Abstract
Tumor cells display a different profile of gene expression than their normal counterparts. Perturbations in the levels of cellular splicing factors can alter gene expression, potentially leading to tumorigenesis. We found that splicing factor SRp20 (SFRS3) is highly expressed in cancers. SRp20 regulated the expression of Forkhead box transcription factor M1 (FoxM1) and two of its transcriptional targets, PLK1 and Cdc25B, and controlled cell cycle progression and proliferation. Cancer cells with RNAi-mediated reduction of SRp20 expression exhibited G2/M arrest, growth retardation, and apoptosis. Increased SRp20 expression in rodent fibroblasts promoted immortal cell growth and transformation. More importantly, we found that SRp20 promoted tumor induction and the maintenance of tumor growth in nude mice and rendered immortal rodent fibroblasts tumorigenic. Collectively, these results suggest that increased SRp20 expression in tumor cells is a critical step for tumor initiation, progression, and maintenance.
Keywords: Cancer, splicing factors, SFRS3, SRp20, G2/M arrest, cell transformation, tumor induction
Introduction
Alternative RNA splicing, a principal molecular event for the gene expression of approximately 70% of all human genes 1,2, increases the coding capacity of the human genome by producing different isoforms from a single pre-mRNA molecule. The regulation of alternative splicing involves interactions between cellular splicing factors and RNA sequences in the pre-mRNA 3,4 and can easily be perturbed by relatively small changes in the levels of splicing factors 5,6. Although alternative splicing events have recently emerged as an important focus in molecular and clinical oncology 7-9, the contribution of alternative splicing to cancer development is poorly understood.
SRp20, recently renamed as SFRS3 10, is a splicing factor that affects alternative splicing by interacting with RNA cis-elements in a concentration- and cell differentiation-dependent manner 11,12. It is the smallest member of the SR protein family 13. In addition to its regulation of RNA splicing, SRp20 plays important roles in cellular functions including termination of transcription 14, alternative RNA polyadenylation 15, RNA export 16,17, and protein translation 18. Mouse embryos lacking SRp20 do not form a blastocyst 19. SRp20 expression is higher than normal in ovarian cancers 20; however, the effect of this increased expression is unclear. Overexpressed SRp20 might alter the RNA splicing and other events of many genes in mammalian cells, thereby substantially affecting the expression levels of various protein isoforms 21. Here we provide evidence that SRp20 is overexpressed in many cancer types and that the increase in SRp20 is essential for cancer cell survival and oncogenesis. In addition, we found that SRp20 is critical for controlling the cell G2/M phase transition and for preventing cell apoptosis.
Materials and methods
Plasmids, cells, and tissue lysates
A T7-SRp20 expression vector (plasmid pJR17) was constructed by swapping the T7-SRp20 coding region from a pCG-T7-SRp20 expression vector (Tom Misteli of NCI and Javier Caceres of Edinburgh) into the pRevTRE vector to put T7-SRp20 under the control of tetracycline.
Immortal rodent fibroblast NIH 3T3 and MEF 3T3 tet-off cells (Clontech) and human C33A, 786-O, U2OS, HeLa, CaSki, WI-38, and MRC-5 cells were grown in Dulbecco's modified Eagle medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS) or calf serum (HyClone), 2 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. The B-cell lymphoma-derived cell lines BCBL-1, JSC-1, and SUDHL-6 were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS. Primary human bronchial epithelial cells (HBEpiC) and primary human renal epithelial cells (HREC) were obtained and grown in medium from ScienCell Research Laboratories. HBEpiC were grown in bronchial epithelial cell medium, and HREC were grown in DMEM supplemented with 10% FBS and Epithelial Cell Growth Supplement (ScienCell Research Laboratories). Primary newborn human foreskin keratinocytes (HFKn) were purchased from Invitrogen and grown in calcium-free Medium 154CF plus human keratinocyte growth supplement (Invitrogen). Peripheral blood mononuclear cells were obtained from healthy blood donors at the NIH Clinical Center blood bank. MEF 3T3 tet-off cells were transfected with plasmid pJR17 (T7-SRp20) by Lipofectamine 2000 (Invitrogen), and selected with 200 μg/mL hygromycin and 1 μg/mL doxycycline for stable transfection. A cell line stably transfected with the empty vector, pRevTRE, was also established as a control. To express T7-SRp20, the stably transfected MEF3T3 tet-off cells were grown in doxycycline-free DMEM growth medium.
Tissue lysates of various pairs of tumor and matched normal tissues from different organs were purchased from Protein Biotechnologies.
RNAi
Synthetic, double-stranded siRNAs were obtained from Dharmacon, Inc. SRp20 and hnRNP U siRNAs were purchased as an siGenome SMARTpool (human SRp20, cat. No. M-030081-00; human hnRNP U, cat. No. J-013501-00). Human YB-1 siRNA is a mixture of siRNAs 393, 394, and oJR1, as described 11. The nonspecific (NS) siRNA has 52% GC content (cat. No. D-001206-08-20). The SRp20 siRNA s12732, targeting a splice junction of SRp20 exon 2 and exon 3, was purchased from Ambion and SRp20 siRNA D-03 was obtained separately from Dharmacon. RNAi was conducted by two or three separate transfections, at intervals of 48 h, with 10 nM (Ambion) or 40 nM (Dharmacon) siRNA in the presence of Lipofectamine 2000. BCBL-1 and JSC-1 lymphoma cell lines were transfected with 40 nM siRNA using the HiPerfect transfection reagent (Qiagen) by three separate transfections in accordance with the instructions of the manufacturer. The cells with knocked-down SRp20, YB-1, or hnRNP U expression were then analyzed for cell number by trypan blue exclusion, for cell cycle by flow cytometry, for RNA splicing by RT-PCR, for protein expression by Western blotting, and for tumor induction by nude mouse injection.
HEKn cells at 2 × 105 cells per well in 12-well plates were transfected 4 h after passage 3 (day 0) with 40 nM SRp20 siRNA or NS siRNA by using Lipofectamine 2000. Cells were transfected again on days 2 and 4 without passage and were counted on day 6. HBEpiC cells at 2.5 × 105 cells per well in 6-well plates were transfected on days 1 and 3 with siRNA as described above for HFKn and were counted on day 5. HREC cells at 2 × 104 cells per well in 24-well plates were transfected on days 1 and 3 with siRNAs as described above for HFKn and were counted on day 5.
Western blot
Protein samples in 2× SDS sample buffer were denatured by boiling for 5 min, separated by NuPAGE Bis-Tris gel electrophoresis (Invitrogen), transferred onto a nitrocellulose membrane, and blotted with the following antibodies: mouse monoclonal antibodies against SRp20 (7B4, American Type Culture Collection), beta-tubulin (BD PharMingen), hnRNP K (Santa Cruz Biotech), and PARP (Calbiochem), or rabbit antibodies against PLK1 (Millipore/Upstate), N-terminal FoxM1 (K19), and Cdc25B (Santa Cruz Biotech).
RNA preparation and RT-PCR
Total cell RNA was prepared from the cells using TRIzol (Invitrogen) following the manufacturer's instructions. After DNase I treatment, 1 μg of RNA was reverse transcribed at 42oC using random hexamers and then amplified using the following gene-specific primer pairs: oJR85 (5'-CGGCTGCCCTACCTACGGA-3') and oJR87 (5'-GGGAGGGCAGCTATTAGGA-3') for human PLK1; oJR104 (5'-CGGCTGCCCTACCTACGAACG-3') and oJR105 (5'- GGCACCAGTCCGAAGGAGAGA-3') for mouse PLK1; oJR111 (5'-TGAAGCCACTGCTACCACG-3') and oJR112 (5'-AGGGCTCTCCACTTTGATGG-3') for human FoxM1; oJR113 (5'-TCGGCCCCGCGTGGAGCAGAC-3') and oJR114 (5'-TAACCCGATTCTGCTCCAGGTGAC-3') for mouse FoxM1; oJR100 (5'- GGGCAAGTTCAGCAACATCGTGGA-3') and oJR101 (5'-GTAGCCGCCTTTCAGGATATACATC-3') for both human and mouse Cdc25B 22.
