Abstract
Genotyping studies show a polarized geographic distribution of Trypanosoma cruzi lineages in humans. Here, we assessed their distribution along Latin America through an immunological approach we designated Western blot (WB) assay with Trypomastigote small-surface antigen (TSSA) I and TSSA II (TSSA-WB). These antigens are expressed by T. cruzi I (TCI; now TcI) and T. cruzi II (TCII; reclassified as TcII to TcVI) parasites. TSSA-WB showed good concordance with genotyping tests. An unexpected frequency of TSSA II recognition was observed in Colombia, Venezuela, and Mexico (northern region of Latin America). In Argentina and Paraguay (southern region), immunophenotyping confirmed the already reported TCII (TcII to TcVI) dominance. The lineage distribution between these regions showed significant difference but not among countries within them (except for Colombia and Venezuela). TSSA-WB shows TCII emergence in the northern region where TCI was reported as dominant or even as the unique T. cruzi lineage infecting humans.
Introduction
Trypanosoma cruzi, the etiological agent of Chagas disease, exhibits a multiclonal structure given its mainly clonal pattern of evolution and little genetic exchange.1,2
The variability of T. cruzi isolates together with the heterogeneity of human populations could be responsible for diverse clinical forms of the infection, which range from asymptomatic to gastrointestinal and heart involvement. They are also variable in different geographic regions.3–5
Multiple studies on the above-mentioned diversity of T. cruzi strains led to their classification into two highly divergent phylogenetic lineages named T. cruzi I (TCI) and T. cruzi II (TCII).6 Further studies described TCII as divided into five discrete subgroups: TCIIa–e.7 Great efforts have also been made to elucidate the genetic structure of the T. cruzi population and relate these data with the described parasite subgroups.8,9 In a recent meeting, an expert committee revised the available information about T. cruzi divergence, reclassified parasite strains by splitting them into six groups and renamed them as discrete typing units (DTUs) designated T. cruzi I to T. cruzi VI.10 According to this proposal, TCI is now named TcI, whereas TCIIa is TcIV, TCIIb is TcII, TCIIc is TcIII, TCIId is TcV, and TCIIe is TcVI.10
Genotyping studies showed that TcI (TCI) is highly dominant in the sylvatic and domestic cycles of transmission from the Amazonas River, in Brazil, northwards. It was detected causing human infections in Colombia, Mexico, and Central America and in a few autochthonous cases in the United States.11–18 In the southern cone of South America, this group has been mainly associated with the sylvatic transmission, whereas TcII, TcV, and TcVI DTUs (included in TCII) show a high prevalence in the domestic cycle, causing the vast majority of infections in humans and other mammalian hosts.19–22
Most typing studies have been performed by employing T. cruzi isolates obtained from blood samples or maintained by serial passages in cultures. Parasite composition of these extracts can differ from that involved in the host infection, because these strategies are known to lead to parasite subpopulation selection,23 thus underestimating the original parasite diversity of the sample. The recent introduction of new direct genotyping tools helps to minimize their underestimation; nevertheless, the low parasitemia that characterizes the chronic phase of the infection remains as a limitation.24 These facts highlight the relevance of developing new strategies to identify parasite subpopulations.
The trypomastigote small-surface antigen (TSSA) is expressed by the circulating forms of T. cruzi and belongs to the group III of the mucin superfamily (TcMUC). TSSA I and TSSA II antigens (cloned from TcI and TcVI parasites, respectively) were originally described as encoded by two alleles (tssa I and tssa II) that are exclusive of the previously denominated TCI and TCII genomes, respectively.25 The detection of anti-TSSA antibodies on serum samples was proposed as an immunological marker of parasite populations involved in T. cruzi infections.25 Recently, a greater diversity of the tssa gene among the new DTUs was reported, showing a high sequence homology among TcII, TcV, and TcVI alleles, whereas TcIII and TcIV genes share features with TcI.26 However, serologic assays that allow us to know whether those polymorphisms correlate with differential humoral immune responses are lacking.
The description of T. cruzi lineages and their distribution along the endemic area provide an adequate reference to analyze the involvement of parasite subpopulations in human disease that have not been clearly defined to date. In this work, lineage typing of the parasites causing human infections in endemic countries (Argentina, Paraguay, Colombia, Venezuela, and Mexico) was performed by detecting antibodies directed to the recombinant antigens TSSA I and TSSA II in Western-blotting assays (TSSA-WB).
Materials and Methods
Human specimens for T. cruzi lineage distribution assessment in Latin America.
A total of 690 serum samples were analyzed by TSSA-WB. They were collected from people living in countries of the endemic area: Mexico (82 seronegative and 83 seropositive), Venezuela (53 seronegative and 103 seropositive), Colombia (42 seronegative and 157 seropositive), Argentina (37 seronegative and 69 seropositive), and Paraguay (21 seronegative and 43 seropositive). Paraguayan samples include sera from 51 Amerindians living in palm tree homes of the Chaco region in contact with wild animals burrowing among wood piles next to their houses where TcI-infecting T. infestans have been detected and characterized; there is an 80% prevalence of T. cruzi infection in this region.27
Conventional T. cruzi diagnosis in human serum samples obtained from different countries of Latin America.
Conventional serological tests routinely used to diagnose T. cruzi infection were run in the laboratories of origin. As general criteria, samples reacting in two serologic tests were scored as infected.
Two serum panels from Mexican individuals were subjected to homemade serologic tests. The first one was evaluated by enzyme-linked immunosorbent assay (ELISA) and WB with total epimastigote extracts (Queretaro strain) as antigen. The second panel was assessed with antigens of the Ninoa Mexican strain: intact epimastigotes were used in indirect immunofluorescence (IIF) assays, and total protein extracts were used in ELISAs.28,29 Colombian samples were analyzed by IIF and ELISA using Dm7, MG8, and Cas 15 T. cruzi strain antigens.30 In Venezuela, diagnosis was defined by the consensus results obtained in two laboratories. One of them carried out IIF, indirect hemagglutination (IHA), and ELISA, as previously reported.31 The other applied two ELISAs: a commercially available kit (BIOSChile, Santiago, Chile) and a homemade test with antigens prepared from metacycle-like forms of the Y T. cruzi strain.32 Sera from Paraguayan individuals were subjected to a homemade ELISA and an IIF test.33 Sera from Argentinean people were assessed using commercial tests: IHA (Laboratory Polychaco, Buenos Aires, Argentina), ELISA (either from Wiener, Rosario, Argentina or Chagatek, Biomerieux, Argentina), and particle agglutination (Serodia, Fujirebio, Japan).24,34
Specimens from non–T. cruzi-infected patients.
