Summary
Clostridium difficile is a leading cause of nosocomial infections. The major virulence factors of this pathogen are the multidomain toxins TcdA and TcdB. These toxins contain a cysteine protease domain (CPD) that autoproteolytically releases a cytotoxic effector domain upon binding intracellular inositol hexakisphosphate. Currently there are no known inhibitors of this protease. Here we describe the rational design of covalent small molecule inhibitors of TcdB CPD. We identified compounds that inactivate TcdB holotoxin function in cells and solved the structure of inhibitor-bound protease to 2.0Å. This structure reveals the molecular basis of CPD substrate recognition and informed the synthesis of activity-based probes for this enzyme. The inhibitors presented here will guide the development of therapeutics targeting C. difficile, and the probes will serve as tools for studying the unique activation mechanism of bacterial toxin CPDs.
Keywords: Protease, activity-based probes, Clostridium difficile, toxin, TcdB, inhibitor
Introduction
The Gram-positive anaerobic bacterium Clostridium difficile is a major cause of hospital-acquired diarrhea and the severe gastrointestinal illness pseudomembraneous colitis (Kelly and LaMont, 2008; Rupnik, et al., 2009). Although infection rates have risen dramatically in the last decade, there is currently a lack of therapeutics to treat C. difficile infection (Halsey, 2008; Kelly and LaMont, 2008). This is in large part due to the organism’s resistance to most classes of antibiotics. A viable strategy for combating C. difficile and other prominent bacterial pathogens is to target virulence factors instead of essential enzymes (Clatworthy, et al., 2007; Puri and Bogyo, 2009). This method limits the selective pressure on the organism to develop resistance to treatment, extending the effective lifespan of the drug. The large glucosylating toxins TcdA and TcdB are ideal targets for this approach because they are the primary virulence factors of C. difficile (Genth, et al., 2008; Jank and Aktories, 2008). TcdB in particular has been shown to be critical for virulence and is found in all clinical isolates (Lyras, et al., 2009; Rupnik, et al., 2009).
Both TcdA and TcdB cause cell death through an orchestrated sequence of events (Jank and Aktories, 2008). These multi-domain toxin proteins first enter cells by triggering receptor-mediated endocytosis (Frisch, et al., 2003; Rolfe and Song, 1993); acidification of toxin-containing endosomal compartments subsequently initiates translocation of the N-terminal cytotoxic glucosyltransferase domain and presumably the cysteine protease domain (CPD) into the cytosol (Just, et al., 1995; Pfeifer, et al., 2003; Qa’Dan, et al., 2000). The CPD is activated by the eukaryotic-specific small molecule inositol hexakisphosphate (InsP6) (Egerer, et al., 2007; Reineke, et al., 2007). This activation catalyzes the autoproteolytic release of the toxin’s cytotoxic glucosyltransferase domain from the endosomal membrane (Egerer, et al., 2007; Pfeifer, et al., 2003). The liberated effector domain then monoglucosylates small Rho family GTPases (Just, et al., 1995), resulting in loss of cell-cell junctions and ultimately cell death (Genth, et al., 2008; Gerhard, et al., 2008; Qa’Dan, et al., 2002).
CPD-mediated autoprocessing of TcdB is a critical step during target cell intoxication. Genetic inactivation of the CPD has been shown to reduce the overall function of TcdB in target cells (Egerer, et al., 2007). A homologous CPD also autoproteolytically regulates the Multifunctional Autoprocessing RTX (MARTX) toxins (Prochazkova, et al., 2009; Sheahan, et al., 2007; Shen, et al., 2009), an otherwise unrelated family of toxins produced by Gram-negative bacteria (Satchell, 2007). Structural analyses of the CPD of both families of toxins have demonstrated that the protease is allosterically regulated by the small molecule InsP6 (Lupardus, et al., 2008; Prochazkova, et al., 2009; Pruitt, et al., 2009). These analyses have also revealed that the CPD is a clan CD protease whose closest known structural homolog is human caspase-7 (Lupardus, et al., 2008). Despite their disparate mechanism of activation, V. cholerae MARTX CPD exhibits similarities in substrate recognition to the caspases (Shen, et al., 2009), except that the CPD cleaves exclusively after a leucine instead of an aspartate residue. In contrast, the molecular details of TcdB CPD substrate recognition remain uncharacterized.
In this study we used a combination of chemical synthesis and structural analyses to probe the substrate recognition and inhibitor sensitivity of the TcdB cysteine protease domain. By screening a focused library of substrate-based CPD inhibitors, we identified several compounds capable of blocking holotoxin function in cell culture. We also solved the structure of TcdB CPD bound to one of these inhibitors. Combined with the structure-activity relationship series derived from our inhibitor analyses, these results provide a foundation for the development of therapeutics targeting this important virulence factor. We further used this information to develop activity-based probes (ABPs) specific for TcdB CPD that will permit the molecular dissection of its unique allosteric activation mechanism. The information presented here may also be valuable for the study of protease domains in other bacterial toxins.
Results
Inhibitor Design and Screening
The use of peptide-based inhibitors is an effective strategy for selectively inactivating proteases through mimicry of natural substrates (Berger, et al., 2006; Kato, et al., 2005; Powers, et al., 2002). Given the importance of the CPD in regulating C. difficile glucosylating toxin function (Egerer, et al., 2007; Reineke, et al., 2007), we sought to identify inhibitors of the TcdB CPD protease. We first tested whether inhibitors specific for a related CPD found in V. cholerae MARTX (MARTXVc) toxin (Shen, et al., 2009) could also inhibit TcdB CPD function (Figure 1). These inhibitors contain tripeptide sequences coupled to either an aza-epoxide or acyloxymethyl ketone (AOMK) reactive electrophile (Figure 1A and S1). Inhibitor potency against TcdB CPD was determined using a gel-based autocleavage assay, in which inhibitor concentration was varied in the presence of the activator inositol hexakisphosphate (InsP6). The autocleavage substrate TcdB(1-804) used in this assay consists of TcdB’s N-terminal 804 amino acids and contains the gIucosyltransferase and CPD domains and the natural autoprocessing site. The aza-epoxide MARTXVc CPD inhibitors were only weakly inhibitory, with both Cbz-LLaL-EP and the related Cbz-EAaL-EP exhibiting observed IC50’s greater than 100 μM (Figure 1B). In contrast, the Cbz-EAL-AOMK inhibitor was significantly more potent, exhibiting an observed IC50 of 7.2 ± 0.7 μM. Because the primary difference between the Cbz-EAaL-EP and Cbz-EAL-AOMK inhibitors is the electrophilic reactive group, we reasoned that the AOMK electrophile is more optimal for TcdB CPD inhibition. Therefore, we synthesized a focused library of covalent AOMK inhibitors based on the natural substrate cleavage sequence of the C. difficile TcdB cysteine protease domain (CPD) (Figure 2A). These inhibitors consist of dipeptide or tripeptide sequences coupled to the dimethylbenzoic acid AOMK, an electrophilic group that has been described as optimal for targeting the structurally related clan CD proteases, the caspases (Kato, et al., 2005; Thornberry, et al., 1994). For all inhibitors, the P1 position (the residue N-terminal to the scissile bond) was held constant as leucine, since this is the primary substrate specificity determinant of bacterial CPDs (Egerer, et al., 2007; Prochazkova, et al., 2009; Shen, et al., 2009). The P2 and P3 positions, as well as the N-terminal capping group, were varied in the library.
