Abstract
Humans are exposed to radiation through the environment and in medical settings. To deal with radiation-induced damage, cells mount complex responses that rely on changes in gene expression. These gene expression responses differ greatly between individuals1 and contribute to individual differences in response to radiation2. Here we identify regulators that influence expression levels of radiation-responsive genes. We treated radiation-induced changes in gene expression as quantitative phenotypes3,4, and conducted genetic linkage and association studies to map their regulators. For more than 1,200 of these phenotypes there was significant evidence of linkage to specific chromosomal regions. Nearly all of the regulators act in trans to influence the expression of their target genes; there are very few cis-acting regulators. Some of the transacting regulators are transcription factors, but others are genes that were not known to have a regulatory function in radiation response. These results have implications for our basic and clinical understanding of how human cells respond to radiation.
In the past 20 years there has been a large increase in the use of radiation in medical diagnostic procedures and treatment protocols. Radiation is genotoxic and induces DNA damage in human cells. To ensure genomic integrity, cells mount complex responses that depend on changes in gene expression. It has long been known that individuals vary in their sensitivity to radiation. This individual variability is also observed at the gene expression level1. We and others have used genetic studies to identify chromosomal regions and genetic variants that influence expression levels of many genes in human cells at the baseline5–9. Here we extend our analysis by genetic mapping of regulatory elements that influence radiation-induced changes in gene expression.
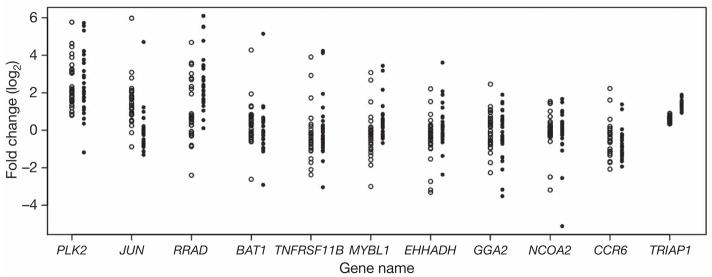
We used microarrays to measure the expression levels of genes in irradiated immortalized B cells from members of 15 Centre d’Étude du Polymorphisme Humain (CEPH) Utah pedigrees10. Data were collected for cells at baseline and at 2 and 6 h after exposure to 10 Gy of ionizing radiation. Of the 10,174 genes on the microarrays that are expressed in immortalized B cells, we focused on 3,280 ‘ionizing-radiation-responsive’ genes that showed at least a 1.5-fold change in gene expression levels at 2 h and/or 6 h after irradiation relative to the baseline. For each of these 3,280 genes we calculated the ratio of expression level at 2 h after irradiation relative to the expression level at the baseline, and the same for the 6 h time point. The log2 of these ratios is the quantitative trait that we mapped to chromosomal locations by genome scans. From here onwards we will refer to the log-transformed expression ratios as the ‘2-h-after-irradiation’ and ‘6-h-after-irradiation’ expression phenotypes. As shown in Fig. 1, the fold change at 2 h and/or 6 h for some of these radiation-responsive genes varies greatly between individuals.
Figure 1. Individual variation in gene expression response to ionizing radiation.
Fold change in 11 radiation-responsive genes; data for each individual are shown as a circle (open circles for 2-h-after-irradiation expression phenotypes; filled circles for 6-h-after-irradiation expression phenotypes). TRIAP1 (TP53-regulated inhibitor of apoptosis 1) is shown as an example of a gene that showed little individual variation in response to ionizing radiation. Other genes show extensive individual variation in gene expression.
Because cellular responses rely in part on changes in gene expression, the extent to which the radiation-responsive genes are induced or repressed influences how cells deal with radiation exposure2. For instance, one of the variable radiation-induced expression phenotypes is the expression level of JUN (Fig. 1). We compared the cellular survival in individuals with low and high JUN induction after exposure to 10 Gy of irradiation. The results showed that in individuals with low JUN induction, cell survival is higher and cell death is lower than in individuals with high JUN induction (Supplementary Fig. 1). To identify the regulators that influence these individual differences in radiation-induced levels of JUN and other responsive genes, we carried out genetic analyses. Supplementary Fig. 2 shows a flow chart of our analyses.