We analyzed serum samples from patients not infected by T. cruzi but with cutaneous leishmaniasis (N = 20, from Paraguay), malaria (N = 7, from Brazil), toxoplasmosis (N = 18, from Argentina), syphilis (N = 10, from Paraguay), idiopathic megaviscera or cardiopathy (N = 22, from Brazil), systemic lupus erythematosus (N = 5), rheumatoid arthritis (N = 1), myositis (N = 1), or mixed connective tissue disease (N = 1). Samples from 16 healthy individuals without epidemiological risk of T. cruzi infection were also included.
Specimens subjected to both genotyping and immunophenotyping.
Molecular and inmunological typing were performed in samples from 66 patients living in Argentina, Bolivia, Colombia, and Paraguay.
For genotyping, DNA was purified from 500 µL of peripheral blood, as previously reported.24 Of 66 patients, 14 were subjected to heart transplant; sera obtained before transplantation were processed. Paraffin-embedded heart explant samples were processed to obtain DNA using the QIAmp tissue kit (Qiagen, Valencia, CA) as reported.34 T. cruzi genotyping was carried out by polymerase chain reaction (PCR) strategies targeted to the intergenic region of spliced leader genes (SL-IR). Three independent reactions (SL-IR I with primers UTCC and TC2, SL-IRac with primers UTCC and Tcac, and SL-IR II with primers UTCC and TC1) allowed classification of T. cruzi into three groups: TcI, TcIII/IV, and TcII/V/VI, respectively.24 Within the last group, some DTUs were identified by PCR targeted to the D7 domain of 24S α rDNA genes and the A-10 fragment to discriminate among TcII, TcV, and TcVI DTUs, as reported.24 All sera were immunophenotyped by TSSA-WB as described below.
Expression and purification of TSSA antigens.
A fragment of tssaI gene (GGATCCGTTACAGCGAATGGTGGGTCTACTAGTTCTACCCCACCTGGTAAGGACAAGAAAACAGCTGCAGGGGGAACTCCATCTCCATCGGGAGCTTCTTCAGGTGAAGCAGAAGCCTCCTCAAAATCGAATTC) from the Dm28c strain (TCI, now TcI) or tssaII gene (GGATCCGTTACAGCGAATGGTGGGTCTACTAGTTCTACCCCACCTTCTGGTACGGAAAATAAACCAGCTACAGGGGAAGCTCCATCTCAACCGGGGGCTTCTTCAGGTGAAGCAGAAGCCTCCTCAAAAATCACTAGTGAATTC) from the CL Brener strain (TCII, presently TcVI) was cloned into pGEX-2T plasmid (BamHI/EcoRI sites are underlined; GE Healthcare).25 Their encoded peptides, namely glutathione S-transferase-TSSA I (GST-TSSA I) and GST-TSSA II, were produced in Escherichia coli and purified by using GSTrap columns (GE Healthcare).25 Recombinant GST was also purified to detect sera background reactivity.
Western blotting with TSSA antigens (TSSA-WB).
GST, GST-TSSA I, and GST-TSSA II proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (Hybond P; GE Healthcare). Diluted sera (1:100) were adsorbed to nitrocellulose-immobilized GST overnight at 4°C and then incubated for 2 hours at room temperature on membranes containing the three separated antigens. Rabbit anti-total human immunoglobulin G (IgG; γ-specific) conjugated to horseradish peroxidase (DAKO, Denmark) was used. Hydrogen peroxide and 3,3′-diaminobenzidine (Sigma) were used for the chromogenic visualization of antigen–antibody specific interaction. Figure 1 shows examples of TSSA I, II and I-II recognition patterns obtained in WB assays by using T. cruzi patients serum samples.
Figure 1.
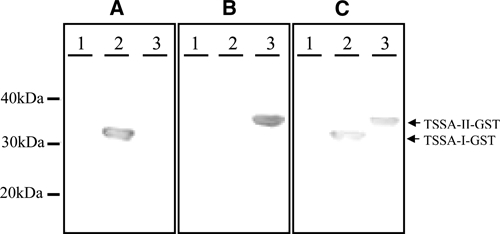
Examples of TSSA I, TSSA II, and TSSA I–II recognition patterns obtained in Western blot assays (A, B, and C, respectively). GST, recombinant TSSA I, and TSSA II (lanes 1, 2, and 3, respectively) were loaded in SDS-PAGE gels and transferred to PVDF membranes to perform TSSA-WB with patients sera. Representation of molecular weight pattern is depicted at the left. GST = glutathione S-transferase; TSSA = trypomastigote small-surface antigen.
Statistical analysis.
Statistical comparisons were performed by the χ2 or Fisher exact tests. Reliability between immunophenotyping and genotyping methods was also determined by the Cohen's κ coefficient.
Ethics statement.
This study was conducted in accordance with the Declaration of Helsinki, under approval of the local ethical committees of the participating institutions from the different countries as well as of the Ethics Review Committee of the World Health Organization. All patients provided written informed consent for sample collection and analysis.
Results
Evaluation of non–T. cruzi-infected human serum samples by TSSA-WB.
The reactivity to the recombinant TSSA antigens of 101 samples from patients suffering different pathologies (cutaneous leishmaniasis, toxoplasmosis, autoimmune diseases, idiopathic megaviscera or cardiopathy, or syphilis) and healthy individuals was analyzed by TSSA-WB. Only one malaria patient showed reactivity against TSSA II.
T.cruzi lineage identification by immunophenotyping and genotyping.
To check the ability of TSSA-WB to identify T. cruzi lineages, we performed a comparative study using genotyping markers as a reference (Table 1). Of 66 samples analyzed, 31 yielded positive PCR results. Among the latter, 17 showed fully coincident genotyping and immunophenotyping determinations. A partial coincidence was observed for two patients: one serum showed mixed TSSA reactivity when genetic markers identified only TcI, whereas the other one recognized TSSA II but was genotyped as a mixed infection. However, 12 samples did not render concordant results; 3 of these samples were genotyped as TcI but were reactive to TSSA II, 7 samples did not recognize any of the TSSA antigens, and the remaining 2 samples did not display conclusive results in TSSA-WB (Table 1).
Table 1.