Figure 1. Chemical inhibition of TcdB CPD.
(A) MARTXVc CPD inhibitors Cbz-EAL-AOMK and Cbz-EAaL-EP, which contain the AOMK and aza-Leu epoxide electrophilic warheads, respectively. Data represents the mean of three experiments ± standard deviation
(B) Gel-based TcdB(1–804) autocleavage assay. Inhibitor concentrations were titrated and the resulting blockade of recombinant toxin autocleavage was assessed by SDS-PAGE (left). The relative cleavage amounts were then quantified and globally fit to determine observed IC50 values for each compound (right).
See also Figure S1.
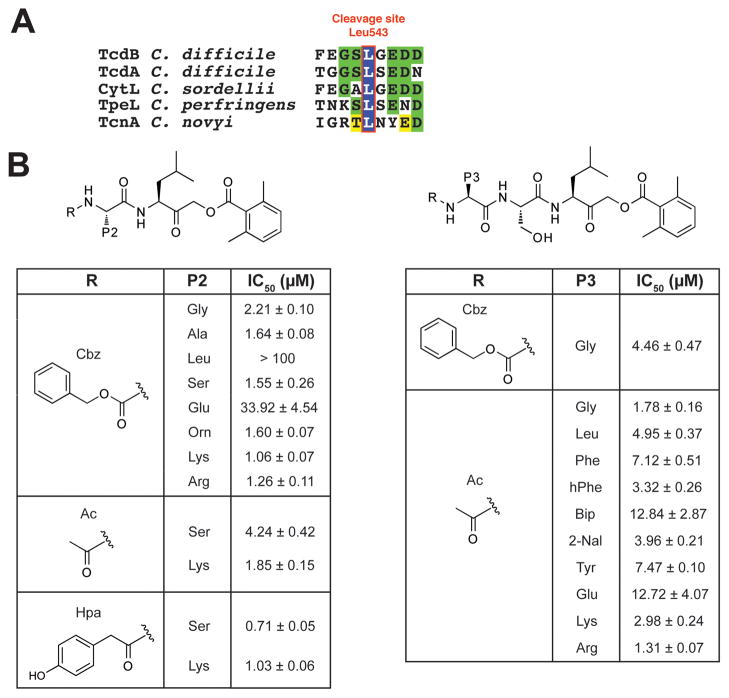
Figure 2. TcdB CPD rational inhibitor design and screening.
(A) Conserved substrate autocleavage site of C. difficile TcdB and related bacterial toxins. The toxin CPD cleaves after the highlighted leucine residue.
(B) Focused screen of capped di- and tripeptide covalent TcdB CPD inhibitors. Observed IC50 values were determined using the autocleavage assay for covalent AOMK inhibitors with diverse P2 (left) and P3 (right) residues. These compounds were N-terminally capped with carboxybenzyl (Cbz), acetyl (Ac), or hydroxyphenyl acetyl (Hpa) groups. The dipeptide inhibitor Hpa-SL-AOMK was found to be the most potent compound. Data represents the mean of three experiments ± standard deviation.
See also Figure S1.
TcdB CPD Inhibitor Structure-Activity Relationship (SAR) Profile
We first probed TcdB CPD P2 specificity using a diverse set of carboxybenzyl (Cbz) capped dipeptide compounds (Figure 2B). Acidic and branched aliphatic amino acids were poorly tolerated in the P2 position, with calculated IC50’s for Cbz-EL-AOMK and Cbz-LL-AOMK of 30 and 100 μM, respectively. Inhibitors containing smaller residues such as alanine and serine were more potent, with IC50’s for Cbz-AL-AOMK and Cbz-SL-AOMK of 1.64 ± 0.08 μM and 1.55 ± 0.26 μM, respectively. Unexpectedly, compounds with basic residues in the P2 position were also potent, with an IC50 of 1.06 ± 0.07 μM observed for Cbz-KL-AOMK. We examined whether the enhanced potency of Cbz-KL-AOMK was specific to this residue or whether other amino acids with a positive charge could recapitulate this effect. Consistent with the latter interpretation, compounds with arginine and ornithine in the P2 position were also potent inhibitors (Figure 2B).
We next assessed the contribution of the carboxybenzyl (Cbz) cap to inhibitor recognition by synthesizing acetyl (Ac) and hydroxyphenyl acetyl (Hpa) capped analogs of Cbz-KL-AOMK and Cbz-SL-AOMK. While the smaller Ac group decreased potency compared to the Cbz cap for both compounds, the Hpa cap increased potency, resulting in an observed IC50 of 0.71 ± 0.05 μM for Hpa-SL-AOMK (Figure 2B). These results indicate that hydrophobic bulk in the dipeptide cap/P3 binding position contributes to TcdB CPD inhibitor potency. Notably, the most potent compound (Hpa-SL-AOMK) contains amino acids found in the natural TcdB CPD substrate (Figure 2A).
Based on these results, we surveyed the P3 specificity of the protease domain using the same technique. We synthesized a focused library of acetyl-capped tripeptide AOMK inhibitors containing the natural leucine and serine residues fixed in the P1 and P2 positions, respectively. The P3 position was varied using a diverse set of amino acids (Figure 2B). Additionally, given the favorable contribution of aromatic bulk in the dipeptide cap site, three non-natural amino acids with an aromatic side chain were included in the P3 position (Figure 2, Figure S1B). Ac-capped tripeptide inhibitors containing small or basic residues in the P3 position were the most potent, while an acidic residue in the P3 position was poorly tolerated (Ac-ESL-AOMK, Figure 2B). More diversity was tolerated in the P3 position of inhibitors containing the natural SL cleavage sequence in the P2/P1 positions. Bulky residues such as leucine were tolerated, with an IC50 of 4.95 ± 0.37 μM observed for Ac-LSL-AOMK. Homophenylalanine (hPhe) was the most potent of the inhibitors containing aromatic P3 residues, with an observed IC50 of 3.32 ± 0.26 μM. This is possibly because its methylene extension at the β-carbon position affords more flexibility in assuming productive interactions with the protease domain (Figure S1B). Conversely, aromatic bulk in the P4 position decreased inhibitor potency, since the Cbz-capped GSL-AOMK inhibitor was ~3-fold less potent than the analogous inhibitor carrying the smaller Ac cap (4.46 ± 0.47 μM vs. 1.78 ± 0.16 μM). None of the compounds in the P3 library, however, were more potent than the most potent dipeptide inhibitor, Hpa-SL-AOMK.
Crystal Structure of Inhibitor-bound TcdB CPD
In order to rationalize the results of the SAR analyses and to gain structural insight into substrate recognition by TcdB CPD, we co-crystallized and solved the structure of InsP6-bound CPD covalently inhibited with the Ac-GSL-AOMK inhibitor at 2.0 Å (Table 1). Although this inhibitor was not the most potent compound identified in the screen, it reflects the natural substrate cleavage site and exhibited improved solubility over dipeptide compounds carrying aromatic caps. The overall structure of inhibitor-bound, activated TcdB CPD (Figure 3A and B) is similar to the previously solved InsP6-bound structure of TcdA CPD, which shares ~60% identity with TcdB CPD (rsmd of ~1Å) (Pruitt, et al., 2009).
Table 1.