Quantitative RT-PCR (qRT-PCR)
Total RNA purified from cells with or without SRp20 knockdown was quantified by qRT-PCR by using Applied Biosystems TaqMan probes for human PLK1 (#HS00983227) and 18S rRNA (#4333760F) in accordance with the manufacturer's instructions. qRT-PCR was performed in a Cepheid Smart cycler. Briefly, 1 μg of total RNA treated with RNase-free DNase I was reverse transcribed at 42oC using random hexamers, followed by a 20-μL PCR reaction that included 2 μL RT product, 1 μL TaqMan Gene Expression Assays (20×), and 10 μL TaqMan Universal PCR Master mix (2×). The PCR reactions were performed at 50̊C for 2 min and 95̊C for 10 min, followed by 40 cycles of 95̊C for 15 s and 60̊C for 10 min. The relative expression level of PLK1 in each sample was calculated by the 2-ΔΔCT or 2-ΔCT method 23 after it was normalized to 18S rRNA.
Immunohistochemical staining
Cervical, soft tissue, and epithelial tumors and normal tissue sections were purchased from US BioMax. Immunohistochemistry was performed with a Vectastain ABC kit (Vector Laboratories) in accordance with the manufacturer's protocol. Sections were deparaffinized, rehydrated, and microwaved for 15 min in the presence of 1× Antigen Retrieval Citra Plus buffer (BioGenex). Endogenous peroxidases were quenched using 3% hydrogen peroxide for 15 min. Sections were incubated with an anti-SRp20 7B4 antibody overnight at 4̊C, followed by a secondary antibody for 30 min and ABC reagent for 30 min. The specific signal was developed by using a DAB substrate kit (Vector Laboratories).
Colony formation assay
NIH 3T3 cells transiently transfected with plasmid pT7-SRp20 (pJR17) or empty vector pFLAG-CMV5.1, or MEF 3T3 tet-off cells stably transfected with plasmid pJR17 or empty vector pRevTRE, were harvested, adjusted to 1 × 104 cells in DMEM containing 0.35% agar and 10% doxycycline-free FBS, and then laid onto a bottom layer containing 0.5% agar and 10% FBS in DMEM in 6-well plates. The plates were stained 3-4 weeks later with 0.005% crystal violet for colony counting.
Tumor induction in nude mice
For tumor induction with HeLa cells, cells transfected twice with SRp20 siRNA or nonspecific siRNAs were collected 24 h after the second transfection and implanted by dorsal subcutaneous inoculation of 1 × 106 cells into both sides of nude mice, with 10 mice in each group. Tumor sizes were measured at 14, 17, and 19 days after implantation. Tumor weight was recorded when the animals were sacrificed on day 22.
For tumor induction with MEF/3T3 tet-off cells, 3 × 106 cells stably transfected with a T7-SRp20 vector or with the empty vector (pRevTRE) were grown in doxycycline-free medium and implanted as described above. Tumor sizes were measured every 4 to 5 days, and tumor weight was recorded when the animals were sacrificed on day 50.
Flow cytometric cell cycle analysis
Cell cycle distribution was determined by flow cytometry. U2OS or HeLa cells transfected twice with SRp20 siRNA or NS siRNA at an interval of 48 h were trypsinized 96 h after the first siRNA transfection, washed twice with PBS, resuspended in Vindelov's propidium iodide buffer, and analyzed with a CYAN MLE cytometer (Dako-Cytomation). Ten thousand events were collected per sample, and data were analyzed with Modfit LT software (Verity Software House).
To analyze the synchronized cells for cell cycle progression, MEF 3T3 tet-off cells stably transfected with T7-SRp20x were seeded in a 6-well plate at 5 × 104 cells per well in DMEM containing 10% doxycycline-free FBS overnight, synchronized by starvation for 48 h in DMEM containing 0.1% FBS, and then collected at the indicated times after serum stimulation (with 10% FBS) for cell cycle analysis by flow cytometry as described above.
Oncomine cancer microarray database analyses
The Oncomine cancer microarray database [http://www.oncomine.com; 24] was used to analyze expression profiles of SRp20 in a variety of human cancerous and normal tissues and in association with tumor progression (grade) and 5-year survival rate. Statistics from individual studies were also obtained from the Oncomine cancer database (January 15 or April 15, 2010, version) and were combined for a Fisher's meta-analysis. Overexpression of SRp20 was defined as significantly higher expression (P < 0.05) in tumor tissues than in the corresponding normal tissues, in high-grade tumors than in low-grade tumors, or in shorter-living (<5 years) cancer patients than in longer-living (>5 years) cancer patients.
Southern blot and semi-quantitative PCR analysis
Southern blot analysis was conducted by using ~5 μg of EcoRI-digested genomic DNA extracted from paired normal and cancerous lung tissues (BioChain Institute, Hayward, CA) and hybridized with an SRp20 DNA probe, a 649-bp PCR fragment amplified with a 5' primer (oJR53, 5'-AAGCCGTCCCGATCCTTCTC-3') and a 3' primer (oJR57, 5'-GACTGCTTGTTCAACTATAGCTGCA-3') from HeLa genomic DNA and randomly labeled with 32P. The blot was reprobed for cyclophilin as a loading control by using a cyclophilin probe, a 660-bp fragment derived from a Hind III-digested 1.1-kb PCR product amplified with a 5' primer (oSB21, 5'-CCAAAGCATTGTACCGCAGAG-3') and a 3' primer (oSB22, 5'-TTGCATATACTGCCTTCTCTTTATC-3') from HeLa genomic DNA and randomly labeled with 32P.
For semi-quantitative PCR analysis of SRp20 gene amplification in paired normal and cancerous cervical or lung tissues, genomic DNA isolated from the cervical or lung tissues was serially diluted and analyzed by PCR by using a primer pair of oJR56 (5'-TCTCTTGAAACAGTGACACAAAGGTG-3') and oJR57 for SRp20 gene detection. Cyclophilin PCR with a primer pair of oSB21 and oSB22 served as loading control.
Statistical analysis
Statistical data for the paired microarray datasets in Fig. 3, were obtained directly from the Oncomine cancer database (www.oncomine.com). Statistics from individual studies with significantly higher expression of SRp20 (P < 0.05) in tumor tissues than in the corresponding normal tissues were also obtained from the Oncomine cancer database (January 15 or April 15, 2010, version) and were combined for Fisher's meta-analysis. All two-group statistical comparisons of means in Fig. 6 and Fig. 10 were calculated with two-tailed student's t test using Excel (Microsoft).
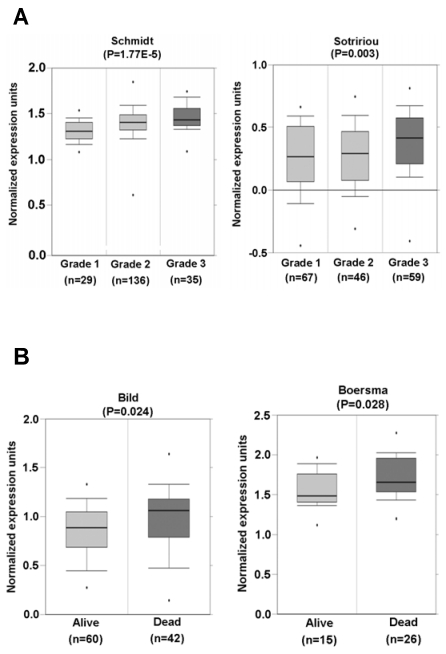
Fig 3.
SRp20 association with tumor progression and prognosis. (A) Increased expression of SRp20 correlates with tumor grade in breast cancer in studies by Schmidt et al. [left; 25] and Sotiriou et al. [right; 26] as obtained from the Oncomine cancer database (January 15, 2010, version). (B) Breast cancer patients who survived less than 5 years expressed a higher level of SRp20 in their tumor tissues than those who lived longer. Data and statistics were compiled from the studies of Bild et al. [left; 27] and Boersma et al. [right; 28], also obtained from the Oncomine cancer database (January 15, 2010, version).
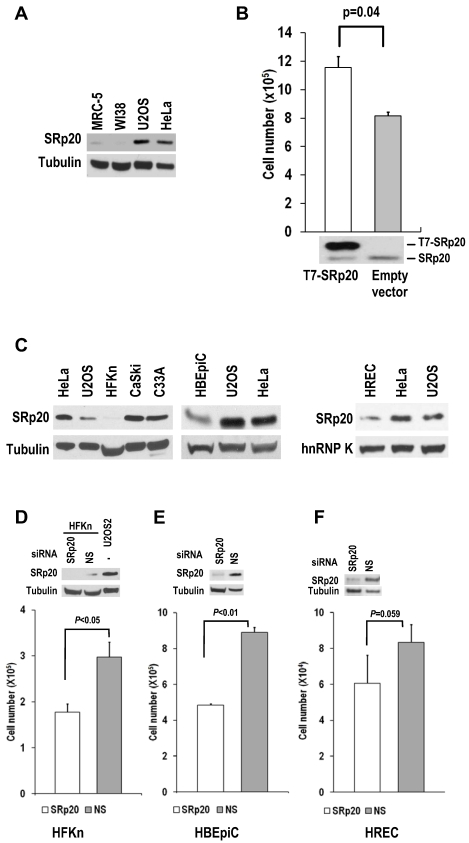
Fig 6.
