Abstract
PBP (peroxisome-proliferator-activated receptor-binding protein) [Med1 (mediator 1)/TRAP220 (thyroid-hormone-receptor-associated protein 220)] is essential for mammary gland development. We established a mammary epithelial cell line with a genotype of PBPLoxP/LoxP by expressing an active form of Notch4. Null mutation of PBP caused severe growth inhibition of the Notch4-immortalized mammary cells. We found that truncated PBP without the two LXXLL motifs could reverse the growth inhibition due to the deficiency of endogenous PBP, indicating that signalling through nuclear receptors is unlikely to be responsible for the growth inhibition as the result of PBP deficiency. Loss of PBP expression was shown to completely ablate the expression of SOX10 [Sry-related HMG (high-mobility group) box gene 10]. The re-expression of SOX10 was capable of reversing the growth inhibition due to PBP deficiency, whereas suppressed expression of SOX10 inhibited the growth of Notch4-immortalized mammary cells. Further studies revealed PBP is directly recruited to the enhancer of the SOX10 gene, indicating that SOX10 is a direct target gene of PBP. We conclude that PBP is essential for the growth of Notch4-immortalized mammary cells by activating SOX10 expression, providing a potential molecular mechanism through which PBP regulates the growth of mammary stem/progenitor cells.
Keywords: mammary gland development, mammary progenitor cell, mediator complex, Notch4, peroxisome-proliferator-activated receptor (PPAR)-binding protein (PBP), Sry-related HMG box gene 10 (SOX10)
INTRODUCTION
PBP [PPAR (peroxisome-proliferator-activated receptor)-binding protein; also known as Med1 (mediator 1)/TRAP220 (thyroid-hormone-receptor-associated protein 220)] is a nuclear receptor coactivator which is a component of the TRAP–DRIP (vitamin D receptor-interacting protein)–Med complex [1–4]. This complex, which contains more than 25 polypeptides, is recruited to nuclear receptors via the subunit PBP. The complex may act by recruiting general transcription machinery to transcription starting sites to facilitate the initiation of transcription [5,6]. In addition to nuclear receptors, PBP also serves as a coactivator for GATA and GABP (GA-binding protein) [7–9].
The knockout of PBP is embryonic lethal [10,11]. Studies with a conditional knockout in mammary glands revealed that PBP-deficient mammary glands exhibited the retarded elongation of ducts during puberty and severely decreased density of alveoli during pregnancy [12,13]. The lactating PBP-deficient mammary glands failed to produce milk to nurture pups, although PBP-deficient mammary epithelial cells expressed abundant milk protein genes. PBP-deficient mammary epithelial cells could not generate mammospheres, which are clonally derived from mammary progenitor/stem cells, indicating that impaired mammary progenitor/stem cells might be responsible for the phenotype. PBP was also found to be amplified and overexpressed in some breast cancers and might play a role in tumorigenesis [14].
The SOX [Sry-related HMG (high-mobility group) box] transcription factor family consists of 20 members, which are widely involved in the control of development [15,16]. Subfamily E has three members: SOX8, SOX9 and SOX10. A point mutation in SOX10 causes Waardenburg-Shah syndrome, which is dominantly inherited and characterized by hypopigmentation of skin, heterochromia irides, deafness and usually Hirschsprung’s disease (aganglionic megacolon) [17,18]. SOX10 plays a critical role in the development of NCCs (neural crest cells), which are multipotent progenitor cells migrating from the dorsal neural tube to various sites in the body [19]. The NCC derivatives include skeletogenic fates, such as craniofacial cartilage and bone, and non-skeletogenic fates, such as melanocytes and neurons and glia of the PNS (peripheral nervous system).
The Notch signalling pathway is widely used for controlling development through stem cell maintenance, cell fate determination and differentiation [20,21]. Notch was found to be preferentially activated in the ductal luminal progenitor epithelium of mammary glands [22]. The persistent activated form of Notch4 induced mammary adenocarcinomas in a transgenic model [23].
The analysis of molecular mechanism by which PBP functions is complicated by the fact that the cells in mammary glands are heterogeneous, consisting of stem/bi-progenitor cells, luminal and myoepithelial cells at their different developmental stages, plus stromal cells. In the present study we report the establishment of a PBPLoxP/LoxP mammary epithelial cell line by expressing the active form of Notch4. We found that null mutation of PBP caused severe growth inhibition of the Notch4-immortalized mammary cells and truncated PBP without the two LXXLL motifs could reverse the growth inhibition. Loss of PBP expression completely ablated the expression of SOX10. The re-expression of SOX10 was capable of reversing the growth inhibition due to PBP deficiency, whereas suppressed expression of SOX10 inhibited the growth of Notch4-immortalized mammary cells. Further studies revealed PBP is directly recruited to the enhancer of the SOX10 gene, indicating that SOX10 is a direct target gene of PBP required for the growth of Notch4-immortalized mammary cells.
MATERIALS AND METHODS
Plasmids and antibodies
The 1.7 kb Notch4 fragment encoding the intracellular domain was amplified from mouse spleen Marathon-ready cDNA (Clontech) using the primers 5′-AGCTAGGAATTCTGGACTCCTGCTGCGC-3′ and 5′-AGCTAGGTCGACCGGAGTCTTCCAGACT-3′. The PCR product was digested with EcoRI and SalI and inserted into PLIB-Bla, which carries a selection gene conferring blasticidian resistance. Partial PBP cDNAs were inserted into the HpaI/ClaI sites of pLNCX to generate pLNCX-PBP1 (amino acids 1–535) and pLNCX-PBP2 (amino acids 1–680). pcDNA3.1-PBP was produced by inserting full-length human PBP cDNA into NotI/XbaI sites of pcDNA3.1(+). pCMV-sport6-SOX10 is an IMAGE clone from Open Biosystem. Luciferase reporter vectors containing conserved regulatory elements were described previously [24]. Anti-SOX10 antibody was purchased from Santa Cruz Biotechnology and kindly provided by Dr Wegner (University Erlangen-Nurnberg, Germany) [25].
Isolation of mammary epithelial cells and retroviral infection
The inguinal glands were collected from wild-type mice. Mammary epithelial cells were separated from stromal cells by collagenase digestion and Percoll gradient centrifugation [26]. Phoenix-Eco packaging cells (5×106) were transfected with 15 µg of PLIB-Bla-Notch4 or PLIB-Bla through the calcium phosphate precipitation method. At 48 h after the transfection, viral supernatant was added to 5×105 mammary epithelial cells with 8 µg/ml polybrene. After 5 h of incubation, the medium with retrovirus was replaced with fresh medium.
Knockout of PBP gene
Mammary cells were infected with retroviruses expressing Cre-ERtam or control viruses [27]. The cells were selected with 1 µg/ml puromycin. The cells were then treated with tamoxifen (1 µM) for 2 days.
Soft agar colony formation assay
The assay was performed as described in [28]. Briefly, Notch4 immortalized cells (2×103) were suspended in a 1 ml mixture containing 0.35% (w/v) agar and DMEM (Dulbecco’s minimal Eagle’s medium) supplemented with 10% FBS (fetal bovine serum), and layered on a 1 ml base of 0.7%(w/v) agar in DMEM with 10% FBS in a six-well plate. Colonies were photographed 2 weeks after plating.
Cell proliferation assay
Cells were seeded into six-well plates at a density of 1×104 cells/well with fresh DMEM plus 10% FBS. Viable cells were counted after Trypan Blue staining once a day for 4 days.
BrdU (bromodeoxyuridine) staining
BrdU was added to cell medium to a final concentration of 10 µM. After 1 h, the cells were washed with PBS, and treated with 4% formaldehyde. The cells were incubated in 2 M HCl solution for 1 h. The cells were then neutralized with 0.1 M borate buffer (pH 8.5). BrdU immunostaining was performed with the Cell Proliferation kit (Amersham Life Sciences) according to the manufacturer’s instructions. The percentage of BrdU-positive cells was estimated by counting 1000 cells of two slides of each genotype.
RNA isolation and RT (reverse transcription)–PCR
Total RNAs were prepared using TRIzol® (Invitrogen). RT–PCR was performed with SuperScript one-step RT–PCR kit from Invitrogen. Each assay was repeated at least once. Representative results are presented.