Genotypes for 4,600 single nucleotide polymorphism (SNP) markers were obtained with a standard SNP-based linkage panel. We used the computer program S.A.G.E. v. 5.4 (http://darwin.cwru.edu/) to carry out genome-wide linkage analysis for each of the 3,280 2-h-after-irradiation and 6-h-after-irradiation expression phenotypes in 15 CEPH families. The analysis gives the strength of the evidence for linkage at each map position in the form of a t value, with an associated pointwise significance level11. We selected expression phenotypes for further analysis by using a threshold of t = 4 from the S.A.G.E. analysis; in our sample of families, this corresponds to a P value of 4 × 10−5 (lod score about 3.4) and about P = 0.05 genome-wide12. We found 1,275 (39%) 2-h-after-irradiation phenotypes and 1,298 (40%) 6-h-after-irradiation phenotypes that exceeded this threshold. With a genome-wide threshold of 0.05, among the 3,280 phenotypes we expect 164 at each time point to show linkage evidence anywhere in the genome with a P value this extreme by chance. We found more than 1,250 phenotypes with linkage significant at this level, so we concluded that false positive findings are at most a small fraction of the results. Some of the expression phenotypes have significant evidence of linkage far beyond the t = 4 threshold. In Table 1 we show the expression phenotypes with the most significant linkage results. These include FAM57A and GADD45B, which are known to influence radiation response through the regulation of cell cycle and apoptosis13–15. Figure 2 shows examples of genome scan results for several expression phenotypes.
Table 1.
Phenotypes with the most significant evidence of linkage
| P | Expression phenotype | Location | Cis/trans |
|---|---|---|---|
| 2-h-after-irradiation phenotypes | |||
| <10−11 | ZNF493 | 11q14–q22 | Trans |
| <10−10 | FAM57A | 17q25 | Trans |
| <10−10 | OCLN | 9q34 | Trans |
| <10−10 | HDGF2 | 18q21–22 | Trans |
| <10−9 | PHYH | 15q14–q25 | Trans |
| 6-h-after-irradiation phenotypes | |||
| <10−11 | EDG1 | 3p21–p14 | Trans |
| <10−10 | GADD45B | 17q12–q21 | Trans |
| <10−10 | IFNAR2 | 6q25–q27 | Trans |
| <10−10 | CP110 | 16p13–p12 | Cis |
| <10−9 | SLC25A15 | 13q13–14 | Cis |
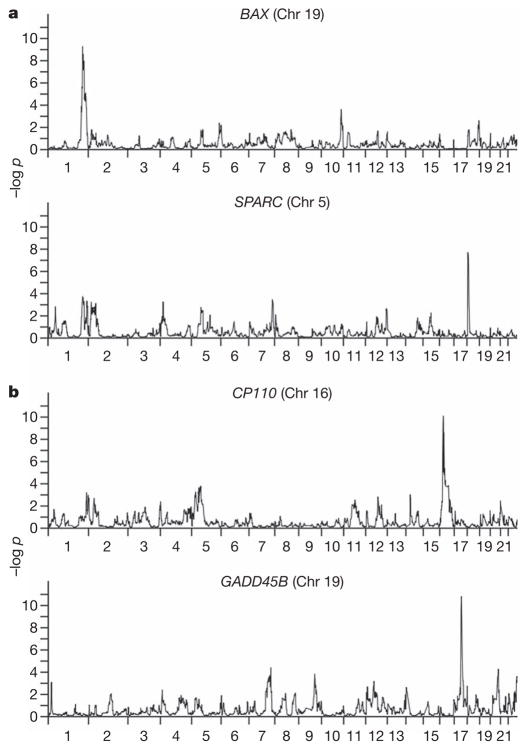
Figure 2. Genome scans for four expression phenotypes.
The chromosomal location of the target gene is given in parentheses. a, Example of 2-h-after-irradiation expression phenotypes. b, Example of 6-h-after-irradiation expression phenotypes.
In comparison with baseline expression, the cis-regulatory and trans-regulatory landscape for radiation-induced gene expression is quite different. We considered cis regulators to be those that were mapped within 5 megabases (Mb) of the target gene5, and all other significant linkage findings to represent trans regulators. Of the 1,275 2-h-after-irradiation phenotypes with significant linkage anywhere (t > 4), only 9 (less than 1%) were cis regulated. Similarly, among the 1,298 6-h-after-irradiation phenotypes, 12 (less than 1%) were cis regulated. The remaining phenotypes were trans regulated. In contrast, for the baseline gene expression phenotypes, we found that about 20% of the phenotypes had a cis-acting regulator and about 80% had a trans-acting regulator5,16.
Hotspots are genomic regions that probably contain regulators influencing the expression levels of many genes4,5,17. To identify these regions we divided the autosomal genome into 554 windows of 5 Mb each and determined the number of regulators mapping to each window. We examined the regulators for the 1,275 2-h-after-irradiation phenotypes and the 1,298 6-h-after-irradiation phenotypes with P < 4 × 10−5. We found several windows that contained many more ‘hits’ than would be expected by chance. If the regulators were randomly distributed across the genome, the probability of 18 or more ‘hits’ within a 5-Mb window would be less than 3 × 10−5. Instead, we found four hotspots with 18 or more hits for the 2-h-after-irradiation phenotypes, and two such hotspots for the 6-h-after-irradiation phenotypes. Table 2 shows the phenotypes that mapped to each of these regions. Because these hotspots are 5 Mb in size, it is possible that they contain more than one regulator of gene expression. The target genes whose regulators mapped to the same hotspots seem to have similar functions or are located very close to each other. For example, among the 19 phenotypes whose expression levels map to the regulatory region on chromosome 2 (the 35–40-Mb window) are three genes (RASL11B, RAP1GDS1 and RASA1) that encode proteins belonging to the RAS family. Two of the target genes in the regulatory region on chromosome 9 (the 0–5-Mb window), CD70 and TNFSF14, are adjacent to each other on chromosome 19. These observations suggest co-regulation of genes that have similar functions or are closely linked.