Human diploid fibroblasts and primary human epithelial cells express minimal amounts of SRp20. (A) MRC-5 and WI-38 fibroblasts express less SRp20 than U2OS and HeLa cells by Western blot analysis. Tubulin served as a control for sample loading. (B) Ectopic expression of SRp20 in WI-38 cells promotes cell growth. WI-38 cells were transfected during cell passage on days 0, 2, and 4 with 2 μg of T7-SRp20 plasmid and were counted on day 6. Shown immediately below the corresponding bar graph are cell lysates blotted with SRp20 antibody 7B4. Data are shown as means ± SE from two separate experiments, each performed in duplicate. (C) Western blot comparison of primary HFKn, HBEpiC, and HREC cells with HeLa, U2OS, CaSki, and C33A cells for SRp20 expression by Western blot. Tubulin or hnRNP K served as a control for sample loading. (D-F) Knockdown of SRp20 expression in HFKn (D), HBEpiC (E), and HREC (F) cells affects cell growth. Western blots above each bar graph show the knockdown efficiency of SRp20 by siRNA. The graph show means ± SE from at least two separate experiments, each in duplicate.
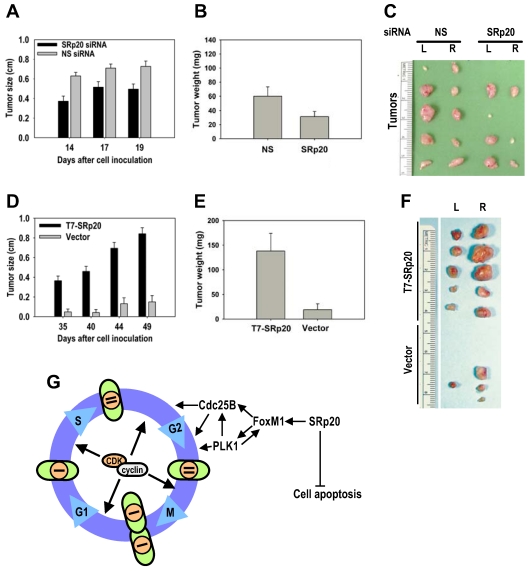
Fig 10.
SRp20 overexpression is tumorigenic in nude mice. (A-B) HeLa cells with reduced SRp20 expression are less competent at inducing tumors. HeLa cells (1 × 106) with or without SRp20 knockdown were implanted subcutaneously, and the tumor diameter (A) was measured on the indicated days. The tumor weight (B) was recorded when the animals were sacrificed on day 22. Data are means ± SE from 10 animals in each group (P < 0.01 for tumor size at each time point and P < 0.05 for tumor weight, t test). (C) Tumors from HeLa cells treated with NS or SRp20 siRNAs and implanted into both sides of the mice (L, left; R, right). Tumors from five animals from each group are shown. (D-E) MEF 3T3 tet-off cells with SRp20 overexpression are tumorigenic. MEF 3T3 tet-off cells (3 × 106) stably transfected with an empty vector only or a T7-SRp20 vector were grown in doxycycline-free medium and then implanted subcutaneously, and the tumor diameter (D) and weight (E) were measured as described above. Data are means ± SE from 5 animals in each group (P < 0.001 for both tumor size at each time point and weight). (F) Tumors from MEF3T3 tet-off cells with or without T7-SRp20 expression implanted into the left or right side. (G) A model for SRp20 regulation of cell cycle progression and tumorigenesis. SRp20 regulates the expression of FoxM1, Cdc25B, and PLK1 and controls cell apoptosis. Green ovals: cells; orange circles: nuclei; bars: genomes.
Results
Increased SRp20 expression in epithelial carcinomas and mesenchymal tissue-derived sarcomas
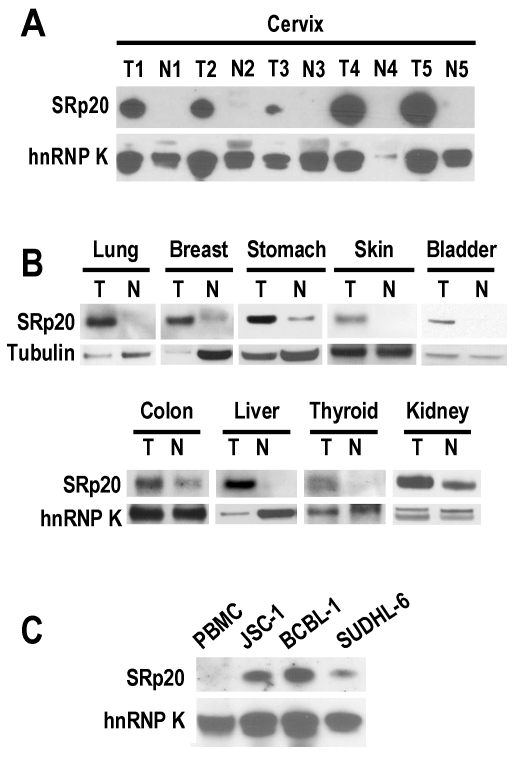
In looking at the role of SRp20 in human papillomavirus (HPV) RNA splicing 11, we found a remarkable increase of SRp20 expression in cervical cancer tissues (Fig. 1A). However, this increase was not limited to cancers caused by HPV infection. We also observed variable increases of SRp20 expression in cancers of the lung, breast, stomach, skin, bladder, colon, liver, thyroid, and kidney (Fig. 1B), as well as in B-cell lymphoma cells (JSC-1 [KSHV+/EBV+], BCBL1 [KSHV+], and SUDHL-6; Fig. 1C).
Fig 1.
SRp20 expression in tumor (T) and normal (N) tissues by Western blot analysis. Tissue samples (A and B) or lymphoma B cells (C) were immunoblotted with an anti-SRp20 7B4 antibody; hnRNP K and tubulin served as controls for sample loading. PBMC, peripheral blood mononuclear cells.
Tissue-array immunohistochemistry demonstrated increased expression of SRp20 not only in epithelial carcinomas (Fig. 2), but also in mesenchymal tissue-derived tumors, including rhadbomyosarcoma, hemangioendothelioma, hemangiopericytoma, neurofibroma, neurilemmoma, liposarcoma, leiomyosarcoma, histiocytoma, and synovial sarcoma (Supplementary information, Fig. S1). By searching the Oncomine cancer microarray database (http://www.oncomine.com), we found a significant increase (P < 0.05) of SRp20 expression in tumor tissues over the expression in corresponding normal tissues in 96 of 190 studies. Fisher's meta analysis indicated that the observed increase in those paired studies was significant (P <0.001). We also found that the increased SRp20 expression correlated with breast cancer progression in 13 of 26 studies (P<0.001) as represented in Fig. 3A 25,26, and with 5-year overall survival in 3 of 6 studies (P = 0.001), as represented in Fig. 3B 27,28.
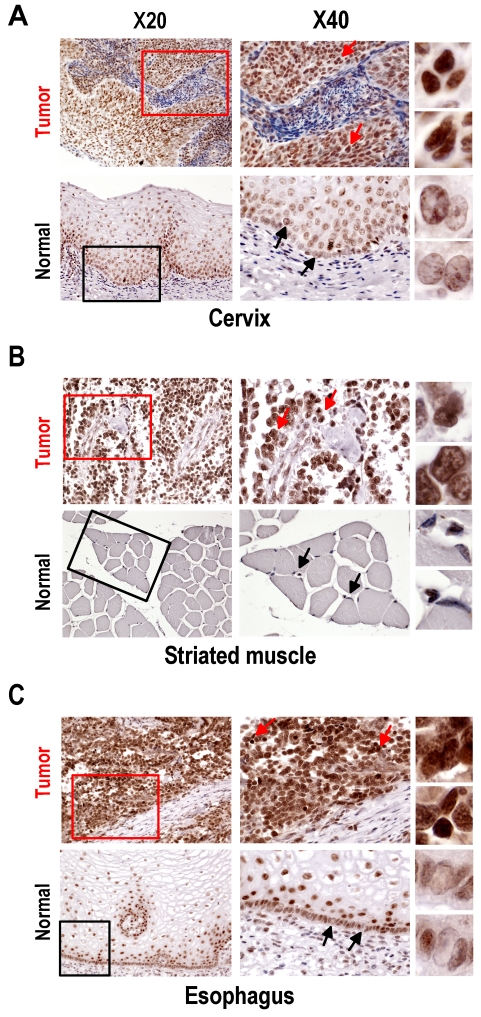
Fig 2.