Data collection and refinement statistics for the TcdB CPD in complex with InsP6 and Ac-GSL-AOMK
| TcdB CPD + Ac-GSL-AOMK | |
|---|---|
| Data collection | |
| Space group | C 2 |
| Cell dimensions | |
| α, β, χ(Å) | 128.3, 45.5, 87.2 |
| α, β, γ(°) | 90, 103.5, 90 |
| Wavelength (Å) | 1.0 |
| Resolution (Å) | 50–2.0 (2.11-2.0) |
| Rmerge | 0.116 (0.534) |
| I/sI | 9.2 (2.9) |
| Completeness (%) | 100.0 (100.0) |
| Redundancy | 6.7 (5.8) |
| Refinement | |
| Resolution (Å) | 50.0-2.0 |
| No. reflections (total/test) | 31654/1692 |
| Rwork/Rfree | 18.8/23.3 |
| No. atoms | |
| Protein | 3886 |
| InsP6 | 72 |
| Calcium | 1 |
| Sodium | 2 |
| Inhibitor | 44 |
| Water | 286 |
| B-factors | |
| Protein | 34.6 |
| InsP6 | 29.1 |
| Calcium | 29.0 |
| Sodium | 34.8 |
| Inhibitor | 43.4 |
| Water | 41.0 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.304 |
Highest resolution shell is in parentheses
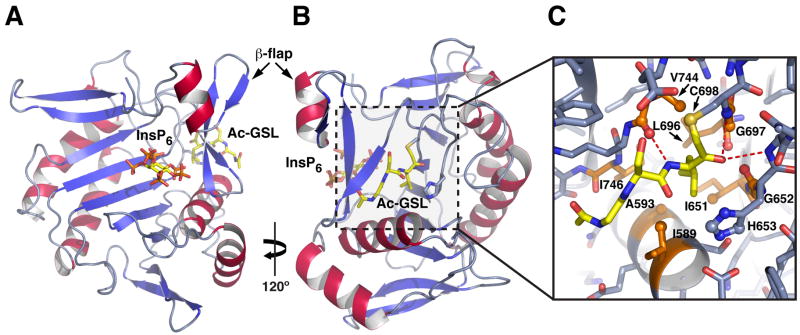
Figure 3. Structure of activated TcdB CPD bound to Ac-GSL-AOMK inhibitor.
(A) Ribbon structure of TcdB CPD in complex with InsP6 viewed from above the InsP6 binding site. InsP6 is shown as a stick model.
(B) A view of the structure rotated ~120º to show inhibitor bound in the active site. InsP6 and the inhibitor are shown as stick models. The β-flap hairpin that separates the InsP6 binding and active site is indicated.
(C) Close-up view of the substrate binding pocket. Hydrophobic residues in the S1 binding pocket are shown as orange sticks, and the inhibitor is shown as yellow sticks. Sidechains that interact with the P1 leucine are shown; hydrogen bonds are indicated by dotted lines.
See also Figure S2.
As with most proteases, the substrate binding pocket of TcdB CPD can be subdivided into multiple subsites. The catalytic residues are positioned between the S1 and S1’ subsites; the numbering of the subsites reflects the corresponding substrate residue recognized, with the prime subsites interacting with substrate residues C-terminal to the scissile bond. The most striking feature of the inhibitor structure is the insertion of the P1 leucine within a deep hydrophobic S1 pocket (Figure 3C). Eight residues, seven of which are nonpolar, are within Van der Waals (4.4 Å) bonding distance of the P1 leucine sidechain. Ile589 and Ala593 are contributed by helix 1 (Figure S2); Ile651 and Gly652 are contributed by strand D; Leu696 and Gly697 are contributed by strand E; and Val744 is contributed by strand G1 of the β-flap, a functional region that is involved in both InsP6 recognition and substrate binding (Lupardus, et al., 2008; Prochazkova, et al., 2009; Pruitt, et al., 2009). Ile746 from the G1 strand of the β-flap forms the distal side of the P1 pocket away from the active site, yet falls just outside van der Waals bonding distance in our structure. It likely also contributes to P1 recognition in vivo, since the inhibitor may be pulled in slightly towards the catalytic cysteine due to the covalent nature of the modification. Since most of these residues are also conserved in the related MARTXVc CPD, which binds to and cleaves after a P1 leucine (Prochazkova, et al., 2009; Shen, et al., 2009), bacterial CPDs would appear to share a common mechanism for recognition of this primary substrate specificity determinant.
Whereas many residues in the S1 subsite participate in recognition of the P1 Leu, residues in the other subsites make minimal interactions with the inhibitor (Figure 3C). In the S2 subsite, the main chain carbonyl of Val744 forms a hydrogen bond with the P2 serine backbone amide of the inhibitor, while on the prime side the main chains of Gly654 and Cys698, which form the “oxyanion hole,” hydrogen bond with the carbonyl formed after inhibitor reaction (Figure 3C). The P2 serine sidechain points towards Glu743; in contrast, the P3 glycine is oriented such that the P3 sidechain would be exposed to solvent. Thus, the inhibitor structure of TcdB CPD reveals the mechanistic basis for substrate recognition.
Inhibitor Docking Findings
To gain additional insight into the inhibitor specificity requirements of the active site, we used docking simulations to replace the Ac-GSL-AOMK in the TcdB CPD crystal structure with other compounds from the focused library. The structure of the modeled inhibitor and all sidechains within 4.5 Å were energy minimized using the default parameter of the Molecular Operating Environment (MOE) software. The docking simulations suggest an explanation for why compounds containing basic amino acids such as lysine or arginine in the P2 position were particularly potent. In these analyses, the positively charged P2 residue forms an electrostatic interaction with the acidic Glu743 residue, which helps to form the S2 pocket (Figure 4). This negatively charged subsite likely also explains why the Cbz-EL-AOMK was not as potent, as this inhibitor cannot form the same favorable electrostatic interactions during binding (Figure 4). Instead, the acidic glutamate of the inhibitor is predicted to interact with Arg745; this interaction, however, may be unfavorable given that Arg745 likely stabilizes the activated conformation of the CPD through a π-cation interaction with the highly conserved Trp761 (Pruitt, et al., 2009).
Figure 4. Molecular docking analyses of TcdB CPD with AOMK inhibitors.
Close-up view of the substrate binding pocket shown with electrostatic surface potential. Blue denotes positively charged surface; red denotes negatively charged surface. Inhibitors are shown as yellow sticks, and relevant residues are indicated. The Ac-GSL-AOMK image is derived from the crystal structure of the inhibitor-bound enzyme. Binding of the Cbz-EL-AOMK is predicted to bind TcdB CPD differently from the other inhibitors.
The docking studies also rationalize the increased potency of the dipeptide inhibitors capped with bulky Cbz or Hpa groups. These caps are predicted to fit into a hydrophobic groove formed between Ile746 and Ile589 such that the hydroxyl of the Hpa cap serves as the donor in a hydrogen bond with Ile746 (Figure 4). This may help favorably orient the inhibitors during binding and subsequent reaction with the catalytic Cys698.