Table 2.
Hotspots and the phenotypes whose regulators mapped to these regions
| Region | Phenotypes |
|---|---|
| 2-h-after-irradiation phenotypes | |
| Chromosome 9: 0–5 Mb | ASPM, STK17B, DOCK10, C4ORF29, PIK3R1, MEF2C, BAT3, RFX3, RXRA, CSTF3, SLC37A4, LOC55652, BCL7A, PFAAP5, TMCO3, ACTN1, TIPIN, SKIP, MYH10, CD70, TNFSF14 |
| Chromosome 20: 55–60 Mb | HOOK1, DNAJC6, PDE4B, RETSAT, RAPGEF2, BXDC2, PRPF4B, C6ORF32, MLLT3, CTSC, DRAM, CKAP4, LHFP, MEIS2, EFTUD1, PBEF1, MGC14376, CDK5R1, TADA2L, KLHL12 |
| Chromosome 2: 35–40 Mb | FUBP1, CXCR4, SLC4A7, WDR48, RASL11B, RAP1GDS1, RASA1, MAN1A1, TWISTNB, STAU2, NACAP1, REXO2, FAM62A, RNASE6, C14ORF111, CENPB, RPS21, SLC5A3, ATP6AP2 |
| Chromosome 2: 225–230 Mb | MR1, ZFP36L2, RY1, LAMP3, LETM1, KIAA0922, POLR3G, PRPF4B, RRS1, STIP1, PDGFD, MLL, ITPR2, TNFSF11, TRAC, NQO1, ARHGDIA, C18ORF24, APOBEC3G |
| 6-h-after-irradiation phenotypes | |
| Chromosome 20: 5–10 Mb | C2ORF3, NCAPG, CENTD1, IL8, HLA-DPA1, ASCC3, ZMIZ2, CSTB, RAD54B, ADM, KLRA1, SMUG1, PTPRU, CPM, ING1, XYLT1, TADA2L, KNT2C, CYP4F3 |
| Chromosome 10: 0–5 Mb | KIAA0492, STK17B, NEK1, EST, MCM4, GTPBP4, TSPAN14, ARHGAP19, DEAF1, EP400, USP10, NPTN, ST3GAL2, WIPF2, ZNP133 |
Next we followed up results of the linkage scans by family-based association analysis, to confirm the linkage findings and obtain much finer resolution within the regions of linkage. Because there are many significant linkage findings, we focused on phenotypes that showed the largest fold induction or repression in response to ionizing radiation. We selected the expression phenotypes that showed at least a twofold change in expression at 2 h and/or 6 h after irradiation compared to the baseline and had at least one marker with significant evidence (t = 4, P < 4 × 10−5) of linkage. There were 182 2-h-after-irradiation and 164 6-h-after-irradiation expression phenotypes that met these criteria.
Of these 346 (182 + 164) phenotypes, 6 were cis regulated. We tested each of these cis-regulated genes for association by the quantitative transmission disequilibrium test (QTDT)18, using SNP markers within the target genes and 5 kilobases (kb) upstream and downstream. The QTDT results showed evidence (nominal P ≤ 0.05) for association (and linkage) for five of these six expression phenotypes, thus supporting the linkage findings that these phenotypes are cis regulated. The five phenotypes with cis regulation are the 2-h-after-irradiation phenotypes of NDUT15, PMAIP1 and PTGER4, and the 6-h-after-irradiation phenotypes of CP110 and PHLDA3.
For the remaining 340 phenotypes (178 at 2 h and 162 at 6 h), the linkage evidence suggests that their expression levels are regulated by trans-acting regulators, giving rise to ‘trans-peaks’. One of these phenotypes is the radiation-induced expression level of BAX (Fig. 2a), which has a role in apoptosis. We found a highly significant linkage peak on chromosome 1 (P <10−9). This candidate region contains the gene TP53BP2, a known regulator of BAX19. We confirmed the linkage finding by family-based association analysis, which showed significant evidence (P < 0.02) for the combined presence of linkage and association at several markers within and near TP53BP2. These results illustrate that genetic analysis allows the identification of polymorphic trans-acting regulators of gene expression. This is important because few trans-acting regulators have previously been identified in studies of the genetics of human gene expression.