Expression of SRp20 in tumor and normal tissues by immunohistochemistry. All tissue sections were stained with the SRp20 7B4 antibody and counterstained with Mayer's hematoxylin. Boxes in the lower-magnification images (×20) indicate locations where the higher-magnification images (×40) were taken. Arrows in the ×40-magnification images indicate individual cells further magnified on the right. (A) Squamous cell carcinoma (grade III) of cervix and normal cervical tissues. Normal cervical tissues express a low level of SRp20 in terminally differentiated cells, but a medium level of SRp20 in dividing basal or parabasal cells. In contrast, all cervical cancer cells show abundant SRp20 expression, with strong nuclear staining of SRp20 (see individual cells on the right). (B) Alveolar rhabdomyosarcoma of right shoulder and normal striated muscle. Rhabdomyosarcoma cells express high levels of SRp20, but normal striated muscle tissues show no detectable SRp20. (C) Esophageal small-cell carcinoma and normal esophagus. All cancer cells show strong nuclear staining for SRp20, but only the dividing basal or parabasal cells in normal esophagus expressed SRp20 at a medium level.
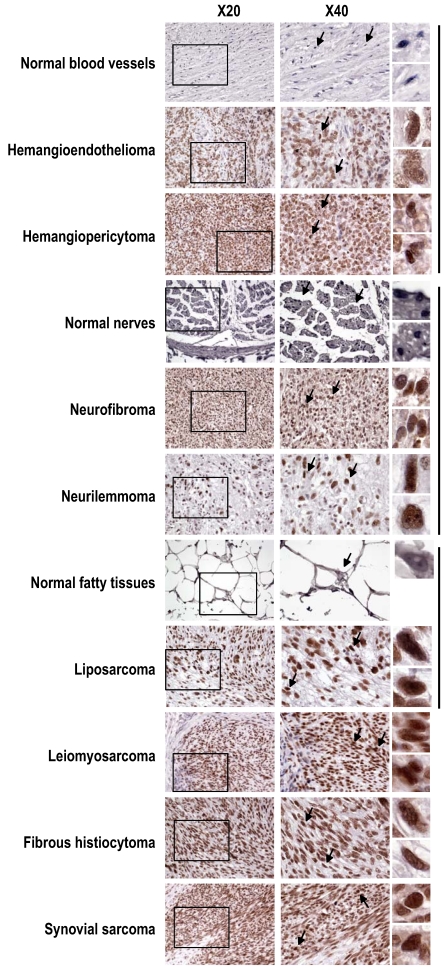
Figure S1.
Increased SRp20 expression in other soft tissue tumors shown by immunohistochemistry with the SRp20 7B4 antibody. SRp20 expression was compared in paired normal and tumor tissues including blood vessels, nerves, and fatty tissues, grouped by the vertical lines on the right. Leiomyosarcoma, fibrous histiocytoma, and synovial sarcoma with strong SRp20 expression are also included as additional examples. See other details in Figure 2.
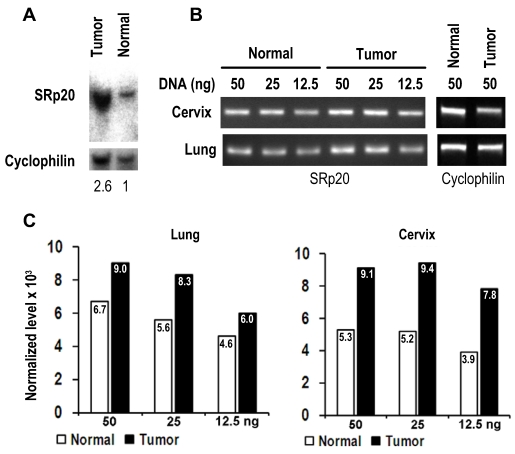
As the gene SFRS3 which encodes SRp20 is located on chromosome 6p21, a common region of DNA amplification seen in many cancers 29, we examined whether gene amplification would be a cause for increased SRp20 expression in cancer tissues. As shown in Fig. 4, we verified SFRS3 gene amplification in lung cancer by Southern blotting and semi-quantitative PCR and in cervical cancers by semi-quantitative PCR, demonstrating that SFRS3 gene amplification could be a cause of increased SRp20 expression in at least a subset of these cancers.
Fig 4.
SRp20 gene amplification in tumor tissues. (A) Southern blot analysis of SRp20 gene amplification in paired normal and cancerous lung tissues. The level of SRp20 relative to cyclophilin DNA is expressed below the blot as a ratio, with the ratio set to 1 for the normal tissue. (B) Semi-quantitative PCR analysis of SRp20 gene amplification in paired normal and cancerous cervical and lung tissues. (C) The relative intensity (x 103) of each PCR amplicon in (B) was normalized to cyclophilin as a control for sample loading.
SRp20 is required for cell proliferation because it promotes the G2/M transition
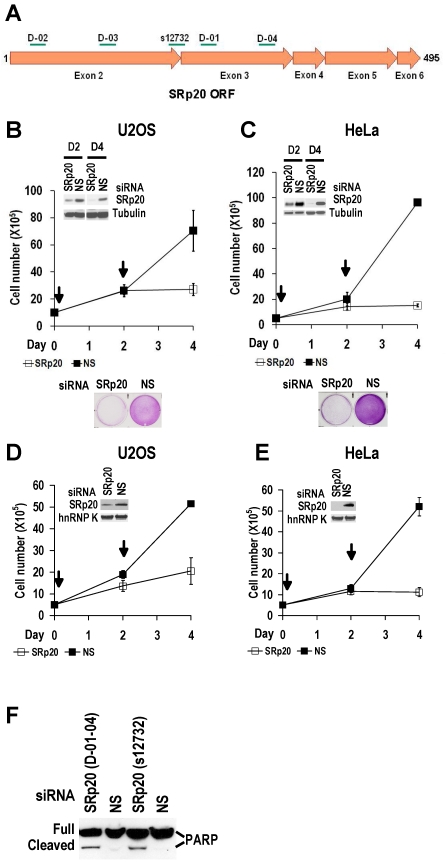
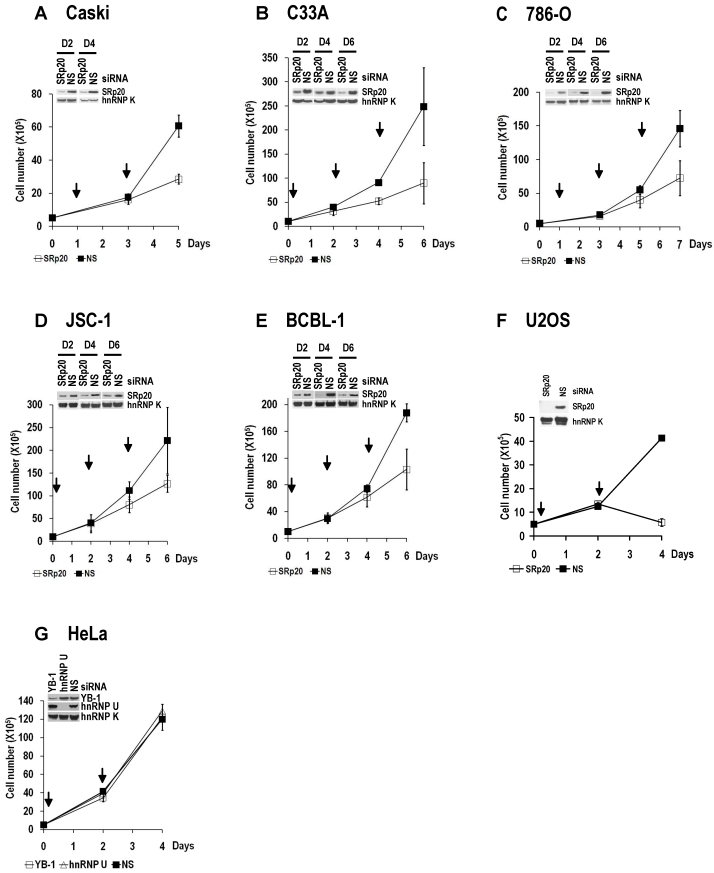
Increased SRp20 could potentially induce the production of mRNA isoforms that encode proteins favoring oncogenesis. We therefore hypothesized that increased SRp20 expression might contribute to maintenance of the carcinogenic phenotype, rather than being a consequence of it. To test this possibility, we examined whether reduced SRp20 expression would affect cancer cell growth. Each of seven cancer cell lines (U2OS, HeLa [HPV18+], CaSki [HPV16+], C33A, 786-O, JSC-1, and BCBL-1) exhibited reduced proliferation after SRp20 expression was knocked down by an siRNA pool targeting different regions of SRp20 mRNA (Fig. 5A-C, Supplementary information, Fig. S2A-E). To exclude any possible off-target effect of the pooled SRp20 siRNAs, we performed similar experiments in U2OS and HeLa cells by using a separate siRNA, siRNA s12732, that targets a splice junction of SRp20 exon 2 and exon 3 (Fig. 5A) and observed the same results (Fig. 5D-E). Using siRNA D-03 alone in U2OS cells also gave the same result (Supplementary information, Fig. S2F). By contrast, knockdown of YB-1 or hnRNP K had no effect on HeLa cell growth (Supplementary information, Fig. S2G). Further studies showed that knockdown of SRp20 expression in U2OS cells led to cell apoptosis, as indicated by apoptotic cleavage of PARP (Fig. 5F). These data indicate that increased SRp20 expression is necessary for the indefinite growth of cancer cells and prevents cancer cell apoptosis.
