Abstract
Reticulons are integral membrane proteins that partition into and shape the tubular endoplasmic reticulum (ER). We propose that reticulons use a membrane-insertion mechanism to generate regions of high membrane curvature in the ER. Reticulon contains two short hairpin transmembrane domains (TMD), which could generate membrane curvature by increasing the area of the cytoplasmic leaflet. Here, we test whether the short length of these hairpin TMDs is required for reticulon membrane-shaping functions in mammalian cells. We lengthened the TMDs of Reticulon 4 to resemble a typical bi-pass TMD that spans both leaflets. We find that TMD mutants oligomerize like wt, however they are not immobilized, do not partition into tubules, do not constrict tubules, and no longer suppress peripheral ER cisternae. Therefore, short hairpin TMD length is required for reticulon protein partitioning and membrane shaping functions. Another membrane protein with a short hairpin TMD is caveolin. We demonstrate that an ER-retained caveolin construct also partitions within the ER in a manner that is dependent on it containing a short hairpin TMD. These data suggest that a short hairpin TMD may be a general feature used by membrane-shaping proteins to partition into and shape regions of high membrane curvature.
Introduction
The ER is an essential eukaryotic organelle required for secretory and membrane protein synthesis, lipid synthesis, and calcium signaling (1). The ER has an elaborate and extensive structure that includes three major domains that are easily resolved by fluorescence microscopy including: 1) the nuclear envelope (NE), 2) the peripheral ER cisternae, and 3) the tubular ER (2). The NE is composed of a double membrane bilayer that includes the inner nuclear membrane (INM) and outer nuclear membrane (ONM) (3). The INM and ONM are stacked over each other separated by a lumen called the perinuclear space (PNS) and are connected to each other at the nuclear pores. The ONM is continuous with the membrane of the peripheral ER, which is composed of the peripheral ER cisternae and peripheral ER tubules. Both the lumen and the membrane of all three of these major ER domains are continuous with each other and yet these domains have very different structures. The shape of these domains varies to a large extent because the membrane curvature of these domains varies. For example, the membrane of the NE is relatively flat and has low membrane curvature everywhere except at the nuclear pores. Likewise, the peripheral ER cisternae also have low membrane curvature except at their edges. In contrast, the tubular ER has high membrane curvature in cross-section along the length of the tubule.
The various domains of the ER are likely to be structured by specific types of membrane shaping proteins that can generate or maintain membrane curvature. It is not known how peripheral ER cisternae are shaped; it could be that the lack of membrane curvature at ER cisternae and the NE is due to the absence of proteins that generate ER domains of high curvature. In particular, one family of ER membrane shaping proteins, the reticulons, specifically partition into regions of high membrane curvature in the ER, like the tubules and edges of cisternae, and these proteins are absent from membrane domains that lack curvature like the NE and the plane of the cisternae (4–9). Several lines of evidence have established the reticulon proteins as membrane shaping proteins that are responsible for generating tubular ER shape. Increasing the concentration of specific reticulons in several eukaryotes leads to longer, thinner, and unbranched tubules while their depletion decreases levels of tubular ER (5, 6, 8, 10, 11). Furthermore, in vitro studies show that purified yeast Rtn1 and Yop1 reconstituted into proteo-liposomes generates membrane tubules (11). Therefore, these proteins partition into the domain of the ER that they shape.
The reticulons are a highly conserved eukaryotic protein family whose distinguishing feature is the reticulon homology domain (RHD). The RHD is an ~200 amino acid domain located at the C-terminus of the protein and is composed of two short hairpin TMDs and the three surrounding cytoplasmic soluble domains, an N-terminal domain, the Nogo-66 domain (the highly conserved central soluble domain), and the C-terminal domain (See Figure 1A for model) (12–14). Reticulon proteins are quite variable at their N-terminus; this domain can be alternatively spliced and up to 80 kDa long for different reticulons. The RHD alone partitions to ER tubules showing that this conserved domain is sufficient for localization to tubules. In addition, the RHD crosslinks into a ladder of complexes showing that reticulons form oligomers via the RHD domain alone (15). Furthermore, all reticulons tested, including the RHD alone, are immobilized in the ER membrane bilayer in FRAP experiments (15). The immobility of reticulons is a conserved property that implies that these proteins could form a scaffold on the tubular ER to shape it by a mechanism similar to that of other oligomeric membrane shaping proteins (16–18).
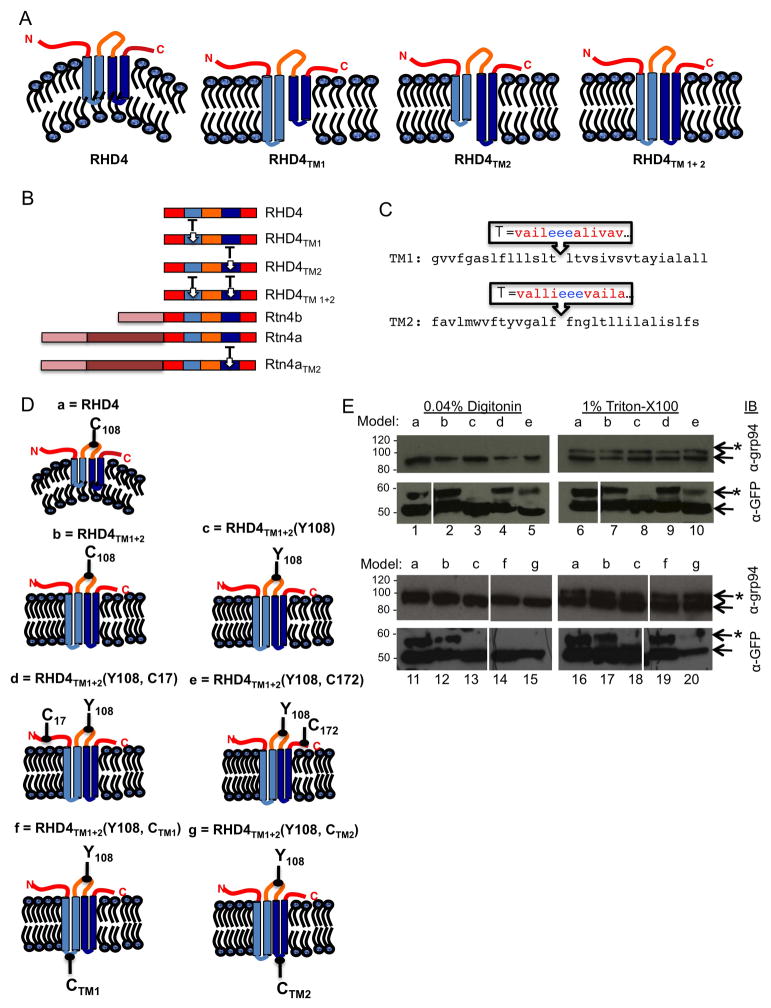
Figure 1.
Sequence and topology of Rtn4 and TM domain mutants generated. (A) Predicted membrane topology of human RHD4 and RHD4 lengthened TM domain mutants: RHD4TM1, RHD4TM2, RHD4TM1+2. (B) Diagram of RHD4, Rtn4b, and Rtn4a demonstrating regions where insertions were made into TM domains. The RHD4 is common to all Rtn4 isoforms, blue domains indicate natural TM domains, insertion sites labeled as T. (C) The sequence of the Rtn4 TM domains (TM1 and TM2) are shown with endogenous residues in black letters, and the inserted hydrophobic and acidic residues are shown in red and blue letters, respectively. (D) Model topology and point cysteine mutations tested (a) GFP-RHD4, (b) GFP-RHD4TM1+2, (c) Cysteine-less mutant GFP-RHD4TM1+2Y108, (d) Single N-terminal cysteine GFP-RHD4TM1+2Y108, C17, (e) single C-terminal cysteine GFP-RHD4TM1+2Y108, C172, (f) single cysteine in lumenal domain of TM1 GFP-RHD4TM1+2Y108, CTM1, and (g) single cysteine in lumenal domain of TM2 GFP-RHD4TM1+2Y108, CTM2. (E) Cos-7 cells transiently transfected with GFP-RHD4TM1+2 cysteine mutants. Cells were treated with 5KDa maleimide-PEG to modify free cysteines. In cells permeabilized with 0.04% digitonin, cysteines in each of mutant GFP-RHD4TM1+2 soluble domains is modified, second and fourth rows, lanes 2, 4, 5, and 12. While cysteine-less mutant GFP-RHD4TM1+2Y108 is not modified, second and fourth rows, lane 3 and 13. Cysteines added to predicted lumenal domains were not modified showing they do not face the cytoplasmic surface, fourth row, lanes 14 and 15. Importantly, lumenal ER protein with one free cysteine grp94 is not modified, first and third rows, lanes 1–5 and 11–15, demonstrating that the ER membrane is not permeabilized. In cells permeabilized with TritonX-100, all GFP-RHD4TM1+2 constructs with free cysteines are modified as well as grp-94, first and third row, lanes 6–10 and 16–20. * denotes modified bands.