The primers used were as follows: β-actin: 5′-CCATCTACGAGGGCTATGCT-3′ and 5′-GCAAGTTAGGTTTTGTCAAAGA-3′; PBP: 5′-TGTATCTGGCTCTCCAATCC-3′ and 5′-AGTGATGAGTTCATACAGGGG-3′; SOX10: 5′-TGCTGCTATTCAGGCTCACT-3′ and 5′-TGCCGAGTAGCTACCCACA-3′; keratin 18: 5′-TCCGCGGCGGAACTCCTGTT-3′ and 5′-TCATCTACCACCTTGCGGAG-3′; SMA (smooth muscle actin): 5′-ATCAGGAAGGATCTCTATGCT-3′ and 5′-CAAAGCTTTGGGCAGGAATG-3′; keratin 5: 5′-GTGGCTTTGGCTTTGGAGGA-3′ and 5′-GCCGTGGTACGCTTGTTGAT-3′; keratin 6 (homologous to both keratin 6a and keratin 6b): 5′-GCCTATGTTTGAGCAGTACA-3′ and 5′-TCAGCCCGACTTCTCTGAG-3′.
Generation of stable cell lines expressing human PBP or SOX10
The Notch4-immortalized cells expressing Cre-ERtam were transfected with pcDNA3.1-hPBP (10 µg) or pcDNA3.1 (10 µg) for PBP expression, and pCMV-Sport6-SOX10 (9 µg) or pCMV-Sport6 (9 µg) plus pcDNA3.1 (1 µg) for SOX10 expression using Lipofectamine™ 2000 (Invitrogen). After selection with G418 (300 µg/ml), the individual clones were combined, expanded and examined for transgene expression.
Microarray hybridization
Notch4-immortalized epithelial cells or Notch4-immortalized epithelial cells expressing Cre-ERtam were treated with tamoxifen (1 µM) for 2 days. The cells were harvested for the preparation of total RNA using TRIzol®. Two preparations of RNA for each type of cells were used for microarray hybridization. RNA samples were further purified with the RNeasy MinElute Cleanup kit (Qiagen). Integrity of the RNA was confirmed using the Agilent 2100 BioAnalyzer. RNA (500 ng) was used to perform in vitro transcription in the presence of biotin-UTP using the Illumina TotalPrep RNA amplification kit. The amplified RNA was then hybridized to an Illumina Whole-Genome mouse Expression BeadChip and detected according to the Illumina user manual. Data normalization and analysis was performed as instructed with Illumina software. Genes whose expression was increased or decreased with a fold of ≥1.5 (P < 0.01) were identified as differentially expressed genes.
siRNA (small interfering RNA) transfection
Two siRNAs against mouse SOX10 and control siRNA were purchased from Sigma. Notch4-immortalized cells were transfected with control siRNA or SOX10 siRNAs using Lipofectamine™ 2000 reagent. After 36 h, total RNA was isolated using TRIzol® reagent. Real-time RT–PCR was performed on ABI 7300 system (Applied Biosystems) using the SYBR Green Supermix (Applied Biosystems) according to the manufacturer’s protocol. One sample of the cells was subjected to proliferation assay as described above.
Enhancer analysis
Notch4-immortalized wild-type or PBP-deficient cells (1×105) were plated in DMEM containing 10% FBS in six-well plates and were cultured for 24 h before transfection. Transfections were performed using Lipofectamine 2000™ reagent with MCSs (multiple species conserved sequences)-LUC or empty e1B-LUC (1.8 µg) and pCMV-β (0.2 µg) as an internal control for transfection efficiency. Cell extracts were prepared 36 h after transfection and were assayed for luciferase and β-galactosidase activities.
ChIP (chromatin immunoprecipitation)
Notch4-immortalized cells were maintained in DMEM containing 10% FBS. Cells were cross-linked with 1% formaldehyde for 10 min at room temperature (22 °C). After cells were collected, ChIP was performed as previously described [29] using antibodies against PBP or Med6 (Santa Cruz Biotechnology). The primers used were: 5′-ATAAATCAGAGAGGGGCTGG-3′ and 5′-AGTGCGTGCCAGCACTCTTT-3′ for SOX10-MCS4; 5′-TTTCCCCACCTCCTCTCCC-3′ and 5′-GGGATGCTCACTGGGATGG-3′ for SOX10-MCS5; 5′-TGGGGTTAAATCTCTCCCTG-3′ and 5′-GAGTAGGGCAGGAGGCGTG-3′ for SOX10-MCS7; 5′-GTGCTCACTCACAGCACTC-3′ and 5′-TTCACTCCCTCTTTCTTCCC-3′ for SOX10-MCS9. In all cases, PCR was performed with a serial dilution of input and various cycles (29–35 cycles) to ensure that amplification was maintained in the linear range. Each assay was repeated three times. Representative results are presented.
RESULTS
Immortalization of primary PBPLoxP/LoxP mammary epithelial cells by an active form of Notch4
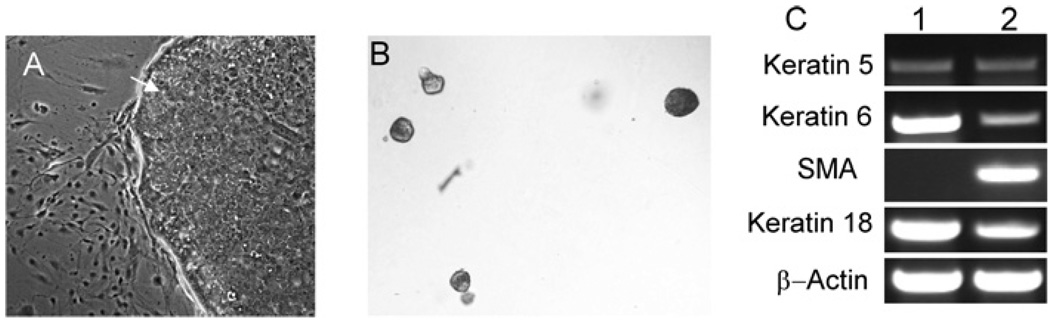
Our conditional knockout model established that PBP is essential for the mammary gland development [13]. To define themolecular mechanism through which PBP controls the mammary gland development, we decided to establish a mammary epithelial cell line with the genotype PBPLoxP/LoxP. Transgenic studies revealed that expression of the intracellular domain of Notch4 led to mammary tumours [23]. This active Notch4 domain could also cause anchorage-independent growth of immortalized mammary epithelial cells [30]. To ascertain if the active Notch4 domain can directly immortalize primary mammary epithelial cells and can be used to establish mammary epithelial cell lines, we infected primary PBPLoxP/LoxP mammary epithelial cells with a retrovirus expressing the constitutively activated form of mouse Notch4. A few days after retroviral infection, colonies emerged and rapidly expanded (Figure 1A). We found that cells from these colonies could proliferate indefinitely. To test if the mammary epithelial cells immortalized by constitutively active Notch4 were transformed, the cells were inoculated into soft agar. The active Notch4-expressing cells demonstrated growth in soft agar (Figure 1B). However, we did not see the formation of tumours in nude mice 3 months after injecting subcutaneously Notch4-immortalized cells into nude mice (results not shown). These cells expressed keratin 18 but not SMA, indicating that the cells were derived from the luminal epithelial cells (Figure 1C). In addition, the cells also expressed keratin 5/6, which appears to be preferentially expressed in mammary progenitor cells (Figure 1C) [31,32].
Figure 1. Immortalization of PBPLoxP/LoxP mammary epithelial cells with active Notch4.
(A) Formation of foci consisting of rapidly proliferating cells after transduction of the active form of Notch4 (arrow). (B) Growth of active Notch4-immortalized cells in soft agar. (C) Active Notch4-immortalized cells (lane 1) expressed keratin 18, keratin 5 and keratin 6, but not SMA, as revealed by RT–PCR. Lane 2: mouse mammary gland as the control.