Under most of the trans-peaks, unlike that for BAX, there are no obvious candidates. Often the linkage peaks are megabases in size and include several genes that are possible candidates. To limit the number of potential regulators for follow-up analyses, we used Chilibot20 to look for co-occurrence of the names of the putative regulator and target genes in the literature and for interactive relationships between them. We used this text-mining program to search for interactive relationships in any cell types and conditions; we did not limit the searches to immortalized B cells or to radiation response. The literature search resulted in 73 potential regulators for 32 expression phenotypes. We performed QTDT analysis using SNP markers within the putative regulators and 5 kb upstream and downstream of these ‘regulatory’ genes. We further limited our analysis to genes with at least one ‘informative’ SNP (defined as 30 informative probands for a SNP). This reduced our QTDT analysis from 73 to 58 regulators for 29 expression phenotypes. The results showed evidence (nominal P < 0.05) for combined linkage and association for 13 of these 29 phenotypes (2 unlinked regulators for expression of FAS 6 h after irradiation). Table 3 shows the linkage and association results for these 13 phenotypes and their corresponding trans regulators. We also regressed the expression levels of these 13 expression phenotypes onto SNP markers in their corresponding regulators. Despite the small sample size (30 individuals), significant evidence of association was found between some radiation-induced expression phenotypes and SNP alleles in their trans regulators (see Supplementary Table 1). These results show that polymorphisms in the trans regulators contribute to individual differences in radiation-induced gene expression.
Table 3.
Family-based association analysis in candidate regions identified by linkage scans
| Radiation-induced gene expression phenotype | Time after irradiation (h) | Potential regulator | GO function | Linkage (P) | Chromosome | QTDT SNP ID | QTDT (P) |
|---|---|---|---|---|---|---|---|
| ARHGDIA | 2 | SERPINE2 | Serine peptidase inhibitor | 3.21 × 10−7 | 2 | rs6734100 | 0.0002 |
| HRAS | 2 | RB1 | Transcription factor | 5.40 × 10−6 | 13 | rs198584 | 0.0033 |
| JUN | 2 | LCP2 | Transmembrane receptor signalling pathway | 1.46 × 10−9 | 5 | rs10068619 | 0.0213 |
| BIRC4 | 2 | CDK9 | Cyclin-dependent kinase | 6.80 × 10−6 | 9 | rs1002095 | 0.0305 |
| TNFSF9 | 2 | CD44 | Receptor | 1.38 × 10−5 | 11 | rs353615 | 0.0321 |
| HMMR | 2 | CDC2 | Cyclin-dependent kinase | 1.10 × 10−6 | 10 | rs1871445 | 0.0385 |
| RFX3 | 2 | HIVEP2 | Regulation of transcription | 6.00 × 10−7 | 6 | rs533173 | 0.0442 |
| TRAF4 | 6 | FAS | Signal transduction | 4.00 × 10−7 | 10 | rs9658786 | 0.0044 |
| TNFSF4 | 6 | LTA4H | Zinc ion binding | 3.38 × 10−7 | 12 | rs7296106 | 0.0057 |
| SPARC | 6 | VDR | Transcription factor activity | 1.85 × 10−7 | 12 | rs2254210 | 0.0077 |
| RFX3 | 6 | DYNC2LI1 | Motor activity | 4.00 × 10−7 | 2 | rs2278356 | 0.01 |
| NT5E | 6 | NDUFB6 | Mitochondrial electron transport | 5.48 × 10−9 | 9 | rs628425 | 0.0179 |
| FAS | 6 | SSB | mRNA binding | 1.20 × 10−6 | 2 | rs7589845 | 0.0215 |
| FAS | 6 | UBA52 | Ribosome/protein modification | 1.13 × 10−8 | 19 | rs2057649 | 0.0296 |
To confirm the cis-linkage and association findings, we conducted a differential allelic gene expression analysis. One of the cis-regulated phenotypes is expression of CP110 (Table 1), which encodes a protein involved in centrosome duplication. Disruption of CP110 leads to unscheduled centrosome duplication21, radiation-induced chromosomal instability and cell death22. To confirm that the radiation-induced level of CP110 is cis regulated, we used quantitative PCR to measure allele-specific changes in CP110 expression levels in irradiated cells from 12 unrelated CEPH individuals who are heterozygous at an exonic SNP (rs179050) in CP110. We found that on average the radiation-induced expression of the T allele at rs179050 was 12% (range 5–40%) more than that of the C allele.