Proteins that shape membrane bilayers often have a direct mechanism for generating membrane curvature (16). We asked what structural features of reticulons might generate membrane curvature in the tubular ER. A common mechanism used by well-characterized membrane-shaping proteins (for example epsin and dynamin) is to generate membrane curvature by the insertion of an amphipathic helix into the cytoplasmic leaflet of the membrane bilayer (16, 19). Reticulons do not have amphipathic helices however they do contain two approximately 33 amino acid long short hairpin transmembrane domains (TMDs) that are predicted to be too long to span the membrane bilayer once, and too short to fully span the membrane bilayer twice. The experimentally determined topology places all reticulon soluble domains in the cytoplasm which means that the short hairpin TMDs would occupy more area in the outer leaflet of the membrane bilayer (6, 8). By this mechanism, reticulon short hairpin TMDs could expand the area of the outer leaflet relative to the inner leaflet of the bilayer to generate the membrane curvature found in an ER tubule. Here we test whether the reticulon short hairpin TMDs are important for reticulon function by testing the effect of lengthening the short hairpin TMDs of human Rtn4 to resemble typical bi-pass TMDs, which fully span both leaflets (see model in Figure 1A).
Results
Mutants Generated
We generated several mutant constructs to test whether short hairpin TMD length is important for reticulon function. We used the RHD of human Rtn4 (RHD4), which has previously been shown to be sufficient for oligomerization, immobilization, and partitioning of Rtn4 into tubules (15). To generate lengthened TMD mutants, we performed site-directed mutagenesis on a previously characterized construct encoding N-terminal monomeric GFP-tagged human RHD4, GFP-RHD4 (15). Insertional mutagenesis generated mutants GFP-RHD4TM1, GFP-RHD4TM2, and GFP-RHD4TM1+2, which had the first TMD lengthened, the second TMD, or both, respectively (Figure 1A and 1B). Each of the short hairpin TMDs was lengthened by inserting ten hydrophobic residues interrupted by three acidic hydrophilic residues into the middle of the short hairpin TMD (Figure 1A, 1B, and 1C). The final length of hydrophobic residues was designed to change the Rtn4 TMDs to a more typical bi-pass TMD length to span both leaflets of the bilayer (see Figure 1C). The acidic residues were added to help pull the middle of the TM hairpin all the way into the ER lumen (see diagram in Figure 1A and 1C).
We used a previously described cysteine modification protocol to confirm that one example of these TMD mutants, GFP-RHD4TM1+2, was still adopting the correct topology in the membrane bilayer (6). The topology of wt RHD4 has all soluble domains on the cytoplasmic side of the membrane bilayer (see topology in Figure 1A) (6). To determine the topology of GFP-RHD4TM1+2, we transiently transfected this construct into Cos-7 cells and incubated the cells with a concentration of the detergent digitonin (0.04%) to permeabilize the plasma membrane but leave the ER membrane intact. Digitonin-permeabilized cells were then incubated with maleimide-PEG (5kDa) to covalently modify free cysteines, and modified samples were analyzed by SDS-PAGE and immuno-blotting with anti-GFP. In 0.04% digitonin the PM is permeabilized but the ER membrane is not, therefore, cytoplasmic cysteines should be modified by Mal-PEG and lumenal cysteines should not. Modification is indicated by a 5kDa mobility shift in the protein by SDS-PAGE. Wild-type GFP-RHD4 has one available cysteine, C108, in the Nogo-66 domain, and it is modified in both GFP-RHD4 and mutant GFP-RHD4TM1+2 demonstrating that this domain faces the cytoplasm (Figure 1D and 1E, models a and b, lanes 1, 2, 11, and 12). When we mutated the endogenous RHD4 Nogo-66 cysteine on GFP-RHD4TM1+2 to a tyrosine [GFP-RHD4TM2(Y108)], the protein is no longer shifted, demonstrating that this cysteine is indeed being modified in GFP-RHD4TM1+2 (Figure 1D and 1E, model c, lane 3 and 13). We then checked the topology of the N- and C- termini of GFP-RHD4TM1+2 by separately mutating a residue to a cysteine in each of these soluble domains GFP-RHD4TM1+2(Y108) mutant. These mutants were designed to now contain a single free cysteine in either the N or C terminus, since the GFP-RHD4TM1+2(Y108) mutant no longer has a cysteine in the Nogo-66 domain. We determined that both the N- and C-terminal constructs were modified by Mal-PEG in digitonin-permeabilized cells demonstrating that both of these domains also face the cytoplasm (Figure 1D and 1E, models d and e, lanes 4 and 5). We also confirmed that cysteines added to the predicted lumenal sequence (EEE to EECE) of GFP-RHD4TM1+2(Y108) were not modified by Mal-PEG in digitonin permeabilized cells showing that this domain does not face the cytoplasm (Figure 1D and 1E, models f and g, lanes 14 and 15) confirming our predicted protein topology as shown in Figure 1A. To confirm that the ER membrane was not being permeabilized under our conditions by digitonin, we simultaneously looked for modification of a lumenal ER protein, grp94, which contains one free cysteine. Grp94 is not modified by Mal-PEG (5kDa) in the digitonin-permeabilized cells (Figure 1E, lanes 1–5 and 11–15, first and third row). In contrast, all proteins that contain a cysteine are modified by Mal-PEG (5kDa) after incubation with a detergent that will permeabilize all cellular membranes, 1% Triton X-100 (Figure 1E, lanes 6–10 and 16–20). These data show that all endogenous soluble domains of GFP-RHD4TM1+2 are in the cytoplasm like wild-type GFP-RHD4 and retain the correct topology.
Short hairpin TM domains localize reticulons to the tubular ER
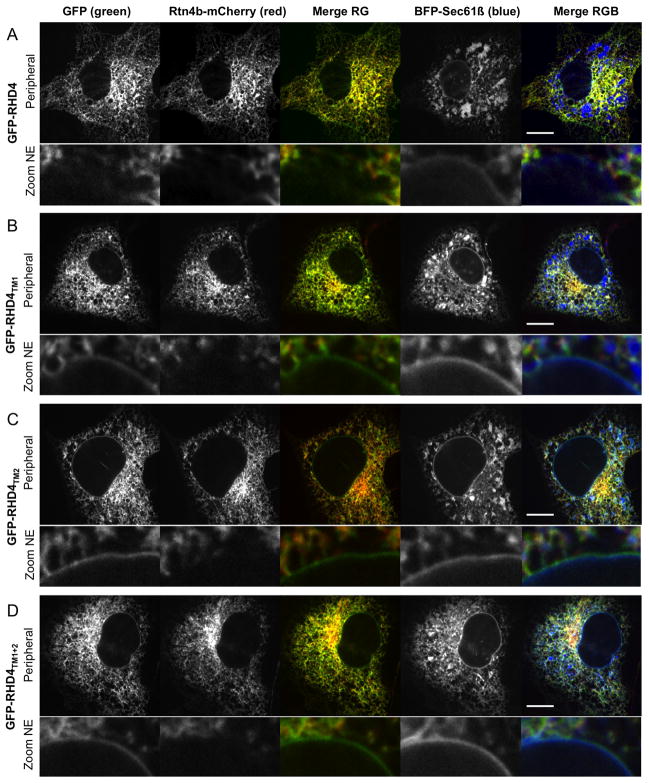
A distinguishing feature of reticulon proteins and the RHD is that they partition into ER tubules and are excluded from the flat sheets of the NE and peripheral ER cisternae (6, 8, 15). We asked whether lengthening the TMDs of RHD4 affected its ability to partition in the ER membrane bilayer. We used confocal fluorescence microscopy to visualize the localization of GFP-RHD4 mutants with lengthened TMDs. Cos-7 cells were transiently co-transfected with a C-terminal mCherry-tagged Rtn4 isoform, Rtn4b-mCherry to visualize tubular ER (Figure 2, second column from left), general ER marker BFP-Sec61β to visualize all ER domains (Figure 2, fourth column from left), and with either GFP-RHD4 or the lengthened TMD mutants (Figure 2, first column). As expected, GFP-RHD4 colocalizes well with Rtn4b-mCherry and is absent from the NE and peripheral ER sheets (Figure 2A, merge RG). In contrast, all TMD mutants (GFP-RHD4TM1, GFP-RHD4TM2, and GFP-RHD4TM1+2, Figure 2B–2D, merge RG) do not partition exclusively to tubules and can be seen at the NE, although not as strongly as BFP-Sec61β (Figure 2B–2D, merge RGB, and zoom to see localization to the NE). These data show that lengthening either one or both of the TMDs of RHD4 reduces its ability to partition exclusively into ER tubules.
