucleomorphs. Genetica. 2002;115:13–28. doi: 10.1023/a:1016011812442. [DOI] [PubMed] [Google Scholar]
- Gilson PR, Su V, Slamovits CH, Reith ME, Keeling PJ, McFadden GI. Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature’s smallest nucleus. Proc Natl Acad Sci USA. 2006;103:9566–9571. doi: 10.1073/pnas.0600707103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SB, Sommer MS, Kroth PG, Gile GH, Keeling PJ, Maier UG. Nucleus-to-nucleus gene transfer and protein retargeting into a remnant cytoplasm of cryptophytes and diatoms. Mol Biol Evol. 2006;23:2413–2422. doi: 10.1093/molbev/msl113. [DOI] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- Gray MW. Origin and evolution of organelle genomes. Curr Opin Genet Dev. 1993;3:884–890. doi: 10.1016/0959-437x(93)90009-e. [DOI] [PubMed] [Google Scholar]
- Haas AL, Siepmann TJ. Pathways of ubiquitin conjugation. FASEB J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JT, Keeling PJ. Nucleus-encoded, plastid-targeted glyceraldehyde-3-phosphate dehydrogenase (GAPDH) indicates a single origin for chromalveolate plastids. Mol Biol Evol. 2003;20:1730–1735. doi: 10.1093/molbev/msg195. [DOI] [PubMed] [Google Scholar]
- Harper JT, Waanders E, Keeling PJ. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int J Syst Evol Microbiol. 2005;55:487–496. doi: 10.1099/ijs.0.63216-0. [DOI] [PubMed] [Google Scholar]
- Hempel F, Bullmann L, Lau J, Zauner S, Maier UG. ERAD-derived preprotein transport across the second outermost plastid membrane of diatoms. Mol Biol Evol. 2009;26:1781–1790. doi: 10.1093/molbev/msp079. [DOI] [PubMed] [Google Scholar]
- Hempel F, Felsner G, Maier UG. New mechanistic insights into pre-protein transport across the second outermost plastid membrane of diatoms. Mol Microbiol. 2010;76:793–801. doi: 10.1111/j.1365-2958.2010.07142.x. [DOI] [PubMed] [Google Scholar]
- Hicke L. A new ticket for entry into budding vesicles-ubiquitin. Cell. 2001;106:527–530. doi: 10.1016/s0092-8674(01)00485-8. [DOI] [PubMed] [Google Scholar]
- Hirakawa Y, Gile GH, Ota S, Keeling PJ, Ishida K. Characterization of periplastidal compartment-targeting signals in chlorarachniophytes. Mol Biol Evol. 2010;27:1538–1545. doi: 10.1093/molbev/msq038. [DOI] [PubMed] [Google Scholar]
- Huang TT, D'Andrea AD. Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol. 2006;7:323–334. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- Idone V, Tam C, Andrews NW. Two-way traffic on the road to plasma membrane repair. Trends Cell Biol. 2008;18:552–559. doi: 10.1016/j.tcb.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki J, Fujita Y, Hase T, Yamamoto Y. Protein translocation within chloroplast is similar in Euglena and higher plants. Biochem Biophys Res Commun. 2000;277:436–442. doi: 10.1006/bbrc.2000.3702. [DOI] [PubMed] [Google Scholar]
- Janouskovec J, Horak A, Obornik M, Lukes J, Keeling PJ. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci USA. 2010;107:10949–10954. doi: 10.1073/pnas.1003335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalanon M, Tonkin CJ, McFadden GI. Characterization of two putative protein translocation components in the apicoplast of Plasmodium falciparum. Eukaryot Cell. 2009;8:1146–1154. doi: 10.1128/EC.00061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnataki A, Derocher A, Coppens I, Nash C, Feagin JE, Parsons M. Cell cycle-regulated vesicular trafficking of Toxoplasma APT1, a protein localized to multiple apicoplast membranes. Mol Microbiol. 2007a;63:1653–1668. doi: 10.1111/j.1365-2958.2007.05619.x. [DOI] [PubMed] [Google Scholar]
- Karnataki A, Derocher AE, Coppens I, Feagin JE, Parsons M. A membrane protease is targeted to the relict plastid of Toxoplasma via an internal signal sequence. Traffic. 2007b;8:1543–1553. doi: 10.1111/j.1600-0854.2007.00637.x. [DOI] [PubMed] [Google Scholar]