TcdB CPD Inhibitor Blocks Full-length Toxin Function
In order to determine if the inhibitors identified in vitro were functional in cells, we assessed their ability to block the intoxication of primary human foreskin fibroblasts (HFFs) by TcdB holotoxin. Pre-treating cells with the most potent inhibitors prevented the cytopathic effects induced by recombinant TcdB holotoxin (Figure 5A). Cell rounding was quantified for select compounds by counting the number of rounded cells visible per field upon inhibitor titration (Figure 5B). The results corroborate the potency rankings observed in the initial autocleavage screen. Two of the most potent compounds, Hpa-SL-AOMK and Hpa-KL-AOMK, both prevented cell rounding. Hpa-SL-AOMK completely inhibited toxin function by 100 μM, with an observed IC50 of approximately 20 μM, while the Hpa-KL-AOMK was slightly less potent. The difference in potency between Hpa-SL-AOMK and Hpa-KL-AOMK may be due to the increased cell permeability of the P2 serine relative to the positively charged lysine. Cell permeability of the inhibitors is likely to determine their potency because CPD inhibition can only occur after toxin entry into cells, when the CPD can become activated by InsP6. In contrast, the negative controls Cbz-EL-AOMK and the epoxide-based pan-cathepsin inhibitor JPM-OEt (Bogyo, et al., 2000; Greenbaum, et al., 2000) both failed to inhibit toxin function just as they poorly inhibit CPD activity (Figure 2B and S1). Surprisingly, Cbz-KL-AOMK and to a lesser extent Cbz-SL-AOMK were found to be cytotoxic (Figure S3). Given that no toxicity was observed for the Hpa-capped analogs, these findings implicate the carboxybenzyl cap in affecting cell viability.
Figure 5. TcdB CPD inhibitors block holotoxin function.
(A) Primary human foreskin fibroblasts (HFFs) pretreated with Hpa-SL-AOMK are protected from holotoxin-mediated cell rounding. HFFs treated with the catalytically inactivated C698A holotoxin or left untreated exhibit minimal cell rounding.
(B) Quantification of inhibitor effects on holotoxin-mediated HFF cell rounding. Pre-treatment with Hpa-SL-AOMK or Hpa-KL-AOMK was protective, while Cbz-EL-AOMK and the pan-cathepsin inhibitor JPM-OEt had no effect on toxin function. Data represents the mean of three experiments ± standard deviation.
(C) Addition of wildtype TcdB holotoxin to HFFs results in complete glucosylation of the Rho GTPase Rac1, as seen by Western blot with the glucosylation-sensitive α-Rac1 monoclonal antibody mAb102. Pre-treating the cells with Hpa-SL-AOMK or Hpa-KL-AOMK protected HFFs from toxin effector domain activity, while inhibiting cathepsin activity with JPM-OEt did not.
See also Figure S3.
We confirmed that inhibition of the TcdB CPD directly prevented holotoxin effector domain activity by monitoring glucosylation of its cellular target Rac1, a Rho GTPase (Genth, et al., 2006; Yang, et al., 2008). Addition of wildtype toxin to cells resulted in complete glucosylation of Rac1, while pretreatment with either Hpa-SL-AOMK or Hpa-KL-AOMK was protective (Figure 5C). Importantly, inhibition of cathepsins using JPM-OEt had no effect on toxin function; JPM-OEt was used as a control since P1 Leu AOMKs have previously been shown to weakly cross-react with the cathepsins (Kato, et al., 2005).
Probe Design to Monitor Toxin Activation
The activation of the CPD by the eukaryotic-specific small molecule InsP6 is a critical step in regulating the function and trafficking of C. difficile glucosylating toxins (Egerer and Satchell, 2010; Shen, 2010). In order to facilitate more detailed studies of this important process, we created fluorescently-tagged and biotin-tagged probe versions of the SL-AOMK inhibitor (AWP19 and AWP15, respectively) to visualize toxin activation (Figure 6A). Both AWP19 and AWP15 covalently label the recombinantly produced TcdB(1-804) autocleavage substrate in response to small molecule activation of the protease by InsP6 (Figure 6B). This substrate represents the toxin region predicted to translocate into the cytosol during intoxication. Notably, the cleavage product TcdB(544-804) formed after proteolytic cleavage was more active than the full-length substrate, as shown by the increase in activity-based probe (ABP) labeling relative to the amount of protein present (Figure 6B and C). This observation suggests that the isolated CPD domain TcdB(544-804) is either more accessible or more reactive with the probe than the full-length substrate. As expected, the ABPs failed to label the catalytically inactive C698A mutant of TcdB(1-804) in the presence or absence of InsP6.
Figure 6. Probe labeling of recombinant TcdB.
(A) Structures of Cy5-labeled AWP19 and biotin-labeled AWP15 TcdB CPD probes.
(B) Labeling of either wildtype or catalytic mutant C698A TcdB(1-804) (0.5 μM) by probes (10 μM) in the presence (25 μM, +) or absence (−) of InsP6. Fluorescence scanning was used to detect AWP19 labeling, while streptavidin-HRP blotting was used to detect AWP15 labeling; total protein was detected by Coomassie staining. A small fraction of cleaved TcdB544–804 is detectably labeled by both probes even though it is not detectable by Coomassie staining.
(C) Labeling of holotoxin produced in E. coli (wildtype and catalytic mutant C698A) and Bacillus megaterium by probes (10 μM) in the presence (25 μM, +) or absence (−) of InsP6. The amount of protein loaded is indicated. “Pre” refers to B. megaterium-produced holotoxin was pre-treated with 25 μM InsP6 to induce autoprocessing for 1 hr at 37 °C after which 10 μM of probe was added to the sample. Fluorescence scanning was used to detect AWP19 labeling, while streptavidin-HRP blotting was used to detect AWP15 labeling. Total protein was visualized using Coomassie staining, while His6-tagged holotoxin was visualized using anti-His antibody Western blotting.
See also Figures S4 and S5.
We also determined that the ABPs could label functional recombinant holotoxin, which was either produced in E. coli or in the Bacillus megaterium expression system (Pruitt, et al., 2010; Yang, et al., 2008) (Figure 6C). The B. megaterium system produces a His6-tagged TcdB holotoxin that is more pure than the native toxin purified from C. difficile culture supernatants (Genth, et al., 2006; Yang, et al., 2008). Both probes could sensitively detect active CPD within the toxin, since the cleaved TcdB(544-2366)-His6 fragment could be labeled by the probe even though it is not detectable by either Coomassie or anti-His6 antibody-conjugated horseradish peroxidase. Titration of probe labeling confirmed the potency of the probes, with AWP19 labeling active holotoxin at probe concentrations below 50 nM (Figure S4). This level of sensitivity will be valuable for tracking CPD activation during toxin trafficking, as it permits the labeling of ensembles of toxin while minimizing the risk of completely inhibiting toxin function.
Monitoring toxin trafficking is further enabled by the cell permeability of AWP19’s near-infrared fluorescent cyanine 5 fluorescent tag, which allows the probe to be used in intact cells. In order to check for off-targets of this probe, we incubated both primary HFFs and the RAW macrophage cell line with AWP19. In both cell types, the only off-target the probe labeled was cathepsin B, as confirmed by comparison with a pan-cathepsin ABP (Figure S5) (Paulick, 2010). Similarly, pre-treatment of both cell types with Cbz-SL-AOMK prevented cathepsin B labeling by the pan-cathepsin probe, but not cathepsin L or X labeling. Thus, cathepsin B would appear to be the primary off-target TcdB CPD inhibitors. Nevertheless, cathepsin B inhibition was not sufficient to reduce TcdB toxin function, since the pan-cathepsin inhibitor JPM had no effect on toxin-induced cell rounding or Rac1 glucosylation (Figure 5). This result strongly suggests that the observed reduction in TcdB glucosylating activity in target cells upon treatment with CPD inhibitors is due to inhibition of CPD function.