Fig 5.
Increased SRp20 is essential for tumor cell growth. (A) Schematic illustration of siRNA target sites in the SRp20 open reading frame (ORF). Arrows represent individual exons and translation directions. Bars above arrows are relative positions of siRNA target sites along with the siRNA names. D-01, D-02, D-03, and D-04 are four different siRNAs in an siRNA pool from Dharmacon. siRNA s12732, which spans the exon 2/exon 3 junction of SRp20 mRNA was from Ambion. (B-E) Knockdown of SRp20 expression affects cancer cell proliferation. SRp20 was knocked down in U2OS cells (an osteosarcoma cell line with wild-type p53 and pRb) and HeLa cells (an HPV18+ cervical cancer cell line) by the Dharmacon siRNAs (40 nM, B-C) or Ambion siRNA s12732 (10 nM, D-E). Insets show SRp20 knockdown efficiency in monolayer cultures by Western blot on day 2 (D2, B-C) and day 4 (D4, B-E) after RNAi. Arrows indicate the days the cells received siRNA treatments. Below the line graphs of B and C are crystal violet staining of the corresponding cells after RNAi. NS, nonspecific siRNA. (F) SRp20 knockdown-mediated cell apoptosis. Poly(ADP-ribose) polymerase (PARP) cleavage was analyzed by Western blot as an apoptotic biomarker 61 in U2OS cells after SRp20 knockdown with the indicated siRNAs in A.
Figure S2.
SRp20 expression is essential for tumor cell growth. SRp20 was knocked down by RNAi as described in Figure 5B, and cell growth was measured in cervical cancer cell lines CaSki (A) and C33A (B), kidney cancer cell line 786-O (C), B-cell lymphoma cell lines JSC-1 (D) and BCBL-1 (E), and U2OS cells (F). The knockdown efficiency in individual cell lines on the indicated days (D2, D4, and D6) in A-E or on day 4 in F was estimated by Western blot and is shown on the top left of each line graph. hnRNP K served as a control for sample loading. Arrows indicate days after cell passage when RNAi was performed. Pooled SRp20 siRNAs D-01 to D-04 (Figure 5A) were used at a final concentration of 40 nM in A-E, and a single D-03 SRp20 siRNA (Figure 5A) was used at a final concentration of 20 nM in F. Data are shown as means ± SD from two separate experiments, each in duplicate. (G) YB-1 or hnRNP U knockdown had no effect on HeLa cell growth.
WI-38 and MRC-5 cells, two human diploid lung fibroblast cell lines with a finite lifetime of about 50 population doublings, naturally express much less SRp20 than U2OS and HeLa cells (Fig. 6A).
When SRp20 was transiently introduced into WI-38 cells, the cell growth increased (Fig. 6B). Interestingly, human primary epithelial cells (human newborn foreskin keratinocytes [HFKn], human bronchial epithelial cells [HBEpiC], and human renal epithelial cells [HREC]), which have limited doubling times in culture, also express less SRp20 than cancer cell lines (Fig. 6C). Knocking down SRp20 expression in these cells also slowed their growth (Fig. 6D-F). Altogether, these data indicate that SRp20 is also essential for normal cell proliferation.
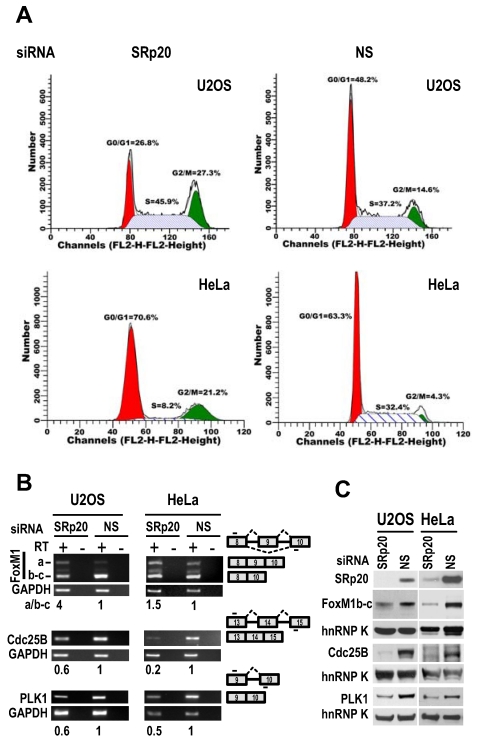
Flow cytometry showed that both U2OS and HeLa cells with siRNA-mediated reduction of SRp20 displayed prominent G2/M arrest (Fig. 7A), accompanied by increased expression of FoxM1a and decreased expression of FoxM1b-c, Cdc25B 30, and polo-like kinase-1 (PLK1; 31 at the mRNA level (Fig. 7B, Supplementary information, Fig. S3). We noticed that U2OS cells which contain a functional G1 check-point 32, but not HeLa cells which lack a functional G1 check-point due to HPV18 E7 degradation of pRb 33, also appeared a slight increase of cell number at S phase after knocking down SRp20 expression (Fig. 7A). Western blotting analyses showed reduced protein expression of FoxM1b-c, Cdc25B, and PLK1 (Fig. 7C), which are all involved in the G2/M transition 34-36. Because the physiological significance of FoxM1a is not clear and it is not generally detectable by Western blot 37,38, we conclude that the increased expression of SRp20 in cancer cells promotes cell cycle progression by affecting the expression of G2/M transition regulators, thus contributing to the indefinite growth of the tumor cell lines in monolayer culture.
Fig 7.
SRp20 regulates expression of FoxM1, Cdc25B, and PLK1 and is essential for cell cycle progression. (A) U2OS and HeLa cells with SRp20 knocked down by RNAi were collected for flow cytometry. NS, non-specific siRNA. One representative of two separate experiments is shown. (B and C) SRp20 regulates expression of FoxM1, Cdc25B, and PLK1. U2OS and HeLa cells with SRp20 knockdown as described in Fig. 3 were examined for FoxM1, Cdc25B, and PLK1 expression at the RNA level by RT-PCR (B) and at the protein level by immunoblot (C). Diagrams to the right of B show RNA splicing directions of FoxM1, Cdc25B, and PLK1 and primer positions (short lines above or below exons [boxes]) for RT-PCR; relative expression levels of each RNA transcript after being normalized to GAPDH are indicated underneath the gels. RT, reverse transcription. Protein levels of SRp20, FoxM1, Cdc25B, and PLK1 were also measured by Western blot (C); GAPDH (B) and hnRNP K (C) served as controls for sample loading in each assay.