- Karnataki A, DeRocher AE, Feagin JE, Parsons M. Sequential processing of the Toxoplasma apicoplast membrane protein FtsH1 in topologically distinct domains during intracellular trafficking. Mol Biochem Parasitol. 2009;166:126–133. doi: 10.1016/j.molbiopara.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW. The tree of eukaryotes. Trends Ecol Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Kilian O, Kroth PG. Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J. 2005;41:175–183. doi: 10.1111/j.1365-313X.2004.02294.x. [DOI] [PubMed] [Google Scholar]
- Kishore R, Muchhal US, Schwartzbach SD. The presequence of Euglena LHCPII, a cytoplasmically synthesized chloroplast protein, contains a functional endoplasmic reticulum-targeting domain. Proc Natl Acad Sci USA. 1993;90:11845–11849. doi: 10.1073/pnas.90.24.11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Maier UG, Leister D. DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol. 2009;60:115–138. doi: 10.1146/annurev.arplant.043008.092119. [DOI] [PubMed] [Google Scholar]
- Kouranov A, Chen X, Fuks B, Schnell DJ. Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J Cell Biol. 1998;143:991–1002. doi: 10.1083/jcb.143.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacheva S, Bedard J, Patel R, Dudley P, Twell D, Rios G, Koncz C, Jarvis P. In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J. 2005;41:412–428. doi: 10.1111/j.1365-313X.2004.02307.x. [DOI] [PubMed] [Google Scholar]
- Leister D. Chloroplast research in the genomic age. Trends Genet. 2003;19:47–56. doi: 10.1016/s0168-9525(02)00003-3. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Lazarowitz SG. Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc Natl Acad Sci U S A. 2010;107:2491–2496. doi: 10.1073/pnas.0909080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HM, Chiu CC. Protein transport into chloroplasts. Annu Rev Plant Biol. 2010;61:157–180. doi: 10.1146/annurev-arplant-042809-112222. [DOI] [PubMed] [Google Scholar]
- Lukes J, Leander BS, Keeling PJ. Cascades of convergent evolution: the corresponding evolutionary histories of euglenozoans and dinoflagellates. Proc Natl Acad Sci USA. 2009;106 Suppl 1:9963–9970. doi: 10.1073/pnas.0901004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Brinkmann H, Savonna C, Cerff R. Evidence for a chimeric nature of nuclear genomes: eubacterial origin of eukaryotic glyceraldehyde-3-phosphate dehydrogenase genes. Proc Natl Acad Sci USA. 1993;90:8692–8696. doi: 10.1073/pnas.90.18.8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Herrmann RG. Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI. Plastids and protein targeting. J Eukaryot Microbiol. 1999;46:339–346. doi: 10.1111/j.1550-7408.1999.tb04613.x. [DOI] [PubMed] [Google Scholar]
- McFadden GI, van Dooren GG. Evolution: red algal genome affirms a common origin of all plastids. Curr Biol. 2004;14:R514–R516. doi: 10.1016/j.cub.2004.06.041. [DOI] [PubMed] [Google Scholar]
- Miller J, Gordon C. The regulation of proteasome degradation by multi-ubiquitin chain binding proteins. FEBS Lett. 2005;579:3224–3230. doi: 10.1016/j.febslet.2005.03.042. [DOI] [PubMed] [Google Scholar]
- Moore RB, Obornik M, Janouskovec J, Chrudimsky T, Vancova M, Green DH, Wright SW, Davies NW, Bolch CJ, Heimann K, Slapeta J, Hoegh-Guldberg O, Logsdon JM, Carter DA. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451:959–963. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Mullin KA, Lim L, Ralph SA, Spurck TP, Handman E, McFadden GI. Membrane transporters in the relict plastid of malaria parasites. Proc Natl Acad Sci USA. 2006;103:9572–9577. doi: 10.1073/pnas.0602293103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassoury N, Cappadocia M, Morse D. Plastid ultrastructure defines the protein import pathway in dinoflagellates. J Cell Sci. 2003;116:2867–2874. doi: 10.1242/jcs.00517. [DOI] [PubMed] [Google Scholar]