Discussion
The rising rate of C. difficile infections necessitates the development of new classes of therapeutics to combat this pathogen. Because of its natural antibiotic resistance, there has been increased focus on targeting the glucosylating toxins TcdA and TcdB for direct therapeutic intervention, as they are the primary mediators of C. difficile pathogenesis (Halsey, 2008; Kelly and LaMont, 2008; Rupnik, et al., 2009). In this study we present the first validation that the TcdB cysteine protease domain (CPD) is a druggable target. Although inhibition of this protease active site is difficult because the small molecule is competing with an intramolecular autoproteolytic event, our findings are encouraging for the development of competitive inhibitors for the TcdB CPD. The most potent compound in our library is the 499 Da capped dipeptide inhibitor Hpa-SL-AOMK (Figure 2), which is within the size constraints generally accepted for therapeutics (Lipinski, 2000). In addition, the minimal interaction between the protease and inhibitor peptide backbone (Figure 4) suggests that inhibitors with non-peptidic scaffolds can be developed to bypass the pharmacokinetic shortfalls of peptidic compounds.
Our rational approach to probing the inhibitor sensitivity of the CPD active site using structural analysis and a focused library of substrate-based compounds yielded multiple inhibitors capable of blocking holotoxin function (Figure 5). These analyses produced the unexpected observation that inhibitors containing basic P2 or P3 residues and bulky hydrophobic N-terminal caps could potently block TcdB autoprocessing. This SAR profile provides a starting point for the development of compounds suitable for therapeutic applications. We note that relying solely on substrate specificity profiling for these domains may not have produced such promising results, since potent substrates do not always translate into viable inhibitors (Drag, et al., 2010). Furthermore, fluorogenic substrate cleavage assays lack sensitivity in detecting bacterial CPD activity because these autoprocessing enzymes exhibit poor transcleavage efficiency (Babe and Craik, 1997; Lupardus, et al., 2008). Our attempts to develop optimized substrates for TcdB CPD produced substrates with poor Km values (~1 mM, data not shown). The approach described here may also prove applicable to other protease domain-containing bacterial toxins (Lebrun, et al., 2009).
Information about the inhibitor sensitivity profile of TcdB CPD was bolstered by our crystal structure of the enzyme bound to the Ac-GSL-AOMK inhibitor (Figure 3). This structure permitted the molecular docking studies that helped rationalize the increased potency of inhibitors with basic P2 residues and bulky hydrophobic N-terminal caps (Figure 4). Furthermore, given the overall similarity between the inhibitor structure of InsP6-bound TcdB CPD presented here and InsP6-bound TcdA CPD (Pruitt, et al., 2009), many of our findings may be translatable to inhibiting this closely related toxin. Our most efficacious inhibitor Hpa-SL-AOMK will likely exhibit similar potency against TcdA due to its identical P1/P2 substrate sequence (Figure 2A), increasing its value as a C. difficile virulence-targeting agent.
This crystal structure also permits comparisons to be made with the inhibitor-bound structure of the related CPD of Vibrio cholerae MARTX toxin (Shen, et al., 2009). Both inhibitor structures reveal that the primary substrate specificity determinant for these proteases is the P1 leucine, and residues involved in recognizing this leucine are conserved in both proteases. The inhibitor structures differ in that MARTXVc CPD makes more backbone interactions with its inhibitor than TcdB CPD and in the S1′ subsite, the region that recognizes residues C-terminal to the scissile bond. For MARTXVc CPD, this region is relatively flat and featureless, whereas in TcdB CPD, Asp656 and Glu657 directly extend into this region and may thus occlude substrate or inhibitor binding (Figure S6). This acidic extension may explain why such a large difference in potency was observed between the Cbz-EAL aza-epoxide and AOMK derivatives for TcdB CPD (Figure 1), while both warheads inhibit MARTXVc CPD to a similar extent (Shen, et al., 2009).
The CPDs of both the C. difficile large gluosylating and MARTX toxin families appears to “sense” the eukaryotic cell environment and activate toxin function accordingly (Egerer and Satchell, 2010; Shen, 2010). However, this unique allosteric activation process is difficult to study using traditional biochemical approaches because it is post-translationally regulated. Our activity-based probes (ABPs) overcome this problem because they afford the direct visualization of CPD activation by InsP6 in complex mixtures and possibly in vivo. Furthermore, the probes provide a robust read-out for CPD activity that could be used in screening applications for competitive (Schneider and Craik, 2009) or allosteric inhibitors (Lee and Craik, 2009) of the CPD, in lieu of fluorogenic substrates (which react poorly with the protease) or autocleavage assays (which are less sensitive).
Because of the covalent nature of these probes, they provide a direct readout of when the toxin has encountered InsP6. This is valuable within the context of studies directed at dissecting toxin trafficking and the molecular mechanisms underlying the allosteric regulation of toxin function by InsP6 (Giesemann, et al., 2008; Jank and Aktories, 2008). For example, it is notable that the cleaved form of TcdB CPD is more effectively labeled by the ABP than the full-length protein (Figure 6). This may imply that the CPD is held in an inhibitory conformation within the native holotoxin and that autoproteolytic cleavage relieves this inhibition. Alternatively, the active site may be more accessible to the ABP following cleavage, which would suggest that the conformation of full-length TcdB in vitro occludes substrate binding. C. difficile glucosylating toxins undergo significant conformational rearrangements during the pH-dependent toxin translocation process (Pruitt, et al., 2010; Qa’Dan, et al., 2000). Based on our observation that cleaved TcdB toxin (aa 544-2366) was also more readily labeled by the probe in the presence of InsP6 (Figure 6C), it is tempting to speculate that the CPD is subject to additional regulation at the level of toxin conformation. Further studies using these promising tools will provide a more detailed understanding of the regulation of the CPD by InsP6 and in the context of the full-length toxin.
Significance
The large glucosylating toxins TcdA and TcdB are the primary virulence factors of the antibiotic-resistant bacterium Clostridium difficile. These toxins are autoproteolytically activated by an internal cysteine protease domain (CPD) in a step that is critical for toxin function. We synthesized a focused library of substrate-based compounds in order to determine a structure-activity relationship for TcdB CPD inhibitors and then gained further insight by co-crystallizing the domain with one of these inhibitors. This rational approach yielded compounds potent enough to inhibit toxin function in cell culture, validating the clostridial glucosylating toxins as druggable targets. Our results provide a promising starting point for the development of therapeutics that minimize the selective pressure on C. difficile to develop resistance. We also used the inhibitor data to develop covalent activity-based probes (ABPs) that can directly measure the allosteric activation of the protease by the small molecule inositol hexakisphosphate. Because these ABPs monitor the post-translational activation of the toxin, they will be useful in studies directed at understanding this unique regulatory mechanism in both biochemical and cell-based assays.
Experimental Procedures
Compound Synthesis
The aza-leu epoxide inhibitors were synthesized in solution using standard chemistry as previously described (Asgian, et al., 2002). The AOMK inhibitors were synthesized using solid-phase synthesis as previously described (Kato, et al., 2005). The activity-based probe AWP19 was synthesized by combining H2N-aminohexanoic-SL-AOMK (1 equiv.) with Cy5-NHS (0.9 equiv.) and DIEA (5 equiv.) in DMSO for one hour and then purifying directly by HPLC. The identity and purity of all compounds was characterized using HR-MS and LCMS.