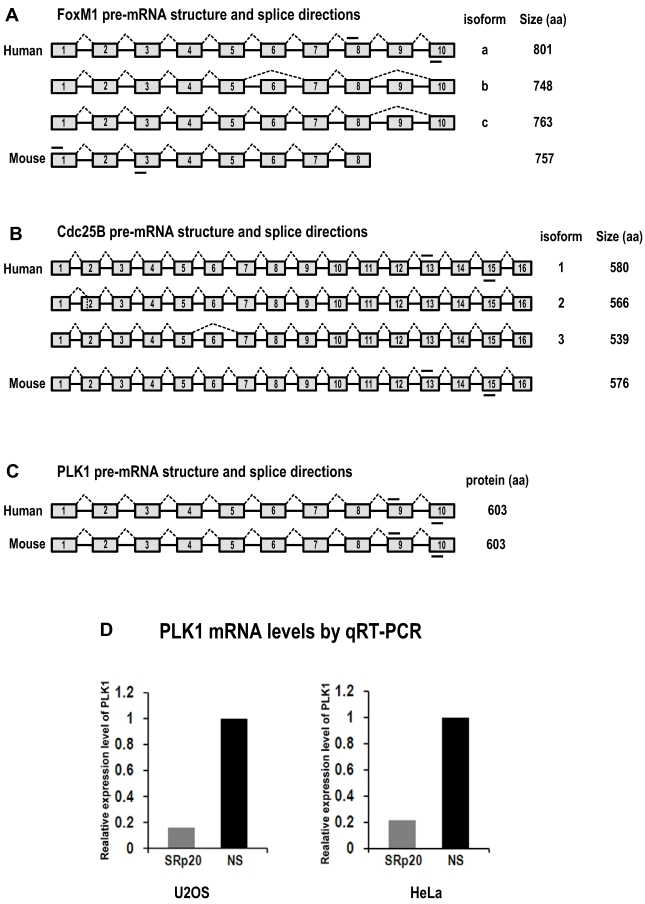
Figure S3.
Diagrams of FoxM1, Cdc25B and PLK1 pre-mRNA structures and splice directions and SRp20 regulation of PLK1 expression by qRT-PCR analysis. (A) Human FoxM1 expresses 3 RNA isoforms by alternative RNA splicing involving exons 6 and 9. FoxM1a, with 10 exons (boxes) and 9 introns (lines between boxes), is a full-length RNA encoding minimal or no detectable protein 37,38. Mouse FoxM1 expresses a major RNA isoform with 8 exons. Mouse FoxM1 has no equivalent sequences to human FoxM1 exon 9 and its exon 8 is a fusion of the human FoxM1 exon 8 and 10 sequences 62. (B) Human Cdc25B express 3 RNA isoforms by alternative RNA splicing involving exons 2 and 6 63. The isoform 2 of Cdc25B, with 16 exons, is a major isoform containing an alternatively spliced exon 2. Mouse Cdc25B expresses a major RNA isoform with 16 exons. (C) Both human and mouse PLK1 contain 10 exons with no reported alternative RNA splicing. Short lines above or below exons are the primers used for RT-PCR analyses of FoxM1, Cdc25B and PLK1 mRNAs. Drawings are not to scale. (D) SRp20 regulates expression of PLK1. Total RNAs were purified from U2OS cells with or without SRp20 knockdown by using Ambion siRNA s12732, or from HeLa cells with or without SRp20 knockdown by using pooled Dharmacon siRNAs. TaqMan probes for human PLK1 and 18S rRNA detection were used for qRT-PCR.
Increased SRp20 expression promotes immortal cell growth and transformation
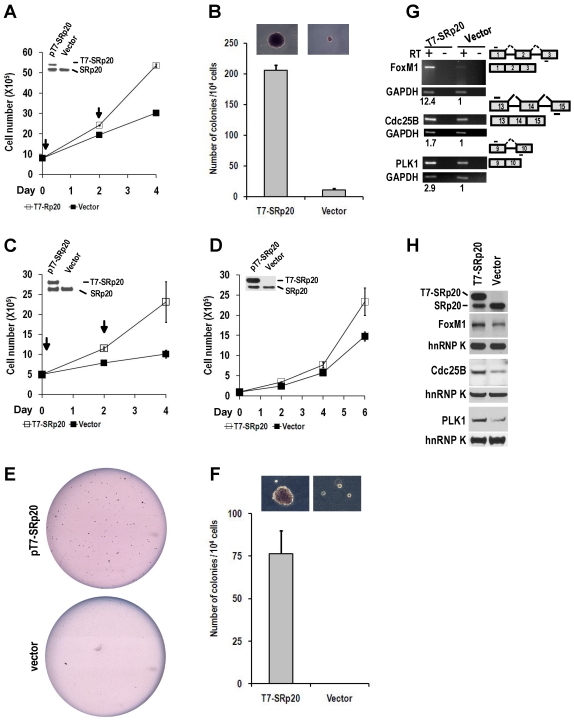
When ectopically expressed in immortal NIH 3T3 fibroblasts in the presence of endogenous SRp20, T7-tagged human SRp20 (T7-SRp20) conferred a substantial growth advantage to cells in monolayer culture (Fig. 8A) and anchorage-independent growth to cells in soft agar (Fig. 8B). This experiment was repeated in another immortal cell line, Swiss MEF (mouse embryonic fibroblast) 3T3 cells. The cells with transient (Fig. 8C) or stable expression (Fig. 8D) of T7-SRp20 displayed a two-fold increase in growth rate; colony formation in soft agar also increased (Fig. 8E-F).
Fig 8.
Overexpression of SRp20 promotes cell growth and transformation. (A and B) Transient expression of T7-SRp20 in the presence of endogenous SRp20 in NIH 3T3 cells promotes cell growth (A) and colony formation in soft agar (B), with the colonies at the top seen under a stereomicroscope at the same magnification for both photos. Arrows indicate the days the cells were transfected with a T7-SRp20 or empty vector. (C-F) Transient (C) or stable (D-F) expression of T7-SRp20 in the presence of endogenous SRp20 in MEF 3T3 tet-off cells promotes cell growth (C and D) and colony formation (E and F). Insets in A, C, and D are expressed T7-SRp20 and endogenous SRp20 by Western blot on day 4. Representative images of colonies in soft agar (E) show two stable cell lines with (top) or without (bottom) T7-SRp20 expression. Colony counts in two separate experiments from two different stable cell lines with or without T7-SRp20 expression are graphed (F), with the colonies at the top seen under a stereomicroscope at the same magnification for both photos. (G and H) stable expression of T7-SRp20 in the presence of endogenous SRp20 in MEF 3T3 tet-off cells promotes the expression of FoxM1, Cdc25B, and PLK1 as detected with RT-PCR for RNAs (G) and with immunoblotting for proteins (H). Diagrams on the right of G show the primer positions (short lines above or below exons [boxes]). RT, reverse transcription. GAPDH (G) and hnRNP K (H) served as controls for sample loading in each assay.
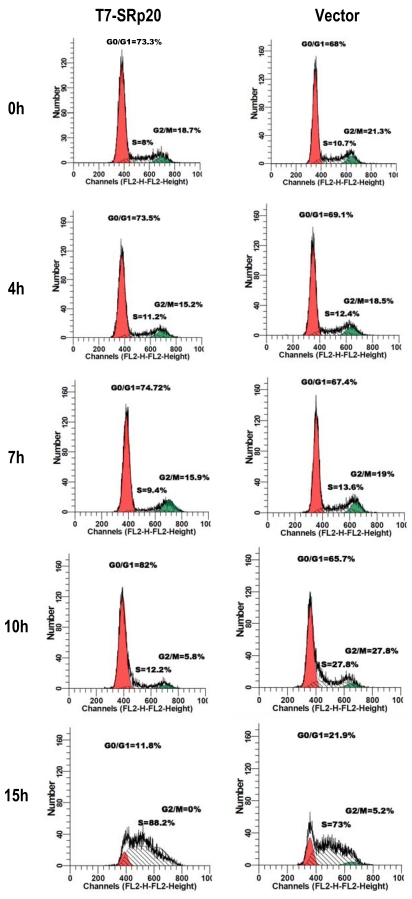
Knockdown of SRp20 expression in cancer cells reduced the expression of the cell cycle regulators FoxM1, Cdc25B, and PLK1 and prevented cell cycle progression through G2/M phase (Fig. 7); in contrast, expression of T7-SRp20 in stably transfected MEF 3T3 cells in the presence of endogenous SRp20 enhanced the expression of FoxM1, Cdc25B, and PLK1 both at the RNA (Fig. 8G) and protein (Fig. 8H) levels and accelerated cell cycle progression of synchronized MEF 3T3 tet-off cells. As shown in Fig. 9, overexpression of T7-SRp20 in synchronized MEF 3T3 tet-off cells pushed the cells quickly go through G2/M phase (compare cell numbers at G2/M phase from 7 h to 10 h) and accelerated the cell cycle transition from G0/G1 to S phase (compare cell numbers at G0/G1 and S phase from 10 h to 15h), over the vector controls.