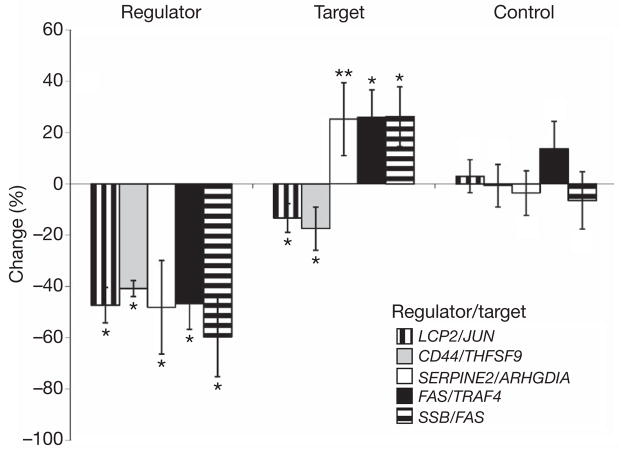
To confirm the findings of trans-acting regulators, we used short interfering RNA (siRNA) to knock down the potential regulators and then measured the expression of the corresponding target genes. We assessed the effect of the knockdown by measuring the expression of the regulators and the corresponding target genes in irradiated cells. Results from our association studies had identified 14 potential regulators for 13 radiation-induced expression phenotypes. Successful knockdown was achieved for 11 of the 14 regulators. The expression levels of the regulators decreased by about 30% to about 70% after the genes were silenced, whereas no changes in expression of the regulators were observed when siRNAs with no sequence homology to the regulators were used. We then measured the expression levels of the target genes and found corresponding changes in expression levels of five of them (Fig. 3). Expression levels of the target genes did not change after knockdown of GAPDH (Supplementary Fig. 3a), indicating that the changes in expression levels of the target genes were specific effects of silencing their regulators. These results provide molecular support for the regulator–target-gene relationships identified by our genetic analyses.
Figure 3. Results of knockdown of five regulators of radiation-expression phenotypes.
Regulators of expression levels of radiation-responsive genes were knocked down by siRNAs. Changes in expression levels of the regulators and their corresponding target genes after knockdown of the regulators are shown, as are changes in expression levels of a control gene (encoding glyceraldehyde-3-phosphate dehydrogenase) after knockdown of the regulators. Data are shown as means ± s.e.m. for four or more independent transfections. Asterisk, P < 0.05; two asterisks, P = 0.07; no symbol, P > 0.05.
The five regulator–target-gene pairs that we validated molecularly were: LCP2 (also known as SLP-76) as regulator of expression of JUN, CD44 as regulator of TNFSF9, FAS as regulator of TRAF4, SERPINE2 as regulator of ARHGDIA, and SSB as regulator of FAS (Fig. 3). Of these five regulators, only FAS is a known regulator of signal transduction in response to ionizing radiation. Others such as SERPINE2 and SSB are not previously known to regulate cellular response to radiation.
Changes in expression of the target genes after knockdown of the regulators support the regulator–target-gene relationships identified by genetic mapping. However, the lack of changes in target gene expression does not argue against the regulatory relationship. The expression levels of the regulators were only partly decreased by the siRNAs; depending on the regulatory mechanism, partial expression may be sufficient to influence the response of a target gene to radiation. In addition, because gene expression response to radiation exposure is critical for survival, there are probably backup mechanisms that will influence the target gene expression even after the main regulator is silenced.
We began with gene expression response to radiation because cellular responses rely on changes in gene expression. We also performed an association analysis to test for allelic association with radiation-induced cell death. We focused on the regulators that were confirmed by QTDT; these included 5 cis regulators, and the 14 trans regulators in Table 3. We measured cell death in irradiated cells from 30 unrelated individuals (parents of the 15 CEPH families) using cytotoxicity and caspase assays, and then tested for evidence of association of these measurements with SNPs within and 5 kb upstream and downstream of the regulators by linear regression. Of the 19 regulators, there were 5 with SNP alleles that were significantly (P < 0.05) associated with cytotoxicity and caspase levels (Table 4). This shows that DNA variants in these polymorphic regulators not only affect gene expression but also cellular response to radiation.
Table 4.
SNPs significantly associated with radiation-induced cytotoxity and caspase levels
| Regulator | Cis/trans | SNP | Major allele | Phenotype | Average activity* |
R2 | P | ||
|---|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | |||||||
| PHLDA3 | Cis | rs3888929; GA | G | Caspase | 2.7 | 3.0 | 4.4 | 0.15 | 0.033 |
| Cytotoxicity | 2.2 | 2.7 | 2.6 | 0.16 | 0.028 | ||||
| LCP2 | Trans | rs4867592; CA | C | Caspase | 2.4 | 3.1 | 4.0 | 0.33 | 0.001 |
| Cytotoxicity | 2.9 | 4.4 | 4.3 | 0.23 | 0.008 | ||||
| LTA4H | Trans | rs7970524; TC | T | Caspase | 3.2 | 2.4 | NA | 0.18 | 0.018 |
| Cytotoxicity | 2.8 | 1.7 | NA | 0.18 | 0.021 | ||||
| NDUFB6 | Trans | rs12003093; AG | A | Caspase | 2.6 | 3.2 | 4.3 | 0.23 | 0.007 |
| Cytotoxicity | 3.1 | 4.1 | 5.5 | 0.26 | 0.004 | ||||
| VDR | Trans | rs4760658; AG | A | Caspase | 2.8 | 2.9 | 4.5 | 0.18 | 0.018 |
| Cytotoxicity | 1.9 | 2.8 | 3.2 | 0.16 | 0.027 | ||||
Average activity compared with unirradiated cells; A represents the major allele, and B the minor allele.