Null mutation of PBP causes growth inhibition of active Notch4-immortalized cells
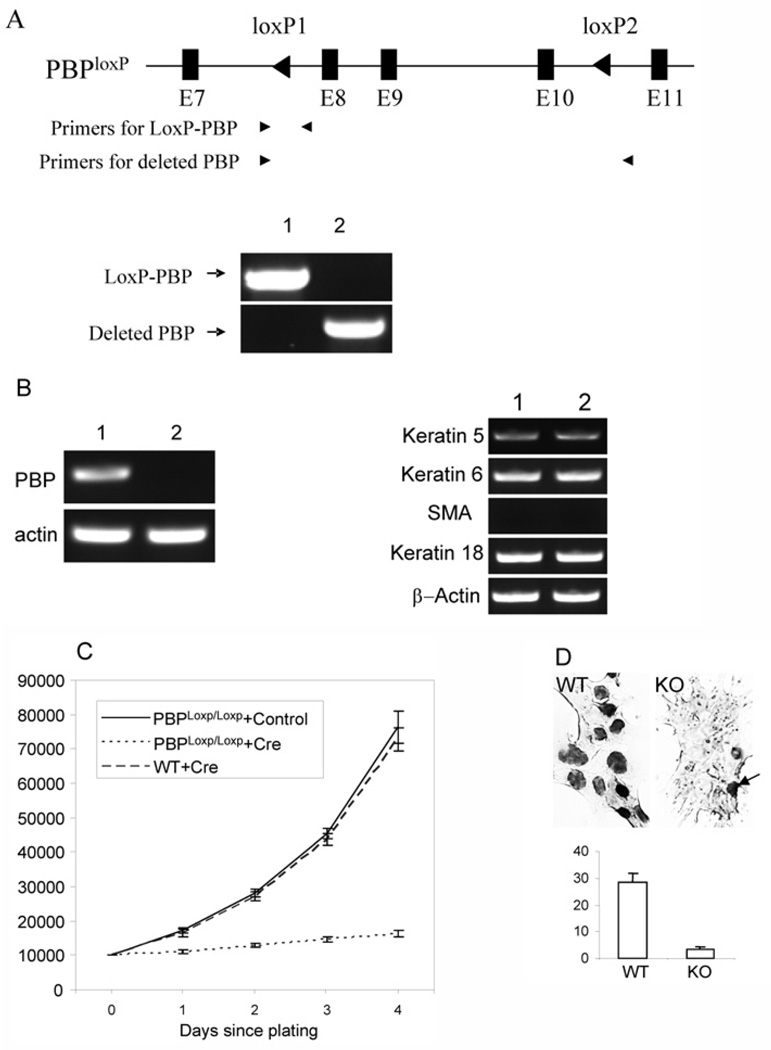
To understand the role of PBP in regulating the growth of mammary epithelial cells, Notch4-immortalized cells were infected with retroviruses expressing Cre-ERtam or control viruses. The cells infected with retrovirus were selected with puromycin and then treated with tamoxifen to mutate the PBP gene. PCR using primers specific for the LoxP-PBP allele or the mutated PBP allele showed that cells infected with Cre-expressing viruses carried mutated PBP genes (Figure 2A). RT–PCR revealed that PBP mRNA was absent in cells infected with Cre-ERtam-expressing viruses (Figure 2B). However, we found that loss of PBP expression did not alter the expression of keratin 18, keratin 5/6 and SMA (Figure 2B), indicating that the identity of cell lineage is not affected. In comparison with the cells infected with control viruses, PBPLoxP/LoxP cells infected with retroviral Cre-ERtam showed severe growth inhibition (Figure 2C). The growth inhibition was not caused by the expression of Cre, as the wild-type cells infected by Cre-ERtam retroviruses exhibited a similar growth rate to that of PBPLoxP/LoxP cells infected by control viruses. To find if PBP-deficient cells enter S-phase and proceed through DNA replication, we examined the incorporation of BrdU. PBP-deficient cells exhibited a dramatic reduction in BrdU-positive cells (Figure 2D). Although approx. 28% of nuclei were labelled in wild-type cells, only approx. 3% of PBP-deficient cells were positive for BrdU staining, indicating that PBP-deficient cells become quiescent.
Figure 2. Requirement of PBP for the growth of Notch4-immortalized cells.
(A) Generation of PBP mutation. Notch4-immortalized cells with PBPLoxP/LoxP genotype were infected with retroviral Cre-ERtam or control viruses. After selection with puromycin and treatment with tamoxifen for 2 days, genomic DNA was prepared. PCR was performed with primers specific for the LoxP-PBP transgene (lane 1, +control; lane 2, +Cre-ERtam) and the deleted PBP gene. (B) Left panel: Loss of PBP mRNA expression in cells infected with retroviral Cre-ERtam. Lane 1, +control; lane 2, +Cre-ERtam. RT–PCR was performed to examine PBP expression. Right panel: the expression of markers for cell lineages was not altered due to loss of PBP expression, as revealed by RT–PCR. Lane 1, wild-type cells; lane 2, PBP-deficient cells. (C) Growth inhibition of PBP-deficient cells. PBPLoxP/LoxP Notch4-immortalized cells infected with retroviral Cre-ERtam or control viruses or wild-type Notch4-immortalized cells infected with retroviral Cre-ERtam were treated with tamoxifen for 2 days and then plated at 1×104 cells per well. Viable cells (y axis) were counted by Trypan Blue staining at the indicated time points after initial seeding. The proliferation of PBP-deficient cells was markedly inhibited (P < 0.001). (D) Markedly decreased nuclei involved in DNA synthesis in PBP-deficient cells, as revealed by BrdU labelling (top panel). WT, wild-type cells (PBPLoxP/LoxP cells infected with control viruses); KO, PBP-deficient cells. The arrow points to one of the few BrdU-labelled cells in the PBP knockout cells. Bottom panel: Quantification of BrdU staining in wild-type (WT) and PBP-deficient (KO) cells.
Growth inhibition is caused solely by PBP deficiency
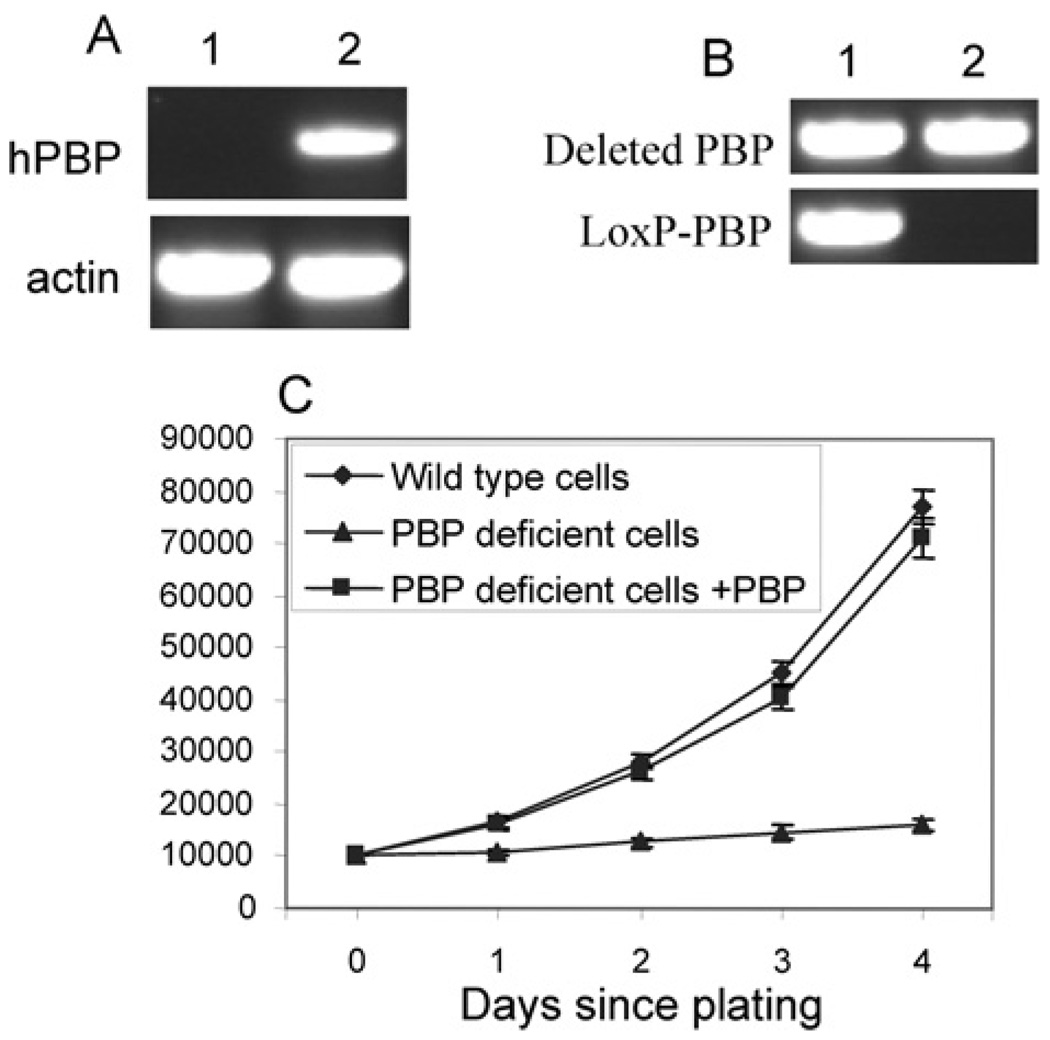
To exclude other factors, such as Cre expression which might contribute to the phenotype in PBP knockout cells, we performed a rescue experiment to demonstrate that PBP deficiency alone was the cause for severe growth retardation in the conditional knockout cells. Cells expressing Cre-ERtam were transfected with human PBP expression vectors or control empty vector. G418-resistant clones were combined and analysed for PBP expression. RT–PCR verified the expression of human PBP (Figure 3A). Interestingly, when the genomic DNA was prepared and the genotype analysed, we found that cells expressing exogenous PBP only carried the mutated PBP gene, whereas cells transfected with control vector still contained the floxed PBP gene (Figure 3B). Cre-ERtam possessed a low level of recombinase activity even without tamoxifen treatment [33]. This residual Cre activity probably constantly mutated the PBP gene, which decreased the number of wild-type cells and generated exogenous PBP-expressing knockout cells capable of proliferating the same as wild-type cells. The whole population of cells expressing exogenous PBP turned into PBP-knockout cells during the 2 weeks of selection by G418. As expected, only the cells expressing exogenous PBP demonstrated vigorous growth after tamoxifen treatment (Figure 3C).