Fig 9.
Stable expression of T7-SRp20 in MEF 3T3 tet-off cells promotes cell cycle progression. MEF 3T3 tet-off cells stably transfected with a T7-SRp20 vector or an empty vector were grown in doxycycline-free medium, synchronized by starvation for 48 h, and collected after the indicated durations of serum stimulation for cell cycle flow cytometry. One representative of two separate experiments is shown.
Increased SRp20 expression is required for tumor induction and maintenance in nude mice
Given the cell growth and transformation potential of SRp20, we conducted a series of nude mouse studies on SRp20 oncogenesis. Much lower tumor induction was seen when HeLa cells with siRNA-reduced SRp20 expression were implanted into nude mice than when HeLa cells receiving a nonspecific siRNA were implanted (Fig. 10A-C). When implanted into nude mice, MEF 3T3 cells with stable T7-SRp20 expression in the presence of endogenous SRp20 exhibited large tumors, whereas the cells stably transfected with the control vector were not competent for tumorigenesis (Fig. 10D-F). Together, these data indicate that increased SRp20 expression in immortalized, untransformed mammalian cells is also oncogenic in nude mice.
Discussion
Although various efforts have been made to understand the molecular basis of tumorigenesis, the mechanisms that lead to tumor-specific gene expression remain largely unknown. Altered expression of splicing factors had been described in various tumor types 9,20,39-41, but the effect of increased expression of splicing factors on tumorigenesis remained to be investigated. To our knowledge, there has been no report on the role of SRp20 in the development of cancer. In this study, we demonstrated a direct cause-effect relationship between increased SRp20 expression and tumor formation with the following evidence: First, all cancer types examined exhibited increased SRp20 expression. SRp20 expression in breast cancer tissues increased with cancer progression and correlated with patients' 5-year survival. Second, cancer cells with reduced SRp20 expression appear to grow slowly, undergo apoptosis as reported while our manuscript was in preparation 42, and are less tumorigenic in nude mice. Third, overexpression of SRp20 in immortal 3T3 cells caused cell transformation and induced tumor formation in nude mice. We characterize SRp20 as a novel proto-oncogene because SRp20 is a normal cellular gene that, when expressed at its physiological level, is essential for normal cell proliferation in cultures (Fig. 6). The cells in the basal layers of cervix and esophagus, which are dividing cells, appear to express SRp20 relatively more than non-dividing, terminally differentiated cells (Fig. 2). Together with the finding that splicing factor SF2/ASF is overexpressed in lung and colon cancers and is oncogenic in nude mice 9, our data provide compelling evidence that altered expression of SR proteins is an important contributor to the development of cancer.
SRp20 that becomes oncogenic appears only when its expression from chromosome 6p is increased, presumably by gene amplification. This chromosome region bearing the SFRS3 (SRp20) gene is commonly amplified in many cancers 29. Our preliminary investigation has confirmed that gene amplification could be a cause of SRp20 overexpression in lung and cervical cancers (Fig. 4). Other oncogenic signaling pathways might also trigger SRp20 expression. One such pathway might be the Wnt signaling pathway, which is implicated in driving the formation of various human cancers and recruits cytosolic β-catenin as a transcriptional coactivator 43 to transactivate SRp20 expression 44. Although the specific cause of increased SRp20 expression in human cancers remains to be investigated, and there may be several possible causes, SFRS3 gene amplification in chromosome 6p as reported in many other studies 29 may lead to increased SRp20 expression in at least a subset of these cancers.
The dysregulated expression of splicing factors can change general RNA splicing and other events essential for the proper expression of the targeted genes, consequently causing diseases and tumor formation 8,45-47. In this report, we identified cellular FoxM1, PLK1, and Cdc25B as three prominent targets of SRp20 in the regulation of cell proliferation and oncogenesis (Fig. 10G). FoxM1, known previously as HFH-11, WIN, MPP-2, FKHL-16, or Trident 48, is a transcription factor of the Forkhead family that regulates expression of PLK1, Cdc25B, and CENP-F 34,49-51. FoxM1 is expressed in proliferating cells in a cell cycle-dependent manner 37,52-54 and plays crucial roles in the G2/M transition 49,50 and in chromosome stability and segregation during mitosis 34. The gene encoding FoxM1 on chromosome 12p13.3 produces a primary transcript that can be alternatively spliced to create three RNA species by inclusion (FoxM1a and 1c) or exclusion (FoxM1b) of exon 6 and inclusion (FoxM1a) or exclusion (FoxM1b and 1c) of exon 9 (Supplementary information, Fig. S3); these three RNA species produce three protein isoforms, FoxM1a, 1b, and 1c, with only FoxM1b and 1c being transcriptionally active 48. Two FoxM1 targets, PLK1 and Cdc25B, are crucial for entry of the cell cycle into mitosis 31,55,56, are all overexpressed in many cancers and oncogenic 48,55,57,58 and were all expressed at reduced levels in U2OS and HeLa cells with SRp20 knockdown, causing the cells to arrest at G2/M. Because PLK1-dependent phosphorylation of FoxM1 is required for G2/M progression 59, reduced PLK1 expression in the cells would result in nonfunctional FoxM1, thereby exaggerating the FoxM1 deficiency caused by SRp20 knockdown. In contrast, overexpression of SRp20 in 3T3 cells stimulated the expression of FoxM1 and its targets PLK1 and Cdc25B and promoted cell proliferation and tumorigenesis.
Although the mechanism by which SRp20 promotes FoxM1 expression remains to be determined, we assume that SRp20 regulation of FoxM1 expression is both transcriptional and posttranscriptional. Because SP1 binding to the FoxM1 promoter regulates FoxM1 expression 60, downregulation of SP1 expression in cells with reduced SRp20 levels 11 may be responsible for FoxM1 reduction at the transcriptional level. Human FoxM1 exon 9 is not conserved in the mouse genome, and human FoxM1a with the exon 9 inclusion does not encode a functional FoxM1a protein; the enhancement of mouse FoxM1 expression by SRp20 therefore presumably occurs by transcription through an indirect mechanism. On the other hand, the enhancement of human FoxM1 expression by SRp20 might occur through the regulation of RNA splicing, most likely by promoting the exclusion of FoxM1 exon 9. Nevertheless, the effects of SRp20 on FoxM1, PLK1, and Cdc25B expression are one mechanism by which SRp20 regulates cell proliferation and oncogenesis.
Acknowledgments
This research was supported by the Intramural Research Program of the Center for Cancer Research, NCI, NIH. We thank Javier Caceres and Tom Misteli for providing T7-SRp20 expression vector, Robert Yarchoan, Douglas Lowy, and Adrian Krainer for their critical comments and encouragement in the course of this study and for their critical reading of the manuscript; Curtis Harris and Izumi Horikawa for their critical reading of the manuscript; and Calvin Chan for his initial analysis of the Oncomine database.