Figure 2.
Localization of RHD4 and lengthened TM domain mutants differs. Cos-7 cells co-expressing ER marker Rtn4b-mCherry (second column in A–D), general ER marker BFP-Sec61β (fourth column in A–D), and either (A) GFP-RHD4, (B) GFP-RHD4TM1, (C) GFP-RHD4TM2, or (D) GFP-RHD4TM1+2 (all GFP localization shown in first column in A–D). Merged images are shown for GFP-tagged proteins (green) and Rtn4b-mCherry (third column in A–D, Merge RG) and for all three channels (BFP-Sec61β in blue, fifth column in A–D, Merge RGB). Whole cell images in top panels were taken with a confocal microscope to image the peripheral ER. Lower row images (Zoom NE) are to highlight relative protein distribution in the NE. Note that all the TM domain mutants are no longer excluded from the NE like GFP-RHD4. Scale bar = 10μM.
Mobility of the RHD is increased in lengthened short hairpin TM domain mutants
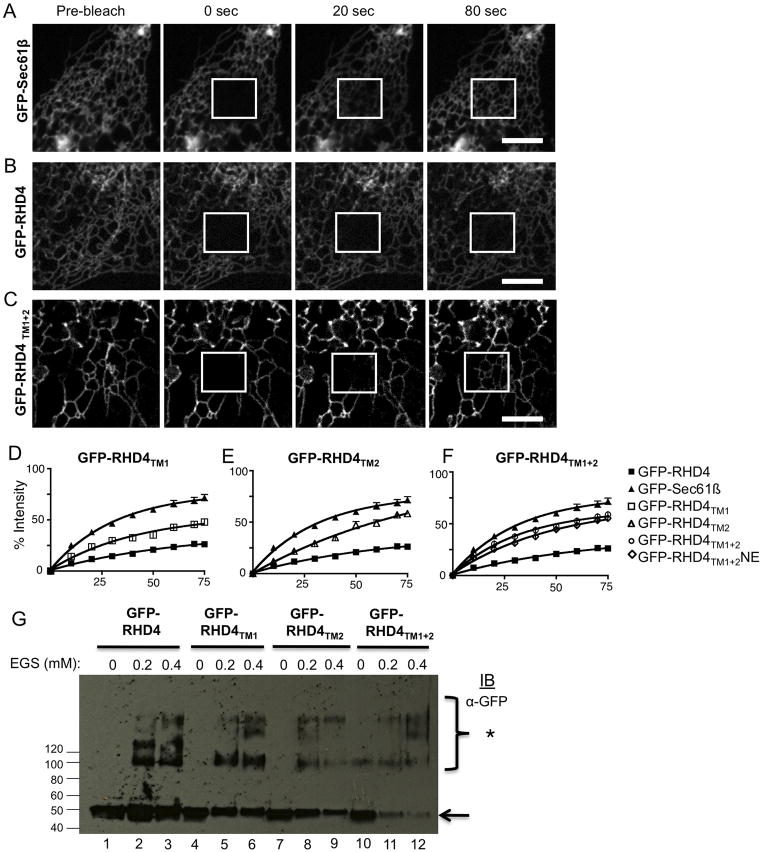
The RHD of both yeast Rtn1 and of human Rtn4 (RHD4) form complexes that are immobile in the membrane bilayer when compared to general ER membrane proteins in fluorescence recovery after photobleaching (FRAP) experiments (15). Therefore, we asked whether lengthening the TMD of RHD4 changes its diffusional mobility in the membrane bilayer. We used FRAP to measure the diffusional mobility of GFP-RHD4 and all TMD mutants after transient transfection of these constructs into Cos-7 cells (Figure 3). A small region of the peripheral ER in these transfected cells was then photobleached by a high power laser in a 5.6 × 5.6μm box and images were taken each second for 80 seconds following the bleaching event. As expected, GFP-RHD4 has a visually obvious slow diffusional mobility when compared to the mobile membrane protein GFP-Sec61β (Figure 3A and B). In contrast, all GFP-RHD4 TMD mutants have a much faster recovery than GFP-RHD4 (see example for GFP-RHD4TM1+2 in Figure 3C). We quantified the rate of recovery for all proteins by averaging normalized fluorescence intensity measurements for several FRAP experiments. These data reveal that GFP-RHD4 recovers slowly while general ER marker, GFP-Sec61β, has a fast mobility. TMD mutants (GFP-RHD4TM1, GFP-RHD4TM2, and GFP-RHD4TM1+2) instead have an intermediate phenotype recovering faster than GFP-RHD4 but not quite as fast GFP-Sec61β (Figure 3D–F). We also tested the pool of GFP-RHD4TM1+2 localized to the NE and saw no difference in mobility when compared to the pool localized to the tubular ER (Figure 3F). Taken together, these data show that RHD4 mutants with lengthened TMDs show an increase in their mobility on the ER membrane bilayer.
Figure 3.
Short hairpin TM domains are required for immobilization of RHD4. (A–C) Cos-7 cells were transiently transfected with GFP tagged proteins (A) GFP-Sec61β, (B) GFP-RHD4 and (C) GFP-RHD4TM1+2, and were imaged live by confocal microscopy. Typical FRAP data is shown for an image pre-bleaching event and then post bleaching for times indicated. Bleached region is in the white box. (D–F) Quantification of FRAP analysis. Each graph contains the averaged curves for GFP-Sec61β (black triangle, N=11) and GFP-RHD4 (black square, N=23). Since curves for TM domain mutants are similar, we separated each into individual graphs, (D) GFP-RHD4TM1 (open squares, N=10), (E) GFP-RHD4TM2 (open triangle, N=14), and (F) GFP-RHD4TM1+2 (open circle, N=13) and GFP-RHD4TM1+2NE (open diamond, N=23). (G) Cos-7 cells were transiently transfected with GFP-RHD4TM mutants as labeled. Cells were then gently permeabilized using 0.04% digitonin to maintain protein-protein interactions. Increasing amounts of crosslinker EGS were added and cells were incubated for an hour. Cells were then further permeabilized with 1% Triton X-100 and protein samples were analyzed by SDS-PAGE and immuno-blotting for GFP. * denotes crosslinked bands. Scale bar = 5μM.
We performed EGS crosslinking to test if the change in diffusional mobility reflected a change in the oligomerization state of the RHD4. Previous work shows that RHD4 can oligomerize into a complex that can be crosslinked by EGS (15). We transiently transfected Cos-7 cells with GFP-RHD4 or each of the GFP-tagged TMD mutants. Cells were then solubilized with digitonin detergent (0.04%) and then incubated with EGS (0 mM, 0.2 mM, and 0.4 mM). After quenching with 5mM DTT, samples were then further solubilized with 1% TritonX-100 and analyzed by SDS-PAGE immuno-blotting with anti-GFP (Figure 3G). The GFP-RHD4 protein gave a similar crosslinking pattern to GFP-RHD4TM1, GFP-RHD4TM2, GFP-RHD4TM1+2. Our crosslinking data demonstrate that TMD mutants are able to form complexes of a similar size to wild-type GFP-RHD4. Therefore, the oligomeric complexes for wild type and TM mutants may each contain similar numbers of reticulon molecules. However, the organization and shape of the reticulon oligomers might be very different for the wild type protein with the short hairpin TM domains. These short hairpins may cause the reticulon subunits to interact at a different angle. This angle may allow the proper immobile reticulon oligomer to form that is able to deform the membrane bilayer. Together, these data show that although short hairpin TMDs are not required for GFP-RHD4 to crosslink into protein complexes, short hairpin TMDs are required for proper partitioning into tubular ER and for it to form an immobilized structure on the tubules.