Figure 3. Restoration of PBP reversed the growth inhibition.
(A) Expression of PBP in PBP conditional knockout cells by transfection. PBPLoxP/LoxP Notch4-immortalized cells expressing Cre-ERtam were transfected with a control vector (lane 1) or PBP expression vector (lane 2). RT–PCR was performed to show the expression of human PBP (hPBP). (B) The PBP gene was mutated in cells transfected with the PBP expression vector but not in cells transfected with the control vector. PCR was performed with primers specific for the LoxP-PBP transgene and deleted PBP gene (Lane 1, +control; lane 2, +pcDNA3.1-PBP). (C) Vigorous growth of PBP knockout cells expressing exogenous PBP. Cells transfected with PBP expression vector (PBP deficient cells + PBP), cells transfected with the control vector (PBP deficient cells) or wild-type Notch4-immortalized cells were treated with tamoxifen for 2 days and then plated and counted at the indicated time points. Unlike PBP-deficient cells, the proliferation of PBP-deficient cells expressing exogenous PBP was not inhibited (P < 0.001).
Truncated PBP without LXXLL motifs restored the growth of cells lacking endogenous PBP
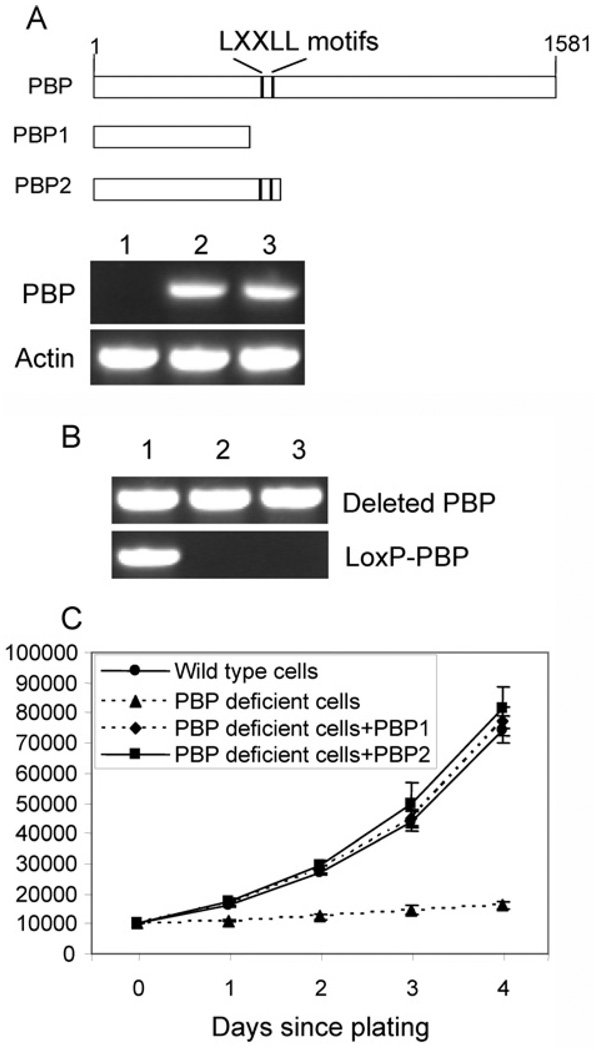
PBP is a nuclear receptor coactivator which interacts with nuclear receptors through two LXXLL motifs. To explore if signalling through nuclear receptors contributes to the function of PBP in the growth of Notch4-immortalized cells, we created two retroviral vectors expressing truncated PBPs (Figure 4A). Truncated PBP1 did not contain any LXXLL motifs, whereas truncated PBP2 carried both motifs. Notch4-immortalized cells expressing Cre-ERtam were infected with viral vector expressing PBP1, PBP2 or a control vector. Clones resistant to G418 were combined and examined for transgene expression (Figure 4).RT–PCR confirmed that both PBP1 and PBP2 were expressed. When the genomic DNA was genotyped, we found that both cells expressing PBP1 and PBP2 only carried the mutated PBP gene (Figure 4B), similar to cells expressing full-length PBP. In contrast, cells infected with the control vector still contained the floxed PBP gene. After the cells were treated with tamoxifen, cells expressing either PBP1 or PBP2 showed robust growth similar to that of wild-type cells, whereas cells infected with the control vector showed severe growth inhibition (Figure 4C).
Figure 4. Truncated PBP without LXXLL motifs reversed the growth inhibition.
(A) Expression of truncated PBPs.PBPLoxP/LoxP Notch4-immortalized cells expressing Cre-ERtam were infected with retroviruses expressing truncated PBPs or control viruses. Two truncated PBPs (PBP1 and PBP2) were created (top panel). The two LXXLL motifs are indicated. After being selected with G418, the expression of truncated PBP was revealed by RT–PCR (bottom panel). Lane 1, cells infected with a control retroviral vector; lane 2, cells infected with retroviral PBP1; lane 3, cells infected with retroviral PBP2. (B) The PBP gene was mutated in cells expressing PBP1 or PBP2, but not in cells transfected with the control vector. PCR was performed with primers specific for the LoxP-PBP transgene and deleted PBP gene (Lane 1, +control; lane 2, +PBP1; lane 3, +PBP2). (C) Vigorous growth of PBP knockout cells expressing either PBP1 or PBP2. Cells expressing PBP1 or PBP2, cells infected with the control vector or wild-type Notch4-immortalized cells were treated with tamoxifen for 2 days and then plated and counted at the indicated time points. The proliferation of PBP knockout cells expressing PBP1 or PBP2 was not inhibited (P < 0.001).
The expression of SOX10 in Notch4-immortalized cells is completely dependent on PBP
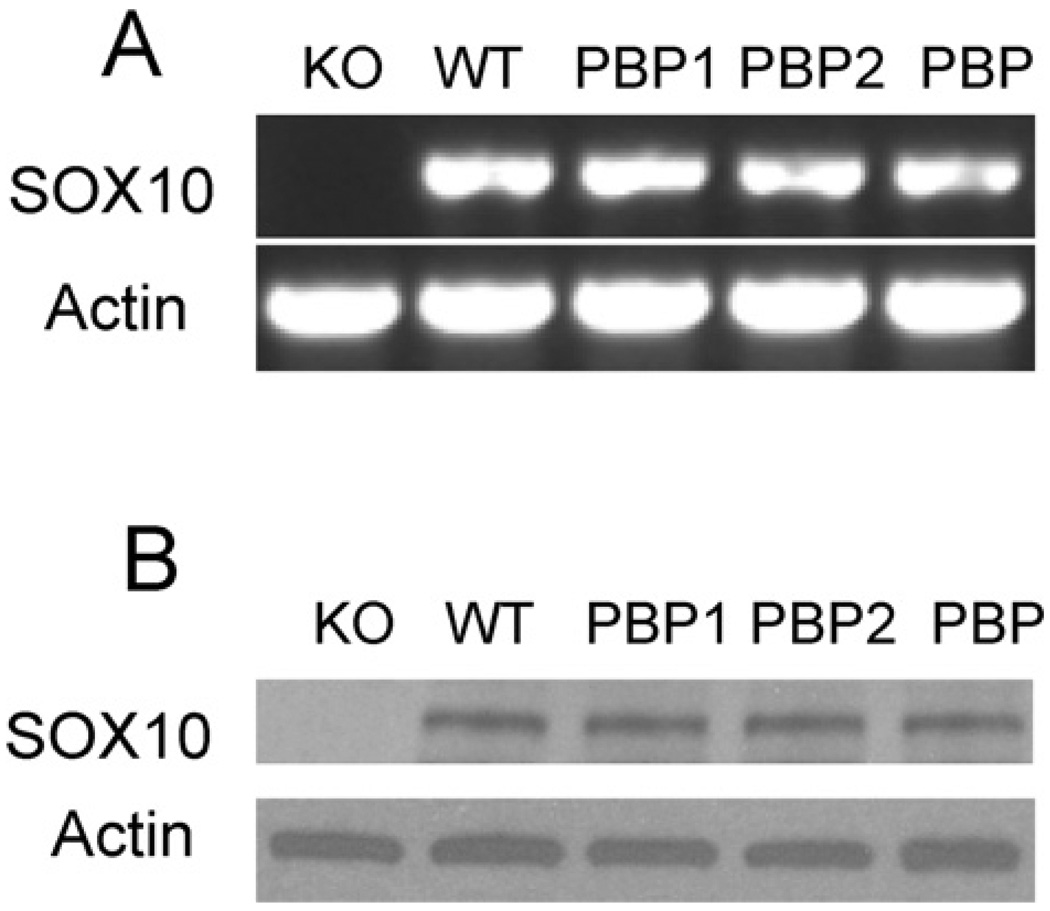
To further understand the mechanism by which PBP regulates mammary cell growth, we performed a microarray analysis to identify potential PBP target genes. The microarray was performed with wild-type and PBP−/− Notch4-immortalized mammary cells. We identified a total of 24 genes which were down-regulated with the loss of PBP (≤1.5-fold; P < 0.01) (Table 1) and a total of 52 genes up-regulated with the loss of PBP (≥1.5-fold; P < 0.01) (Table 2). The expression of keratin 14 was increased approx. 40-fold with PBP deficiency. Among the down-regulated genes is SOX10, which is reduced approx. 30-fold due to PBP deficiency. RT–PCR showed that SOX10 was expressed in wild-type cells and not expressed in PBP-deficient cells (Figure 5A), which is confirmed by Western blotting (Figure 5B). We also found that the cells expressing full-length PBP, partial PBP1 or PBP2 expressed SOX10.