Lineage identification by immunophenotyping and genotyping assays
| Immunophenotyping | Genotyping | ||
|---|---|---|---|
| TCI | TCII | TCI + TCII | |
| TSSA I | 3 | 0 | 0 |
| TSSA II | 3 | 13* | 1 |
| TSSA I–II | 1 | 0 | 1† |
| Inconclusive | 2 | 0 | 0 |
| Negative | 5 | 2 | 0 |
| Total | 14 | 15 | 2 |
In some cases, DTUs among TCII infections were identified by genotyping. P < 0.0001 (κ coefficient).
Four patients with TcV, one patient with TcVI, and one patient with TcII and/or TcVI bloodstream parasites.
One patient with TcI and TcV mixed infection.
Measure of agreement between genotyping and immunophenotyping results was assessed by the Cohen's κ coefficient (inconclusive and negative results were excluded from the analysis). As seen in Table 1, genotyping and immunophenotyping data were highly concordant (P < 0.0001).
Of the 35 samples non-reactive by genotyping methods, 26 recognized TSSA II, and 3 reacted with both TSSA I and TSSA II. Two sera did not render conclusive results, and four samples were not reactive in the immunophenotyping assay (data not shown).
TSSA-WB and conventional serology reactivity of serum samples.
TSSA-WB and tests routinely used for T. cruzi diagnosis (conventional serology [CS]) were assayed in samples from individuals living in Mexico, Colombia, Venezuela, Paraguay, and Argentina.
When co-reactivity between TSSA-WB and CS was assessed, co-positivity and co-negativity values were 61.6% and 85.2%, respectively. Parameters detailed for each country in Table 2 show higher co-reactivity between samples from Argentina and Paraguay than for patients from Mexico and Colombia/Venezuela.
Table 2.
TSSA-WB and CS co-reactivity of samples from Latin American patients
| Sample origin | TSSA I/TSSA II vs. CS | |
|---|---|---|
| Co-positivity (%) | Co-negativity (%) | |
| Mexico | 36.1 | 78.0 |
| Colombia | 62.0 | 63.4 |
| Venezuela | 42.7 | 82.0 |
| Argentina | 97.1 | 100.0 |
| Paraguay | 93.0 | 71.4 |
Co-positivity = 100 × (frequency of TSSA-WB reactive samples/frequency of CS positive samples). Co-negativity = 100 × (frequency of TSSA-WB unreactive samples/frequency of CS negative samples).
T.cruzi immunophenotyping in patients from Latin American countries.
A comparative analysis among different countries was done based on TSSA-WB and CS co-positive serum samples. Table 3 shows details of recombinant TSSA antigens recognition.
Table 3.
T. cruzi immunophenotyping in patients from Latin America
| Sample origin | TSSA reactivity | Total | ||
|---|---|---|---|---|
| I | II | I–II | ||
| Mexico | 10 (33%) | 11 (37%) | 9 (30%) | 30 |
| Colombia | 16 (17%) | 41 (44%) | 36 (39%) | 93 |
| Venezuela | 14 (34%) | 19 (46%) | 8 (19%) | 41 |
| Argentina | 0 (0%) | 66 (99%) | 1 (1%) | 67 |
| Paraguay | 0 (0%) | 39 (98%) | 1 (2%) | 40 |
| Total | 40 | 176 | 55 | 271 |
Samples co-positive by TSSA-WB and CS were considered. Data represent the lineage distribution analysis for countries, with comparisons between TSSA reactivity (I vs. II; II vs. I–II; I vs. I–II) being performed with the χ2 or Fisher exact tests. Overall difference was P < 0.00001 (χ2 test, degrees of freedom (df) = 8). Comparisons of TSSA reactivity between southern countries (Argentina and Paraguay) were not significant. Comparisons of TSSA reactivity among northern countries (Mexico, Venezuela, and Colombia) remained insignificant except for TSSA I vs. TSSA I–II reactivity between Venezuela and Colombia (P < 0.025; χ2 test, df = 1). Comparison for lineage distribution on dividing into northern (Mexico, Venezuela, and Colombia) and southern (Paraguay and Argentina) countries: overall difference P < 0.00001 (χ2, df = 2); TSSA I vs. TSSA II: P < 0.0001 (χ2, df = 1); TSSA II vs. TSSA I–II: P < 0.0001 (χ2, df = 1). Comparisons of TSSA I vs. TSSA I/II reactivity were not significant.
Similar proportions of Mexican patients were found to recognize solely TSSA I, TSSA II, or both (mixed infections) (Table 3). Among Colombian samples, single TSSA II and mixed reactivity showed high frequencies (41 and 36 of 93 patients, respectively), whereas a small prevalence of exclusive TSSA I reactivity was detected (16/93). In Venezuela, around one-half of the samples contained antibodies to TSSA II (19/41 cases), but additionally, TSSA I antibodies were developed (14/41); 8 of the 41 samples showed mixed reactivity.
All TSSA-WB positive samples from Argentina (N = 67) showed reactivity to TSSA II; one of them also recognized TSSA I (mixed infection). All reactive samples from Paraguayan people showed anti-TSSA II antibodies in single (39/40) or mixed (1/40) infections.
We compared the prevalence of human infections caused by the previously denominated TCI, TCII, or both based on TSSA-WB data among the countries under study. The overall statistical analysis showed significant differences in T. cruzi lineage distribution (P < 0.00001). For further analysis, we made paired comparisons of peptide recognition (TSSA I versus TSSA II; TSSA II versus TSSA I–II; TSSA I versus TSSA I–II) between countries within the north (Mexico, Colombia, and Venezuela) and south (Argentina and Paraguay) of Latin America. No differences were found, except for that Colombia and Venezuela (Table 3) related to single TSSA I and mixed reactivity (P < 0.025).
Then, we made pair-wise comparisons of the TSSA-WB results of serum samples from one country in the north and one in the south of Latin America. Overall differences for the distribution of TSSA I, TSSA II, and TSSA I–II reactivities were highly significant (P < 0.00001) (Table 3). Moreover, differences were significant for TSSA I versus TSSA II and TSSA II versus TSSA I–II reactivities (P < 0.0001) (Table 3). However, TSSA I versus TSSA I–II comparisons did not render significant differences, probably because of the absence of reactivity to only TSSA I and the very low number of samples recognizing both antigens in the southern countries.
Immunophenotyping to assess T. cruzi lineage distribution in Latin American regions.