References
- 1.Venter JC, Adams MD, Myers EW. et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 2.Johnson JM, Castle J, Garrett-Engele P. et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 3.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 4.Zheng ZM. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and Mammalian gene expression. J Biomed Sci. 2004;11:278–294. doi: 10.1159/000077096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Y, Maroney PA, Denker JA. et al. Dynamic regulation of alternative splicing by silencers that modulate 5' splice site competition. Cell. 2008;135:1224–1236. doi: 10.1016/j.cell.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heinzen EL, Ge D, Cronin KD. et al. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venables JP, Klinck R, Bramard A. et al. Identification of alternative splicing markers for breast cancer. Cancer Res. 2008;68:9525–9531. doi: 10.1158/0008-5472.CAN-08-1769. [DOI] [PubMed] [Google Scholar]
- 8.Christofk HR, Vander Heiden MG, Harris MH. et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 9.Karni R, de Stanchina E, Lowe SW. et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins) Genes Dev. 2010;24:1073–1074. doi: 10.1101/gad.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia R, Liu X, Tao M. et al. Control of the papillomavirus early-to-late switch by differentially expressed SRp20. J Virol. 2009;83:167–180. doi: 10.1128/JVI.01719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavaloc Y, Bourgeois CF, Kister L. et al. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zahler AM, Lane WS, Stolk JA. et al. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 14.Cui M, Allen MA, Larsen A. et al. Genes involved in pre-mRNA 3'-end formation and transcription termination revealed by a lin-15 operon Muv suppressor screen. Proc Natl Acad Sci U S A. 2008;105:16665–16670. doi: 10.1073/pnas.0807104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lou H, Neugebauer KM, Gagel RF. et al. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol Cell Biol. 1998;18:4977–4985. doi: 10.1128/mcb.18.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Gattoni R, Stevenin J. et al. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 17.Hautbergue GM, Hung ML, Golovanov AP. et al. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc Natl Acad Sci U S A. 2008;105:5154–5159. doi: 10.1073/pnas.0709167105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bedard KM, Daijogo S, Semler BL. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007;26:459–467. doi: 10.1038/sj.emboj.7601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jumaa H, Wei G, Nielsen PJ. Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr Biol. 1999;9:899–902. doi: 10.1016/s0960-9822(99)80394-7. [DOI] [PubMed] [Google Scholar]
- 20.He X, Ee PL, Coon JS. et al. Alternative splicing of the multidrug resistance protein 1/ATP binding cassette transporter subfamily gene in ovarian cancer creates functional splice variants and is associated with increased expression of the splicing factors PTB and SRp20. Clin Cancer Res. 2004;10:4652–4660. doi: 10.1158/1078-0432.CCR-03-0439. [DOI] [PubMed] [Google Scholar]
- 21.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 22.Guo J, Kleeff J, Li J. et al. Expression and functional significance of CDC25B in human pancreatic ductal adenocarcinoma. Oncogene. 2004;23:71–81. doi: 10.1038/sj.onc.1206926. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Rhodes DR, Yu J, Shanker K. et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt M, Bohm D, von Torne C. et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 26.Sotiriou C, Wirapati P, Loi S. et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 27.Bild AH, Yao G, Chang JT. et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 28.Boersma BJ, Reimers M, Yi M. et al. A stromal gene signature associated with inflammatory breast cancer. Int J Cancer. 2008;122:1324–1332. doi: 10.1002/ijc.23237. [DOI] [PubMed] [Google Scholar]
- 29.Santos GC, Zielenska M, Prasad M. et al. Chromosome 6p amplification and cancer progression. J Clin Pathol. 2007;60:1–7. doi: 10.1136/jcp.2005.034389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varmeh-Ziaie S, Manfredi JJ. The dual specificity phosphatase Cdc25B, but not the closely related Cdc25C, is capable of inhibiting cellular proliferation in a manner dependent upon its catalytic activity. J Biol Chem. 2007;282:24633–24641. doi: 10.1074/jbc.M703105200. [DOI] [PubMed] [Google Scholar]
- 31.van Vugt MA, Bras A, Medema RH. Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol Cell. 2004;15:799–811. doi: 10.1016/j.molcel.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 32.Guan KL, Jenkins CW, Li Y. et al. Growth suppression by p18, a p16INK4/MTS1- and p14INK4B/MTS2-related CDK6 inhibitor, correlates with wild-type pRb function. Genes Dev. 1994;8:2939–2952. doi: 10.1101/gad.8.24.2939. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez SL, Stremlau M, He X. et al. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J Virol. 2001;75:7583–7591. doi: 10.1128/JVI.75.16.7583-7591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laoukili J, Kooistra MR, Bras A. et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 35.Lammer C, Wagerer S, Saffrich R. et al. The cdc25B phosphatase is essential for the G2/M phase transition in human cells. J Cell Sci. 1998;111( Pt 16):2445–2453. doi: 10.1242/jcs.111.16.2445. [DOI] [PubMed] [Google Scholar]
- 36.Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 37.Ye H, Kelly TF, Samadani U. et al. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17:1626–1641. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu M, Dai B, Kang SH. et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–3602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- 39.Stickeler E, Kittrell F, Medina D. et al. Stage-specific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene. 1999;18:3574–3582. doi: 10.1038/sj.onc.1202671. [DOI] [PubMed] [Google Scholar]
- 40.Fischer DC, Noack K, Runnebaum IB. et al. Expression of splicing factors in human ovarian cancer. Oncol Rep. 2004;11:1085–1090. [PubMed] [Google Scholar]
- 41.Ghigna C, Moroni M, Porta C. et al. Altered expression of heterogenous nuclear ribonucleoproteins and SR factors in human colon adenocarcinomas. Cancer Res. 1998;58:5818–5824. [PubMed] [Google Scholar]
- 42.He X, Arslan AD, Pool MD. et al. Knockdown of splicing factor SRp20 causes apoptosis in ovarian cancer cells and its expression is associated with malignancy of epithelial ovarian cancer. Oncogene. 2010 doi: 10.1038/onc.2010.426. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barker N, Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat Rev Drug Discov. 2006;5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 44.Goncalves V, Matos P, Jordan P. The beta-catenin/TCF4 pathway modifies alternative splicing through modulation of SRp20 expression. RNA. 2008;14:2538–2549. doi: 10.1261/rna.1253408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettigrew CA, Brown MA. Pre-mRNA splicing aberrations and cancer. Front Biosci. 2008;13:1090–1105. doi: 10.2741/2747. [DOI] [PubMed] [Google Scholar]
- 47.Grosso AR, Martins S, Carmo-Fonseca M. The emerging role of splicing factors in cancer. EMBO Rep. 2008;9:1087–1093. doi: 10.1038/embor.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 49.Krupczak-Hollis K, Wang X, Kalinichenko VV. et al. The mouse Forkhead Box m1 transcription factor is essential for hepatoblast mitosis and development of intrahepatic bile ducts and vessels during liver morphogenesis. Dev Biol. 2004;276:74–88. doi: 10.1016/j.ydbio.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Kiyokawa H, Dennewitz MB. et al. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci U S A. 2002;99:16881–16886. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang IC, Chen YJ, Hughes D. et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korver W, Roose J, Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 1997;25:1715–1719. doi: 10.1093/nar/25.9.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao KM, Sha M, Lu Z. et al. Molecular analysis of a novel winged helix protein, WIN. Expression pattern, DNA binding property, and alternative splicing within the DNA binding domain. J Biol Chem. 1997;272:19827–19836. doi: 10.1074/jbc.272.32.19827. [DOI] [PubMed] [Google Scholar]
- 54.Leung TW, Lin SS, Tsang AC. et al. Over-expression of FoxM1 stimulates cyclin B1 expression. FEBS Lett. 2001;507:59–66. doi: 10.1016/s0014-5793(01)02915-5. [DOI] [PubMed] [Google Scholar]
- 55.Galaktionov K, Lee AK, Eckstein J. et al. CDC25 phosphatases as potential human oncogenes. Science. 1995;269:1575–1577. doi: 10.1126/science.7667636. [DOI] [PubMed] [Google Scholar]
- 56.Karlsson C, Katich S, Hagting A. et al. Cdc25B and Cdc25C differ markedly in their properties as initiators of mitosis. J Cell Biol. 1999;146:573–584. doi: 10.1083/jcb.146.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takai N, Hamanaka R, Yoshimatsu J. et al. Polo-like kinases (Plks) and cancer. Oncogene. 2005;24:287–291. doi: 10.1038/sj.onc.1208272. [DOI] [PubMed] [Google Scholar]
- 58.Park HJ, Carr JR, Wang Z. et al. FoxM1, a critical regulator of oxidative stress during oncogenesis. EMBO J. 2009;28:2908–2918. doi: 10.1038/emboj.2009.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fu Z, Malureanu L, Huang J. et al. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol. 2008;10:1076–1082. doi: 10.1038/ncb1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petrovic V, Costa RH, Lau LF. et al. Negative regulation of the oncogenic transcription factor FoxM1 by thiazolidinediones and mithramycin. Cancer Biol Ther. 2010;9:1008–1016. doi: 10.4161/cbt.9.12.11710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tewari M, Quan LT, O'Rourke K. et al. Yama/CPP32 beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 62.Ma RY, Tong TH, Cheung AM. et al. Raf/MEK/MAPK signaling stimulates the nuclear translocation and transactivating activity of FOXM1c. J Cell Sci. 2005;118:795–806. doi: 10.1242/jcs.01657. [DOI] [PubMed] [Google Scholar]
- 63.Baldin V, Cans C, Superti-Furga G. et al. Alternative splicing of the human CDC25B tyrosine phosphatase. Possible implications for growth control? Oncogene. 1997;14:2485–2495. doi: 10.1038/sj.onc.1201063. [DOI] [PubMed] [Google Scholar]