Table 1.
Genes with significantly decreased expression in PBP-deficient mammary cells
| Gene | Fold(PBP+/+/PBP−/−) |
|---|---|
| SOX10 | 29.6 |
| Renin-binding protein | 9.5 |
| Scavenger receptor class A, member 3 (Scara3) | 7.6 |
| Casein κ | 6.9 |
| Transcription elongation factor A (SII)-like 8 | 6.6 |
| Syndecan binding protein (syntenin) 2 | 6.4 |
| Epithelial mitogen | 5.4 |
| CYP2D22 | 5.0 |
| Csnk | 4.9 |
| PGLYRP1 | 4.5 |
| Claudin 1 | 4.2 |
| CD59a | 3.7 |
| Phosphatase and actin regulator 1 | 3.6 |
| CTTNBP2N-terminallike | 3.4 |
| Plekhb1 | 3.3 |
| Wnt7a | 3.3 |
| ATPase, Na+/K+ transporting, β1 polypeptide | 3.2 |
| GRB10 | 2.9 |
| Interleukin 24 | 2.7 |
| Profilin 2 | 2.7 |
| Transmembrane protein 117 | 2.6 |
| Keratin 23 | 2.6 |
| Surfactant-associated protein D | 2.6 |
| Plasminogen activator, tissue | 2.5 |
Table 2 .
Genes with significantly increased expression in PBP-deficient mammary cells
| Gene | Fold (PBP+/+ /PBP−/−) |
|---|---|
| Keratin 14 | 0.025 |
| Cadherin 15 | 0.055 |
| Crystallin βA4 | 0.12 |
| Homeobox C9 | 0.17 |
| Matrix Gla protein | 0.17 |
| Proline arginine-rich end leucine-rich repeat | 0.17 |
| Interleukin 33 | 0.19 |
| Aldehyde dehydrogenase family 3, subfamily A1 | 0.19 |
| Lama3 | 0.20 |
| IL18R1 | 0.20 |
| LOXL1 | 0.21 |
| Aquaporin 1 | 0.21 |
| Calcium- and integrin-binding family member 3 | 0.22 |
| Kruppel-like factor 2 | 0.22 |
| 5'-Nucleotidase domain-containing 2 | 0.24 |
| WNT1-inducible signalling pathway protein 2 | 0.25 |
| Stabilin 1 | 0.25 |
| SPARC-related modular calcium-binding 1 | 0.26 |
| B-cell leukaemia/lymphoma 11B (Bcl11b) | 0.26 |
| AnnexinA8 | 0.27 |
| Syndecan 3 | 0.27 |
| Kelch-like 30 | 0.30 |
| Suppressor of cytokine signalling 3 | 0.30 |
| Adrenergic receptor, α2a | 0.30 |
| Lipopolysaccharide-binding protein | 0.30 |
| Lymphocyte antigen 6 complex, locus C1 | 0.30 |
| 5' -Nucleotidase, ecto | 0.30 |
| Diacylglycerol O-acyltransferase 2 | 0.31 |
| Procollagen, type VI, α1 | 0.31 |
| Fascin homologue 1, actin-bundling protein | 0.31 |
| Ly6/Plaur-domain-containing 3 | 0.31 |
| Dysferlin | 0.32 |
| Phosphodiesterase 1B, Ca2+/calmodulin dependent | 0.32 |
| Interleukin 13 receptor, α2 | 0.32 |
| Carbonic anyhydrase 12 | 0.32 |
| Keratin 6B | 0.33 |
| Adamts2 | 0.33 |
| Phospholipase A1 member A | 0.35 |
| Tripartite motif protein 29 | 0.35 |
| Procollagen, type V, α1 | 0.35 |
| NUAK family, SNF1-like kinase 2 | 0.36 |
| Slc24a3 | 0.36 |
| Musculin | 0.37 |
| P2rx1 | 0.37 |
| Stearoyl-CoA desaturase 1 | 0.37 |
| Leucine-rich-repeat-containing 26 | 0.37 |
| Lipoprotein lipase | 0.37 |
| FXYD domain-containing ion transport regulator 3 | 0.38 |
| Scinderin | 0.39 |
| Endothelin receptor type A | 0.39 |
| Regenerating islet-derived 1 | 0.39 |
| Prominin 2 | 0.40 |
Figure 5. PBP deficiency led to the complete loss of SOX10 expression.
(A) RT–PCR was performed to reveal SOX10 mRNA. WT, wild-type cells; KO, PBP−/− cells; PBP, PBP−/− cells expressing exogenous full-length PBP; PBP1, PBP−/− cells expressing the PBP1 fragment; PBP2, PBP−/− cells expressing the PBP2 fragment. (B) Western blotting was performed to reveal SOX10 protein expression.
The growth inhibition due to PBP deficiency can be reversed by SOX10
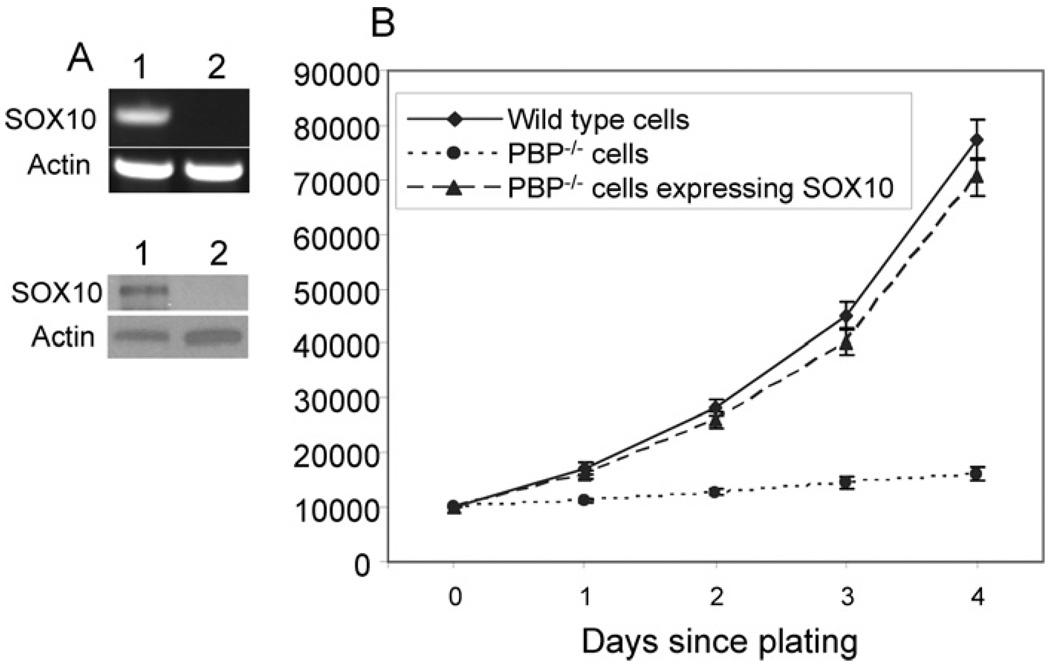
To test if SOX10 can compensate the loss of PBP expression, Notch4 immortalized cells expressing Cre-ERtam were transfected with the SOX10 expression vector or control vector. The expression of SOX10 in these cells was revealed by RT–PCR and Western blotting (Figure 6A). We compared the proliferation of wild-type cells, PBP-deficient cells and PBP-deficient cells expressing SOX10. The cells were inoculated at equal number and counted every day for 4 days. The PBP-deficient cells expressing SOX10 and wild-type cells showed similar rate of proliferation, whereas the growth of PBP-deficient cells was severely inhibited (Figure 6B).