In this study we used genetic linkage and association analyses to identify DNA variants that influence the expression levels of genes in irradiated cells. The results allowed us to characterize the global regulatory landscape of gene expression response to radiation and will facilitate the development of genetic tools for radiosensitivity. First, we found that the regulatory landscape of gene expression in irradiated cells is quite different from that of cells at the baseline. Of the gene expression phenotypes in cells at the baseline, at least 20% are cis regulated5,16,23. In contrast, less than 1% of the radiation-induced expression phenotypes are cis regulated; almost all of the phenotypes are regulated by trans-acting factors. The large number of trans-acting regulators provides cells with multiple mechanisms to mount responses to different types of stressors, and because trans regulators can influence the expression of several genes, it also permits coordinated responses. In C. elegans24 and yeast25, trans regulators are also found more frequently than cis regulators in influencing gene expression response to external stimuli, suggesting that this may be a phenomenon that is seen across different organisms.
Second, by combining results from genetic mapping and molecular validation studies, we have identified regulatory regions and regulators that influence radiation-induced changes in gene expression. The results allowed us to uncover polymorphic regulators of gene expression response to radiation. One might expect many of the trans-acting regulators to be transcription factors; indeed, three of them (RB1, HIVEP2 and VDR) are. The remaining regulators include a cell-surface receptor (CD44) and genes that have few known functions (DYNC2LI1 and UBA52). These results indicate that many genes other than transcription factors have a function in regulating gene expression. Functions of a large number of human genes remain unknown; the genetics of gene expression is a powerful approach for identifying those that have a regulatory role.
Last, our results have medical implications. Individuals differ in their response to radiation. The regulatory variants identified in this study will enable the genetic prediction of individual sensitivity to radiation. In addition, the identification of genes involved in regulating radiation response will enable the development of radiosensitizers that increase the sensitivity of tumours to radiation. Together, information on patients’ sensitivity to radiation and methods of sensitizing malignant cells to radiation will improve outcomes of radiotherapies. Thus, the results of studies of genetics of human gene expression expand our understanding of gene regulation and have clinical implications.
METHODS SUMMARY
The data were from parents and a mean of 8 offspring per sibship (range 7–9) of 15 CEPH families (CEPH 1333, 1341, 1346, 1362, 1408, 1416, 1420, 1421, 1423, 1424, 1444, 1447, 1451, 1454 and 1582). For the expression analysis, immortalized B cells were grown at a density of 5 × 105 cells ml−1 and irradiated at 10 Gy in a 137Cs irradiator. Cells were harvested before irradiation and at 2 h and 6 h after exposure to ionizing radiation. RNA was extracted from the cells, labelled and hybridized onto Affymetrix Human U133A 2.0 arrays. For the genetic studies, low-density genotypes for 4,600 autosomal SNP markers were obtained with the Illumina Linkage Panel (v. 3) and high-density genotypes were obtained from the HapMap Project26 and by inference27. Multipoint genome-wide linkage analysis was done by SIBPAL in S.A.G.E. (http://darwin.cwru.edu/) using the ‘W4’ option28. Family-based association analysis with SNP markers within, and 5 kb upstream and downstream of the target genes (cis) or candidate regulators (trans) was carried out with QTDT software, using the orthogonal (ao) model and variance component options (wega)18.
Various cellular and molecular analyses were performed. Cell death was measured 24 h after irradiation by using the multitox-fluor multiplex cytotoxicity assay (Promega) and caspase-glo 3/7 assay (Promega). Allele-specific quantitative PCR was performed with complementary DNA samples as templates and TaqMan SNP genotyping assay for rs179050 (Applied Biosystems). For knockdown assays, immortalized B cells were transfected with Accell siRNA pools (Dharmacon) against candidate regulators or non-target control in accordance with the manufacturer’s instructions. After transfection, the cells were irradiated at 10 Gy in a 137Cs irradiator and RNA was harvested at the indicated times after irradiation. The effect of siRNA on gene expression was analysed by quantitative PCR.
METHODS
CEPH samples and expression phenotyping
The data were from parents and a mean of eight offspring per sibship (range seven to nine) of 15 CEPH families (CEPH 1333, 1341, 1346, 1362, 1408, 1416, 1420, 1421, 1423, 1424, 1444, 1447, 1451, 1454 and 1582). For the expression analysis, immortalized B cells (lymphoblastoid cells) were grown at a density of 5 × 105 cells ml−1 in RPMI 1640 with 15% fetal bovine serum, 2 mM L-glutamine and 100 U ml−1 penicillin/streptomycin and irradiated at 10 Gy in a 137Cs irradiator. Cells were harvested before irradiation and at 2 h and 6 h after exposure to ionizing radiation. RNA was extracted from the cells and hybridized onto Affymetrix Human U133A 2.0 arrays. We used a random number generator to determine the order in which the cells were grown, and array hybridizations were performed; cells from family members were not processed together except by chance. The baseline, 2-h-after-irradiation and 6-h-after-irradiation samples for each individual were processed together. The cRNA samples were prepared in a total of four batches (about 96 samples per batch). Hybridizations were performed in batches of 48 samples. Expression intensity was scaled to 500 using the global scaling method implemented in the Expression Console software (Affymetrix) and transformed by log2. We defined ‘radiation-responsive’ genes as those that showed at least a 1.5-fold change in gene expression levels in 6 or more of 30 unrelated individuals. We focused on these 3,280 radiation-responsive genes in this study.