Short hairpin TM domains are also required for partitioning Rtn4a
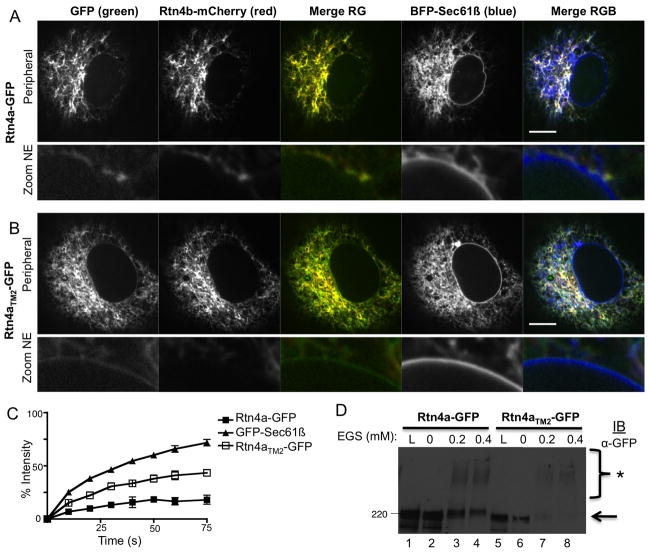
Rtn4a (aka Nogo A) is a vertebrate reticulon isoform that contains a long cytoplasmic N-terminal domain in addition to the RHD4 (see diagram in Figure 1). This isoform has been shown to generate ER tubules both in vivo and in vitro (6, 11). We next tested whether lengthening the second TMD of Rtn4a-GFP to generate Rtn4aTM2-GFP would similarly change its properties in the ER membrane bilayer (see diagram in Figure 1). Lengthening TM2 of GFP-RHD4 was sufficient to alter the localization and mobility of the RHD4 (Figures 1, 2, and 3), so we only lengthened TM2 of Rtn4a-GFP to make Rtn4aTM2-GFP (Figure 1B). We used insertional mutagenesis to lengthen TM2 of Rtn4a-GFP to make Rtn4aTM2-GFP. We first compared the ER localization of Rtn4aTM2-GFP with Rtn4a-GFP. Cos-7 cells were transiently co-transfected with Rtn4b-mCherry to show typical reticulon distribution, along with BFP-Sec61β to label all ER membranes, and with either Rtn4a-GFP or Rtn4aTM2-GFP (Figure 4A and 4B). Transfected cells were imaged live by confocal fluorescence microscopy. As expected, the Rtn4a-GFP protein partitioned exclusively to the tubular ER with perfect co-localization with Rtn4b-mCherry (Figure 4A, Merge RG) and the Rtn4a-GFP did not colocalize with the BFP-Sec61β signal in the NE and ER cisternae (Figure 4A, and zoom NE). In contrast, Rtn4aTM2-GFP did not overlap perfectly with Rtn4b-mCherry (Figure 4B, Merge RG). While Rtn4aTM2-GFP was indeed enriched in ER tubules, it could also be found localizing with the BFP-Sec61β in the NE and in peripheral ER cisternae (Figure 4B and zoom NE). Together these data show that lengthening a single short hairpin TMD in the full-length isoform, Rtn4a, also decreases its ability to partition exclusively to tubular ER.
Figure 4.
Short hairpin TM domains are required for Rtn4a localization and immobilization. A representative image of a Cos-7 cell co-expressing a general ER marker BFP-Sec61β (blue, forth column), tubular ER marker Rtn4b-mCherry (red, second column), and either (A) Rtn4a-GFP or (B) Rtn4aTM2-GFP (green, first column). Note that Rtn4a (in A) co-localizes well with Rtn4b-mCherry and does not co-localize with BFP-Sec61β (Merge RGB) in the NE, whereas Rtn4aTM2-GFP can be seen in the NE (compare Zoom NE images for A and B). (C) Quantification of FRAP analysis. Graph contains the averaged curves for GFP-Sec61β (filled triangles, N=11), Rtn4a-GFP (filled squares, N=10), and Rtn4aTM2-GFP (open squares, N=11). (D) Cos-7 cells were transiently transfected with GFP-RHD4TM mutants as labeled. Cells were then gently permeabilized using 0.04% digitonin to maintain protein-protein interactions. Increasing amounts of crosslinker EGS were added and cells were incubated for an hour. Cells were then further permeabilized with 1% Triton X-100 and protein samples were analyzed by SDS-PAGE and immuno-blotting for GFP. * denotes crosslinked bands. Scale bar = 10μM.
Short hairpin TM domains immobilize Rtn4a
FRAP data shows that Rtn4a is immobilized in the ER membrane similar to that of the RHD4 alone and similar to that of a known membrane scaffolding protein lamin B receptor (15). We tested whether the mobility of the Rtn4a isoform would also increase, simply as a result of lengthening its second TMD despite its longer N-terminal cytoplasmic domain. Cos-7 cells were transiently co-transfected with either Rtn4a-GFP or Rtn4aTM2-GFP and FRAP experiments were performed as described for Figure 3. As expected, Rtn4a-GFP recovers more slowly by FRAP than ER membrane protein GFP-Sec61β (Figure 4C, compare filled squares to filled triangles, respectively). In contrast, Rtn4aTM2-GFP has a mobility that is intermediate between Rtn4a-GFP and GFP-Sec61β (Figure 4C, open squares). These results are similar to those observed for GFP-RHD4 TMD mutants and demonstrate that TMD length has a dramatic effect on the diffusional mobility of the Rtn4a complexes.
We tested whether Rtn4aTM2-GFP can still form higher molecular weight complexes in the membrane bilayer that can be crosslinked by EGS and visualized by SDS-PAGE (Figure 4D). Rtn4a-GFP or Rnt4aTM2-GFP were transiently transfected into Cos-7 cells, cells were then solubilized with 0.04% digitonin and treated with the EGS concentrations indicated. The reaction was quenched with 5mM DTT. Then crosslinked samples were further solubilized in 1% TritonX-100 and analyzed by SDS-PAGE and immuno-blotting. We find that both Rtn4a-GFP and Rtn4aTM2-GFP crosslink in a similar pattern demonstrating that both protein forms can establish protein-protein interactions. Taken together, these FRAP and crosslinking data reveal that although short hairpin TMDs may not be important for Rtn4a complex formation alone, short hairpin TMDs are important for organizing the Rtn4 protein complex into the structural shape that is properly immobilized and partitioned within the membrane bilayer.
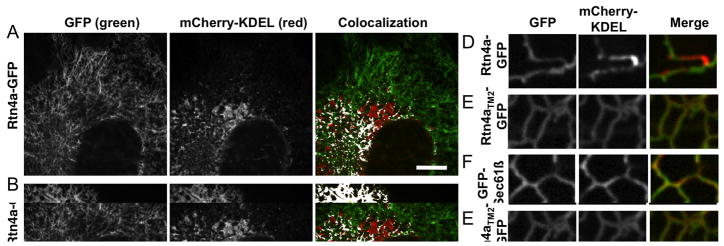
Short hairpin TM domains of Rtn4a are required to constrict ER tubules
Previous work has shown that overexpression of reticulon proteins can constrict ER tubules to a smaller diameter; this alteration in tubule diameter can affect the distribution and diffusion of lumenal ER proteins in the peripheral ER (5, 11). Overexpression of Rtn4a in Cos-7 cells can even “squeeze” lumenal ER proteins out of the peripheral ER tubules and concentrate these proteins into the remaining lumen within more perinuclear ER (11). We next wanted to address whether the short hairpin TMDs are important for the ability of Rtn4a to constrict tubular ER and limit lumenal protein diffusion. Cos-7 cells were transiently transfected with a lumenal ER marker mCherry-KDEL along with either Rtn4a-GFP or Rtn4aTM2-GFP. Cells were then imaged live 24 hours post-transfection by confocal fluorescence microscopy in order to visualize the amount of co-localization between each of these constructs and the lumenal ER protein. Our prediction was that Rtn4a-GFP should constrict the tubular ER and exclude the lumenal ER protein from regions of high Rtn4a expression. In contrast, if the lengthened TMD proteins could no longer generate membrane curvature and constrict ER tubules it should not displace lumenal mCherry-KDEL. As predicted, Rtn4a overexpression displaced lumenal mCherry-KDEL from the tubular ER (Figure 5A). When we zoomed in on individual tubules, we could clearly see that areas of high Rtn4a-GFP had low levels of lumenal mCherry-KDEL and vice versa (Figure 5D). This exclusion property is specific to Rtn4a, since overexpression of a general ER membrane protein, GFP-Sec61β, does not exclude the lumenal protein from ER tubules (Figure 5C and F). In contrast to wt Rtn4a-GFP, overexpression of Rtn4aTM2-GFP did not exclude the co-expressed mCherry-KDEL from the tubular ER (Figure 5B). Rtn4aTM2-GFP and mCherry-KDEL also colocalized along the length of the tubule similar to GFP-Sec61β and mCherry-KDEL while Rtn4a-GFP clearly partitions relative to mCherry-KDEL (Figure 5E).