Figure 6. Expression of SOX10 overcame the growth inhibition due to PBP deficiency.
(A) Expression of SOX10 in cells transfected with SOX10, as revealed by RT–PCR (top panel) andWestern blot (bottom panel). Lane 1, PBP-deficient cells transfected with SOX10; lane 2, PBP-deficient cells (Cre-ERtam-expressing PBPLoxP/LoxP cells with a 2 day treatment with tamoxifen). (B) SOX10-expressing PBP-deficient cells exhibited a similar growth rate to that of wild-type cells. PBP-deficient cells expressing exogenous SOX10, PBP-deficient cells or wild-type cells were plated at 1×104 cells per well. Viable cells were counted by Trypan Blue staining at different times. Similar to wild-type cells, PBP-deficient cells expressing SOX10 proliferated far faster than PBP-deficient cells (P < 0.001).
Suppression of SOX10 expression inhibits the proliferation of Notch4-immortalized cells
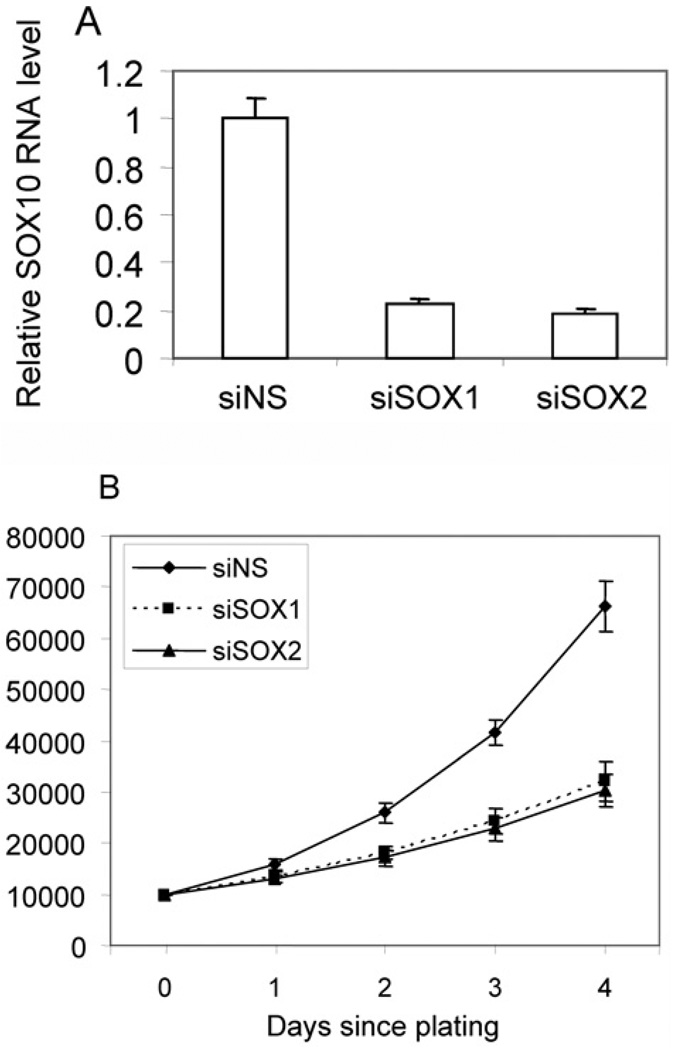
To confirm that SOX10 is required for the proliferation ofNotch4-immortalized cells, the cells were transfected with two siRNAs targeting SOX10, each of which reduced SOX10 transcript by approx. 80% compared with controls (Figure 7A). We found that cells with reduced SOX10 expression showed inhibited proliferation compared with control cells (Figure 7B).
Figure 7. Suppression of SOX10 inhibits the proliferation of Notch4-immortalized cells.
Notch4-immortalized cells were transfected with two siRNAs targeting SOX10 (siSOX1 and siSOX2) or control siRNA (siNS). (A) The suppression of SOX10 was demonstrated by real-time PCR. (B) The cells (1×104) were plated and counted at indicated time points after initial seeding. The cells expressing siRNAs against SOX10 proliferated slower than cells expressing control siRNA (P < 0.01).
Conserved regulatory regions 4, 7 and 9 at SOX10 display high enhancer activity in Notch4-immortalized Cells
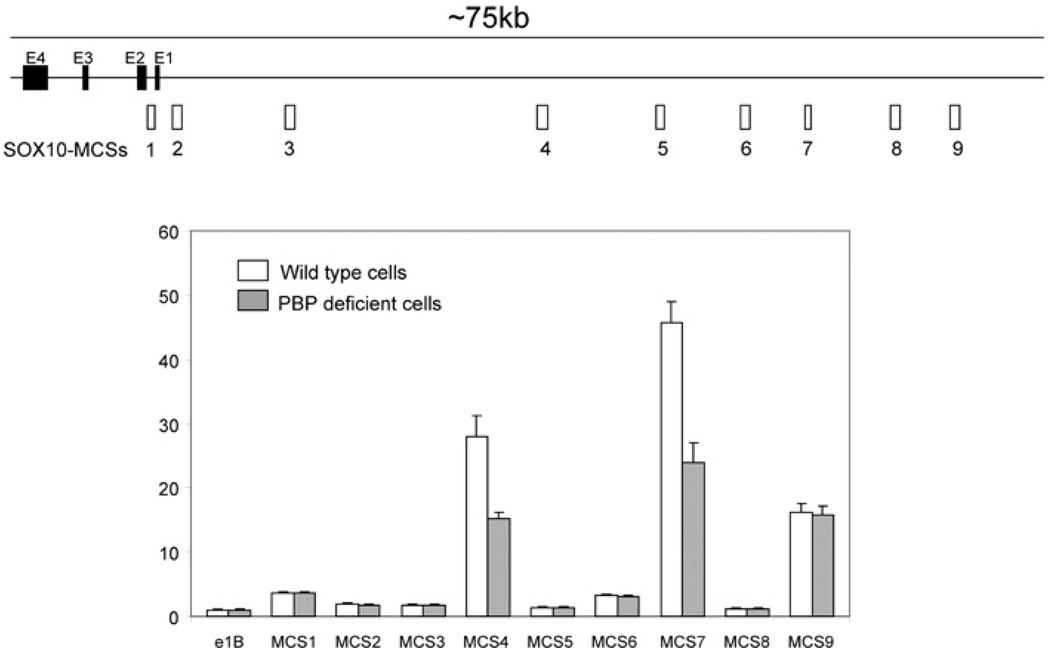
The SOX10 gene is under complicated regulation with regulatory elements distributed in more than 75 kb of DNA [24,34,35] (Figure 8). A total of nine MCSs, which show a high conservation among different species, have been identified through sequence comparison [23]. The enhancer activities of these MCSs have been assessed in melanocytes and Schwann cells. To gain insight into the mechanism by which the transcription of SOX10 is regulated in Notch4-immortalized mammary cells, the nine individual regulatory regions were cloned into a luciferase report gene with a minimal promoter. These vectors were transfected into wild-type cells or PBP-knockout cells. We found that fragments 4, 7 and 9 confer a high expression of luciferase (more than 10-fold over the control) in Notch4-immortalized cells (Figure 8). Although the SOX10 gene is not expressed in PBP-deficient cells, these regulatory regions were active in these cells. However, fragments 4 and 7 showed decreased activity in PBP-knockout cells, although the difference is much smaller compared with that of SOX10 mRNA expression.
Figure 8. Conserved regions 4, 7 and 9 are highly active in Notch4-immortalized cells.
The 75 kb SOX10 gene including its exons (E1 to E4) and its upstream sequence are presented along with the nine MCSs which show high conservation among different species (top panel). The SOX10 MCSs inserted into the upstream of a minimal promoter-driven luciferase reporter were transfected along with the β-gal expression vector into Notch4-immortalized wild-type (open bars) or PBP-deficient cells (grey bars) (bottom panel). E1B represents the minimal promoter-driven luciferase reporter. Luciferase activity was normalized to β-gal activity.