The results described above about the distribution of parasite groups in different countries prompted us to perform a global comparative analysis based on TSSA I and TSSA II markers. For this purpose, we defined three areas: Southern region (south of South America; Argentina and Paraguay), Northern region (north of South America; Colombia and Venezuela), and Mexico. Figure 2 shows a graphic representation of TSSA-WB data within each of these regions. In line with data presented in Table 3, lineage prevalence analysis indicates significant differences in the Southern region versus Northern region and Southern region versus Mexico. However, no differences were found between Mexico and the Northern region (P < 0.44). Comparisons with combined data from the Northern region and Mexico versus the results from the Southern region also indicate a differential distribution of T. cruzi parasite lineages between the southern and northern areas of Latin America. The comparison of the distribution of TSSA I and mixed infection cases remained non-significant, probably because of the low number of samples from Argentina and Paraguay falling into these categories.
Figure 2.
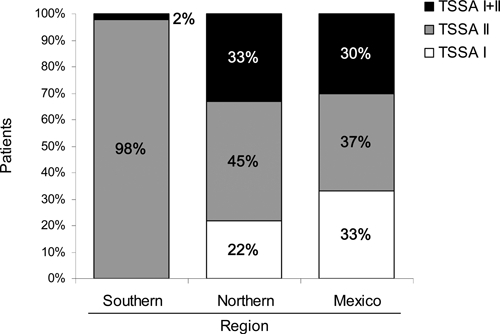
Prevalence of T. cruzi lineages in human infection in Latin America based on TSSA I and TSSA II reactivity: Southern region (Argentina and Paraguay), Northern region (Colombia and Venezuela), and Mexico. Overall difference is P < 0.00001 (χ2 test, degrees of freedom (df) = 4). Overall comparison between Southern and Northern region is P < 0.00001 (χ2 test, df = 2). Comparison between Southern region and Mexico for TSSA I and TSSA II reactivities is P < 0.00001 (Fisher exact test, df = 1). Overall comparison between Northern region and Mexico is P < 0.44 (χ2 test, df = 2). Overall comparison between Northern region and Mexico vs. Southern region is P < 0.00001 (χ2 test, df = 2).
Discussion
Given the relevance of T. cruzi persistence as responsible for the pathogenesis of the infection, parasite variability may be a key factor determining the clinical outcome of Chagas disease.34,35 Then, the distribution of T. cruzi genotypes may explain the regional variations in the manifestations of the chronic infection.
The immunological marker used along this work allows the classification of T. cruzi in two main groups, TCI and TCII.6,25 Recently, TCII was reclassified into five DTUs (TcII to TcVI) based on genetic markers.10 In this new context, no conflict arises when interpreting TSSA I recognition as related to TcI infections. However, for the other DTUs, we can assert that TSSA II-WB recognizes TcVI infections, because the TSSA II peptide was cloned from CL Brener (TcVI). As expected, TSSA II is also recognized by antibodies raised in mice infected with other TcVI strains (RA and Cvd) and in humans harboring TcVI bloodstream parasites (Table 1).36 Moreover, we also detected TSSA II-specific antibodies in serum samples from patients with TcII and TcV infections (Table 1) as well as in samples from mice infected with TcII (Br strain).36 These experimental data are in agreement with those data published by Bhattacharyya and others,26 who showed the potential use of TSSA II to detect TcII, TcV, and TcVI based on the analysis of the sequences of the antigenic region.26 However, Bhattacharyya and others26 suggest the incapability of TSSA II to recognize TcIII and TcIV DTUs, mainly associated with non-human infections, and a possible cross-reaction with TSSA I based on their predicted aminoacid sequence.26,37,38 Unfortunately, the absence of serological assays impedes the confirmation of these speculations. Proper studies must be carried out to verify them.
Herein, we immunocharacterized, for the first time, the T. cruzi infection of Mexican, Colombian, and Venezuelan people through TSSA markers. These antigens have already been used in an ELISA assay to analyze serum samples of patients from the southern cone of South America.25 Because we applied a WB-based assay that improves the detection of anti-TSSA I antibodies in the human infection, patients from the south of South America were also included.
We tested serum samples from patients undergoing other infectious or non-infectious illnesses (malaria, leishmaniasis, syphilis, and megasyndromes autoimmune disorders) and samples obtained from healthy individuals. The recognition of TSSA antigens only in one patient with malaria shows the specificity of TSSA-WB, even when assaying samples from patients infected with Leishmania spp., which frequently causes cross-reactivity in serological tests.39,40
A good concordance between immunophenotyping and direct genotyping was observed. Full concordance was obtained in 17 of 31 samples (Table 1). Partially coincident determinations may be because of variations in parasitic load, differential parasite tissue tropism, and/or the variability proper of human populations, among others, which also account for the lack of concordance for three patients (Table 1). Positive TSSA-WB results of negative genotyped samples show the usefulness of this immunological tool to characterize T. cruzi populations during indeterminate or chronic human infections when low parasitemia makes direct genotyping difficult. In those cases, although laboratory amplification of parasites is an alternative to obtaining enough DNA, it is detrimental for assessing the original complexity of the infecting parasite population. Moreover, the immunological characterization is also independent of parasite tissue tropism.
TSSA-WB findings obtained in human serum samples from different countries were analyzed in combination with the serological diagnosis of the infection. Although high co-reactivity arises from the comparisons of both approaches for Argentinean and Paraguayan patients (Table 2), lower agreement was detected for patients from Mexico and the north of South America (Colombia and Venezuela) (Table 2). This phenomenon may be caused by the use of the recombinant TSSA antigens obtained from parasites isolated in the south of South America, to a dissimilar immunogenicity of TSSA I and TSSA II that was already reported for other mammalian species as well as the variable features of the human populations under study.25 In addition, the use of individual recombinant antigens in serologic tests is known to reduce their sensitivity.