- Patron NJ, Waller RF, Archibald JM, Keeling PJ. Complex protein targeting to dinoflagellate plastids. J Mol Biol. 2005;348:1015–1024. doi: 10.1016/j.jmb.2005.03.030. [DOI] [PubMed] [Google Scholar]
- Patron NJ, Waller RF, Keeling PJ. A tertiary plastid uses genes from two endosymbionts. J Mol Biol. 2006;357:1373–1382. doi: 10.1016/j.jmb.2006.01.084. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Rusten TE, Stenmark H. Protein sorting into multivesicular endosomes. Curr Opin Cell Biol. 2003;15:446–455. doi: 10.1016/s0955-0674(03)00080-2. [DOI] [PubMed] [Google Scholar]
- Ralph SA, Foth BJ, Hall N, McFadden GI. Evolutionary pressures on apicoplast transit peptides. Mol Biol Evol. 2004a;21:2183–2194. doi: 10.1093/molbev/msh233. [DOI] [PubMed] [Google Scholar]
- Ralph SA, van Dooren GG, Waller RF, Crawford MJ, Fraunholz MJ, Foth BJ, Tonkin CJ, Roos DS, McFadden GI. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol. 2004b;2:203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Richter S, Lamppa GK. A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc Natl Acad Sci USA. 1998;95:7463–7468. doi: 10.1073/pnas.95.13.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers MB, Gilson PR, Su V, McFadden GI, Keeling PJ. The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol Biol Evol. 2007;24:54–62. doi: 10.1093/molbev/msl129. [DOI] [PubMed] [Google Scholar]
- Schleiff E, Soll J, Kuchler M, Kuhlbrandt W, Harrer R. Characterization of the translocon of the outer envelope of chloroplasts. J Cell Biol. 2003;160:541–551. doi: 10.1083/jcb.200210060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber F, Soldati-Favre D. Metabolic pathways in the apicoplast of apicomplexa. Int Rev Cell Mol Biol. 2010;281:161–228. doi: 10.1016/S1937-6448(10)81005-6. [DOI] [PubMed] [Google Scholar]
- Siepmann TJ, Bohnsack RN, Tokgoz Z, Baboshina OV, Haas AL. Protein interactions within the N-end rule ubiquitin ligation pathway. J Biol Chem. 2003;278:9448–9457. doi: 10.1074/jbc.M211240200. [DOI] [PubMed] [Google Scholar]
- Slavikova S, Vacula R, Fang Z, Ehara T, Osafune T, Schwartzbach SD. Homologous and heterologous reconstitution of Golgi to chloroplast transport and protein import into the complex chloroplasts of Euglena. J Cell Sci. 2005;118:1651–1661. doi: 10.1242/jcs.02277. [DOI] [PubMed] [Google Scholar]
- Soll J, Schleiff E. Protein import into chloroplasts. Nat Rev Mol Cell Biol. 2004;5:198–208. doi: 10.1038/nrm1333. [DOI] [PubMed] [Google Scholar]
- Sommer MS, Gould SB, Lehmann P, Gruber A, Przyborski JM, Maier UG. Der1-mediated preprotein import into the periplastid compartment of chromalveolates? Mol Biol Evol. 2007;24:918–928. doi: 10.1093/molbev/msm008. [DOI] [PubMed] [Google Scholar]
- Spork S, Hiss JA, Mandel K, Sommer M, Kooij TW, Chu T, Schneider G, Maier UG, Przyborski JM. An unusual ERAD-like complex is targeted to the apicoplast of Plasmodium falciparum. Eukaryot Cell. 2009;8:1134–1145. doi: 10.1128/EC.00083-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl A, Nilsson S, Lundberg P, Bhushan S, Biverstahl H, Moberg P, Morisset M, Vener A, Maler L, Langel U, Glaser E. Two novel targeting peptide degrading proteases, PrePs, in mitochondria and chloroplasts, so similar and still different. J Mol Biol. 2005;349:847–860. doi: 10.1016/j.jmb.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Staub O, Rotin D. Role of ubiquitylation in cellular membrane transport. Physiol Rev. 2006;86:669–707. doi: 10.1152/physrev.00020.2005. [DOI] [PubMed] [Google Scholar]
- Sulli C, Fang Z, Muchhal U, Schwartzbach SD. Topology of Euglena chloroplast protein precursors within endoplasmic reticulum to Golgi to chloroplast transport vesicles. J Biol Chem. 1999;274:457–463. doi: 10.1074/jbc.274.1.457. [DOI] [PubMed] [Google Scholar]