Protein Expression and Purification
An overnight culture of pET28a-TcdB1–804 was diluted 1:500 into 4 L 2YT media and grown shaking at 37ºC. When an OD600 of 0.6–0.9 was reached, IPTG was added to 250 μM, and cultures were grown for 3 hr at 225 rpm at 30ºC. Cultures were pelleted, resuspended in 60 mL lysis buffer [500 mM NaCl, 50 mM Tris-HCl, pH 7.5, 15 mM imidazole, 10% glycerol] and flash frozen in liquid nitrogen. Lysates were thawed, then lysed by sonication and cleared by centrifugation at 15,000 × g for 30 minutes. C-terminally His6-tagged TcdB(1-804) was affinity purified by incubating the lysates in batch with 2.0 mL Ni-NTA Agarose beads (Qiagen) with shaking for 3 hr at 4ºC. The binding reaction was pelleted at 1,500 × g, and the pelleted Ni-NTA agarose beads were washed 3 x with lysis buffer. His6-tagged CPD was eluted from the beads by the addition of 400 μL high imidazole elution buffer [500 mM NaCl, 50 mM Tris-HCl, pH 7.5, 175 mM imidazole, 10% glycerol]. The elution was repeated three times; the eluate was pooled, buffer exchanged in gel filtration buffer [200 mM NaCl, 10 mM Tris pH 7.5, 5% glycerol), and concentrated to 750 μL. The concentrated prep was pelleted at 13,000 × g for 10 min at 4 °C prior to loading on a HiPrep S200 16/60 Sephacryl column (GE Healthcare). Purified His6-tagged CPD was concentrated and stored at −20ºC in gel filtration buffer.
TcdB Autocleavage Assay
Recombinant TcdB(1-804) was diluted to a final concentration of 0.5 μM in assay buffer [60 mM NaCl, 20 mM Tris pH 7.5, 250 mM sucrose] in a 96-well plate. 0.5 μL of a 100X inhibitor stock in DMSO was then added to each well in triplicate, and the samples were incubated at 37 °C for 30 minutes. Inositol hexakisphosphate (0.5 μL, InsP6, Calbiochem) was then added to a final concentration of 25 μM, and the reaction was incubated at 37 °C for 1 hour. Samples were then diluted in 4X SDS-PAGE loading buffer and resolved by SDS-PAGE on 12% gels. Cleavage reactions were visualized by Coomassie staining and quantified using the program ImageJ (http://rsb.info.nih.gov/ij/). For each sample, the amount of autocleaved protein relative to the total protein amount was plotted versus the concentration of inhibitor and globally fit using the sigmoidal function in KaleidaGraph.
Protein purification, crystallization, and data collection
An overnight culture of pET22b-TcdB544–797 was diluted 1:500 into 3 L 2YT media and grown shaking at 37ºC. When an OD600 of 0.6–0.9 was reached, IPTG was added to 250 μM, and cultures were grown for 12–16 hr (225 rpm) at 18–20ºC. Cultures were pelleted, resuspended in 50 mL lysis buffer [500 mM NaCl, 50 mM Tris-HCl, pH 7.5, 15 mM imidazole, 10% glycerol] and flash frozen in liquid nitrogen. Lysates were thawed, then lysed by sonication and cleared by centrifugation at 15,000 × g for 30 minutes. TcdB544–797 was purified as described above except that it was concentrated to 1 mM, and the gel filtration buffer was 150 mM NaCl, 10 mM Tris pH 7.5. Gel filtration purified TcdB(544-797) was treated with 2 mM InsP6 and 2 mM Ac-GSL-AOMK (inhibitor stock was at 200 mM in DMSO). The inhibitor was added slowly due to poor solubility; reaction with the protease improved inhibitor solubility. The reaction was allowed to proceed for 1 hr at room temperature after which excess inhibitor was pelleted by centrifuging the reaction at 13,000 × g for 10 min at 4 °C. Crystallization screening was carried out using the sitting drop vapor-diffusion method and initial hits were observed in 0.1 M Tris HCl pH 8.0 and 30% PEG2000 MME as the precipitant. Crystals used for data collection were grown in 1 μL drops by mixing equal volumes of protein with mother liquor and appeared only after ~45–60 days. Crystals were cryoprotected in 45% PEG2000 MME, flash frozen in liquid nitrogen, and data collected under cryo-cooled conditions at 100 K at beamline 8.2.1 at the Advanced Light Source (UC-Berkeley). Diffraction data were processed using MOSFLM (Leslie) and SCALA (Potterton, et al., 2003), and processing statistics are listed in Table 1.
Structure determination and refinement
Initial phases were obtained by molecular replacement with PHASER (McCoy, et al., 2007) using the structure of the TcdA CPD (PDB ID 3HO6) as a search model (Pruitt, et al., 2009). Using the molecular replacement phases, the TcdB CPD was built by ARP/wARP (Perrakis, et al., 1999) to approximately 85% completeness, followed by rounds of model building and adjustment with COOT (Emsley and Cowtan 2004) and refinement with PHENIX (Adams, et al., 2002). Restraints for the Ac-GSL molecule were obtained from the PRODRG server (Schuttelkopf and van Aalten, 2004). The final model underwent restrained and translation/libration/screw refinement in REFMAC5 (Murshudov, et al., 1997), resulting in a final R/Rfree values of 18.8% and 23.3%. Ramachandran analysis of model geometry by Molprobity (http://molprobity.biochem.duke.edu/) indicates that 99.0% of residues reside in the most favorable regions, with none in the disallowed regions. Refinement statistics can be found in Table 1. Structural figures were prepared with Pymol (DeLano, 2002). The final model contains two TcdB CPD molecules in the asymmetric unit (ASU), with each bound to one InsP6 molecule, one Ac-GSL inhibitor molecule, two sodium ions and a calcium ion bridging the crystal contact between the molecules in the ASU. Chain A of the Tcd CPD is used for all figures in the paper.
Docking Simulations
The different homology models of TcdB CPD were built from the crystal structure of the protease bound of Ac-GSL-AOMK using the default parameters of the Molecular Operating Environment (MOE) software. In each model, the covalent linkage to Cys698 and the position of leucine in the P1 pocket were initially fixed as those of the crystal structure. The N-terminal cap and P2 side chain were manually built into the active site of the protease. Energy minimization was performed first on the modified region of the inhibitor and all side chains within 4.5 Å, and second on the entire inhibitor and all side chains within 4.5 Å.
Cell Rounding Assay
Primary human foreskin fibroblasts (HFFs) were seeded in 96-well plates at a density of 1–2 × 104 cells/well in DMEM supplemented with 10% FBS. Prior to the assay the cells were washed three times with 100 μL DMEM alone. One microliter of a 100X inhibitor stock in DMSO was then added to each well in triplicate, and the cells were incubated at 37 °C for 30 minutes. Recombinant TcdB holotoxin expressed in E. coli was then added to the cells at a final concentration of 0.3 pM. The samples were incubated at 37 °C for 2 hours and then imaged using a 20X objective on an inverted microscope. Four fields were imaged per well, and the average number of rounded cells per field was calculated.