In the southern cone of South America, TcII, TcV, and TcVI are proposed as the main parasite groups circulating in the domiciliary cycle of transmission; TcI was observed in sylvatic areas, where TcIII and TcIV are confined.19–22,24,41 However, in the north of South America, Central America, and Mexico, currently available data show a clear dominance of TcI in both transmission cycles.11–18
Our results from Argentinean and Paraguayan patients indicate almost exclusive anti-TCII reactivity, even in the particular situation of the Amerindians in the Chaco Region of Paraguay where the detection of TcI may be expected because of the lack of barrier between the sylvatic and domestic areas. T. cruzi I was only observed in a few cases of mixed infection, which coincides with the results that Di Noia and others25 report. These findings and those communicated for humans, dogs, and vectors confirm the predominance of TcII, TcV, and TcVI in the domestic cycle, whereas TcI is more closely related to sylvatic mammals.19,22,41–45
In northern countries, TSSA-WB delineates a distribution pattern that coincides only partially with previous descriptions based on molecular and biochemical markers. Interestingly, we observed single and mixed infections in patients from Venezuela where only single infections had been described for human and dog populations.11,46 In Colombia, a high proportion of samples showed single TSSA II and mixed TSSA I–II recognition, thus contrasting with previous data that describe single TcI as the main genotype both in vectors and mammals.13,17,18,47–50 Finally, similar proportions of single and mixed (TSSA I–II) infections were observed in patients from Mexico, where genotyping reports describe TcI as the unique parasite DTU involved in human infections.14,51 Indeed, our findings point, for the first time, to the involvement of TcII/V/VI parasites in human Chagas disease in Mexico.
Despite the reported dominance of TcI, few cases of TcII, TcIV, TcV, and TcVI infections in humans, dogs, primates, and vectors have been recently found in Venezuela, Colombia, Guatemala, and the Brazilian Amazonia; most of those found were detected using modified typing strategies.11,17,46,52–54 The introduction of new procedures shows the emergence of different T. cruzi populations in the human infection in this region. Our results obtained using a strategy not dependent on parasitemia, culture isolation, or tissue tropism are in line with this new picture for the geographical distribution of T. cruzi lineages.
The comparative analysis of the results among the defined regions—south of South America (Paraguay and Argentina), north of South America (Venezuela and Colombia), and Mexico—showed statistically significant differences in the distribution of TCI (TcI) and TCII (TcII, TcV, and TcVI) between the first region and the other two regions. Moreover, TCII (TcII, TcV, and TcVI) is preferentially associated with human infections in the southern cone, whereas both parasite groups are widely distributed in the north of Latin America.
A proper description of T. cruzi geographic distribution will help link the parasite genotype with clinical features in humans and evaluate new prophylactic and therapeutic strategies necessary to succeed in controlling the infection.
ACKNOWLEDGMENTS
The authors are grateful to the Laboratory of Molecular Biology of Chagas Disease (LabMECh) at Institute of Research in Genetic Engineering and Molecular Biology (INGEBI)- National Council of Science and Technology (CONICET) for facilitating the infrastructure for genotyping studies. Also, we thank Dr. C. A. Buscaglia for critically reading the manuscript and Drs. J. Blejer and S. Angel for providing serum samples used as controls. Laboratory assistance by I. Colaianni is also appreciated. A.G.S., O.A.B., and M.S.L. are members of the National Council of Science and Technology Researcher's Career. M.G.R., P.A.S., and J.M.B. are fellows of the CONICET.
Footnotes
Financial support: Major financial support was provided by Grant A40278 from the World Health Organization and Grant PICT 33103 from the National Agency of Science and Technology (to M.S.L.). Partial support was provided by Grants FONACIT G2000001530 and LOCTI P935015 (to L.B.), Estrategia de Sostenibilidad Universidad de Antioquia (to O.T.C.), and Grants PICT 33955 and PIP 112 CONICET (to A.G.S.).
Authors' addresses: Marikena G. Risso, Paula A. Sartor, and Maria Susana Leguizamón, Departamento de Microbiología, Facultad de Medicina, Universidad de Buenos Aires, Buenos Aires, Argentina, E-mails: mkrisso@yahoo.com, psartor@fmed.uba.ar, and sleguiza@fmed.uba.ar. Juan M. Burgos and Alejandro G. Schijman, Laboratorio de Biología Molecular de la Enfermedad de Chagas (LaBMECh), Instituto de Investigaciones en Ingeniería Genética y Biología Molecular (INGEBI), Buenos Aires, Argentina, E-mails: jburgos@dna.uba.ar and schijman@dna.uba.ar. Luis Briceño, Instituto de Biomedicina, Facultad de Medicina, Universidad Central de Venezuela, San Nicolás a Providencia, Sector Hospital Vargas, Caracas, Venezuela, E-mail: lbricenozoppi@gmail.com. Eva M. Rodríguez, Centro de Investigación en Salud Pública “Jacinto Convit,” Facultad de Medicina, Universidad Central de Venezuela, Estado Lara, Venezuela, E-mail: evamary@gmail.com. Felipe Guhl, Centro de Investigaciones en Parasitología Tropical (CIMPAT), Universidad de los Andes, Bogotá, Colombia, E-mail: fguhl@uniandes.edu.co. Omar Triana Chavez, Instituto de Biología, Grupo de Chagas, Sede de Investigación Universitaria (SIU), Universidad de Antioquia, Medellín, Colombia, E-mail: otriana@gmail.com. Berta Espinoza, Departamento de Inmunología, Instituto de Investigaciones Biomédicas, Ciudad Universitaria, México, E-mail: besgu@biomedicas.unam.mx. Victor M. Monteón, Centro de Investigaciones en Enfermedades Tropicales, Universidad Autónoma de Campeche, Campeche, México, E-mail: victormonteon@yahoo.com.mx. Graciela Russomando, Departamento de Biología Molecular y Genética, Instituto de Investigaciones en Ciencias de la Salud, Universidad Nacional de Asunción, Río de la Plata y Lagerenza, Asunción, Paraguay, E-mail: grusso@rieder.net.py. Oscar A. Bottasso, Instituto de Inmunología, Facultad de Ciencias Médicas, Universidad Nacional de Rosario, Santa Fe, Argentina, E-mail: oscarbottasso@yahoo.com.ar.
Reprint requests: Maria Susana Leguizamón, Departamento de Microbiología, Facultad de Medicina, Universidad de Buenos Aires, Argentina, Paraguay 2155 Piso 13, (1121) Buenos Aires, Argentina, E-mail: sleguiza@fmed.uba.ar.