- Tawk L, Chicanne G, Dubremetz JF, Richard V, Payrastre B, Vial HJ, Roy C, Wengelnik K. Phosphatidylinositol 3-phosphate, an essential lipid in Plasmodium, localises to the food vacuole membrane and the apicoplast. Eukaryot Cell. 2010 doi: 10.1128/EC.00124-10. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomova C, Geerts WJ, Muller-Reichert T, Entzeroth R, Humbel BM. New comprehension of the apicoplast of Sarcocystis by transmission electron tomography. Biol Cell. 2006;98:535–545. doi: 10.1042/BC20060028. [DOI] [PubMed] [Google Scholar]
- Tomova C, Humbel BM, Geerts WJ, Entzeroth R, Holthuis JC, Verkleij AJ. Membrane contact sites between apicoplast and ER in Toxoplasma gondii revealed by electron tomography. Traffic. 2009;10:1471–1480. doi: 10.1111/j.1600-0854.2009.00954.x. [DOI] [PubMed] [Google Scholar]
- Tonkin CJ, Roos DS, McFadden GI. N-terminal positively charged amino acids, but not their exact position, are important for apicoplast transit peptide fidelity in Toxoplasma gondii. Mol Biochem Parasitol. 2006a;150:192–200. doi: 10.1016/j.molbiopara.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Tonkin CJ, Struck NS, Mullin KA, Stimmler LM, McFadden GI. Evidence for Golgi-independent transport from the early secretory pathway to the plastid in malaria parasites. Mol Microbiol. 2006b;61:614–630. doi: 10.1111/j.1365-2958.2006.05244.x. [DOI] [PubMed] [Google Scholar]
- van Dooren GG, Schwartzbach SD, Osafune T, McFadden GI. Translocation of proteins across the multiple membranes of complex plastids. Biochim Biophys Acta. 2001;1541:34–53. doi: 10.1016/s0167-4889(01)00154-9. [DOI] [PubMed] [Google Scholar]
- van Dooren GG, Su V, D’Ombrain MC, McFadden GI. Processing of an apicoplast leader sequence in Plasmodium falciparum and the identification of a putative leader cleavage enzyme. J Biol Chem. 2002;277:23612–23619. doi: 10.1074/jbc.M201748200. [DOI] [PubMed] [Google Scholar]
- van Dooren GG, Tomova C, Agrawal S, Humbel BM, Striepen B. Toxoplasma gondii Tic20 is essential for apicoplast protein import. Proc Natl Acad Sci USA. 2008;105:13574–13579. doi: 10.1073/pnas.0803862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dooren GG, Reiff SB, Tomova C, Meissner M, Humbel BM, Striepen B. A novel dynamin-related protein has been recruited for apicoplast fission in Toxoplasma gondii. Curr Biol. 2009;19:267–276. doi: 10.1016/j.cub.2008.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi T, Tsudzuki T, Sugiura M. The genomics of land plant chloroplasts: Gene content and alteration of genomic information by RNA editing. Photosynth Res. 2001;70:107–118. doi: 10.1023/A:1013892009589. [DOI] [PubMed] [Google Scholar]
- Waller RF, Reed MB, Cowman AF, McFadden GI. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 2000;19:1794–1802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller RF, McFadden GI. The apicoplast: a review of the derived plastid of apicomplexan parasites. Curr Issues Mol Biol. 2005;7:57–79. [PubMed] [Google Scholar]
- Wastl J, Maier UG. Transport of proteins into cryptomonads complex plastids. J Biol Chem. 2000;275:23194–23198. doi: 10.1074/jbc.M003125200. [DOI] [PubMed] [Google Scholar]
- Whatley JM, John P, Whatley FR. From extracellular to intracellular: the establishment of mitochondria and chloroplasts. Proc Roy Soc Lond B Biol Sci. 1979;204:165–187. doi: 10.1098/rspb.1979.0020. [DOI] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Bhattacharya D. A single origin of the peridinin-and fucoxanthin-containing plastids in dinoflagellates through tertiary endosymbiosis. Proc Natl Acad Sci USA. 2002;99:11724–11729. doi: 10.1073/pnas.172234799. [DOI] [PMC free article] [PubMed] [Google Scholar]