Rac1 Glucosylation Assay
HFF cells were seeded into 24-well treated plates (7.5 × 105) and grown to 100% confluency overnight, washed once with pre-warmed DMEM, and left in 0.25 mL DMEM per well. The indicated concentration of inhibitor was added as a 1:100 dilution from a DMSO stock and incubated for 30 min. Recombinant TcdB holotoxin purified from E. coli was added to the cells (0.3 pM) and incubated for 90 min at 37 °C. The media was removed and then the cells were lysed in 25 μL 1 X FSB by scraping the cells in concentric circles. Cell lysates were boiled for 5 min at 95 °C, and 15 μL of lysate was resolved on a 14% SDS-PAGE gel and transferred to nitrocellulose. Unglucosylated Rac1 was detected using a 1:1000 dilution of mAb102 (Millipore) and a 1:5000 dilution of anti-mouse IgG HRP (Biorad). Actin was simultaneously visualized using a polyclonal anti-actin antibody at 1:2000 dilution (Sigma), and a 1:5000 dilution of anti-rabbit IgG HRP (Biorad).
Purification of TcdB holotoxin from E. coli
Overnight cultures of pET28a-TcdB wildtype or C698A holotoxin were diluted 1:500 into 3 L 2YT media and grown shaking at 37ºC. C-terminally His6-tagged holotoxin was purified as described for His6-tagged TcdB(1-804) with the exception that β-mercaptoethanol was added to the lysis buffer at 2 mM.
AWP19 and AWP15 labeling of recombinant TcdB(1-804)
Wildtype and C698A TcdB 1-804 (0.5 μM) were incubated with 10 μM of the indicated probe. InsP6 was added to a final concentration of 25 μM (1:100 dilution) in a total volume of 50 μL where indicated, and probe labeling proceeded for 1 hr at 37 °C. Fifteen microliters of 4X final sample buffer was added and then the sample was boiled for 3 min at 95 °C. The samples were resolved on a 10% gel by SDS-PAGE. For AWP19 labeling, fluorescent labeling was visualized using a fluorescent scanner followed by Coomassie staining. For AWP15 staining, the sample was loaded in duplicate, and one set was visualized by Coomassie staining (5 μL sample loaded), while the other set was transferred to nitrocellulose (2.5 μL sample loaded) and blotted using streptavidin-HRP (Sigma) at 1:3000.
AWP19 and AWP15 labeling of recombinant TcdB
For labeling of TcdB holotoxin purified from E. coli, wildtype and C698A TcdB was diluted to 0.3 μM in assay buffer and then the indicated probe was added to 10 μM (1:100 dilution from DMSO stock) in a total volume of 15 μL. InsP6 was added at a final concentration of 25 μM where indicated. For labeling of TcdB holotoxin purified from B. megaterium (a gift from R. Pruitt and D.B. Lacy), 0.5 μM of toxin was diluted in CPD buffer and then 10 μM of the indicated probe was added (1:100 dilution from DMSO stock). InsP6 was then added at a final concentration of 25 μM. Alternatively, InsP6 (25 μM) was added to the B. megaterium produced toxin (0.5 μM) and incubated for 1 hr at 37 °C and then the indicated probe was added at 10 μM final concentration. Following probe addition, the labeling reactions were allowed to proceed for 1 hr at 37 °C after which 5 μL of 4 X FSB was added, and the samples were boiled for 3 min at 95 °C. For AWP19 labeled samples, 15 μL was resolved on a 10% gel and then visualized by fluorescence scanning followed by Coomassie staining. For AWP15 labeled samples, the samples (either 5 or 2.5 μL) were loaded in duplicate and resolved on a 10% gel, then transferred to nitrocellulose. The membranes were probed with Streptavidin-HRP (Sigma) at 1:3000 (5 μL sample loaded) or with an anti-His antibody (Pierce) and anti-rabbit IgG HRP (Biorad) at 1:10,000 (2.5 μL sample loaded).
Supplementary Material
Acknowledgments
We thank D. Borden Lacy and Rory Pruitt (Vanderbilt University) for the generous gift of TcdB toxin produced in B. megaterium and Elaine Hamm and Jimmy Ballard (Oklahoma Health Sciences University) for Clostridium difficile VPI 10463 genomic DNA. A.W.P. is supported by an NSF Graduate Research Fellowship. P.J.L. is a Damon Runyon Fellow, supported by the Damon Runyon Cancer Research Foundation. K.C.G. is supported by the Keck Foundation and the Howard Hughes Medical Institute. M.B. is supported by the Burroughs Wellcome Foundation and NIH grants R01EB005011 and R01AI078947. A.S. is supported by an NIH National Institutes of General Medical Sciences 1-K99-GM092934-01. Coordinates and structure factors have been deposited in the Protein Data Bank (www.rcsb.org) under accession number XXXX.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Asgian JL, James KE, Li ZZ, Carter W, Barrett AJ, Mikolajczyk J, Salvesen GS, Powers JC. Aza-peptide epoxides: a new class of inhibitors selective for clan CD cysteine proteases. J Med Chem. 2002;45:4958–4960. doi: 10.1021/jm025581c. [DOI] [PubMed] [Google Scholar]
- Babe LM, Craik CS. Viral proteases: evolution of diverse structural motifs to optimize function. Cell. 1997;91:427–430. doi: 10.1016/s0092-8674(00)80426-2. [DOI] [PubMed] [Google Scholar]
- Berger AB, Sexton KB, Bogyo M. Commonly used caspase inhibitors designed based on substrate specificity profiles lack selectivity. Cell Res. 2006;16:961–963. doi: 10.1038/sj.cr.7310112. [DOI] [PubMed] [Google Scholar]
- Bogyo M, Verhelst S, Bellingard-Dubouchaud V, Toba S, Greenbaum D. Selective targeting of lysosomal cysteine proteases with radiolabeled electrophilic substrate analogs. Chemistry & biology. 2000;7:27–38. doi: 10.1016/s1074-5521(00)00061-2. [DOI] [PubMed] [Google Scholar]
- Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nature chemical biology. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
- Drag M, Bogyo M, Ellman JA, Salvesen GS. Aminopeptidase fingerprints, an integrated approach for identification of good substrates and optimal inhibitors. The Journal of biological chemistry. 2010;285:3310–3318. doi: 10.1074/jbc.M109.060418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egerer M, Giesemann T, Jank T, Satchell KJ, Aktories K. Auto-catalytic cleavage of Clostridium difficile toxins A and B depends on cysteine protease activity. The Journal of biological chemistry. 2007;282:25314–25321. doi: 10.1074/jbc.M703062200. [DOI] [PubMed] [Google Scholar]
- Egerer M, Satchell KJ. Inositol hexakisphosphate-induced autoprocessing of large bacterial protein toxins. PLoS Pathog. 2010;6:e1000942. doi: 10.1371/journal.ppat.1000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch C, Gerhard R, Aktories K, Hofmann F, Just I. The complete receptor-binding domain of Clostridium difficile toxin A is required for endocytosis. Biochem Biophys Res Commun. 2003;300:706–711. doi: 10.1016/s0006-291x(02)02919-4. [DOI] [PubMed] [Google Scholar]
- Genth H, Dreger SC, Huelsenbeck J, Just I. Clostridium difficile toxins: more than mere inhibitors of Rho proteins. Int J Biochem Cell Biol. 2008;40:592–597. doi: 10.1016/j.biocel.2007.12.014. [DOI] [PubMed] [Google Scholar]
- Genth H, Huelsenbeck J, Hartmann B, Hofmann F, Just I, Gerhard R. Cellular stability of Rho-GTPases glucosylated by Clostridium difficile toxin B. FEBS Lett. 2006;580:3565–3569. doi: 10.1016/j.febslet.2006.04.100. [DOI] [PubMed] [Google Scholar]
- Gerhard R, Nottrott S, Schoentaube J, Tatge H, Olling A, Just I. Glucosylation of Rho GTPases by Clostridium difficile toxin A triggers apoptosis in intestinal epithelial cells. J Med Microbiol. 2008;57:765–770. doi: 10.1099/jmm.0.47769-0. [DOI] [PubMed] [Google Scholar]