Viability, cytotoxicity and caspase activity measurements
Immortalized B-cell lines from 30 CEPH parents and individuals with high (GM07048, GM10834 and GM10861) or low (GM10842, GM10843 and GM12752) radiation induction of JUN expression were irradiated at 10 Gy in a 137Cs irradiator. The cells were then analysed 24 h after irradiation with a multitox-fluor multiplex cytotoxicity assay (Promega) and a caspase-glo 3/7 assay (Promega).
Genotypes
Low-density genotypes for 4,600 autosomal SNP markers were obtained with the Linkage Panel, v. 3 (Illumina). We used PEDSTATS29 to check for Mendelian inconsistencies. This resulted in the removal of 48 genotypes at four distinct SNP markers. High-density genotypes for family-based association (QTDT) were obtained by inference using the low-density genotypes and high-density genotypes on selected individuals27.
Analysis of linkage and association
Multipoint genome-wide linkage analysis was done by SIBPAL in S.A.G.E. v. 5.4 (http://darwin.cwru.edu/) using the ‘W4’ option28. SIBPAL determines evidence for linkage at each SNP from regression of the phenotype difference between siblings on the estimated proportion of marker alleles shared identical-by-descent between siblings; the result is reported as a t value with corresponding significance, as given in the text. Pointwise significance was converted to genome-wide significance12. In permutation analysis by S.A.G.E. of the results for 14 phenotypes (6 from Table 1, and 8 phenotypes with t values close to 5: 4.99–5.4), we found one phenotype with two t values over 5 and one phenotype with seven t values over 5 among 100,000 replicates. For the other 12 phenotypes (including the 6 from Table 1), we did not find any t values over 5 in 100,000 permutations for each phenotype.
Family-based association analysis with SNPs near and within the target genes or candidate regulators was performed with QTDT. We used the orthogonal (ao) model18 and variance component options (wega). For association analysis, after irradiation the expression phenotype, cytotoxicity or caspase level, as dependent variable, was regressed on SNP genotypes (coded 0, 1 or 2). R2 was estimated for each phenotype–SNP combination as the ratio of the regression sum of squares to the total sum of squares.
Hotspots
The autosomal genome was divided into 554 windows of 5 Mb each (the last window of the q arm of a chromosome can be smaller). For each of the 1,275 2-h-after-irradiation phenotypes and 1,298 6-h-after-irradiation phenotypes, we considered all SNPs with t > 4, P < 4 × 10−5. Any window with one or more such SNPs was counted as having one ‘hit’ for that phenotype. Some phenotypes had more than one hit in the genome because some had multiple linkage peaks, or because peaks for some phenotypes were broad and spanned adjacent 5-Mb windows; in this case, each window was counted as having one hit. There were 3,151 such hits for the 2-h-after-irradiation phenotypes and 3,120 hits for the 6-h-after-irradiation phenotypes. We assumed that if the hits were distributed randomly across the genome, their distribution over windows would be approximately Poisson, with a mean of 5.69 and 5.63 for the 2-h and 6-h phenotypes, respectively (3,151/554 and 3,120/554).
SNP genotyping and allele-specific RT–PCR
Allele-specific quantitative PCR was performed with cDNA samples as templates, using TaqMan primer for rs179050 (Applied Biosystems). A standard curve for the quantitative PCR was produced with genomic DNA from individuals homozygous for rs179050. The standard curve showed that genomic DNA from the individuals heterozygous at that SNP contained about 50% of the T and C alleles as expected. The standard curve was then used to quantify the T:C allele in the 12 cDNA samples at the baseline and 6 h after irradiation.
Knockdown of candidate regulators
Immortalized B cells were transfected with Accell siRNAs (Dharmacon) against candidate regulators or non-target control in accordance with the manufacturer’s instructions. For each regulator we used a pool of siRNAs that targeted the regulators, to minimize off-target effects30. After transfection, the cells were irradiated at 10 Gy in a 137Cs irradiator, and RNA was harvested after irradiation. In addition to the siRNAs, we also used short hairpin RNA (shRNA) to knock down two regulators, LCP2 and CD44. Although the shRNAs were not as effective as the siRNAs in the knockdown, similar trends were seen, further supporting the regulator–target-gene relationships (Supplementary Fig. 3b). MISSION shRNA plasmid (Sigma-Aldrich) transfections were performed with the nucleofector cell line nucleofector kit V (Amaxa), in accordance with the manufacturer’s instructions. The effect of siRNA (or shRNA) on gene expression was analysed by quantitative PCR. Expression of β-actin was used as a control for normalization, and changes in expression were calculated relative to cells transfected with non-target control siRNA or shRNA. Sequences of PCR primers, siRNAs and shRNAs are presented in Supplementary Tables 2–4.