References
- 1.Tibayrenc M, Ward P, Moya A, Ayala FJ. Natural populations of Trypanosoma cruzi, the agent of Chagas disease, have a complex multiclonal structure. Proc Natl Acad Sci USA. 1986;83:115–119. doi: 10.1073/pnas.83.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tibayrenc M, Ayala FJ. The clonal theory of parasitic protozoa: 12 years on. Trends Parasitol. 2002;18:405–410. doi: 10.1016/s1471-4922(02)02357-7. [DOI] [PubMed] [Google Scholar]
- 3.Macedo AM, Martins MS, Chiari E, Pena SD. DNA fingerprinting of Trypanosoma cruzi: a new tool for characterization of strains and clones. Mol Biochem Parasitol. 1992;55:147–153. doi: 10.1016/0166-6851(92)90135-7. [DOI] [PubMed] [Google Scholar]
- 4.Macedo AM, Pena SD. Genetic variability of Trypanosoma cruzi: implications for the pathogenesis of Chagas disease. Parasitol Today. 1998;14:119–124. doi: 10.1016/s0169-4758(97)01179-4. [DOI] [PubMed] [Google Scholar]
- 5.Macedo AM, Machado CR, Oliveira RP, Pena SD. Trypanosoma cruzi: genetic structure of populations and relevance of genetic variability to the pathogenesis of chagas disease. Mem Inst Oswaldo Cruz. 2004;99:1–5. doi: 10.1590/s0074-02762004000100001. [DOI] [PubMed] [Google Scholar]
- 6.Anonymous Recommendations from a satellite meeting. Mem Inst Oswaldo Cruz. 1999;94:429–432. doi: 10.1590/s0074-02761999000700085. [DOI] [PubMed] [Google Scholar]
- 7.Brisse S, Dujardin JC, Tibayrenc M. Identification of six Trypanosoma cruzi lineages by sequence-characterised amplified region markers. Mol Biochem Parasitol. 2000;111:95–105. doi: 10.1016/s0166-6851(00)00302-9. [DOI] [PubMed] [Google Scholar]
- 8.Westenberger SJ, Barnabé C, Campbell DA, Sturm NR. Two hybridization events define the population structure of Trypanosoma cruzi. Genetics. 2005;171:527–543. doi: 10.1534/genetics.104.038745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freitas JM, Augusto-Pinto L, Pimenta JR, Bastos-Rodrigues L, Gonçalves VF, Teixeira SM, Chiari E, Junqueira AC, Fernandes O, Macedo AM, Machado CR, Pena SD. Ancestral genomes, sex, and the population structure of Trypanosoma cruzi. PLoS Pathog. 2006;2:e24. doi: 10.1371/journal.ppat.0020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zingales B, Andrade SG, Briones MR, Campbell DA, Chiari E, Fernandes O, Guhl F, Lages-Silva E, Macedo AM, Machado CR, Miles MA, Romanha AJ, Sturm NR, Tibayrenc M, Schijman AG. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz. 2009;104:1051–1054. doi: 10.1590/s0074-02762009000700021. [DOI] [PubMed] [Google Scholar]
- 11.Añez N, Crisante G, da Silva FM, Rojas A, Carrasco H, Umezawa ES, Stolf AM, Ramírez JL, Teixeira MM. Predominance of lineage I among Trypanosoma cruzi isolates from Venezuelan patients with different clinical profiles of acute Chagas' disease. Trop Med Int Health. 2004;9:1319–1326. doi: 10.1111/j.1365-3156.2004.01333.x. [DOI] [PubMed] [Google Scholar]
- 12.Ruíz-Sánchez R, León MP, Matta V, Reyes PA, López R, Jay D, Monteón VM. Trypanosoma cruzi isolates from Mexican and Guatemalan acute and chronic Chagasic cardiopathy patients belong to Trypanosoma cruzi I. Mem Inst Oswaldo Cruz. 2005;100:281–283. doi: 10.1590/s0074-02762005000300012. [DOI] [PubMed] [Google Scholar]
- 13.Salazar A, Schijman AG, Triana-Chávez O. High variability of Colombian Trypanosoma cruzi lineage I stocks as revealed by low-stringency single primer-PCR minicircle signatures. Acta Trop. 2006;100:110–118. doi: 10.1016/j.actatropica.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Sánchez-Guillén M del C, Bernabé C, Tibayrenc M, Zavala-Castro J, Totolhua JL, Méndez-López J, González-Mejía ME, Torres-Rasgado E, López-Colombo A, Pérez-Fuentes R. Trypanosoma cruzi strains isolated from human, vector, and animal reservoir in the same endemic region in Mexico and typed as T. cruzi I, discrete typing unit 1 exhibit considerable biological diversity. Mem Inst Oswaldo Cruz. 2006;101:585–590. doi: 10.1590/s0074-02762006000600002. [DOI] [PubMed] [Google Scholar]
- 15.Teixeira MM, da Silva FM, Marcili A, Umezawa ES, Shikanai-Yasuda MA, Cunha-Neto E, Kalil J, Stolf N, Stolf AM. Trypanosoma cruzi lineage I in endomyocardial biopsy from a north-eastern Brazilian patient at end-stage chronic Chagasic cardiomyopathy. Trop Med Int Health. 2006;11:294–298. doi: 10.1111/j.1365-3156.2006.01575.x. [DOI] [PubMed] [Google Scholar]
- 16.Roellig DM, Brown EL, Barnabé C, Tibayrenc M, Steurer FJ, Yabsley MJ. Molecular typing of Trypanosoma cruzi isolates, United States. Emerg Infect Dis. 2008;14:1123–1125. doi: 10.3201/eid1407.080175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zafra G, Mantilla JC, Valadares HM, Macedo AM, González CI. Evidence of Trypanosoma cruzi II infection in Colombian chagasic patients. Parasitol Res. 2008;103:731–734. doi: 10.1007/s00436-008-1034-0. [DOI] [PubMed] [Google Scholar]