PBP is recruited to the enhancer of SOX10
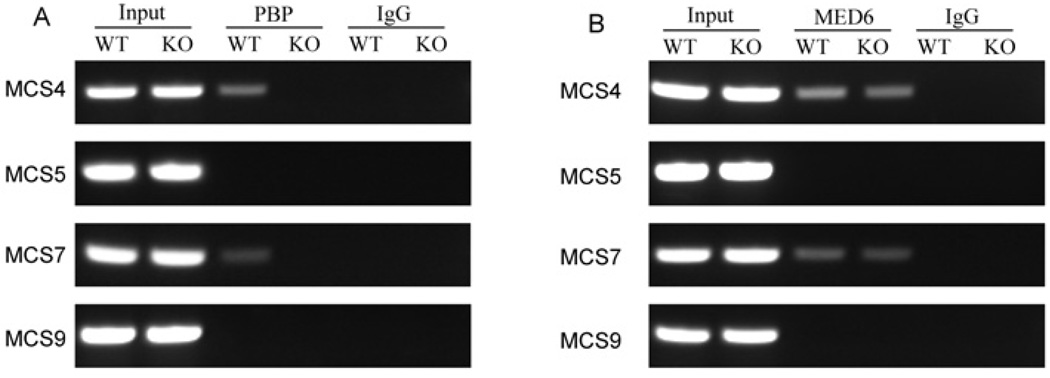
To investigate if PBP is recruited to SOX10 regulatory regions in the nuclei of living cells, we performed a ChIP assay. We examined four regions: MCS4 and MCS7, which are highly active in Notch4-immortalized cells and showed reduced activity with PBP deficiency, MCS9, which is highly active and is not affected by PBP deficiency, and MCS5, which is not active in this cell line. The antibodies against PBP could efficiently precipitate the fragments 4 and 7 of SOX10 gene, but could not pull down fragments 5 and 9 (Figure 9). When PBP-deficient cells were used in the ChIP assays, no PCR products were seen for any of the fragments, confirming the specificity of the antibodies. We also performed ChIP assays with the anti-Med6 antibody; Med6 is a component of TRAP–DRIP–Med complex. We observed the amplification with fragments 4 and 7 but not with fragments 5 and 9 in wild-type cells. In PBP-deficient cells, unlike the anti-PBP antibody, the anti-Med6 antibody precipitated fragments 4 and 7 but not fragments 5 and 9, indicating that the TRAP–DRIP–Med complex without PBP was recruited to these regulatory regions. These results suggested that the TRAP–DRIP–Med complex is recruited to the SOX10 enhancer in Notch4-immortalized cells.
Figure 9. PBP is recruited to enhancer regions of SOX10.
(A) Association of PBP with MCS4 and MCS7. Notch4-immortalized wild-type (WT) or PBP-deficient cells (KO) were fixed with formaldehyde. ChIP assays were performed using anti-PBP antibody or control IgG. Primers located in the MCSs 4, 5, 7 and 9 were used for PCR. (B) Association of the TRAP–Med complex with MCSs 4 and 7 in both wild-type cells (WT) and PBP−/− cells (KO), as revealed by ChIP using anti-Med6 antibody.
DISCUSSION
PBP is essential for the normal mammary gland development, as shown by the retarded elongation of ducts during puberty, severely decreased density of alveoli during pregnancy and the failure to produce milk to nurture pups [13]. Furthermore, PBP-deficient mammary epithelial cells cannot form mammospheres that are generated from mammary progenitor/stem cells. In the present study, we demonstrated that PBP is essential for the proliferation of active Notch4-immortalized cells by activating SOX10 expression. In contrast, we recently found that PBP is not required for the proliferation of Ras-transformed mammary epithelial cells (C. Qi, Y. T. Zhu, Y. Jia and Y. J. Zhu, unpublished work). Our findings suggest that the Notch signalling during normal mammary gland development is disrupted without PBP, which provides potential molecular mechanisms responsible for the phenotypes associated with PBP-deficient mammary glands.
At first, we found that the active form of Notch4 can immortalize the primary luminal epithelial cells, providing an easy way to establish mammary epithelial cell lines. This direct immortalization is in contrast with that by the expression of other oncogenes such as H-rasV12, which takes months to establish a stable cell line and this occurs only after the cells go through a process of senescence followed by apoptosis. Mammary epithelial cells with active Notch signalling express CD61, a marker for luminal progenitor cells [22]. We showed that Notch4-immortalized cells express keratin 5/6, which is considered as a mammary progenitor marker [31,32]. In addition, our microarray data showed that Notch4-immortalized cells do not express any differentiation markers such as β-casein. These data indicated that active Notch4-immortalized cells are similar to luminal progenitor cells.
Loss of PBP expression led to severe growth inhibition in active Notch4-immortalized cells. Interestingly, the PBP fragment without LXXLL motifs can reverse the growth inhibition. As the LXXLL motif is needed for the interaction between nuclear receptors and PBP, this finding suggested that the signalling through nuclear receptors is not critical for the function of PBP in the proliferation of Notch4-immortalized cells.
SOX10 plays a role in the formation of NCCs, maintenance of stem cell multipotency, specification of a subset of NCC fate and differentiation of specific cell types [19]. Its expression is under the control of several extrinsic signals, such as Wnts, BMPs (bone morphogenetic proteins) and FGFs (fibroblast growth factors) [19]. Its expression patterns are complicated, with transient expression in certain cells and persistent expression in others. Spatial and temporal control of tissue-specific SOX10 gene expression is mediated by the cis-regulatory elements, which are located as far as 65 kb upstream of SOX10 gene. It is impressive that the loss of PBP completely ablates the expression of the SOX10 gene. It would be of interest to see if the dependence of SOX10 expression on PBP occurs in all mammary epithelial cells expressing SOX10 or only in certain mammary epithelial cells at a certain developmental stage. It would be interesting to investigate if PBP is also critical for SOX10 expression in NCCs. If it is the case, the mutation on PBP or altered PBP expression affecting the SOX10 expression might be involved in those Waardenburg-Shah syndromes with no identified SOX10 mutations so far.
We found that SOX10 can compensate the growth inhibition caused by loss of PBP expression. Furthermore, suppressed expression of SOX10 inhibited the proliferation of Notch4-immortalized mammary cells. SOX10 is well known for its critical role in neural crest development, but its function in mammary gland development has not been investigated. Our findings imply that SOX10 is an essential gene for the proliferation of luminal progenitor cells. SOX10 has been shown to be expressed in myoepithelial cells of mammary glands [36], indicating that SOX10 could also play a role in these cells.
We found that MCS4 and MCS7 showed differential activities in the wild-type and knockout cells. MCS4 and MCS7 are likely to contain sites for certain transcription factors which require PBP for their activities. This argument is supported by the ChIP assay showing that PBP was directly recruited to MCS4 and MCS7. A number of transcription factors, including SOX9, AP2 (adaptor protein 2) and LEF (lymphoid enhancer factor), have been found to be involved in the regulation of SOX10 [35]. We can propose two scenarios through which PBP controls SOX10 expression. Certain transcription factors could interact with PBP to recruit the TRAP–Med complex to the SOX10 gene. Or PBP is indirectly recruited to the SOX10 gene by other components of the TRAP–Med complex, and PBP is required for the function of the TRAP–Med complex to activate the SOX10 gene. Given that truncated PBP without the last 1100 amino acids is sufficient for SOX10 expression and this fragment shares homology with Med1 from yeast and Drosophila, there is evidence for the second scenario. Intriguingly, the PBP fragment without any LXXLL motifs was found to be sufficient for its function in adipogenesis mediated by PPARγ [37]. To this end, identifying the transcription factor recruiting the TRAP–Med complex will clarify the issue and is our focus for the immediate future.
ACKNOWLEDGEMENTS
We thank Dr Wegner and Dr Cleary (Stanford University, U.S.A.) for providing the anti-SOX10 antibody and retroviral Cre-ERtam vector respectively. We also thank Dr J.K. Reddy for his advice.
FUNDING
This work was supported by the National Institutes of Health [grant number CA 88898 (to Y.-J. Z)]. A. S. M. and M. K. P. were supported by an award from the National Institute of General Medical Sciences [grant number R01GM071648 (to A. S. M.)].