Supplementary Material
Acknowledgments
We thank D. George and W. Ewens for advice and discussion, A. Bruzel, S. Solomon, T. Weber and K. Halasa for technical help, and C. McGarry for manuscript preparation. Some analyses for this paper were performed by using the program package S.A.G.E., which is supported by a grant from the National Center for Research Resources. This work is supported by grants from the National Institutes of Health (to V.G.C. and R.S.S.), by seed grants from the University of Pennsylvania Center for Excellence in Environmental Toxicology (to V.G.C.), by the W. W. Smith Endowed Chair (to V.G.C.) and the Howard Hughes Medical Institute (to V.G.C.).
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information The microarray data are deposited in the GEO database under accession number GSE12626. Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Correa CR, Cheung VG. Genetic variation in radiation-induced expression phenotypes. Am J Hum Genet. 2004;75:885–890. doi: 10.1086/425221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amundson SA, et al. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res. 2008;68:415–424. doi: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
- 3.Cheung VG, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nature Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 4.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 5.Morley M, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung VG, et al. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 8.Dixon AL, et al. A genome-wide association study of global gene expression. Nature Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 9.Stranger BE, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dausset J, et al. Centre d’Étude du Polymorphisme Humain (CEPH): collaborative genetic mapping of the human genome. Genomics. 1990;6:575–577. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- 11.Haseman JK, Elston RC. The investigation of linkage between a quantitative trait and a marker locus. Behav Genet. 1972;2:3–19. doi: 10.1007/BF01066731. [DOI] [PubMed] [Google Scholar]
- 12.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nature Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 13.Pan D, et al. Down-regulation of CT120A by RNA interference suppresses lung cancer cells growth and sensitizes to ultraviolet-induced apoptosis. Cancer Lett. 2006;235:26–33. doi: 10.1016/j.canlet.2005.03.045. [DOI] [PubMed] [Google Scholar]
- 14.Vairapandi M, Balliet AG, Hoffman B, Liebermann DA. GADD45b and GADD45g are cdc2/cyclinB1 kinase inhibitors with a role in S and G2/M cell cycle checkpoints induced by genotoxic stress. J Cell Physiol. 2002;192:327–338. doi: 10.1002/jcp.10140. [DOI] [PubMed] [Google Scholar]
- 15.Selvakumaran M, et al. The novel primary response gene MyD118 and the proto-oncogenes myb, myc, and bcl-2 modulate transforming growth factor β1-induced apoptosis of myeloid leukemia cells. Mol Cell Biol. 1994;14:2352–2360. doi: 10.1128/mcb.14.4.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price AL, et al. Effects of cis and trans genetic ancestry on gene expression in African Americans. PLoS Genet. 2008;4:e1000294. doi: 10.1371/journal.pgen.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rockman MV, Kruglyak L. Genetics of global gene expression. Nature Rev Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 18.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samuels-Lev Y, et al. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell. 2001;8:781–794. doi: 10.1016/s1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- 20.Chen H, Sharp BM. Content-rich biological network constructed by mining PubMed abstracts. BMC Bioinformatics. 2004;5:147. doi: 10.1186/1471-2105-5-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z, et al. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev Cell. 2002;3:339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- 22.Sato N, Mizumoto K, Nakamura M, Tanaka M. Radiation-induced centrosome overduplication and multiple mitotic spindles in human tumor cells. Exp Cell Res. 2000;255:321–326. doi: 10.1006/excr.1999.4797. [DOI] [PubMed] [Google Scholar]
- 23.Pant PV, et al. Analysis of allelic differential expression in human white blood cells. Genome Res. 2006;16:331–339. doi: 10.1101/gr.4559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, et al. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2006;2:e222. doi: 10.1371/journal.pgen.0020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith EN, Kruglyak L. Gene-environment interaction in yeast gene expression. PLoS Biol. 2008;6:e83. doi: 10.1371/journal.pbio.0060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burdick JT, Chen WM, Abecasis GR, Cheung VG. In silico method for inferring genotypes in pedigrees. Nature Genet. 2006;38:1002–1004. doi: 10.1038/ng1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shete S, Jacobs KB, Elston RC. Adding further power to the Haseman and Elston method for detecting linkage in larger sibships: weighting sums and differences. Hum Hered. 2003;55:79–85. doi: 10.1159/000072312. [DOI] [PubMed] [Google Scholar]
- 29.Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 30.Myers AJ, et al. Minimizing off-target effects by using diced siRNAs for RNA interference. J RNAi Gene Silencing. 2006;2:181–194. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.