Figure 5.
Rtn4a short hairpin TM domains are required to constrict ER tubules. (A–C) Representative images of Cos-7 cells co-expressing a lumenal ER protein mCherry-KDEL (second column, red) and either (A) Rtn4a-GFP (first column, green), (B) Rtn4aTM2-GFP (first column, green), or (C) GFP-Sec61β (first column, green). Areas of colocalization are highlighted in white on colocalization images for A–C (last column). (D–F) Zoom on tubules of cells transfected with the same constructs as in (A–C, respectively). (G) Average Pearson’s Coefficient of cells co-expressing mCherry-KDEL similar to those shown in (A–C) (n=9–12 cells each). (G) Average Pearson’s Coefficient values of Cos-7 cells co-expressing mCherry-Sec61β and Rtn4a-GFP, Rtn4aTM2-GFP, or GFP-Sec61β (n=9–12 cells each). Scale bar = 10μm for A–C.
We calculated the Pearson’s Coefficient (PC) of correlation to quantify the degree of colocalization between the lumenal protein mCherry-KDEL and each membrane protein: Rtn4a-GFP, Rtn4aTM2-GFP, or GFP-Sec61β. The PC measures the intensity of fluorescence of each pixel in both channels and compares the channels to determine the amount of correlation. A PC of 1.0 indicates complete colocalization; a PC of 0 means no correlation; and a PC of −1.0 means they are negatively correlated (20). We found that Rtn4a-GFP and mCherry-KDEL are negatively correlated with a PC of −0.21 showing that Rtn4a-GFP is able to limit the distribution of this lumenal ER protein (Figure 5G). In contrast, both Rtn4aTM2-GFP and GFP-Sec61β had high colocalization with mCherry-KDEL with calculated PC values of 0.71 and 0.72, respectively (Figure 5G). These data demonstrate that, unlike wild-type Rtn4a, Rtn4aTM2 behaves more like a general ER membrane protein and is not able to constrict ER tubules and to limit the diffusion of lumenal proteins when overexpressed. We also looked at the colocalization of Rtn4a-GFP and Rtn4aTM2-GFP with the general ER membrane protein, mCherry-Sec61β. Both Rtn4a-GFP and Rtn4aTM2-GFP had positive PC values with mCherry-Sec61β (0.31 and 0.52 respectively), showing that Rtn4a-GFP does not effect the localization of ER membrane proteins (Figure 5H). Rtn4aTM2-GFP and mCherry-Sec61β have a higher PC value than Rtn4a-GFP with mCherry-Sec61β, which would be expected since Rtn4aTM2-GFP no longer partitions exclusively to tubules. As a control, we co-transfected cells with GFP-Sec61β and mCherry-Sec61β and measured the PC to be 0.81 showing high colocalization of two different fluorescently tagged proteins that should co-localize. These data show that short hairpin TMD length has a dramatic effect on the distribution of lumenal ER proteins in tubules. We propose that this effect on lumenal protein distribution reflects a requirement for short hairpin TMDs to cause constriction of ER tubule diameter.
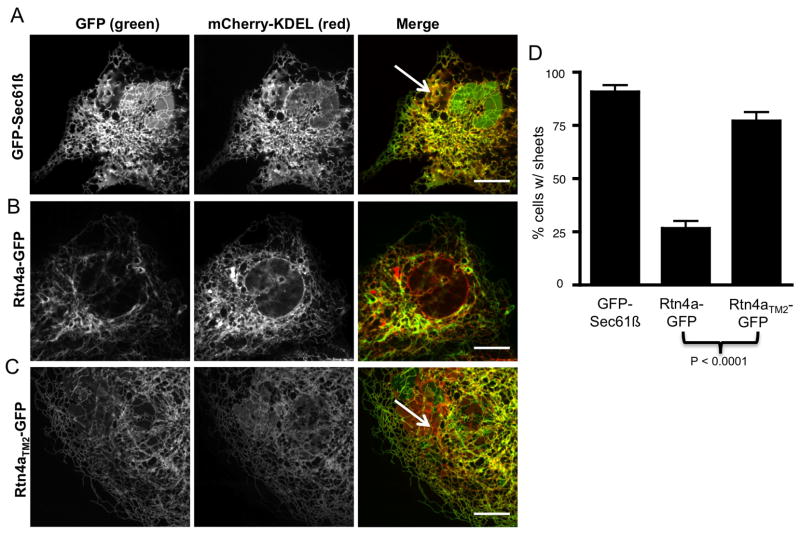
Short hairpin TM domains are required for Rtn4a to convert peripheral ER cisternae into tubules
We next tested whether TMD length affects the ability of Rtn4a to convert peripheral ER cisternae into tubules. The levels of reticulons can change the overall structure and organization of the peripheral ER. Depletion of reticulon proteins leads to cells with mostly cisternal peripheral ER, whereas overexpression of Rtn1 in yeast or Rtn4a in mammalian cells leads to a reduction in peripheral ER cisternae concomitant with an increase in tubular ER (6, 10). We measured whether expression of Rtn4aTM2-GFP can convert peripheral ER cisternae into tubules. Cos-7 cells were transiently co-transfected with mCherry-KDEL (to visualize all ER domains), and with either GFP-Sec61β, Rtn4a-GFP or Rtn4aTM2-GFP (representative images Figure 6A–C). Twenty-four hours after transfection, cells were imaged live by confocal fluorescence microscopy and scored in a Bernoulli trial test for either the presence of cisternae (with the unit 1) or the lack of cisternae (with the unit 0). The upper threshold for the mCherry-KDEL expression levels was set to 2 times the background. Thus, cells that were greatly over-expressing mCherry-KDEL were omitted. After counting many cells (N=87 for GFP-Sec61β, N=162 for Rtn4a-GFP, N=105 for Rtn4aTM2-GFP), we determined that the outcome of the cisternal binomial distribution was independent of the relative GFP fluorescence intensity readings (data not shown). Almost all cells transfected with GFP-Sec61β, 90.8% (SEM= ± 3.1%), had cisternal ER (Figure 6A and 6D). About one fourth of Rtn4a-GFP transfected cells, 26.5% (SEM=± 3.5%), had cisternal ER (Figure 6B and 6D); similar to previous reports (6). The extension of the hairpin loop in Rtn4aTM2-GFP had a significant increase in the occurrence of ER cisternae at 77.1% (SEM=± 4.1%) (Figure 6C and 6D). These data demonstrate that lengthening the short hairpin TMD of just one hairpin loop in Rtn4a is sufficient to inhibit its ability to transform peripheral ER cisternae into tubules.
Figure 6.
Rtn4a short hairpin TM domains are required to convert ER cisternae into tubules. (AC) Representative images of Cos-7 cells co-expressing a lumenal ER protein mCherry-KDEL (second column, red) and (A) a general ER membrane protein GFP-Sec61β (first column, green), (B) Rtn4a-GFP (first column, green), or (C) Rtn4aTM2-GFP (first column, green). Note the presence of peripheral ER cisternae (white arrows) in cells expressing GFP-Sec61β (A) and Rtn4aTM2-GFP (C). (D) The percent of cells containing peripheral ER cisternae for each cell type A–C. (N=87 for GFP-Sec61β, N=162 for Rtn4a-GFP, and N=105 for Rtn4aTM2-GFP). Scale bar = 10μm.