Abbreviations used
- BrdU
bromodeoxyuridine
- ChIP
chromatin immunoprecipitation
- DMEM
Dulbecco’s minimal Eagle’s medium
- DRIP
vitamin D receptor-interacting protein
- FBS
fetal bovine serum
- MCS
multiple species conserved sequence
- Med
mediator
- NCC
neural crest cell
- PBP
peroxisome-proliferator-activated receptor-binding protein
- PPAR
peroxisome-proliferator-activated receptor
- RT
reverse transcription
- siRNA
small interfering RNA
- SMA
smooth muscle actin
- SOX
Sry-related HMG (high-mobility group) box
- TRAP
thyroid-hormone-receptor-associated protein.
Footnotes
AUTHOR CONTRIBUTION
Yi-Jun Zhu designed the study. Megana Prasad and Andrew McCallion provided the MCS reporter vectors. Chao Qi developed the cell line. Liping Hu constructed PBP expression vectors and expressed PBP in Notch4-immortalized cells. Yuzhi Jia performed the microarray analysis. Yiwei Tony Zhu analysed the cell lines and the MCSs of SOX10. Yi-Jun Zhu wrote the manuscript.
REFERENCES
- 1.Zhu Y, Qi C, Jain S, Rao MS, Reddy JK. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J. Biol. Chem. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- 2.Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl. Acad. Sci. U.S.A. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 4.Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Naar AM, Erdjument-Bromage H, Tempst P, Freedman LP. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 5.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Crawford SE, Qi C, Misra P, Stellmach V, Rao MS, Engel JD, Zhu Y, Reddy JK. Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J. Biol. Chem. 2002;277:3585–3592. doi: 10.1074/jbc.M107995200. [DOI] [PubMed] [Google Scholar]
- 8.Stumpf M, Waskow C, Krotschel M, van Essen D, Rodriguez P, Zhang X, Guyot B, Roeder RG, Borggrefe T. The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc. Natl. Acad. Sci. U.S.A. 2006;103:18504–18509. doi: 10.1073/pnas.0604494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Udayakumar TS, Belakavadi M, Choi KH, Pandey PK, Fondell JD. Regulation of Aurora-A kinase gene expression via GABP recruitment of TRAP220/MED1. J. Biol. Chem. 2006;281:14691–14699. doi: 10.1074/jbc.M600163200. [DOI] [PubMed] [Google Scholar]
- 10.Zhu Y, Qi C, Jia Y, Nye JS, Rao MS, Reddy JK. Deletion of PBP/PPARBP, the gene for nuclear receptor coactivator peroxisome proliferator-activated receptor-binding protein, results in embryonic lethality. J. Biol. Chem. 2000;275:14779–14782. doi: 10.1074/jbc.C000121200. [DOI] [PubMed] [Google Scholar]
- 11.Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell. 2000;5:683–693. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- 12.Jia Y, Qi C, Kashireddi P, Surapureddi S, Zhu YJ, Rao MS, Le Roith D, Chambon P, Gonzalez FJ, Reddy JK. Transcription coactivator PBP, the peroxisome proliferator-activated receptor (PPAR)-binding protein, is required for PPARα-regulated gene expression in liver. J. Biol. Chem. 2004;279:24427–24434. doi: 10.1074/jbc.M402391200. [DOI] [PubMed] [Google Scholar]
- 13.Jia Y, Qi C, Zhang Z, Zhu YT, Rao SM, Zhu YJ. Peroxisome proliferator-activated receptor-binding protein null mutation results in defective mammary gland development. J. Biol. Chem. 2005;280:10766–10773. doi: 10.1074/jbc.M413331200. [DOI] [PubMed] [Google Scholar]
- 14.Zhu Y, Qi C, Jain S, Le Beau MM, Espinosa R, 3rd, Atkins GB, Lazar MA, Yeldandi AV, Rao MS, Reddy JK. Amplification and overexpression of peroxisome proliferator-activated receptor binding protein (PBP/PPARBP) gene in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10848–10853. doi: 10.1073/pnas.96.19.10848. Reddy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefebvre V, Dumitriu B, Penzo-Mendez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int. J. Biochem. Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiefer JC. Back to basics: Sox genes. Dev. Dyn. 2007;236:2356–2366. doi: 10.1002/dvdy.21218. [DOI] [PubMed] [Google Scholar]
- 17.Pingault V, Bondurand N, Kuhlbrodt K, Goerich DE, Prehu MO, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, Matthijs G, et al. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat. Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- 18.Bondurand N, Kuhlbrodt K, Pingault V, Enderich J, Sajus M, Tommerup N, Warburg M, Hennekam RC, Read AP, Wegner M, Goossens M. A molecular analysis of the yemenite deaf-blind hypopigmentation syndrome: SOX10 dysfunction causes different neurocristopathies. Hum. Mol. Genet. 1999;8:1785–1789. doi: 10.1093/hmg/8.9.1785. [DOI] [PubMed] [Google Scholar]
- 19.Kelsh RN. Sorting out Sox10 functions in neural crest development. Bioessays. 2006;28:788–798. doi: 10.1002/bies.20445. [DOI] [PubMed] [Google Scholar]
- 20.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 21.Politi K, Feirt N, Kitajewski J. Notch in mammary gland development and breast cancer. Semin. Cancer Biol. 2004;14:341–347. doi: 10.1016/j.semcancer.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 22.Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat ML, Oakes SR, Lindeman GJ, Visvader JE. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Jhappan C, Gallahan D, Stahle C, Chu E, Smith GH, Merlino G, Callahan R. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev. 1992;6:345–355. doi: 10.1101/gad.6.3.345. [DOI] [PubMed] [Google Scholar]
- 24.Antonellis A, Huynh JL, Lee-Lin SQ, Vinton RM, Renaud G, Loftus SK, Elliot G, Wolfsberg TG, Green ED, McCallion AS, Pavan WJ. Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish. PLoS Genet. 2008;4:e1000174. doi: 10.1371/journal.pgen.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maka M, Stolt CC, Wegner M. Identification of Sox8 as a modifier gene in a mouse model of Hirschsprung disease reveals underlying molecular defect. Dev. Biol. 2005;277:155–169. doi: 10.1016/j.ydbio.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Richards J, Guzman R, Imagawa W, Nandi S. Sustained growth in primary culture of normal mammary epithelial cells embedded in collagen gels. Proc. Natl. Acad. Sci. U.S.A. 1980;77:2088–2092. doi: 10.1073/pnas.77.4.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 28.Cifone MA, Fidler IJ. Correlation of patterns of anchorage-independent growth with in vivo behavior of cells from a murine fibrosarcoma. Proc. Natl. Acad. Sci. U.S.A. 1980;77:1039–1043. doi: 10.1073/pnas.77.2.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 30.Robbins J, Blondel BJ, Gallahan D, Callahan R. Mouse mammary tumor gene int-3: a member of the notch gene family transforms mammary epithelial cells. J. Virol. 1992;66:2594–2599. doi: 10.1128/jvi.66.4.2594-2599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sapino A, Macri L, Gugliotta P, Pacchioni D, Liu YJ, Medina D, Bussolati G. Immunophenotypic properties and estrogen dependency of budding cell structures in the developing mouse mammary gland. Differentiation. 1993;55:13–18. doi: 10.1111/j.1432-0436.1993.tb00028.x. [DOI] [PubMed] [Google Scholar]
- 32.Smith GH, Mehrel T, Roop DR. Differential keratin gene expression in developing, differentiating, preneoplastic, and neoplastic mouse mammary epithelium. Cell Growth Differ. 1990;1:161–170. [PubMed] [Google Scholar]
- 33.Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deal KK, Cantrell VA, Chandler RL, Saunders TL, Mortlock DP, Southard-Smith EM. Distant regulatory elements in a Sox10-β GEO BAC transgene are required for expression of Sox10 in the enteric nervous system and other neural crest-derived tissues. Dev. Dyn. 2006;235:1413–1432. doi: 10.1002/dvdy.20769. [DOI] [PubMed] [Google Scholar]
- 35.Werner T, Hammer A, Wahlbuhl M, Bosl MR, Wegner M. Multiple conserved regulatory elements with overlapping functions determine Sox10 expression in mouse embryogenesis. Nucleic Acids Res. 2007;35:6526–6538. doi: 10.1093/nar/gkm727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nonaka D, Chiriboga L, Rubin BP. Sox10: a pan-schwannian and melanocytic marker. Am. J. Surg. Pathol. 2008;32:1291–1298. doi: 10.1097/PAS.0b013e3181658c14. [DOI] [PubMed] [Google Scholar]
- 37.Ge K, Cho YW, Guo H, Hong TB, Guermah M, Ito M, Yu H, Kalkum M, Roeder RG. Alternative mechanisms by which mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor γ-stimulated adipogenesis and target gene expression. Mol. Cell Biol. 2008;28:1081–1091. doi: 10.1128/MCB.00967-07. [DOI] [PMC free article] [PubMed] [Google Scholar]