- Giesemann T, Egerer M, Jank T, Aktories K. Processing of Clostridium difficile toxins. J Med Microbiol. 2008;57:690–696. doi: 10.1099/jmm.0.47742-0. [DOI] [PubMed] [Google Scholar]
- Greenbaum D, Medzihradszky KF, Burlingame A, Bogyo M. Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chemistry & biology. 2000;7:569–581. doi: 10.1016/s1074-5521(00)00014-4. [DOI] [PubMed] [Google Scholar]
- Halsey J. Current and future treatment modalities for Clostridium difficile-associated disease. Am J Health Syst Pharm. 2008;65:705–715. doi: 10.2146/ajhp070077. [DOI] [PubMed] [Google Scholar]
- Jank T, Aktories K. Structure and mode of action of clostridial glucosylating toxins: the ABCD model. Trends Microbiol. 2008;16:222–229. doi: 10.1016/j.tim.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Just I, Selzer J, Wilm M, von Eichel-Streiber C, Mann M, Aktories K. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature. 1995;375:500–503. doi: 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C, Chehade KA, Salvesen GS, Bogyo M. Activity-based probes that target diverse cysteine protease families. Nature chemical biology. 2005;1:33–38. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- Kelly CP, LaMont JT. Clostridium difficile--more difficult than ever. N Engl J Med. 2008;359:1932–1940. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- Lebrun I, Marques-Porto R, Pereira AS, Pereira A, Perpetuo EA. Bacterial toxins: an overview on bacterial proteases and their action as virulence factors. Mini Rev Med Chem. 2009;9:820–828. doi: 10.2174/138955709788452603. [DOI] [PubMed] [Google Scholar]
- Lee GM, Craik CS. Trapping moving targets with small molecules. Science. 2009;324:213–215. doi: 10.1126/science.1169378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AG. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. :26. [Google Scholar]
- Lipinski CA. Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods. 2000;44:235–249. doi: 10.1016/s1056-8719(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Lupardus PJ, Shen A, Bogyo M, Garcia KC. Small Molecule-Induced Allosteric Activation of the Vibrio cholerae RTX Cysteine Protease Domain. Science. 2008;322:265–268. doi: 10.1126/science.1162403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyras D, O’Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, Poon R, Adams V, Vedantam G, Johnson S, et al. Toxin B is essential for virulence of Clostridium difficile. Nature. 2009;458:1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunztleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Paulick MG, Bogyo M. Development of activity-based probes for the characterization and visualization of cathepsin X. 2010 submitted. [Google Scholar]
- Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nature structural biology. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Pfeifer G, Schirmer J, Leemhuis J, Busch C, Meyer DK, Aktories K, Barth H. Cellular uptake of Clostridium difficile toxin B. Translocation of the N-terminal catalytic domain into the cytosol of eukaryotic cells. The Journal of biological chemistry. 2003;278:44535–44541. doi: 10.1074/jbc.M307540200. [DOI] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Powers JC, Asgian JL, Ekici OD, James KE. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- Prochazkova K, Shuvalova LA, Minasov G, Voburka Z, Anderson WF, Satchell KJ. Structural and molecular mechanism for autoprocessing of MARTX toxin of Vibrio cholerae at multiple sites. The Journal of biological chemistry. 2009;284:26557–26568. doi: 10.1074/jbc.M109.025510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt RN, Chagot B, Cover M, Chazin WJ, Spiller B, Lacy DB. Structure-function analysis of inositol hexakisphosphate-induced autoprocessing in Clostridium difficile toxin A. The Journal of biological chemistry. 2009;284:21934–21940. doi: 10.1074/jbc.M109.018929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt RN, Chambers MG, Ng KK, Ohi MD, Lacy DB. Structural organization of the functional domains of Clostridium difficile toxins A and B. Proc Natl Acad Sci U S A. 2010;107:13467–13472. doi: 10.1073/pnas.1002199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri AW, Bogyo M. Using small molecules to dissect mechanisms of microbial pathogenesis. ACS Chem Biol. 2009;4:603–616. doi: 10.1021/cb9001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qa’Dan M, Ramsey M, Daniel J, Spyres LM, Safiejko-Mroczka B, Ortiz-Leduc W, Ballard JD. Clostridium difficile toxin B activates dual caspase-dependent and caspase-independent apoptosis in intoxicated cells. Cell Microbiol. 2002;4:425–434. doi: 10.1046/j.1462-5822.2002.00201.x. [DOI] [PubMed] [Google Scholar]
- Qa’Dan M, Spyres LM, Ballard JD. pH-induced conformational changes in Clostridium difficile toxin B. Infect Immun. 2000;68:2470–2474. doi: 10.1128/iai.68.5.2470-2474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke J, Tenzer S, Rupnik M, Koschinski A, Hasselmayer O, Schrattenholz A, Schild H, von Eichel-Streiber C. Autocatalytic cleavage of Clostridium difficile toxin B. Nature. 2007;446:415–419. doi: 10.1038/nature05622. [DOI] [PubMed] [Google Scholar]
- Rolfe RD, Song W. Purification of a functional receptor for Clostridium difficile toxin A from intestinal brush border membranes of infant hamsters. Clin Infect Dis. 1993;16(Suppl 4):S219–227. doi: 10.1093/clinids/16.supplement_4.s219. [DOI] [PubMed] [Google Scholar]
- Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nature reviews. 2009;7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- Satchell KJ. MARTX, multifunctional autoprocessing repeats-in-toxin toxins. Infect Immun. 2007;75:5079–5084. doi: 10.1128/IAI.00525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider EL, Craik CS. Positional scanning synthetic combinatorial libraries for substrate profiling. Methods Mol Biol. 2009;539:59–78. doi: 10.1007/978-1-60327-003-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Sheahan KL, Cordero CL, Satchell KJ. Autoprocessing of the Vibrio cholerae RTX toxin by the cysteine protease domain. Embo J. 2007;26:2552–2561. doi: 10.1038/sj.emboj.7601700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A. Allosteric regulation of protease activity by small molecules. Mol Biosystems. 2010;6:1431–1443. doi: 10.1039/c003913f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A, Lupardus PJ, Albrow VE, Guzzetta A, Powers JC, Garcia KC, Bogyo M. Mechanistic and structural insights into the proteolytic activation of Vibrio cholerae MARTX toxin. Nature chemical biology. 2009 doi: 10.1038/nchembio.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornberry NA, Peterson EP, Zhao JJ, Howard AD, Griffin PR, Chapman KT. Inactivation of interleukin-1 beta converting enzyme by peptide (acyloxy)methyl ketones. Biochemistry. 1994;33:3934–3940. doi: 10.1021/bi00179a020. [DOI] [PubMed] [Google Scholar]
- Yang G, Zhou B, Wang J, He X, Sun X, Nie W, Tzipori S, Feng H. Expression of recombinant Clostridium difficile toxin A and B in Bacillus megaterium. BMC Microbiol. 2008;8:192. doi: 10.1186/1471-2180-8-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.