The oligomerization and short hairpin TM domain length of caveolin similarly affects its ability to partition in the ER
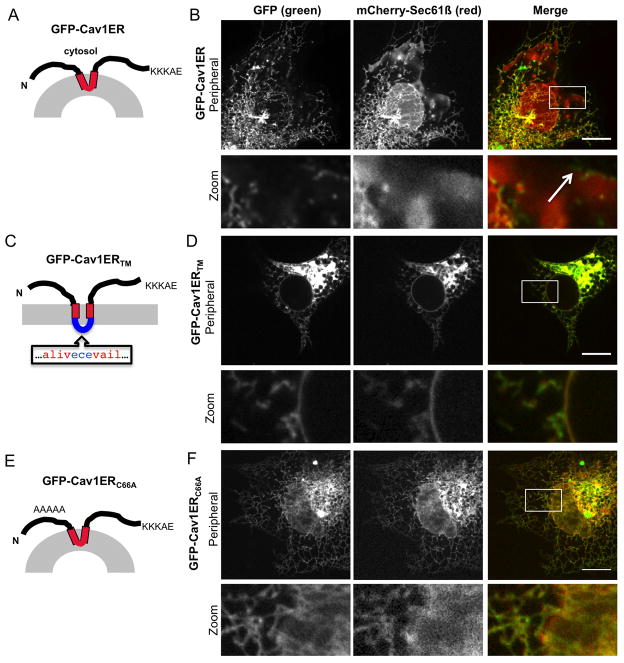
Caveolins localize to and generate the structure of caveolae, regions of high membrane curvature on the plasma membrane (21). There are striking similarities between reticulons and caveolin. Like reticulons, caveolins are integral membrane proteins that form oligomeric structures on the membrane bilayer and generate membrane curvature (17, 21–24). Like reticulons, caveolins also have a short hairpin TMD (see topology in Figure 7A) and it has been suggested that caveolin may generate high curvature vesicles at the plasma membrane (PM) by inserting this short hairpin TMD into the cytoplasmic leaflet of the PM (21, 24, 25). We asked whether these shared features of caveolin (oligomerization and short hairpin TMD length) could also localize caveolin to regions of high membrane curvature in the ER. To test this, we generated a form of caveolin that is retained in the ER membrane by adding the human Rtn3 ER retention signal (KKKAE) onto the C-terminus of a GFP-tagged human Caveolin1 construct to make GFP-Cav1ER (Figure 7A). Cos-7 cells were transiently transfected with GFP-Cav1ER and mCherry-Sec61β and were imaged live by confocal fluorescence microscopy. While mCherry-Sec61β localized to all ER domains, GFP-Cav1ER localizes specifically to ER tubules and is absent from the flat sheets of the NE and peripheral ER cisternae in a manner similar to reticulons (Fig. 7B). We also saw some GFP-Cav1ER at the edges of ER cisternae (Figure 7B, zoom images, and white arrow), which is similar to the localization pattern of reticulon at ER tubules and edges of ER cisternae previously shown (6, 8, 9). We next tested whether GFP-Cav1 with a lengthened TMD (GFP-Cav1TMER) can still partition into tubular ER. We generated this construct by inserting a mostly hydrophobic sequence into the middle of the caveolin TMD (see model in Figure 7C). The mutant with a lengthened TMD, GFP-Cav1ERTM, now localizes to all ER domains including the NE similar to the Rtn4TM mutants (Figure 7D and zoom images).
Figure 7.
An ER-retained Caveolin-1 partitions into tubules. (A) Predicted topology of GFP-tagged Caveolin-1 with a C-terminal ER retention signal (KKKAE). (B) GFP-Cav1ER (first column, green) expressed in Cos-7 cells localizes to the tubular ER and edges of cisternae (zoom image, white arrow) whereas normal ER resident ER membrane protein, mCherry-Sec61β (second column, red), localizes to all ER sub-domains, including the NE and peripheral ER cisternae. (C) Model of GFP-Cav1ERTM with lengthened TM domain. (D) Colocalization images as in (B) for construct in (C) showing localization of GFP-Cav1ERTM at the NE. (E) Model of monomeric ER-retained caveolin, GFP-Cav1ERC66A construct with mutations changing residues 66–70 to alanines (zoom image). (F) Colocalization images as in (B) for construct in (E) showing localization of GFP-Cav1ERC66A at the NE (zoom image). Scale bar = 10μm.
Reticulon mutants that fail to oligomerize redistribute to all ER domains similar to lengthened TMD mutants (15). Thus, we asked whether a monomeric form of GFP-Cav1ER partitions into ER tubules. We changed residues 66–70 into alanines of Caveolin 1 that are required for its oligomerization (22)to make a monomeric ER-retained Cav1 construct, GFP-Cav1ERC66A. The GFP-Cav1ERC66A was co-transfected with mCherry-Sec61B into Cos-7 cells and imaged live by confocal fluorescence microscopy (see model in Figure 7E). This mutant localized to all ER domains including the NE showing that oligomerization is also required for partitioning Cav1 to tubules (Figure 7F and zoom images). Taken together, these data show that another integral membrane shaping protein that also oligomerizes and contains a short hairpin TMD can partition into ER tubules by a mechanism that requires the length of its short hairpin TMD.
Discussion
One distinguishing feature of reticulon proteins is their short hairpin TMDs. The TMDs have been predicted to form a wedge in the outer leaflet of the ER bilayer to expand it relative to the inner leaflet to generate membrane curvature (6). This property could allow reticulons to make ER tubules out of flat membranes. Here, we test this hypothesis by lengthening the short hairpin TMDs of human Rtn4 to resemble typical bi-pass TMDs fully spanning both leaflets of the membrane bilayer and found that Rtn4 mutants with bi-pass TMDs do not function like wild-type Rtn4.
Reticulons interact with other ER membrane shaping proteins including the DP1/REEP/Yop1 family (6, 26). The DP1/REEP/Yop1 family is structurally similar to the reticulons and members also partition into tubular ER. Mal-PEG modification and protease digestion experiments show that all the DP1/REEP/Yop1 family members contain both short hairpin TMDs and the same membrane topology as reticulons (6, 26). The DP1/REEP/Yop1 proteins also crosslink and form immobile oligomers suggesting they may also use a scaffolding mechanism like reticulons (15). Our data suggests, that the short hairpin TMDs of these other family members could also be important for their abilities to partition and shape membrane bilayers. It remains to determined whether short hairpin TMDs contribute to the function of all reticulon orthologs. For example, investigators studying plant reticulons model them with unusually long diagonal TMDs rather than short hairpin TMDs (8, 27). However, topology experiments do not rule out that plant reticulons in fact have four short hairpin TMDs rather than four unusually long diagonal TMDs. Otherwise, if plant reticulons do have unusually long bi-pass TMDs, then the plant reticulons appear to generate membrane curvature by a different mechanism than what has been proposed for animal reticulons (27).
Analogies can be made between the mechanism that reticulon uses to generate membrane curvature and those described for other types of membrane-bending proteins (16, 19, 28–31). Partial insertion of an amphipathic alpha helix to change bilayer symmetry is a mechanism previously described for Sar-1, a protein involved in generating membrane curvature during COPII vesicle formation. When Sar-1 binds to GTP it undergoes a conformational change that exposes the amphipathic helix and triggers membrane insertion into the outer leaflet at ER exit sites to generate membrane curvature (18, 32, 33). Similar to reticulons, purified Sar1 can also convert liposomes into membrane tubules of a ~26 nm diameter and the amphipathic helix is required for this ability (18, 33).
Caveolins share more properties with reticulons than proteins like Sar-1, which transiently associate with the membrane bilayer to generate membrane curvature. Caveolins are integral membrane proteins that generate membrane curvature to make caveolae at the plasma membrane (21). Like reticulons, caveolins form immobile oligomers and this feature is required for caveolin to generate caveolae; mutants that do not oligomerize do not generate caveolae (17, 22). Caveolins also have a single short hairpin TMD similar in length and topology to the TMD of Rtn4. It has not been tested whether the length of this TMD is required for caveolin localization and function at caveolae. The striking similarity to reticulons led us to test whether the short hairpin TMD of caveolin contributes to its ability to localize to regions of membrane curvature. We attached an ER retention signal onto the C-terminus of caveolin to test whether caveolin would also partition within the ER. A caveolin with an ER retention signal localizes to the ER and indeed partitions into ER tubules and is excluded from the nuclear envelope (Figure 7B). Like reticulon, a caveolin mutant that could not form immobile oligomers did not partition into ER tubules (Figure 7F). And yet, the short hairpin TMD and oligomerization are both important, because an ER-retained caveolin mutant that could oligomerize, but now had a lengthened TMD does not partition into ER tubules (Figure 7D). Together these data show two shared and important features that contribute to the ability of both reticulon and caveolin to partition into a region of high membrane curvature: oligomerization and short hairpin TMDs.