- 18.Falla A, Herrera C, Fajardo A, Montilla M, Vallejo GA, Guhl F. Haplotype identification within Trypanosoma cruzi I in Colombian isolates from several reservoirs, vectors and humans. Acta Trop. 2009;110:15–21. doi: 10.1016/j.actatropica.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Buscaglia CA, Di Noia JM. Trypanosoma cruzi clonal diversity and the epidemiology of Chagas' disease. Microbes Infect. 2003;5:419–427. doi: 10.1016/s1286-4579(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 20.Higo H, Miura S, Horio M, Mimori T, Hamano S, Agatsuma T, Yanagi T, Cruz-Reyes A, Uyema N, Rojas de Arias A, Matta V, Akahane H, Hirayama K, Takeuchi T, Tada I, Himeno K. Genotypic variation among lineages of Trypanosoma cruzi and its geographic aspects. Parasitol Int. 2004;53:337–344. doi: 10.1016/j.parint.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Yeo M, Acosta N, Llewellyn M, Sánchez H, Adamson S, Miles GA, López E, González N, Patterson JS, Gaunt MW, de Arias AR, Miles MA. Origins of Chagas disease: Didelphis species are natural hosts of Trypanosoma cruzi I and armadillos hosts of Trypanosoma cruzi II, including hybrids. Int J Parasitol. 2005;35:225–233. doi: 10.1016/j.ijpara.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 22.Cardinal MV, Lauricella MA, Ceballos LA, Lanati L, Marcet PL, Levin MJ, Kitron U, Gürtler RE, Schijman AG. Molecular epidemiology of domestic and sylvatic Trypanosoma cruzi infection in rural northwestern Argentina. Int J Parasitol. 2008;38:1533–1543. doi: 10.1016/j.ijpara.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devera R, Fernandes O, Coura JR. Should Trypanosoma cruzi be called “cruzi” complex? A review of the parasite diversity and the potential of selecting population after in vitro culturing and mice infection. Mem Inst Oswaldo Cruz. 2003;98:1–12. doi: 10.1590/s0074-02762003000100001. [DOI] [PubMed] [Google Scholar]
- 24.Burgos JM, Altcheh J, Bisio M, Duffy T, Valadares HM, Seidenstein ME, Piccinali R, Freitas JM, Levin MJ, Macchi L, Macedo AM, Freilij H, Schijman AG. Direct molecular profiling of minicircle signatures and lineages of Trypanosoma cruzi bloodstream populations causing congenital Chagas disease. Int J Parasitol. 2007;37:1319–1327. doi: 10.1016/j.ijpara.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 25.Di Noia JM, Buscaglia CA, De Marchi CR, Almeida IC, Frasch AC. A Trypanosoma cruzi small surface molecule provides the first immunological evidence that Chagas' disease is due to a single parasite lineage. J Exp Med. 2002;195:401–413. doi: 10.1084/jem.20011433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhattacharyya T, Brooks J, Yeo M, Carrasco HJ, Lewis MD, Llewellyn MS, Miles MA. Analysis of molecular diversity of the Trypanosoma cruzi trypomastigote small surface antigen reveals novel epitopes, evidence of positive selection and potential implications for lineage-specific serology. Int J Parasitol. 2010;40:921–928. doi: 10.1016/j.ijpara.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Cura C, Mejía-Jaramillo M, Duffy T, Burgos JM, Rodriguero M, Cardinal MV, Kjos S, Gurgel Gonçalves R, Blanchet D, De Pablos LM, Da Silva A, Russomando G, Cuba Cuba CA, Aznar C, Abate T, Levin MJ, Osuna A, Gürtler RE, Diosque P, Solari A, Triana-Chávez O, Schijman AG. Trypanosoma cruzi genotypes in different geographic regions and transmission cycles based on a microsatellite motif of the intergenic spacer of spliced leader genes. Int J Parasitol. 2010 doi: 10.1016/j.ijpara.2010.06.006. doi:10.1016/j.ijpara.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sánchez B, Monteón V, Reyes P, Espinoza B. Standardization of ELISA and Western blot for detection of Trypanosoma cruzi. Antibodies using extracts of Mexican strains as antigens. Concordance between laboratories. Arch Med Res. 2001;32:382–388. doi: 10.1016/s0188-4409(01)00303-4. [DOI] [PubMed] [Google Scholar]
- 29.Rangel-Flores H, Sánchez B, Mendoza-Duarte J, Barnabé C, Breniére SF, Ramos C, Espinoza B. Serological and parasitological demonstration of Trypanosoma cruzi infections in an urban central area of Mexico: correlation with electrocardiographic alterations. Am J Trop Med Hyg. 2001;65:887–895. doi: 10.4269/ajtmh.2001.65.887. [DOI] [PubMed] [Google Scholar]
- 30.Guhl F, Nicholls S. Manual de Procedimientos para el Diagnostico de la Enfermedad de Chagas. Bogotá DC: Quebecor Press; 2001. pp. 12–58. [Google Scholar]
- 31.Feliciangeli MD, Sánchez-Martín MJ, Suárez B, Marrero R, Torrellas A, Bravo A, Medina M, Martínez C, Hernandez M, Duque N, Toyo J, Rangel R. Risk factors for Trypanosoma cruzi human infection in Barinas State, Venezuela. Am J Trop Med Hyg. 2007;76:915–921. [PubMed] [Google Scholar]
- 32.Briceño L, Rodriguez EM, Medina M, Campos Y, Leon G, Briceño A, Mosca W. An inexpensive antigen for serodiagnosis Chagas' disease. Invest Clin. 2010;51:101–113. [PubMed] [Google Scholar]
- 33.Otani MM, Vinelli E, Kirchhoff LV, del Pozo A, Sands A, Vercauteren G, Sabino EC. WHO comparative evaluation of serologic assays for Chagas disease. Transfusion. 2009;49:1076–1082. doi: 10.1111/j.1537-2995.2009.02107.x. [DOI] [PubMed] [Google Scholar]
- 34.Diez M, Favaloro L, Bertolotti A, Burgos JM, Vigliano C, Lastra MP, Levin MJ, Arnedo A, Nagel C, Schijman AG, Favaloro RR. Usefulness of PCR strategies for early diagnosis of Chagas' disease reactivation and treatment follow-up in heart transplantation. Am J Transplant. 2007;7:1633–1640. doi: 10.1111/j.1600-6143.2007.01820.x. [DOI] [PubMed] [Google Scholar]
- 35.Revollo S, Oury B, Laurent JP, Barnabé C, Quesney V, Carrière V, Noël S, Tibayrenc M. Trypanosoma cruzi: impact of clonal evolution of the parasite on its biological and medical properties. Exp Parasitol. 1998;89:30–39. doi: 10.1006/expr.1998.4216. [DOI] [PubMed] [Google Scholar]
- 36.Risso MG, Garbarino GB, Mocetti E, Campetella O, Gonzalez Cappa SM, Buscaglia CA, Leguizamon MS. Differential expression of a virulence factor, the trans-sialidase, by the main Trypanosoma cruzi phylogenetic lineages. J Infect Dis. 2004;189:2250–2259. doi: 10.1086/420831. [DOI] [PubMed] [Google Scholar]
- 37.Marcili A, Lima L, Valente VC, Valente SA, Batista JS, Junqueira AC, Souza AI, da Rosa JA, Campaner M, Lewis MD, Llewellyn MS, Miles MA, Teixeira MM. Comparative phylogeography of Trypanosoma cruzi TCIIc: new hosts, association with terrestrial ecotopes, and spatial clustering. Infect Genet Evol. 2009;9:1265–1274. doi: 10.1016/j.meegid.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 38.Llewellyn MS, Lewis MD, Acosta N, Yeo M, Carrasco HJ, Segovia M, Vargas J, Torrico F, Miles MA, Gaunt MW. Trypanosoma cruzi IIc: phylogenetic and phylogeographic insights from sequence and microsatellite analysis and potential impact on emergent Chagas disease. PLoS Negl Trop Dis. 2009;3:e510. doi: 10.1371/journal.pntd.0000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caballero ZC, Sousa OE, Marques WP, Saez-Alquezar A, Umezawa ES. Evaluation of serological tests to identify Trypanosoma cruzi infection in humans and determine cross-reactivity with Trypanosoma rangeli and Leishmania spp. Clin Vaccine Immunol. 2007;14:1045–1049. doi: 10.1128/CVI.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frank FM, Fernández MM, Taranto NJ, Cajal SP, Margni RA, Castro E, Thomaz-Soccol V, Malchiodi EL. Characterization of human infection by Leishmania spp. in the northwest of Argentina: immune response, double infection with Trypanosoma cruzi and species of Leishmania involved. Parasitology. 2003;126:31–39. doi: 10.1017/s0031182002002585. [DOI] [PubMed] [Google Scholar]
- 41.Burgos JM, Begher S, Silva HM, Bisio M, Duffy T, Levin MJ, Macedo AM, Schijman AG. Molecular identification of Trypanosoma cruzi I tropism for central nervous system in Chagas reactivation due to AIDS. Am J Trop Med Hyg. 2008;78:294–297. [PubMed] [Google Scholar]
- 42.Diosque P, Barnabé C, Padilla AM, Marco JD, Cardozo RM, Cimino RO, Nasser JR, Tibayrenc M, Basombrío MA. Multilocus enzyme electrophoresis analysis of Trypanosoma cruzi isolates from a geographically restricted endemic area for Chagas' disease in Argentina. Int J Parasitol. 2003;33:997–1003. doi: 10.1016/s0020-7519(03)00139-5. [DOI] [PubMed] [Google Scholar]
- 43.Marcet PL, Duffy T, Cardinal MV, Burgos JM, Lauricella MA, Levin MJ, Kitron U, Gürtler RE, Schijman AG. PCR-based screening and lineage identification of Trypanosoma cruzi directly from faecal samples of triatomine bugs from northwestern Argentina. Parasitology. 2006;132:57–65. doi: 10.1017/S0031182005008772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freitas JM, Lages-Silva E, Crema E, Pena SD, Macedo AM. Real time PCR strategy for the identification of major lineages of Trypanosoma cruzi directly in chronically infected human tissues. Int J Parasitol. 2005;35:411–417. doi: 10.1016/j.ijpara.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 45.Ceballos LA, Cardinal MV, Vazquez-Prokopec GM, Lauricella MA, Orozco MM, Cortinas R, Schijman AG, Levin MJ, Kitron U, Gürtler RE. Long-term reduction of Trypanosoma cruzi infection in sylvatic mammals following deforestation and sustained vector surveillance in northwestern Argentina. Acta Trop. 2006;98:286–296. doi: 10.1016/j.actatropica.2006.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crisante G, Rojas A, Teixeira MM, Añez N. Infected dogs as a risk factor in the transmission of human Trypanosoma cruzi infection in western Venezuela. Acta Trop. 2006;98:247–254. doi: 10.1016/j.actatropica.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 47.Triana O, Jaramillo N, Moreno J. Genetic variability of Colombian populations of Trypanosoma cruzi and Trypanosoma rangeli. Biol Res. 1999;32:1–10. [PubMed] [Google Scholar]
- 48.Jaramillo N, Moreno J, Triana O, Arcos-Burgos M, Muñoz S, Solari A. Genetic structure and phylogenetic relationships of Colombian Trypanosoma cruzi populations as determined by schizodeme and isoenzyme markers. Am J Trop Med Hyg. 1999;61:986–993. doi: 10.4269/ajtmh.1999.61.986. [DOI] [PubMed] [Google Scholar]
- 49.Cuervo P, Cupolillo E, Segura I, Saravia N, Fernandes O. Genetic diversity of Colombian sylvatic Trypanosoma cruzi isolates revealed by the ribosomal DNA. Mem Inst Oswaldo Cruz. 2002;97:877–880. doi: 10.1590/s0074-02762002000600023. [DOI] [PubMed] [Google Scholar]
- 50.Montilla MM, Guhl F, Jaramillo C, Nicholls S, Barnabe C, Bosseno MF, Breniere SF. Isoenzyme clustering of trypanosomatidae Colombian populations. Am J Trop Med Hyg. 2002;66:394–400. doi: 10.4269/ajtmh.2002.66.394. [DOI] [PubMed] [Google Scholar]
- 51.Bosseno MF, Barnabé C, Magallón Gastélum E, Lozano Kasten F, Ramsey J, Espinoza B, Brenière SF. Predominance of Trypanosoma cruzi lineage I in Mexico. J Clin Microbiol. 2002;40:627–632. doi: 10.1128/JCM.40.2.627-632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Triana O, Ortiz S, Dujardin JC, Solari A. Trypanosoma cruzi: variability of stocks from Colombia determined by molecular karyotype and minicircle southern blot analysis. Exp Parasitol. 2006;113:62–66. doi: 10.1016/j.exppara.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 53.Pennington PM, Paiz C, Grajeda LM, Cordón-Rosales C. Short report: concurrent detection of Trypanosoma cruzi lineages I and II in domestic Triatoma dimidiata from Guatemala. Am J Trop Med Hyg. 2009;80:239–241. [PubMed] [Google Scholar]
- 54.Marcili A, Valente VC, Valente SA, Junqueira AC, da Silva FM, Pinto AY, Naiff RD, Campaner M, Coura JR, Camargo EP, Miles MA, Teixeira MM. Trypanosoma cruzi in Brazilian Amazonia: lineages TCI and TCIIa in wild primates, Rhodnius spp. and in humans with Chagas disease associated with oral transmission. Int J Parasitol. 2009;39:615–623. doi: 10.1016/j.ijpara.2008.09.015. [DOI] [PubMed] [Google Scholar]