All the membrane-shaping mechanisms described here follow the rules of the bilayer couple hypothesis (34). Briefly, in a closed lipid membrane bilayer both leaflets may respond differently to changes but will remain coupled to each other. This mechanism requires that the protein(s) insert a hydrophobic or amphipathic alpha helix into one leaflet of the lipid bilayer to expand it like a wedge. This would expand the area of one leaflet relative to the second leaflet to generate curvature. But how is membrane curvature initially generated and then stabilized? Vesicles and tubules have fairly defined and conserved diameters that could be determined by both membrane insertion and scaffolding mechanisms. Our data suggest that reticulon proteins use both mechanisms to shape the ER membrane. In vitro work shows that many membrane-shaping proteins sense and generate curvature via a membrane-inserted alpha-helix (18, 35, 36). The insertion of an alpha helix into one leaflet of the bilayer may locally induce curvature on a flat membrane, providing a (small) curved membrane template to recruit other membrane-bending proteins. Oligomerization would stabilize membrane curvature further and recruit more proteins to generate vesicles and tubules (6, 17, 18, 37). Our data predict that membrane-bending proteins similar to reticulon may initially bend membrane bilayers by insertion of an alpha helical short hairpin TMD and maintain curvature over a large area by oligomerizing. However, since Rtn4 mutants with long TMDs are still able to crosslink into oligomers, the short hairpin TMDs are likely not the only reticulon domains required for shaping tubular ER. Ultimately, high-resolution structures of reticulon complexes (and other membrane shaping proteins) are needed to address the role of each domain in generating curvature.
Materials & Methods
Mammalian Plasmid Constructs
Rtn4a-GFP (human), GFP-RHD4 (human), and GFP-Sec61β (human) were described previously (15). Cav1ER was cloned by PCR amplifying human Cav1 (NCBI accession number: NM_001753) from Hela cell cDNA (provided by Jens Lykke-Andersen) with primers containing a C-terminal ER retention signal from human Rtn3 (KKKAE) and inserted into pAC-GFP-N1 (Clontech) using the BglII and EcoRI restriction sites at the 5′ and 3′ ends, respectively. Site-directed mutagenesis (Stratagene) was used to change GFP-Cav1ER into GFP-Cav1ERTM and GFP-Cav1ERC66A; to change GFP-RHD4 into GFP-RHD4TM1, GFP-RHD4TM2 and RHD4TM1+TM2; Rtn4a-GFP to Rtn4aTM2-GFP; and to change GFP-RHD4TM1+2 into the cysteine mutants used for topology experiments. mCherry-Sec61β was cloned in sequential steps by PCR amplifying the monomeric Cherry coding region out of a plasmid (kindly provided by Amy Palmer, accession number: AY678264) and insertion into pAcGFP-N1 with restriction sites NheI and BglII at the 5′ and 3′ ends, respectively, followed by in-frame insertion of Sec61β from GFP-Sec61β into restriction sites BglII and EcoRI. mCherry-KDEL was cloned by PCR amplification of the mCherry coding region with a primer containing the N-terminal signal sequence of human BiP (aa #1–18) and a reverse primer that contains the KDEL coding sequence and was inserted into pAcGFP1-C1 with restriction sites NheI and EcoRI at the 5′ and 3′ ends, respectively. Rtn4b-mCherry was cloned in sequential steps by PCR amplifying human Rtn4b (accession number: AAG12177) from HEK293 cDNA and insertion into pAcGFP1-N1 with restriction sites XhoI and KpnI at the 5′ and 3′ ends, respectively, followed by replacement of the mGFP coding region with the mCherry coding region that was PCR amplified from mCherry-KDEL and inserted into the restriction sites: Kpn1 and NotI. BFP-Sec61β was cloned by digesting GFP-Sec61β with BglII and PstI at the 5′ and 3′ ends respectively. This piece was then inserted at the 3′ end of BFP in plasmid pTagBFP-C1 (Evrogen) in the restriction sites BglII and PstI.
Topology and Chemical Crosslinking
Topology experiments were performed as previously described (6). Briefly, Cos-7 cells were transfected with GFP-RHD4 or individual cysteine GFP-RHD4TM1+2 mutant constructs, harvested and rinsed with HCN buffer (50mM Hepes pH 7.5, 150mM NaCl, 2mM CaCl2). Cells were then treated with either 0.04% Digitonin (Sigma) or 1% Triton-X 100 (Fisher) for 30 min on ice. Samples were then incubated with 1mM Maleimide-PEG 5 kDa (Sigma) at 4°C for 1hr. After modification, samples were quenched with 100mM DTT and were further solubilized with 1% Triton X-100 for 15min then analyzed by SDS-PAGE and immuno-blotting with anti-GFP (MBL) or anti-GRP94 (Santa Cruz) and secondary HRP-conjugated antibodies. Chemical crosslinking was performed by transiently transfecting Cos-7 cells with GFP-RHD4 or individual cysteine GFP-RHDTM1+2 mutant constructs. Cells were grown for 18–24 hours after transfection and rinsed with PBS, scraped off the dish, and pelleted for 10,000 rpm for 3 min. Cell pellet was resuspended and lysed in HKME buffer (25mM Hepes, 150mM KOAc, 2.5mM MgOAc, 5mM EDTA, 0.04% digitonin) and incubated on ice for 30 min. Samples were divided and incubated for 1 hour on ice with either buffer alone, 0.2mM or 0.4mM EGS. Samples were then incubated in 1% Digitonin for 1 hour on ice, followed by addition of 5mM DTT and 1% Triton-X100. Crosslinked samples were then analyzed by SDS-PAGE and immuno-blotting with anti-GFP and secondary antibodies.
Tissue Culture, Transfection, Confocal Microscopy of Cos-7 cells
Cos-7 cells (ATCC) were grown at 37°C with 5% CO2 in Dulbecco’s modified Eagle’s medium (Invitrogen) containing 10% fetal bovine serum. Transfection of DNA into Cos-7 cells was performed using Lipofectamine 2000 (Invitrogen) in Optimem media. Cells were split after 5 hours of transfection to microscope imaging dishes (Mattek) and imaging and biochemical assays were performed 18–24 hours later. Imaging of live cells was performed in a heated stage chamber at 37°C (Pathology Devices) with a Nikon TE2000-U inverted fluorescence microscope equipped with: Photometrics Cascade II EM-CCD camera and Yokagowa Spinning disc Confocal (CSU-Xm2) (Nikon Instruments, Inc.) and a 100x NA 1.4 oil objective. Images were acquired using Metamorph (v7.0) then contrasted, merged, and scale bars were added using ImageJ.
Fluorescence Recovery after Photobleaching
Transfected Cos-7 cells were imaged in a heated stage chamber at 37°C by confocal microscopy as described above. Metamorph software was used to image cells for 80s at 1s intervals at 500ms exposure. Using the Mosaic Digital Diaphragm System for Nikon TE2000 Microscope (Photonic Instruments), four images were taken pre-bleaching after which a region of 65 × 65 pixels was photobleached for 500ms (Uniblitz VCM-D1 shutter, Vincent Associates) using an Argon laser with wavelength of 450–515nm (National Laser Company) and images were taken at 1s intervals for the remaining time. Raw data was quantified and extracted using MetaMorph Software looking at 3 regions per cell, the photobleached region (PB), a region outside of the cell for background intensity (BG), and a region within the cell outside of the photobleached region to correct for overall fluorescence loss due to imaging (CR). Microsoft Excel was used to normalize the data. First we subtracted BG fluorescence from all time points, as PB-BG = PBbg and CR-BG = CRbg. CRbg was then plotted and fit to the exponential equation CRbg = CR0e−kt to solve for k. Next k was used in the equation PBf = PBbgekt to adjust for loss in fluorescence of the photobleached region. To eliminate the unbleached pool (10–20%) from further calculations, PBf at the time point immediately following the photobleaching event was subtracted from all PBf corrected intensity values. The fluorescence intensity was converted to a percent by averaging the time points before the bleaching event and setting that value to 100% and the time point after photobleaching to 0%. Several individual FRAP experiments where normalized as such, plotted, and subjected to analysis by GraphPadPrism5.
Pearson’s Coefficient of Correlation
Cos-7 cells were co-transfected with indicated constructs and imaged live using confocal fluorescence microscopy in a heated chamber as previously described. Transfected cells used in subsequent analyses had a signal to noise ratio for both channels that was between 3–4:1. Analyses were done using the Colocalization Finder plugin within the ImageJ MBF “ImageJ for Microscopy” Collection. The PC was quantified in at least 9 cells for each experimental combination and results were then entered into GraphPadPrism5 to determine mean values, standard deviation, and standard error of the mean (SEM) for each data set. The p-values were calculated using non-parametric t-test analysis.
Cisternae counting experiments were performed by co-transfecting Cos-7 cells with mCherry-KDEL, and with either GFP-Sec61β, Rtn4a-GFP, or Rtn4aTM2-GFP. Twenty-four hours after transfection, cells were imaged using confocal microscopy at 500ms per wavelength and were scored as either having peripheral ER cisternae (unit 1) or lacking cisternae (unit 0). Cells that had levels of mCherry-KDEL more than 2X above background were also not counted. Three GFP labeled ER proteins were analyzed in this assay co-transfected with mCherry-KDEL. Binomial results were entered in GraphPad Prism 5 to determine percentage of cells containing cisternae, standard deviation, standard error of the mean, and to graph the results. P-values were determined using a non-parametric t-test analysis.
Acknowledgments
We thank G. Odorizzi, W. Prinz, T. Rapoport, and A. Staehelin for helpful discussions and suggestions; J. Friedman and A. English for critical reading of this manuscript; J. Friedman for construction of the BFP-Sec61β plasmid; A. Palmer for the mCherry plasmid, J. Lykke-Andersen for HeLa cell cDNA. NAZ was supported by predoctoral training grant from the National Institutes of Health (NIH), GM07135. GKV was supported by the Searle Scholar Award and NIH grant GM083977.
References
- 1.Baumann O, Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol. 2001;205:149–214. doi: 10.1016/s0074-7696(01)05004-5. [DOI] [PubMed] [Google Scholar]
- 2.English AR, Zurek N, Voeltz GK. Peripheral ER structure and function. Curr Opin Cell Biol. 2009;21(4):596–602. doi: 10.1016/j.ceb.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hetzer MW, Wente SR. Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev Cell. 2009;17(5):606–616. doi: 10.1016/j.devcel.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Craene JO, Coleman J, Estrada de Martin P, Pypaert M, Anderson S, Yates JR, 3rd, Ferro-Novick S, Novick P. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol Biol Cell. 2006;17(7):3009–3020. doi: 10.1091/mbc.E06-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tolley N, Sparkes IA, Hunter PR, Craddock CP, Nuttall J, Roberts LM, Hawes C, Pedrazzini E, Frigerio L. Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic. 2008;9(1):94–102. doi: 10.1111/j.1600-0854.2007.00670.x. [DOI] [PubMed] [Google Scholar]
- 6.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124(3):573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 7.Audhya A, Desai A, Oegema K. A role for Rab5 in structuring the endoplasmic reticulum. J Cell Biol. 2007;178(1):43–56. doi: 10.1083/jcb.200701139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sparkes I, Tolley N, Aller I, Svozil J, Osterrieder A, Botchway S, Mueller C, Frigerio L, Hawes C. Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell. 2010;22(4):1333–1343. doi: 10.1105/tpc.110.074385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol. 2009;187(4):525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson DJ, Hetzer MW. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J Cell Biol. 2008;182(5):911–924. doi: 10.1083/jcb.200805140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu J, Shibata Y, Voss C, Shemesh T, Li Z, Coughlin M, Kozlov MM, Rapoport TA, Prinz WA. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319(5867):1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- 12.Oertle T, Schwab ME. Nogo and its paRTNers. Trends Cell Biol. 2003;13(4):187–194. doi: 10.1016/s0962-8924(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 13.Yang YS, Strittmatter SM. The reticulons: a family of proteins with diverse functions. Genome Biol. 2007;8(12):234. doi: 10.1186/gb-2007-8-12-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan R, Shi Q, Hu X, Zhou X. Reticulon proteins: emerging players in neurodegenerative diseases. Cell Mol Life Sci. 2006;63(7–8):877–889. doi: 10.1007/s00018-005-5338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem. 2008;283(27):18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7(1):9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 17.Hill MM, Bastiani M, Luetterforst R, Kirkham M, Kirkham A, Nixon SJ, Walser P, Abankwa D, Oorschot VM, Martin S, Hancock JF, Parton RG. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell. 2008;132(1):113–124. doi: 10.1016/j.cell.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long KR, Yamamoto Y, Baker AL, Watkins SC, Coyne CB, Conway JF, Aridor M. Sar1 assembly regulates membrane constriction and ER export. J Cell Biol. 2010;190(1):115–128. doi: 10.1083/jcb.201004132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pucadyil TJ, Schmid SL. Conserved functions of membrane active GTPases in coated vesicle formation. Science. 2009;325(5945):1217–1220. doi: 10.1126/science.1171004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224(Pt 3):213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 21.Richter T, Floetenmeyer M, Ferguson C, Galea J, Goh J, Lindsay MR, Morgan GP, Marsh BJ, Parton RG. High-resolution 3D quantitative analysis of caveolar ultrastructure and caveola-cytoskeleton interactions. Traffic. 2008;9(6):893–909. doi: 10.1111/j.1600-0854.2008.00733.x. [DOI] [PubMed] [Google Scholar]
- 22.Machleidt T, Li WP, Liu P, Anderson RG. Multiple domains in caveolin-1 control its intracellular traffic. J Cell Biol. 2000;148(1):17–28. doi: 10.1083/jcb.148.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez I, Ying Y, Albanesi J, Anderson RG. Mechanism of caveolin filament assembly. Proc Natl Acad Sci U S A. 2002;99(17):11193–11198. doi: 10.1073/pnas.172196599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krajewska WM, Maslowska I. Caveolins: structure and function in signal transduction. Cell Mol Biol Lett. 2004;9(2):195–220. [PubMed] [Google Scholar]
- 25.Hansen CG, Nichols BJ. Molecular mechanisms of clathrin-independent endocytosis. J Cell Sci. 2009;122(Pt 11):1713–1721. doi: 10.1242/jcs.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SH, Zhu PP, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, andatlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest. 2010;120(4):1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tolley N, Sparkes I, Craddock CP, Eastmond PJ, Runions J, Hawes C, Frigerio L. Transmembrane domain length is responsible for the ability of a plant reticulon to shape endoplasmic reticulum tubules in vivo. The Plant Journal. 2010 doi: 10.1111/j.1365-313X.2010.04337.x. [DOI] [PubMed] [Google Scholar]
- 28.Voeltz GK, Prinz WA. Sheets, ribbons and tubules -how organelles get their shape. Nat Rev Mol Cell Biol. 2007;8(3):258–264. doi: 10.1038/nrm2119. [DOI] [PubMed] [Google Scholar]
- 29.Itoh T, De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta. 2006;1761(8):897–912. doi: 10.1016/j.bbalip.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 30.Madsen KL, Bhatia VK, Gether U, Stamou D. BAR domains, amphipathic helices and membrane-anchored proteins use the same mechanism to sense membrane curvature. FEBS Lett. 2010;584(9):1848–1855. doi: 10.1016/j.febslet.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 31.Bauer M, Pelkmans L. A new paradigm for membrane-organizing and -shaping scaffolds. FEBS Lett. 2006;580(23):5559–5564. doi: 10.1016/j.febslet.2006.08.077. [DOI] [PubMed] [Google Scholar]
- 32.Bielli A, Haney CJ, Gabreski G, Watkins SC, Bannykh SI, Aridor M. Regulation of Sar1 NH2 terminus by GTP binding and hydrolysis promotes membrane deformation to control COPII vesicle fission. J Cell Biol. 2005;171(6):919–924. doi: 10.1083/jcb.200509095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee MCS, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R. Sar1p N-Terminal Helix Initiates Membrane Curvature and Completes the Fission of a COPII Vesicle. Cell. 2005;122(4):605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 34.Sheetz MP, Singer SJ. Biological membranes as bilayer couples. A molecular mechanism of drug-erythrocyte interactions. Proc Natl Acad Sci U S A. 1974;71(11):4457–4461. doi: 10.1073/pnas.71.11.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatzakis NS, Bhatia VK, Larsen J, Madsen KL, Bolinger PY, Kunding AH, Castillo J, Gether U, Hedegard P, Stamou D. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat Chem Biol. 2009;5(11):835–841. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- 36.Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, McMahon HT. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25(12):2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farsad K, Ringstad N, Takei K, Floyd SR, Rose K, De Camilli P. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J Cell Biol. 2001;155(2):193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]